Abstract
In comparison to extraarticular ligaments and tendons, the intraarticular ligaments such as the anterior and posterior cruciates exhibit different biochemical, biomechanical, and viscoelastic properties and most importantly, differential abilities to heal after surgical repair. Little is known about the underlying basis for these differences, in large measure due to the paucity of molecular markers distinguishing different classes of tendons and ligaments. To date, there has been no systematic analysis of gene expression differences between different types of connective tissues. We used Affymetrix expression arrays to analyze the differences in gene expression levels between the anterior cruciate, posterior cruciate, and medial collateral ligaments, the patellar and Achilles tendons and the synovium. We have identified five clusters of gene cohorts displaying similar expression patterns. These clusters group into three categories including: (1) genes that are strongly expressed in all connective tissues compared to the synovium control tissue; (2) genes that distinguish intraarticular connective tissues from extraarticular connective tissues; and (3) a group of genes expressed in common by the patellar tendon and the synovium. Our analysis identifies a new marker of tendons and ligaments (fibin2), demonstrates molecular diversity between subtypes of tendons and ligaments, and indicates that the primary molecular subdivision among dense regular connective tissues is intra- versus extraarticular rather than ligament versus tendon.
Keywords: intraarticular, extraarticular, ligaments, tendons, affymetrix, fibin
Tendon and ligament injuries are increasingly common. 1,2 Yet the ability of specific tendons or ligaments to heal themselves after tearing or after surgical transection varies widely. Interestingly, the location of the connective tissue appears to be more significant in this respect than the traditional ligament versus tendon classification. In particular, ligaments and tendons within the joint (intraarticular) do not heal after rupture3 or after surgical repair,4–9 whereas ligaments and tendons outside the joint environment (extraarticular) typically heal uneventfully.10–12 Although intraarticular and extraarticular ligaments reside in very different environments in situ, it has also been demonstrated that fibroblasts generated from the different sources exhibit unique proliferative abilities. This may indicate that the differential properties of intraarticular and extraarticular ligament and tendons may be at least partially inherent as opposed to entirely environmental.13–16
Although it is clear that there is a diversity in the properties of cells of different tendons and ligaments, to date there have been no molecular markers to distinguish between them. In contrast, recent reports have described genes expressed in these connective tissues as a group. One of the most specific markers of adult tendons and ligaments, Scleraxis (Scx), is expressed in all tendon and ligament progenitors from the earliest stage of cellular commitment to adulthood.17–19 Thus, the expression of Scx and of other recently described molecular markers of tendons and ligaments cannot distinguish, either developmentally or functionally, between ligaments and tendons, or between intra- and extraarticular ligaments. Whether this is due to an actual lack of developmental subdivision is currently unclear.
We hypothesized that there would be significant differences between the gene expression of the tendon and ligament cells, as well as between intraarticular and extraarticular tissues. Identification of differences in gene expression among different connective tissues is important for understanding the effect of the different environments on the tissues within them and for understanding the regulation of their distinct properties. To discover potential gene expression differences between different connective tissues and to better understand subdivisions of connective tissues, we carried out an Affymetrix based genome-wide gene expression analysis on adult intraarticular and extraarticular ligaments and tendons. To further characterize the expression of genes, we then performed in situ hybridization analyses on selected genes of interest identified in the gene expression analysis.
MATERIALS AND METHODS
Hybridization of Affymetrix Array
Adult porcine tissue was obtained after euthanasia from animals undergoing other IACUC approved studies. All tissues were dissected immediately after euthanasia and flash frozen in liquid nitrogen. Tissues studied included the anterior cruciate ligament (ACL), posterior cruciate ligament (PCL), medial collateral ligament (MCL), Achilles tendon (AT), patellar tendon (PT), and synovium (Syn). Tissues were cut into smaller pieces with a fresh razor blade and homogenized in Trizol (acidified phenol), using a polytron mixer. Total RNA was purified from the Trizol mixture and cleaned over a Qiagen RNeasy column. RNA integrity and concentration was analyzed on an Agilent 2100 bioanalyzer. Double-stranded cDNA was synthesized from the total RNA and biotin-labeled cRNA was produced by in vitro transcription from the cDNA pool. The cRNA probe was fragmented, spiked with array controls, and hybridized to an Affymetrix Porcine Genome Array (#900623), for 16 h. This array contains 23,937 probe sets recognizing approximately 23,256 transcripts from 20,201 porcine genes. The arrays were washed using an automated Affymetrix GeneChip 400 Fluidics Station and scanned twice on an HP/Affymetrix GeneArray Chip Scanner.
Data Analysis
Raw (*.CEL) files were normalized using the software ArrayAssist lite running the GC-Robust multiarray average (RMA) algorithm20 (using RMA or PLIER normalization algorithms gave similar results). Normalized data sets were compared to each other using the GeneSpring GX program. In our primary analysis, clusters were identified by plotting the ratio of the average normalized expression value for each gene in an individual tissue to the average expression value of the same gene across all tissues. Thus, a plot above 1.0 indicates that a gene is expressed higher than average in the particular tissue. From this plot, clusters were discovered by searching for gene expressions that track with specific peaks or valleys in different samples. Every gene identified in this manner was later individually analyzed to assure against inappropriate assignment to a particular cluster. All information on specific Affymetrix probe sets (annotated ‘‘Ssc.# # ##. 1.A1_at’’), including target sequence, oligonucleotide probes, predicted target genes, and Blast homologies, can be found at the online Affymetrix Netaffx Analysis Center (http://www.affymetrix.com/analysis/index.affx). Our GC-RMA normalized data set is contained in Supplemental data 1.
