Abstract
Background
Detection of melanoma cells in circulation may be important in assessing tumor progression. The objective of this study was to develop a specific, reliable, multimarker quantitative real-time reverse transcription-PCR (qRT) assay for detecting melanoma cells in patients’ blood.
Methods
We developed qRT assays for the mRNA of four melanoma-associated markers: MART-1, GalNAc-T, PAX-3, and MAGE-A3. In optimization studies, we tested 17 melanoma cell lines and 49 peripheral blood leukocyte (PBL) samples from volunteers. We performed RNA and melanoma cell dilution studies to assess the detection limits and imprecision of the assays. We measured the mRNAs in blood specimens from 94 melanoma patients [American Joint Committee on Cancer (AJCC) stage I, n = 20; II, n = 20; III, n = 32; IV, n = 22].
Results
All markers were frequently detected in melanoma cell lines, whereas none of the markers was detected in PBLs from volunteers. The qRT assay could detect 1 melanoma cell in 107 PBLs in the melanoma cell-dilution studies. Markers were detected in 15%, 30%, 75%, and 86% of melanoma patients with AJCC stage I, II, III, and IV disease, respectively. The number of positive markers and AJCC stage were significantly correlated (Spearman correlation coefficient = 0.58; P <0.0001).
Conclusions
Multimarker qRT can detect circulating melanoma cells in blood. Measurement of the studied molecular markers in blood may be useful in detection of metastasis and monitoring treatment response of melanoma patients.
The metastasis of melanoma to distant sites often portends a poor prognosis (1). Assessment of primary and/or metastatic melanoma has been addressed in the new American Joint Committee on Cancer (AJCC)5 staging criteria (2, 3). The staging system, however, does not accurately take into account the disease progression events, particularly ongoing systemic metastasis, at the time patients are seen. The detection of melanoma cells in circulation may be important in assessing tumor progression and potential for metastasis.
The molecular detection of circulating melanoma cells in blood by reverse transcription-PCR (RT-PCR) has been studied previously, and associations with stage and outcome of disease have been reported (4–6). RT-PCR can detect a few tumor cells among millions of peripheral blood leukocytes (PBLs) (7, 8), but there are limitations on the specificity and reliability of assays based on gel electrophoresis systems (9, 10). Recently, quantitative real-time RT-PCR (qRT) assays have offered a more robust, accurate, and less labor-intensive approach that allows rapid and reproducible quantitative analysis for detection of a few tumor cells in tissues (11). Investigators have reported the detection of circulating tumor cells in blood by both single and multimarker qRT, but few studies have assessed the agreement of qRT with evidence of disease progression (12–15). Heterogeneity of the expression of tumor genes and variable performance of the assays have posed major problems for detection of circulating tumor cells in blood. As we first reported, this heterogeneity of marker expression in blood and lymph nodes makes multimarker RT-PCR assays advantageous compared with single-marker assays for detecting circulating malignant cells (7, 16). Although tumor cells may express all markers assessed, the concentrations of expressed marker mRNAs can vary. Because of the detection limits for mRNAs, this difference must be considered in developing sensitive and reliable qRT assays (5, 6, 17, 18).
To date, only a limited number of tumor-associated markers have been identified that are absent in healthy cells but produced in melanoma cells. We have developed a qRT assay for four specific predictive markers for study of primary and metastatic melanoma (11, 19): melanoma antigen recognized by T cells-1 (MART-1), β1→4-N-acetylgalactosaminyltransferase (GalNAc-T), paired box homeotic gene transcription factor 3 (PAX-3), and melanoma antigen gene-A3 family (MAGE-A3) (11). MART-1 is a major melanocyte-differentiation antigen that is frequently produced by melanoma cells and is not produced by nonmelanoma malignancies and lymph nodes from cancer-free patients (11, 20). GalNAc-T (21, 22) has been investigated in melanoma and other tumors (22, 23). PAX-3 (24) is re-expressed in rhabdomyosarcoma, Ewing sarcoma, and melanoma (25, 26). MAGE-A3 is commonly produced by various tumors and not by healthy tissue, except male germline cells and placenta (10, 11). Use of qRT testing for the above four specific markers can aid in the identification of metastatic melanoma cells in paraffin-embedded sentinel lymph node (SLN) samples and thus can upstage patients whose SLNs were negative by immunohistochemistry and on examination of hematoxylineosin-stained tissue (11). In this study, we applied this qRT assay to blood of melanoma patients, using four mRNA markers predictive for the SLNs to detect occult tumor cells.
The major objective of the study was to determine whether the predictive multimarker qRT assay for SLNs could detect metastatic tumor cells in blood. Our hypothesis is that the qRT assay could be useful in detecting metastatic cells in the blood of melanoma patients and be used as a surrogate of disease progression.
Materials and Methods
MELANOMA CELL LINES
Seventeen melanoma cell lines (MA, MB, MC, MD, ME, MF, MG, MH, MI, MJ, MK, ML, MM, MN, MO, MP, and MQ) were established and characterized at the John Wayne Cancer Institute (JWCI). Cells were grown in RPMI 1640 containing 100 mL/L heat-inactivated fetal calf serum and 10 g/L penicillin/streptomycin (Gibco) in a T75-cm2 flask and were used when they reached 70%– 80% confluence.