Because these clusters describe transcript expression levels throughout the entire genome, they can be used to describe the relatedness of specific tissues. For example, two blindly analyzed samples that showed identical genome-wide transcript profiles would be considered identical tissues. A third tissue that was highly similar but exhibited a few transcript expression differences would be considered very highly related. Additionally, two very different tissues (e.g., skeletal muscle vs. cerebellum), would have vastly different expression profiles. By adding smooth muscle and hippocampal tissues to the analysis, we would generate gene expression clusters linking the two neural tissues (containing neural specific genes), and the two muscle tissues. Thus, a collection of tissue samples can be compared among themselves by analyzing gene expression clusters and comparing the number of genes whose expression links them together.
Generation of Chick In Situ Hybridization Probes
To further characterize the expression of genes in all of our clusters we performed in situ hybridization analyses. Because it is impractical to perform in situ hybridization on porcine tissue, we limited our secondary analysis to expression in the embryonic chick. Our rational for using chick tissue is threefold. First, we can obtain large numbers of staged embryos that are easily probed for gene expression. Second, there is a large collection of chicken cDNA sequences allowing us to obtain DNA constructs suitable for generating in situ probes without having to screen libraries for individual genes. Finally, using the chick instead of a more closely related mammalian organism allows us to identify genes within these clusters whose expression in tendons and ligaments is highly conserved between divergent species and thus is more likely to represent a functional expression pattern rather than simply promiscuous and benign transcription.
To generate probes suitable for expression analysis in the chick, the target porcine sequence from the Affymetrix array (identified through the Netaffx analysis center) was used to search the NCBI genebank server using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/BLAST/). The identified closest chick relative of the target porcine gene was then used to identify Expressed Sequence Tag (EST) sequences in the chick EST server (http://www.chick.manchester.ac.uk/).
In Situ Hybridizations
Chick EST in situ probe templates were generated by PCR amplification of the insert using M13 forward and M13 reverse primers flanking the vector inserts [primer sequences are (M13forward: 5′-GTA AAA CGA CGG CCAG-3′; M13reverse: 5′-AAC AGC TAT GAC CATG-3′)]. Antisense digoxigenin labeled cRNA riboprobes were generated using T3 RNA polymerase in the presence of ribonucleotides including digoxigenin-Uridine. Fertilized White Leghorn chick eggs (ordered from Charles River Spafas, N. Franklin, CT) were incubated in a 37°C humidified chamber until reaching the desired stage of development. Adult chicken tissue was harvested fresh from a 1-year-old Rhode Island Red rooster (Mayflower Poultry Company, Cambridge, MA). In situ hybridizations were performed as described in our lab protocols (http://genetics.med.harvard.edu/~cepko/protocol/index.html), with the exception of Proteinase K treatment in whole mount specimens where PK concentration was varied based on the age of the embryo.
RESULTS
Genomic Analysis of Different Connective Tissues