PATIENTS
All patients enrolled in the study had documented physical and medical histories, and their AJCC stage of disease was determined and recorded at the time of blood drawing. Blood was drawn from 94 melanoma patients (20 with stage I, 20 with stage II, 32 with stage III, and 22 with stage IV disease) immediately before they received any treatment at JWCI. All patients signed consents for the use of their blood specimens, and the study was carried out according to the guidelines set forth by the Saint John’s Health Center and the JWCI Institutional Review Board committee.
The study was conducted in a double-blinded fashion: the patients’ disease status was not known to the individuals who performed the PCR assay or analyzed the PCR data, and PCR results were not known to the individuals who recorded disease status.
BLOOD PROCESSING AND RNA EXTRACTION
We collected 10 mL of blood from each patient with melanoma. Blood samples were collected in sodium citrate-containing tubes, and the first several milliliters of blood at the initial venipuncture were discarded to eliminate skin-plug contamination, as described previously (7, 10). Within 2 h after being drawn, blood was processed in a designated blood-processing room. Blood cells were collected by use of Purescript RBC Lysis Solution (Gentra), according to the manufacturer’s instructions.
Tri-Reagent (Molecular Research Center) was used to isolate total cellular RNA from blood samples and cell lines, as described previously (9, 10). All of the RNA extraction procedures were performed in a designated sterile laminar flow hood with RNase-free labware. RNA was quantified and assessed for purity by ultraviolet spectrophotometry. Blood processing, RNA extraction, RT-PCR assay set up, and post-RT-PCR product analysis were carried out in separate designated rooms to prevent cross-contamination (7, 19).
PRIMERS AND PROBES
Primer and probe sequences were designed for the qRT as described previously (11, 19). The fluorescence resonance energy transfer probe sequences were as follows: MART-1, 5′-FAM-TGCAGAACAGTCACCACCACC-BHQ-1-3′; GalNAc-T, 5′-FAM-ATGAGGCTGCTTTCACTATCCGCA-BHQ-1-3′; PAX-3, 5′-FAM-CCAGACTGATTACGCGCTCTCCC-BHQ-1-3′; MAGE-A3, 5′-FAMAGCTCCTGCCCACACTCCCGCCTGT-BHQ-1-3′; and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′-FAM-CAGCAATGCCTCCTGCACCACCAA-BHQ-1-3′, where FAM is 6-carboxyfluorescein and BHQ-1 is Black Hole Quencher 1.
Melanoma cells and 49 PBL samples from healthy donors were used to optimize the assay. The GAPDH gene was used as a control housekeeping gene. Any specimen with insufficient GAPDH mRNA was excluded from the study.
MULTIMARKER qRT ASSAY
Reverse transcription reactions were performed with Moloney murine leukemia virus reverse transcriptase (Promega) with oligo(dT) primer (10, 11). The multimarker qRT assay was performed in an iCycler iQ Real-Time Thermocycler Detection System (Bio-Rad Laboratories). We transferred 5 µL of cDNA from 250 ng of total RNA to a well of a 96-well PCR plate (Fisher Scientific) in which 0.5 µM each primer, 0.3 µM fluorescence resonance energy transfer probe, 1 U of AmpliTaq Gold polymerase (Applied Biosystems), 200 µM each deoxynucleotide triphosphate, 4.5 mM MgCl2, and PCR buffer were added to a final volume of 25 µL. Samples were amplified with a precycling hold at 95 °C for 10 min, followed by 42 cycles of denaturation at 95 °C for 1 min, annealing for 1 min (at 55 °C for GAPDH, 59 °C for MART-1, 62 °C for GalNAc-T and PAX-3, and 58 °C for MAGE-A3), and extension at 72 °C for 1 min. The calibration curve was generated with the threshold cycle (Ct) of 9 serial dilutions of plasmid templates (108–100 copies). The Ct of each sample was interpolated from the calibration curve, and the number of mRNA copies was calculated by the iCycler iQ Real-Time Detection System Software (Bio-Rad Laboratories). Each assay was performed at least twice and included marker-positive (melanoma cell lines) and -negative controls (PBLs of healthy donors) and reagent controls (reagent alone without RNA or cDNA) for qRT assays to verify the results. The mean number of mRNA copies for each gene was used for analysis.
SERIAL DILUTION STUDY OF MELANOMA CELLS IN PBLs
To determine the detection limit for melanoma cells in blood, we performed qRT on serially diluted melanoma cells mixed with PBLs from healthy blood donors. This in vitro model system to some extent mimics the circulating melanoma cells in blood. In the assay, serial dilutions of melanoma cells (100, 10, 5, 2.5, 1, and 0 cells) that expressed all four markers were mixed with 107 donor-derived PBLs and assayed for each marker by qRT. This in vitro assay was performed 10 times to validate the reproducibility and robustness of the assay system.
PLASMID CONTROLS
Specific plasmid controls were synthesized as described previously (19). PCR products generated from MART-1, GalNAc-T, PAX-3, MAGE-A3, and GAPDH were run on 2% agarose gel electrophoresis and extracted by the QIAquick gel extraction method (Qiagen) according to the manufacturer’s instructions. Extracted PCR products were ligated into pCR II-TOPO cloning vector (Invitrogen) and transformed into Escherichia coli DH5-α cells. Plasmids containing the target gene were purified and quantified for use in the quantitative PCR setup. To confirm that the inserted PCR product size was correct, plasmids were digested with specific restriction enzymes, and the products were visualized after gel electrophoresis.