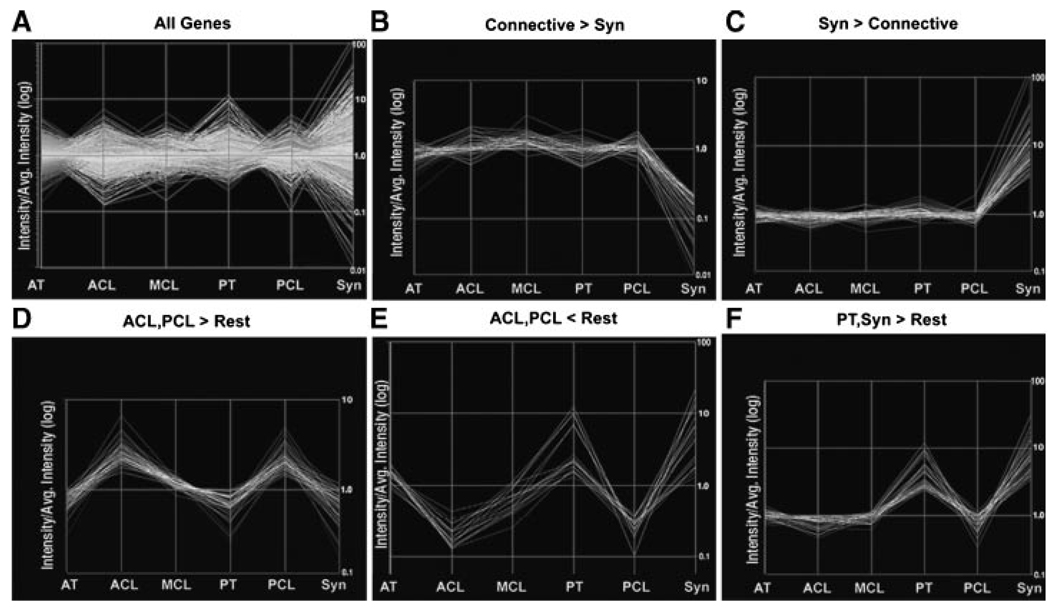
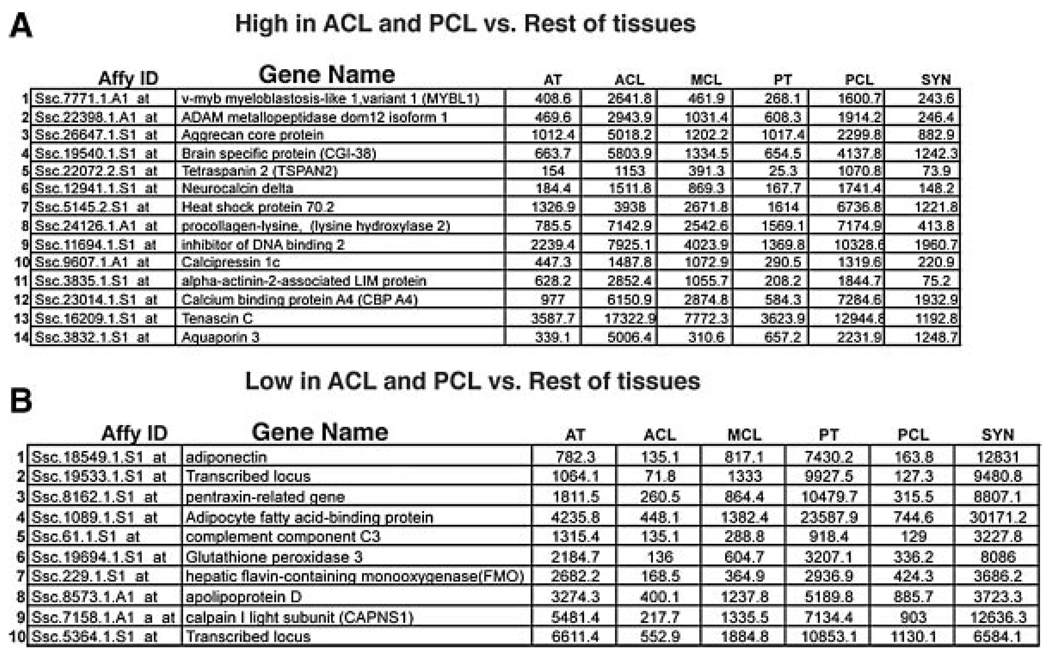
Our comparison identified five unique clusters of genes whose expression patterns were similar to each other across the six tissues (Fig. 1). We discarded as background those genes whose expression was uniformly high (i.e., “housekeeping” genes), or low (nonexpressed genes), and genes that were anomalously assigned to clusters due to chance variation of weak signals below the noise threshold. We defined a cluster as a group of five or more genes with similar relative expression changes across the six tissues. Clusters identified contained: genes that were expressed at a higher level in all connective tissues compared to the synovium (Connective>Syn); genes that were expressed at a higher level in the synovium compared to all of the connective tissues (Syn>Connective); genes that were expressed at a higher level in the ACL and PCL compared to all other tissues examined (ACL, PCL>Rest); genes that were expressed at a lower level in ACL and PCL compared to the other tissues (ACL, PCL<Rest); and genes that were expressed at a higher level in the PT and synovium compared to all the other tissues (PT, Syn>Rest).
Figure 1.
Cluster analysis of gene expression among different connective tissues. Relative values of gene expression in the different panels are plotted on a log scale as the ratio of expression in the indicated tissue to the average expression across all tissues. Thus, two genes in the same cluster may track together with respect to their fold expression changes across tissues but can still vary widely in their absolute expression levels. Tissues plotted are the Achilles tendon (AT), anterior cruciate ligament (ACL), medial collateral ligament (MCL), patellar tendon (PT), posterior cruciate ligament (PCL), and the Synovium of the knee capsule (Syn). The “All Genes” plot demonstrates the ratio of means for all genes analyzed across the six tissues.
Our clusters were then assigned to one of three functional groups. One group contained two separate clusters of genes whose expression patterns distinguished the connective tissues from the control tissue (Connective>Syn, and Syn>Connective). In these two clusters gene expression levels varied up to 100-fold compared to the average expression levels in all tissues (Fig. 1B and C). The second group contained clusters that distinguished intraarticular connective tissues from extraarticular connective tissues (ACL, PCL>Rest, and ACL, PCL<Rest; Fig. 1D and E). Genes in these clusters demonstrated expression differences on the order of 10-fold between different tissues. A third group, represented by a single cluster, indicated an epigenetic relationship between the patellar tendon and the synovium (PT, Syn > Rest; Fig. 1F). Interestingly, there were no definitive clusters distinguishing tendons from ligaments, which would have been indicated by genes differentially expressed in ACL, PCL, and MCL compared to PT and AT. However, we do recognize that within the “ACL, PCL > Rest” and “ACL, PCL < Rest” clusters, the average gene expression levels in the extraarticular ligament (MCL) were no closer to their levels in the two extraarticular tendons (PT and AT) than to the their corresponding levels in the intraarticular ACL or PCL tissues. These genes generally displayed expression levels in the MCL midway between their levels in the interarticular ligaments and the extraarticular tendons (Fig. 1D and E).