STATISTICAL ANALYSIS
We used the Mann–Whitney U-test to compare positive marker detection among AJCC stages. The Pearson correlation coefficient and Cochran–Armitage trend test were used to examine the significance of associations of the number of markers and AJCC stage of disease. Kappa analysis was used to assess the relationship between two markers. Spearman correlation coefficients were used to assess the relationship between multimarker detection and AJCC stage. All two-sided P values ≤0.05 were considered statistically significant.
Results
CALIBRATION CURVES AND ASSAY VARIATION
The calibration curves (Fig. 1 in the Data Supplement that accompanies the online version of this article at http://www.clinchem.org/content/vol51/issue6/) showed the expected linear increase of signal with logarithm of the copy number. PCR efficiency, assessed from the slopes of the curves, was between 90% and 100%. The correlation coefficients for all calibration curves (Ct vs log copy number) in the study were ≥0.99.
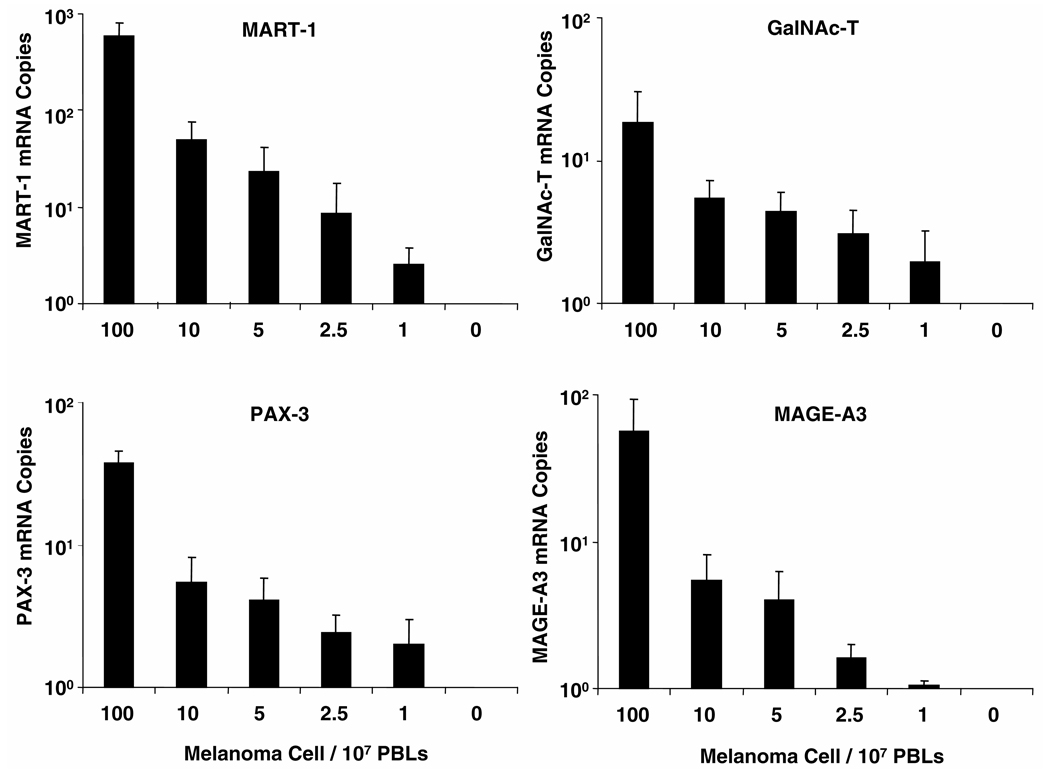
Fig. 1. qRT detection limits for melanoma cells mixed with PBLs from healthy blood donors.
Serially diluted melanoma cells (100, 10, 5, 2.5, 1, and 0) were mixed with 107 PBLs from healthy blood donors. RNA was isolated from the cell mixture, and qRT was performed; the assay was performed 10 times. Mean (SD; error bars) absolute mRNA copies from a representative cell line (MA) are given according to serial dilution.
When qRT was performed in different experiments with mRNA from one melanoma cell line, the imprecision values (CVs) for GAPDH, MART-1, GalNAc-T, PAX-3, and MAGE-A3 were 7.7%, 21%, 14%, 27%, and 34%, respectively between assays (n = 3) and 1.8%–22% for triplicate results (intraassay variation).