Clusters of Connective Tissue Gene Expression Cohorts
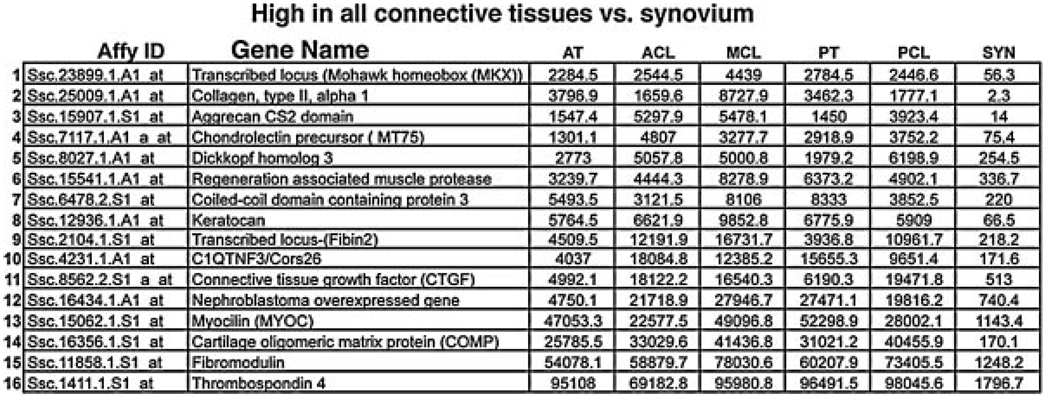
The first cluster we analyzed was one containing genes that were highly expressed in all of the connective tissues (ACL, PCL, MCL, and AT) but not expressed (or expressed at low levels), in the Synovium (Fig. 2). In our normalized data we have empirically determined that expression values below 100 are undetectable, values at 500 are weak, values at 1000 are easily detectable, and values above 5000 are very strong. These genes represent potential tendon and ligament markers. We did, in fact, observe several genes in this cluster that have already been identified as being highly expressed in adult tendons and ligaments including Aggrecan, Connective Tissue Growth Factor (CTGF), Cartilage Oligomeric Matrix Protein (COMP), Fibromodulin, and Thrombospondin 4.21–25 The tendon and ligament markers Scleraxis (Scx) and Tenomodulin (TeM), are not represented in our gene arrays. We were, however, able to determine that probe set I.D. “Ssc.23899.1.A1_at” represents the porcine homolog of the recently described tendon marker Mohawk (Mkx). Mkx is a TALE class homeobox gene, belonging to the Iroquois subfamily of homeobox genes, that has already been described as being expressed in the syndetomal tendon progenitors and its expression persists in later embryonic tendons.26 Our analysis indicates that it remains highly expressed even in adult connective tissue. Because the probe sets represented on this array are developed from Expressed Sequence Tag (EST), libraries, many of them represent 3′ untranslated regions (UTRs) of the targeted porcine genes. Because 3′ UTR sequences diverge rapidly, these sequences are often insufficient to identify the homologous genes in other organisms. Thus, it is possible that some of the other genes identified only as “transcribed locus” on the Affymetrix array represent porcine 3′ UTR sequences for Scx, TeM, or other unidentified genes, and simply cannot be unambiguously identified due to sequence divergence.
Figure 2.
Genesdifferentiallyexpressed between connective tissues and the control synovium. The table lists all genes in the Connective > Syn cluster from Figure 1B. Probe set IDs in the leftmost column can be used to query the Affymetrix data base to retrieve all information about a specific probe set as described in Materials and Methods. Values listed represent the expression levels of the individual genes in each tissue and are normalized from the raw hybridization data using the GC-RMA algorithm.