MULTIMARKER mRNA EXPRESSION IN MELANOMA CELL LINES
All melanoma cell lines showed GAPDH expression with high copy numbers (mean, 1.6 × 107; range, 1.7 × 106 to 5.5 × 107), and expression of the mRNA markers MART-1, GalNAc-T, PAX-3, and MAGE-A3 was detected in 88%, 100%, 100%, and 94%, respectively, of these melanoma cell lines. The numbers of MART-1 mRNA copies ranged from 0 to 8.4 × 106 (mean, 1.2 × 106) per 250 ng of total RNA from individual melanoma lines. The copy numbers of GalNAc-T ranged from 9.4 × 101 to 1.5 × 105 (mean, 2.2 × 104), PAX-3 ranged from 3.2 × 103 to 2.7 × 106 (mean, 2.0 × 105), and MAGE-A3 ranged from 0 to 3.3 × 105 (mean, 7.3 × 104). Fourteen cell lines expressed all 4 markers, and 3 lines expressed 3 markers. No marker expression was detected in PBLs from 49 healthy donors under the optimum conditions established for individual markers.
qRT DETECTION LIMIT OF MARKER EXPRESSION
After establishing potential marker genes for melanoma, we used RNA dilution series to determine the detection limit of each marker. Total RNA was isolated from melanoma cell lines that expressed all 4 markers; we then performed qRT on RNA serially diluted from 2.5 × 10−1 to 10−8 µg for individual markers (Fig. 2 in the online Data Supplement). The assay was performed several times with three different cell lines. Although mRNA concentrations differed among cell lines, the instrument– qRT assay combination detected all markers consistently at picogram concentrations: MART-1, PAX-3, and MAGE-A3 expression from 1 pg of RNA and GalNAc-T from 10 pg of RNA above background. The background subtraction was obtained from the net (inner – outer) fluorescence for each well, and threshold fluorescence for the experiment was set at 10 times the mean of the SD of the fluorescence of each well from cycles 2 to 10 by the iCycler iQ Real-Time Detection System Software. Linear regression analysis of curves from serial RNA dilutions demonstrated that the correlation coefficient was 0.95– 0.998 and slopes were −3.50 to −3.75 for individual markers with r = 0.95– 0.998. GAPDH expression was detected from 0.01 pg of RNA in all cell lines assessed.
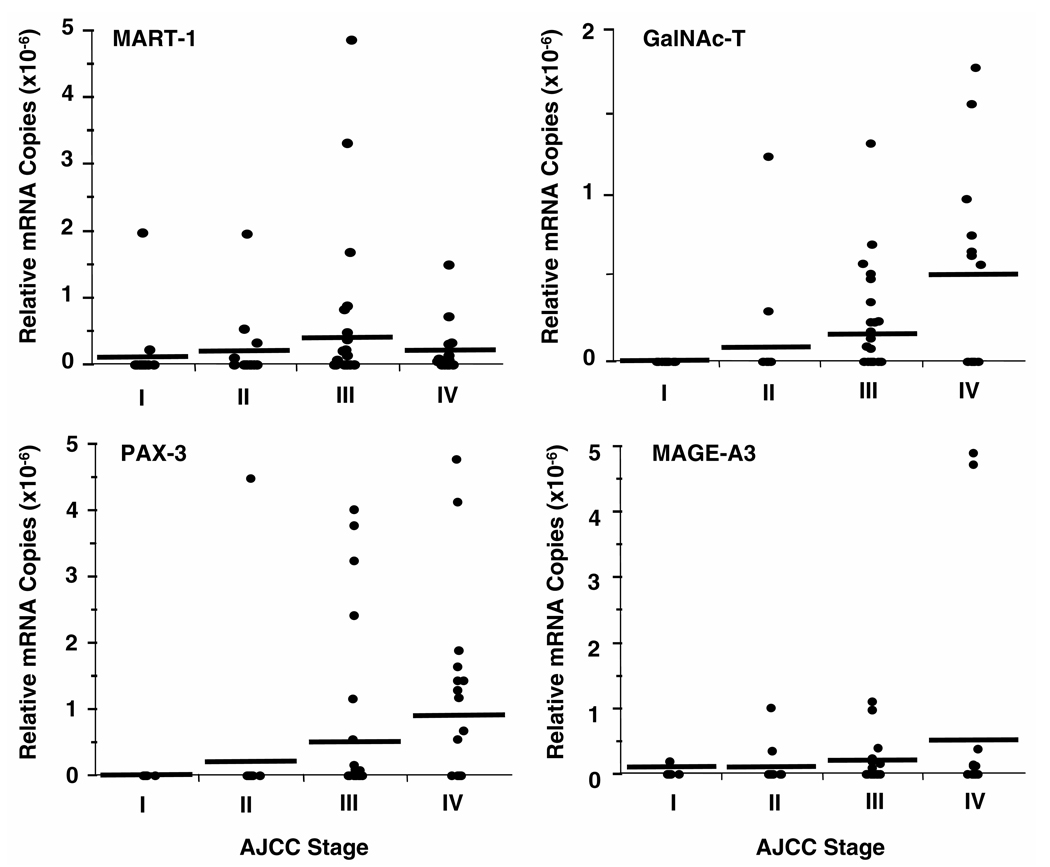
Fig. 2. qRT quantification of cancer markers in peripheral blood of melanoma patients.