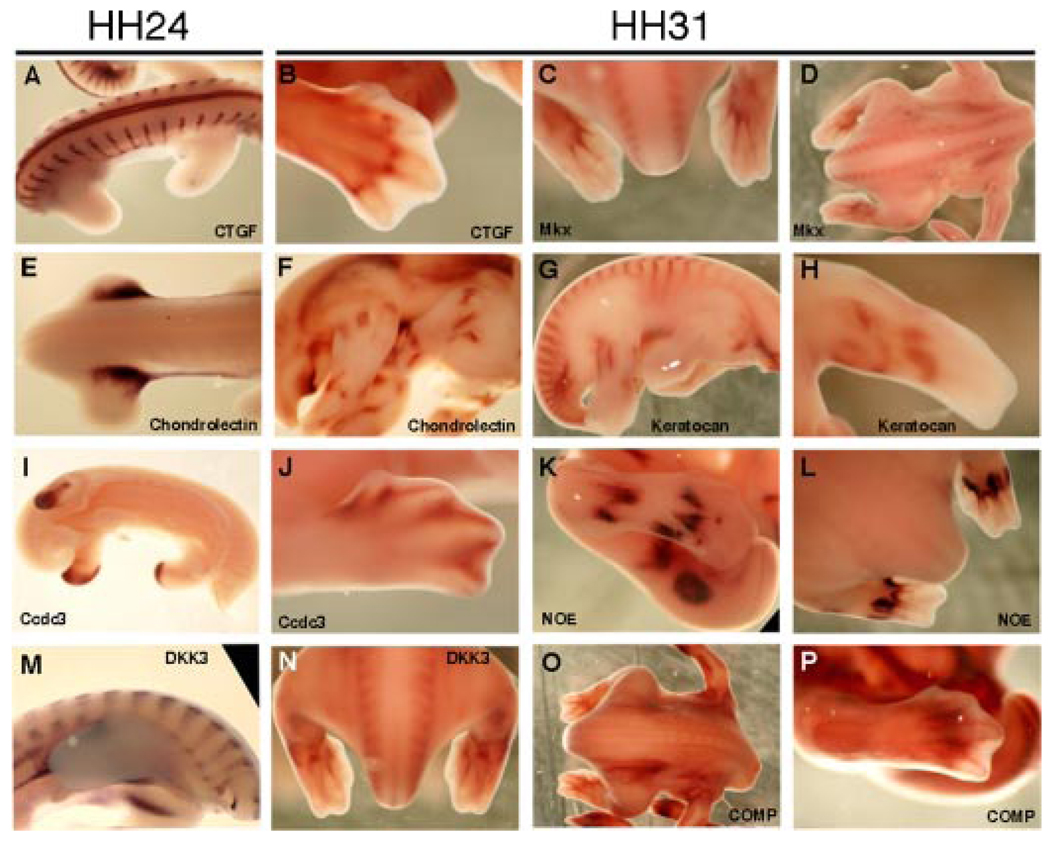
We have also identified in this cluster known genes that have not been previously associated with ligament or tendon development or physiology including Dick-kopf-3 (DKK3), coiled-coil domain protein 3 (Ccdc3), tumor necrosis factor-3 (C1Q/TNF3/Cors26), keratocan, and Nephroblastoma overexpressed (NOE) gene. To support this finding, we performed in situ hybridization to analyze the expression of genes in this cluster. In these analyses, we found that factors known to be expressed in developing tendons and ligaments, specifically Mkx,26 CTGF,22 and COMP23 and the new genes that have not been previously associated with ligament or tendon development or physiology (specifically Keratocan, Coiled- coil domain protein 3, Nephroblastoma overexpressed gene, DKK3, and Chondrolectin) all demonstrate dynamic and spatially restricted expression patterns in the chick embryo at Hamburger-Hamilton stages 24 and 31 (HH24 and HH31),27 of development (Fig. 3) with similarities between the previously identified genes associated with tendon development and the new genes identified in this analysis. For example, CTGF and DKK3 show repetitive expression patterns in the early developing somites that contain cells fated to become mature spinal tendons (as well as precursors to skeletal and muscle cells). Mkx, COMP, Keratocan, and DKK3 all show staining in the lateral aspects of the developing spine, which is reminiscent of Scx staining in the developing spinal tendons (Fig. 3).18,19 Other factors show unique expression patterns in the developing limbs including Mkx, and Dkk3 in the ventral hind limb tendons, and NOE in structures surrounding and outlining developing tendons in the hand plate, and CTGF in specific joints of the autopod.
Figure 3.
In situ hybridization analysis of factors from the Connective > Syn cluster. Darker regions of embryos label regions of specific gene upregulation. Embryos shown are either at Hamburger Hamilton (HH; ref. 27) stage HH24(A, E, I, M), orHH31(B–D,F–H, J–L,N–P). Specific factors probed for are indicated on each panel. Orientation of embryos is as follows: (C, L, N) dorsal posterior view with the head toward the top of the frame; (A, D, E, O), dorsal views with heads to right; (B, J, K, P) closeups of dorsal right hindlimbs with the distal limb to the right; (F, G, I, M) right flank views with head to the right; (H) anterior–ventral view of left forelimb. CTGF is expressed in the developing somites (A), and in the forming joints of the autopod (B). Mkx is expressed in the ventral tendons of the hindlimbs (C), and in tissues of the developing spine (D). Chondrolectin is highly expressed in the proximal girdle of the developing limb bud (E), and at HH31 at unique regions of the developing skeleton (F). Keratocan is expressed in developing somites and in regions of the dorsal hindlimb (G), and in regions of the ventral forelimb (H). Ccdc3 is expressed in the distal tip of the HH24 limb buds (I), and surrounding the condensing digits of the HH31 autopod (J).NOE is expressed in developing limb regions surrounding primary tendon condensation sites (K—dorsal, L—ventral hindlimb). DKK3is expressed in the early somites (M), and atHH31in the developing spine and ventral tendons of the hindlimb (N).COMP is expressed in regions surrounding mesenchymal condensations in the developing spine (O) and autopod (P).
Identification of a Novel Connective Tissue Marker
We have seen that one of the “transcribed locus” genes within the “Connective > Syn” cluster (Ssc.2104.1.S1_at), represents a unique gene that is highly conserved across many species and, like Scx, TeM, and Mkx, is a very early marker of connective tissue differentiation 17–19,26,28–30 The protein product from this locus has no identifiable conserved domains but is similar to the recently described fibin gene in Danio rerio (full sequence and species alignment in Supplemental Data 2). In D. rerio, fibin28 is described as a novel secreted factor that is required for fin bud outgrowth and initiation of T-Box transcription factor-5 (Tbx5) expression and does not seem to be expressed in developing or mature tendons or ligaments. Because our newly identified gene is very strongly expressed in developing and mature connective tissues in the chick (Fig. 4) and in the pig (Figs. 1 and 2), and because of the zebrafish genomic duplication, we do not assume that it is a direct ortholog of the described zebrafish fibin gene. We thus, refer to this gene as fibin2 (fbn2). Like zebrafish Fbn, Fbn2 is predicted to contain a signal peptide based on analysis using PSORT (www.psort.org), indicating that it encodes a secreted protein.
Figure 4.
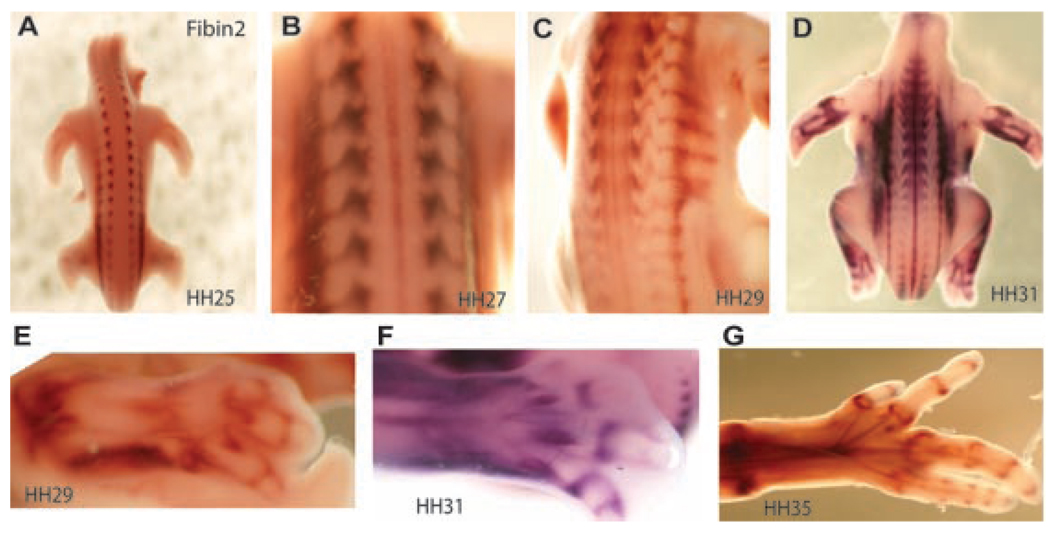
Embryonic expression patterns of Fbn2, a novel gene expressed in developing tendon progenitors. In situ hybridization of Fbn2 (A–G) was performed in chicken embryos at stage of development indicated.27 Expression is indicated by dark brown or purple staining. All panels are imaged from the dorsal side of the embryo (A–D) or limb (E–G) with anterior toward the top. Fbn2 is expressed from the early stages of syndetome specification in the developing somites (A; and 19), and is maintained through the development of axial connective tissue (D). Fbn2 is dynamically expressed in the developing limb in a pattern consistent with developing and mature connective tissues (E–G).
In situ hybridization of Fbn2 (Fig. 4) demonstrated that it is expressed from the earliest stages of tendon development in the syndetome of the developing somites (Fig. 4A).19 As development proceeds, Fbn2 continues to be expressed in a tendon specific manner in both the developing axial tendons (Fig. 4B–D) and the limb tendons and joint capsules (Fig. 4E–G). Thus, Fbn2 is dynamically expressed in the developing limb in a pattern consistent with developing and mature connective tissues (Fig. 4A–G).
Clusters of Gene Expression Cohorts Defining Subsets of Connective Tissues
The next clear distinction among the tissues analyzed was between intraarticular and extraarticular connective tissues. The two intraarticular connective tissues analyzed (ACL, and PCL), grouped together in two separate clusters (ACL, PCL > Rest and ACL, PCL < Rest). These clusters contain candidate genes for the mediation of properties specific to intra- versus extraarticular connective tissues. Among the genes identified here, some are well-established connective tissue genes (Tenascin C), whereas others (including Neurocalcin delta, Aquaporin 3, Adiponectin, and the Pentraxin-related gene), represent genes that have not been described as playing a role in connective tissue function (Fig. 5). Interestingly, although we did not see clusters (five or more), of genes whose expression patterns distinguished tendons from ligaments, we did see a few individual genes within clusters that were generally expressed lower or higher in ligaments compared to tendons. These included Complement component C3, Glutathione peroxidase 3, and Flavin Containing mono-oxygenase 1 (FMO1), which all showed lower expression in ligaments compared to tendons (Fig. 5B). Additionally, Neurocalcin delta, and αActinin-2-associated LIM protein both showed somewhat higher expression levels in ligaments compared to tendons (Fig. 5A). We also did not see a cluster of genes that grouped all intraarticular tissues (ACL, PCL, and synovium) from all extraarticular tissues (MCL, AT, and PT). Again, however, we did see a single gene (Aquaporin 3; Fig. 5A), whose expression was consistently higher in ACL, PCL, and Synovium compared to MCL, AT, and PT.
Figure 5.
Genes differentially expressed between intraarticular and extraarticular connective tissues. The table lists all genes in the (A) ACL, PCL > Rest and (B) ACL, PCL < Rest clusters from Figure1D and E. Values listed represent the normalized expression levels of the individual genes within each tissue.