Blood specimens were collected from patients with different AJCC stages of melanoma. RNA was isolated from total cells in blood, and qRT was performed. Relative mRNA copies are given according to AJCC stage. Pearson correlation analysis indicated significant correlations between AJCC stage and relative mRNA copies for all markers: MART-1, P = 0.0071; GalNAc-T, P = 0.0002; PAX-3, P =0.0001; MAGE-A3, P = 0.014. Horizontal bars indicate mean mRNA copies.
qRT DETECTION LIMIT FOR MELANOMA CELLS MIXED WITH PBLs IN VITRO
The assay detected mRNA for each marker from 1 melanoma cell mixed with 107 PBLs; mRNA copies gradually decreased on serial dilution of melanoma cells (Fig. 1). The number of mRNA copies for individual markers varied. All markers were positive at 10 melanoma cells mixed with 107 PBLs in all 10 experiments (Table 1 in the online Data Supplement). Detection frequencies of MART-1, GalNAc-T, PAX-3, and MAGE-A3 from 1 melanoma cell mixed with 107 PBLs were 90%, 80%, 50%, and 20%, respectively. These rates are equivalent to 1 melanoma cell in several milliliters of blood. When we tested dilutions of 1 melanoma cell in 108 PBLs, we detected expression of MART-1, GalNAc-T, PAX-3, and MAGE-A3 in 6, 4, 2, and 2 of 10 attempts, respectively.
Table 1.
mRNA detection in blood from melanoma patients.
| AJCC stage, n (%) | ||||||
|---|---|---|---|---|---|---|
| I | II | III | IV | Total | ||
| Marker | (n = 20) | (n = 20) | (n = 32) | (n = 22) | (n = 94) | Pa |
| MART-1 | 2 (10) | 4 (20) | 11 (34) | 11 (50) | 28 (30) | 0.003 |
| GalNAc-T | 0 (0) | 2 (10) | 13 (41) | 8 (36) | 23 (24) | 0.0006 |
| PAX-3 | 0 (0) | 1 (5) | 10 (31) | 10 (45) | 21 (22) | <0.0001 |
| MAGE-A3 | 1 (5) | 2 (10) | 7 (22) | 7 (32) | 17 (18) | 0.016 |
Comparison between AJCC stage and each marker detection by Cochran–Armitage trend test.
ASSESSMENT OF MULTIMARKER mRNA DETECTION IN BLOOD FROM MELANOMA PATIENTS
The range of relative mRNA copies (absolute mRNA copies of each marker/absolute mRNA copies of GAPDH) was 10−6 to 10−8 for each marker (Fig. 2). The relative number of mRNA copies was higher at a higher disease stage. Pearson correlation coefficient analysis results were significant between AJCC stage and relative mRNA copies for all markers: r = 0.28 (P = 0.01) for MART-1; r = 0.37 (P = 0.0002) for GalNAc-T; r = 0.28 (P <0.0001) for PAX-3; r = 0.25 (P = 0.014) for MAGE-A3. Relative mRNA copies for patients with stage III/IV disease (metastatic disease) were significantly higher (Mann–Whitney U-test) than for patients with stage I/II disease (localized disease) for all individual markers (MART-1, P = 0.01; GalNAc-T, P = 0.0002; PAX-3, P = 0.0001; MAGE-A3, P = 0.03).
Overall, in blood samples from 94 melanoma patients, MART-1, GalNAc-T, PAX-3, and MAGE-A3 were detected in 30%, 24%, 22%, and 18% of patients, respectively (Table 1), with lower detection rates of marker genes in patients with early-stage disease. The detection rate for each marker was significantly related (Cochran–Armitage trend test) to AJCC stage (MART-1, P = 0.003; GalNAc-T, P = 0.0006; PAX-3, P <0.0001; MAGE-A3, P = 0.016). We found no significant coincidence (kappa test) of marker detection between pairs of marker genes other than MART-1 and MAGE-A3 (P = 0.021).
The number of multimarkers detected in patients was also higher in those with higher AJCC stage (Spearman r = 0.58; P <0.0001; Table 2). Only 3 (15%) of 20 AJCC stage I patients had at least 1 positive marker detected, whereas 19 (86%) of 22 AJCC stage IV patients had at least 1 positive marker and 12 (55%) of 22 AJCC stage IV patients had multiple markers detected. When patients were divided into those with no or 1 positive marker detected and those with 2 or more (multiple) positive markers detected, the Cochran–Armitage trend test indicated a significant increase of patients with multiple markers detected who had advanced stages of disease (P <0.0001). The number of markers detected showed a sharp contrast in distribution between stages I/II and III/IV. In stages I/II, 15%, 7%, 0%, and 0% of patients had 1, 2, 3, and 4 markers detected, respectively. On the other hand, 35%, 28%, 15%, and 2% of patients had 1, 2, 3, and 4 markers detected, respectively, in stage III/IV. There was a significant difference in multimarker detection between AJCC stages I/II and III/IV (P <0.0001).
Table 2.
Number of markers correlated to disease stage.
| AJCC stage, n (%) | ||||||
|---|---|---|---|---|---|---|
| Markers | ||||||
| detected, | I | II | III | IV | Total | |
| n | (n = 20) | (n = 20) | (n = 32) | (n = 22) | (n = 94) | P |
| 0 | 17 (85) | 14 (70) | 8 (25) | 3 (14) | 42 (45) | <0.0001a |
| 1 | 3 (15) | 3 (15) | 12 (37) | 7 (32) | 25 (26) | |
| 2 | 0 (0) | 3 (15) | 7 (22) | 8 (36) | 18 (19) | |
| 3 | 0 (0) | 0 (0) | 5 (16) | 3 (14) | 8 (9) | |
| 4 | 0 (0) | 0 (0) | 0 (0) | 1 (5) | 1 (1) | |
| ≤1 | 20 (100) | 17 (85) | 20 (63) | 10 (45) | 67 (71) | <0.0001b |
| ≥2 | 0 (0) | 3 (15) | 12 (37) | 12 (55) | 17 (29) | |
Spearman correlation coefficient analysis showed a significant correlation between number of positive markers and AJCC stage.