In these clusters that divide the connective tissues into subpopulations we saw that gene expression patterns varied by degrees. Although some of the variations are striking, none demonstrate absolute differential expression.
Additional Clusters
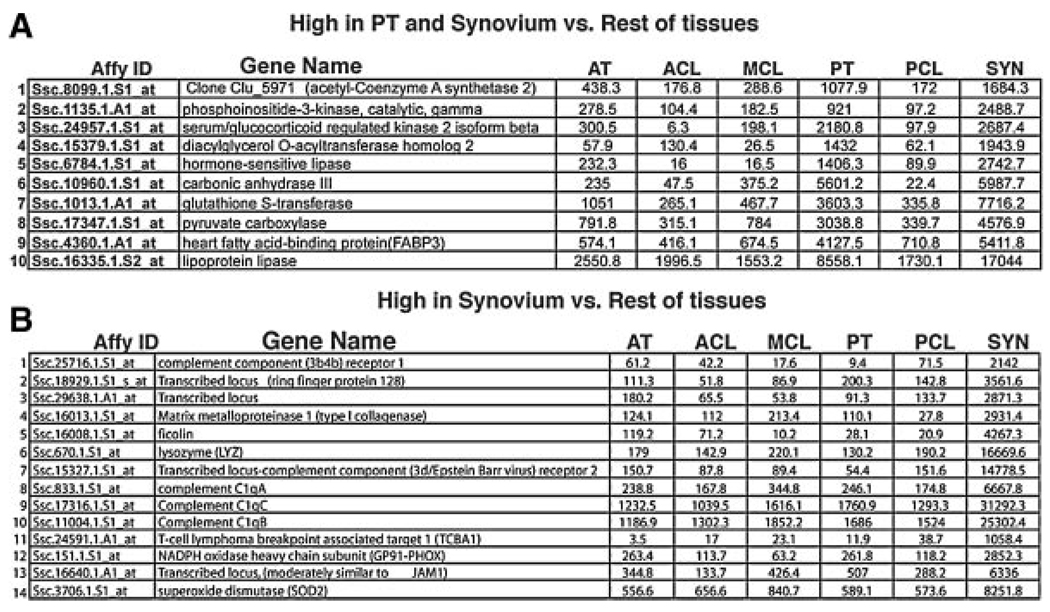
We also see an unexpected cluster that connects, on a gene expression level, patellar tendon with the intra-articular synovium in the knee (Fig. 6A). Although these tissues are widely different from each other, the connection on the molecular level in this analysis is rather strong. In particular genes like Carbonic anhydrase III and Diacylglycerol o-acyltransferase homolog 2, are both expressed strongly in PT and Syn but show little or no expression in the other tissues analyzed.
Figure 6.
Clusters of genes that (A) connect the patellar tendon with the synovium of the knee and (B) are highly expressed in the Synovium compared to the connective tissues analyzed. The table lists all genes in the (A) PT, Syn > Rest and (B) Syn > Connective clusters from Figure 1F and C, respectively. Values listed represent the normalized expression levels of the individual genes within each tissue.
Finally, we see a cluster of genes that are more highly expressed in the synovium compared to the connective tissues analyzed (Fig. 6B). In this case, genes belonging to the cluster could be factors that are important for the function of the developing and or mature Synovium, or genes whose expression is detrimental for the development or function of connective tissues. All of the genes from this cluster displayed very low to no expression when tested in chick embryos by in situ hybridization (data not shown). This is not surprising, because the synovium does not develop until stages later than we analyzed but indicates that these genes are not globally expressed during early development.
DISCUSSION
In this work we have set out to better clarify the epigenetic differences between connective tissues using a genomic expression analysis of ACL, PCL, MCL, AT, PT, and synovium, with our underlying hypothesis being that significant differences would exist specifically between tendon and ligament. Our analysis uncovered several genes that are of potential interest in understanding the development and physiology of connective tissues. In particular, we have identified a unique transcript (fibin2), that exhibits very strong and restricted expression in developing and mature tendons and ligaments. Faithful molecular markers of tendons and ligaments have only recently been identified starting with Scleraxis and Tendin/Tenomodulin in 2001,18,29,30 followed by Mohawk in 2006.26 The identification of molecular markers of tendons and ligaments, particularly of Scleraxis, which is the most faithful to date, has spurred rapid progress and interest in the molecular development of these tissues. Molecular visualization has allowed researchers to track the commitment and fate of tendon progenitors,17,19 describe the effect of systemic mutations on tendon development, and to track the ability of stem cells to develop into tendon precursors in vitro (Pearse et al. unpublished data). The identification of fibin2 provides both an independent marker gene for analysis of tendon and ligament precursors and, as a putative secreted signal, is a candidate gene for cooperative specification and maintenance of tendon and ligament tissues.
Our analysis also highlights several known genes that have not previously been linked to tendon and ligament function. Specifically DKK3, a family member of a group of Wnt inhibitory proteins, Nephroblastoma Overexpressed Gene (NOE), Ccdc3, and Keratocan, were highly expressed in connective tissues relative to control tissues. These particular genes also demonstrated spatially restricted expression in the developing chick implicating a role for them during embryogenesis (Fig. 3). It is, in fact, interesting that most of the genes within this cluster are expressed in patterns that are individually unique from each other but that are all spatially related to aspects of the development of connective tissues, bones and muscles. These expression patterns are compelling because tendons and ligaments will ultimately bind together the complexity of the musculoskeletal system in the adult.