Cochran–Armitage trend test showed a significant increase of patients with more than 2 positive markers detected in advanced stage of disease.
Discussion
To date, assessment of risk factors for tumor progression has been studied on primary and/or metastatic tumors, and tumor–node–metastasis staging has been used for treatment and patient management. Detection of circulating tumor cells is an attractive assessment approach because tumor cells in circulation are essential for the formation of metastatic lesion and because blood analysis can depict real-time tumor status (5, 18, 27). However, the clinical utility of molecular detection of circulating tumor cells in blood continues to be debated, mostly because of inconsistency among the previous findings, indicating the necessity for performing careful characterizations of these tests. In this study, we demonstrated the usefulness of multimarker qRT as a sensitive and specific quantitative assay to detect circulating melanoma cells in blood. In vitro models showed the reproducibility and reliability of the assay and the feasibility of clinical application. The assay detected ~1 melanoma cell in 107–108 PBLs from healthy blood donors. The number of positive markers in blood was significantly higher in patients with advanced-stage melanoma than in patients with early-stage disease.
The qRT assay has high-throughput capacity that can analyze large numbers of samples without PCR product carryover contamination. Moreover, this assay enables accurate and reproducible quantification of mRNA to compare gene expression among samples. This assay system offers an overall logistic advantage because it can detect occult metastatic tumor cells among millions of healthy leukocytes in blood without requiring cell-separation methods such as magnetic beads, separation medium, or other approaches.
The 4 mRNA markers selected in the study were frequently found in melanoma cells but not in healthy donor blood PBLs under the optimized assay conditions. Our previous studies demonstrated that metastatic melanoma tumors are heterogeneous in melanoma-associated marker expression (11, 19). Because MART-1, GM2/GD2, and MAGE-A3 have been demonstrated to be highly immunogenic in humans (28–30), cells expressing these antigens may be deleted by host immunity. However, the combination of markers in the multimarker assay can compensate for individual marker expression; thus, we expect detection of tumor cells to be increased and false-negative results reduced.
In blood specimens from patients with melanoma, the overall detection rate in blood was highest for MART-1, similar for GalNAc-T and PAX-3, and lowest for MAGEA3. Differences in cell line analysis compared with blood may be related to the physiology of cells during circulation in blood or the clonal phenotype (7). Detection rates of the multimarker qRT assay were higher than any of the individual markers alone. These findings support the supposition that a single-marker assay in blood has limited clinical utility (7, 31, 32). We did not use tyrosinase in the study because of previously reported variable detection rates and potential problems with false positives (31, 32). The detection of tumor cells in blood has often been tested directly in correlation with outcome in previously reported studies. However, such analyses must be carried out in defined patient specimens because many variables play a role in tumor metastasis (7). Blood markers can serve both diagnostic and predictive functions. In this study, predictive markers for clinical stage II patients were validated through molecular upstaging of SLNs (11, 19). Previously we have shown the significance of circulating melanoma cells in prediction of disease outcome in patients with stage III/IV melanoma (5, 6). In the present study, we quantitatively demonstrated the differences in blood from patients with AJCC stages I/II and III/IV disease. Because disease relapse in melanoma patients, particularly those with AJCC stage I, II, and III disease, requires long-term follow-up (5, 8) and is influenced by treatment interventions, we did not correlate the patient outcome and long-term clinical significance of the qRT results in this study. A prospective multicenter treatment trial of long-term outcomes is currently being performed for validation of our markers. Potentially this quantitative real-time assay of circulating cells could be useful for assessing patient disease status and guiding treatment management (4, 12).
In summary, melanoma prognosis is currently determined based on tumor and host demographic and static factors, but dynamic factors of ongoing tumor metastasis are also important. Molecular markers in blood can be a very informative indicator of systemic disease progression. Our findings suggest the potential clinical usefulness of the qRT assay for detecting circulating cells in blood of melanoma patients. Future studies involving serial blood analysis and long-term follow-up evaluation of patients may allow a more detailed assessment of the predictive ability of the qRT assay.
Acknowledgments
This study was supported in part by NIH National Cancer Institute P01 Grants CA 29605, Project II, and CA 12528, Project II, and by the Martin H. Weil Laboratory.
Footnotes
Nonstandard abbreviations: AJCC, American Joint Committee on Cancer; RT-PCR, reverse transcription-PCR; PBL, peripheral blood leukocyte; qRT, quantitative real-time reverse transcription-PCR; MART-1, melanoma antigen recognized by T cells-1; GalNAc-T, β1→4-N-acetylgalactosaminyltransferase; PAX-3, paired box homeotic gene transcription factor 3; MAGE-A3, melanoma antigen gene-A3 family; SLN, sentinel lymph node; JWCI, John Wayne Cancer Institute; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; and Ct, threshold cycle.
References
- 1.Greene FL, Fleming ID, Fritz AG, Balch CM, Haller DG, Morrow M. AJCC cancer staging manual. 6th ed. New York: Springer-Verlag; 2002. [Google Scholar]
- 2.Balch CM, Buzaid AC, Soong SJ, Atkins MB, Cascinelli N, Coit DG, et al. Final version of the American Joint Committee on Cancer staging system for cutaneous melanoma. J Clin Oncol. 2001;19:3635–3648. doi: 10.1200/JCO.2001.19.16.3635. [DOI] [PubMed] [Google Scholar]
- 3.Balch CM, Buzaid AC, Soong SJ, Atkins MB, Cascinelli N, Coit DG, et al. New TNM melanoma staging system: linking biology and natural history to clinical outcomes. Semin Surg Oncol. 2003;21:43–52. doi: 10.1002/ssu.10020. [DOI] [PubMed] [Google Scholar]
- 4.Pantel K, Cote RJ, Fodstad O. Detection and clinical importance of micrometastatic disease. J Natl Cancer Inst. 1999;91:1113–1124. doi: 10.1093/jnci/91.13.1113. [DOI] [PubMed] [Google Scholar]
- 5.Hoon DS, Bostick P, Kuo C, Okamoto T, Wang HJ, Elashoff R, et al. Molecular markers in blood as surrogate prognostic indicators of melanoma recurrence. Cancer Res. 2000;60:2253–2257. [PubMed] [Google Scholar]
- 6.Wascher RA, Morton DL, Kuo C, Elashoff RM, Wang HJ, Gerami M, et al. Molecular tumor markers in the blood: early prediction of disease outcome in melanoma patients treated with a melanoma vaccine. J Clin Oncol. 2003;21:2558–2563. doi: 10.1200/JCO.2003.06.110. [DOI] [PubMed] [Google Scholar]
- 7.Hoon DS, Wang Y, Dale PS, Conrad AJ, Schmid P, Garrison D, et al. Detection of occult melanoma cells in blood with a multiple-marker polymerase chain reaction assay. J Clin Oncol. 1995;13:2109–2116. doi: 10.1200/JCO.1995.13.8.2109. [DOI] [PubMed] [Google Scholar]
- 8.Mocellin S, Del Fiore P, Guarnieri L, Scalerta R, Foletto M, Chiarion V, et al. Molecular detection of circulating tumor cells is an independent prognostic factor in patients with high-risk cutaneous melanoma. Int J Cancer. 2004;111:741–745. doi: 10.1002/ijc.20347. [DOI] [PubMed] [Google Scholar]
- 9.Bostick PJ, Chatterjee S, Chi DD, Huynh KT, Giuliano AE, Cote R, et al. Limitations of specific reverse-transcriptase polymerase chain reaction markers in the detection of metastases in the lymph nodes and blood of breast cancer patients. J Clin Oncol. 1998;16:2632–2640. doi: 10.1200/JCO.1998.16.8.2632. [DOI] [PubMed] [Google Scholar]
- 10.Miyashiro I, Kuo C, Huynh K, Iida A, Morton D, Bilchik A, et al. Molecular strategy for detecting metastatic cancers with use of multiple tumor-specific MAGE-A genes. Clin Chem. 2001;47:505–512. [PubMed] [Google Scholar]
- 11.Takeuchi H, Morton DL, Kuo C, Turner RR, Elashoff D, Elashoff R, et al. Prognostic significance of molecular upstaging of paraffinembedded sentinel lymph nodes in melanoma patients. J Clin Oncol. 2004;22:2671–2680. doi: 10.1200/JCO.2004.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stathopoulou A, Gizi A, Perraki M, Apostolaki S, Malamos N, Mavroudis D, et al. Real-time quantification of CK-19 mRNA-positive cells in peripheral blood of breast cancer patients using the LightCycler system. Clin Cancer Res. 2003;9:5145–5151. [PubMed] [Google Scholar]
- 13.Howe JG, Crouch J, Cooper D, Smith BR. Real-time quantitative reverse transcription-PCR for cyclin D1 mRNA in blood, marrow, and tissue specimens for diagnosis of mantle cell lymphoma. Clin Chem. 2004;50:80–87. doi: 10.1373/clinchem.2003.024695. [DOI] [PubMed] [Google Scholar]
- 14.Keilholz U, Goldin-Lang P, Bechrakis NE, Max N, Letsch A, Schmittel A, et al. Quantitative detection of circulating tumor cells in cutaneous and ocular melanoma and quality assessment by real-time reverse transcriptase-polymerase chain reaction. Clin Cancer Res. 2004;10:1605–1612. doi: 10.1158/1078-0432.ccr-0610-3. [DOI] [PubMed] [Google Scholar]
- 15.Schuster R, Max N, Mann B, Heufelder K, Thilo F, Grone J, et al. Quantitative real-time RT-PCR for detection of disseminated tumor cells in peripheral blood of patients with colorectal cancer using different mRNA markers. Int J Cancer. 2004;108:219–227. doi: 10.1002/ijc.11547. [DOI] [PubMed] [Google Scholar]
- 16.Sarantou T, Chi DD, Garrison DA, Conrad AJ, Schmid P, Morton DL, et al. Melanoma-associated antigens as messenger RNA detection markers for melanoma. Cancer Res. 1997;57:1371–1376. [PubMed] [Google Scholar]
- 17.Baker MK, Mikhitarian K, Osta W, Callahan K, Hoda R, Brescia F, et al. Molecular detection of breast cancer cells in the peripheral blood of advanced-stage breast cancer patients using multimarker real-time reverse transcription-polymerase chain reaction and a novel porous barrier density gradient centrifugation technology. Clin Cancer Res. 2003;9:4865–4871. [PubMed] [Google Scholar]
- 18.O’Hara SM, Moreno JG, Zweitzig DR, Gross S, Gomella LG, Terstappen LW. Multigene reverse transcription-PCR profiling of circulating tumor cells in hormone-refractory prostate cancer. Clin Chem. 2004;50:826–835. doi: 10.1373/clinchem.2003.028563. [DOI] [PubMed] [Google Scholar]
- 19.Takeuchi H, Kuo C, Morton DL, Wang HJ, Hoon DS. Expression of differentiation melanoma-associated antigen genes is associated with favorable disease outcome in advanced-stage melanomas. Cancer Res. 2003;63:441–448. [PubMed] [Google Scholar]
- 20.Kawakami Y, Eliyahu S, Delgado CH, Robbins PF, Rivoltini L, Topalian SL, et al. Cloning of the gene coding for a shared human melanoma antigen recognized by autologous T cells infiltrating into tumor. Proc Natl Acad Sci U S A. 1994;91:3515–3519. doi: 10.1073/pnas.91.9.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsuchida T, Saxton RE, Irie RF. Gangliosides of human melanoma: GM2 and tumorigenicity. J Natl Cancer Inst. 1987;78:55–60. doi: 10.1093/jnci/78.1.55. [DOI] [PubMed] [Google Scholar]
- 22.Tsuchida T, Saxton RE, Morton DL, Irie RF. Gangliosides of human melanoma. J Natl Cancer Inst. 1987;78:45–54. doi: 10.1093/jnci/78.1.45. [DOI] [PubMed] [Google Scholar]
- 23.Kuo CT, Bostick PJ, Irie RF, Morton DL, Conrad AJ, Hoon DS. Assessment of messenger RNA of β 1→4-N-acetylgalactosaminyl-transferase as a molecular marker for metastatic melanoma. Clin Cancer Res. 1998;4:411–418. [PubMed] [Google Scholar]
- 24.Walther C, Guenet JL, Simon D, Deutsch U, Jostes B, Goulding MD, et al. Pax: a murine multigene family of paired box-containing genes. Genomics. 1991;11:424–434. doi: 10.1016/0888-7543(91)90151-4. [DOI] [PubMed] [Google Scholar]
- 25.Schulte TW, Toretsky JA, Ress E, Helman L, Neckers LM. Expression of PAX3 in Ewing's sarcoma family of tumors. Biochem Mol Med. 1997;60:121–126. doi: 10.1006/bmme.1997.2567. [DOI] [PubMed] [Google Scholar]
- 26.Scholl FA, Kamarashev J, Murmann OV, Geertsen R, Dummer R, Schafer BW. PAX3 is expressed in human melanomas and contributes to tumor cell survival. Cancer Res. 2001;61:823–826. [PubMed] [Google Scholar]
- 27.Koyanagi K, Ozawa S, Ando N, Kitagawa Y, Ueda M, Kitajima M. Clinical significance of telomerase activity in peripheral blood of patients with esophageal squamous cell carcinoma. Ann Thorac Surg. 2002;73:927–932. doi: 10.1016/s0003-4975(01)03435-x. [DOI] [PubMed] [Google Scholar]
- 28.Irie RF, Matsuki T, Morton DL. Human monoclonal antibody to ganglioside GM2 for melanoma treatment. Lancet. 1989;1:786–787. doi: 10.1016/s0140-6736(89)92606-8. [DOI] [PubMed] [Google Scholar]
- 29.Gaugler B, Van den Eynde B, van der Bruggen P, Romero P, Gaforio JJ, De Plaen E, et al. Human gene MAGE-3 codes for an antigen recognized on a melanoma by autologous cytolytic T lymphocytes. J Exp Med. 1994;179:921–930. doi: 10.1084/jem.179.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawakami Y, Eliyahu S, Sakaguchi K, Robbins PF, Rivoltini L, Yannelli JR, et al. Identification of the immunodominant peptides of the MART-1 human melanoma antigen recognized by the majority of HLA-A2-restricted tumor infiltrating lymphocytes. J Exp Med. 1994;180:347–352. doi: 10.1084/jem.180.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glaser R, Rass K, Seiter S, Hauschild A, Christophers E, Tilgen W. Detection of circulating melanoma cells by specific amplification of tyrosinase complementary DNA is not a reliable tumor marker in melanoma patients: a clinical two-center study. J Clin Oncol. 1997;15:2818–2825. doi: 10.1200/JCO.1997.15.8.2818. [DOI] [PubMed] [Google Scholar]
- 32.Jung FA, Buzaid AC, Ross MI, Woods KV, Lee JJ, Albitar M, et al. Evaluation of tyrosinase mRNA as a tumor marker in the blood of melanoma patients. J Clin Oncol. 1997;15:2826–2831. doi: 10.1200/JCO.1997.15.8.2826. [DOI] [PubMed] [Google Scholar]