We also saw expression level differences between intraarticular and extraarticular connective tissues in 24 different genes. In each of these cases the observed difference was a matter of level of expression rather than of absolute expression. Our studies, in fact, were unable to identify a single gene demonstrating absolute “on or off” expression differences between subtypes of connective tissues regardless of the genome-wide coverage of the expression array used in our analysis. There could be multiple reasons for this. It is possible that there simply are no factors that are absolutely restricted to a specific subtype of connective tissue and that the differential properties that they exhibit are due to the more subtle alterations we see in this analysis. There could still be embryonic factors that are sufficient to specify subtypes of connective tissues but from our analysis of the adult tissue, the resulting differences largely specify an adjustment up or down in levels of specific cohorts of connective tissue factors.
The analysis of these expression patterns indicates, from a broader perspective, that there is a closer molecular relationship between extraarticular ligaments and tendons than there is between intraarticular ligaments and extraarticular ligaments. That is, from a molecular standpoint, the major subdivision of dense regular connective tissues is not tendon versus ligament, it is intra- versus extraarticular. This conclusion is based on the observation that, among the five different connective tissues, the genome-wide expression profiles of the ACL and PCL tissues were linked to each other (but not to the MCL), by two clusters, containing 24 transcripts, whereas there are only five genes (discussed above), whose expression pattern distinguishes tendons and ligaments. These differentially expressed genes, which are all listed in Figure 5A and B, are candidates for effectors of the differential biology of intra- and extraarticular connective tissues. Our observations are consistent with recent work demonstrating that intra- and extraarticular connective tissues undergo very different developmental pathways. Specifically, all intraarticular tissues, including ligaments, cartilage, and synovial lining, are descended from the same GDF5 expressing progenitor cells while extraarticular connective tissues are not.31 Additionally, analysis of mice containing null mutations in both the Sox5 and Sox6 genes demonstrated a lack of intraarticular ligaments but normal development of extraarticular tendons and ligaments (V. Lefebvre personal communication), underscoring the unique developmental requirements of intra- and extraarticular connective tissues. It is worth reiterating, however, that all of the gene cohorts we have analyzed (while tracking together) show a spectrum of expression levels across the tissues analyzed. Thus, there is a gradient of expression levels of these genes from intraarticular ligaments, to MCL, to extraarticular tendons.
This study also found that the gene for collagen Type II alpha I (COL2A1) was expressed by tendon and ligament cells. COL2A1 expression is typically associated with chondrocyte function, but the finding here in fibroblasts is consistent with prior work showing type II collagen production by rat renal fibroblasts,32 as well as chick embryo fibroblasts33 and tenocytes.34 Similarly, the matrix metalloproteinase I (MMP-1) expression noted in the synovium is consistent with prior studies of rat synovial fibroblasts where MMP-1 is noted particularly in the lining layer of the synovium,35 and has been shown to be regulated by IL-1b36,37 and GG2-1.38
One of the weaknesses in this study was the small number of tissues examined. This makes it difficult to determine whether the appropriate distinction is between intra- and extraarticular tissues or intra- and extrasynovial tissues. Future studies including analyses of additional tissues (e.g., the extraarticular flexor tendons and additional extraarticular ligaments) are planned to follow up and expand on the initial findings reported here.
In summary, we have identified the molecular signatures of a set of diverse connective tissues. These data have helped to identify at least one novel transcript, Fbn2, that is expressed very specifically in developing and mature tendons and ligaments. They also demonstrate a stronger molecular connection between tendons and extraarticular ligaments than between intra and extraarticular connective tissues.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a grant from the National Institutes of Health (C.J.T. and NIAMS R01 AR054099 and K02 AR049346 to M.M.M.).
REFERENCES
- 1.Adirim TA, Cheng TL. Overview of injuries in the young athlete. Sports Med. 2003;33:75–81. doi: 10.2165/00007256-200333010-00006. [DOI] [PubMed] [Google Scholar]
- 2.Burt CW, Overpeck MD. Emergency visits for sports-related injuries. Ann Emerg Med. 2001;37:301–306. doi: 10.1067/mem.2001.111707. [DOI] [PubMed] [Google Scholar]
- 3.Murray MM, Martin SD, Martin TL, et al. Histological changes in the human anterior cruciate ligament after rupture. J Bone Joint Surg Am. 2000;82-A:1387–1397. doi: 10.2106/00004623-200010000-00004. [DOI] [PubMed] [Google Scholar]
- 4.Feagin JA, Jr, Curl WW. Isolated tear of the anterior cruciate ligament: 5-year follow-up study. Am J Sports Med. 1976;4:95–100. doi: 10.1177/036354657600400301. [DOI] [PubMed] [Google Scholar]
- 5.Kaplan N, Wickiewicz TL, Warren RF. Primary surgical treatment of anterior cruciate ligament ruptures: a long-term follow-up study. Am J Sports Med. 1990;18:354–358. doi: 10.1177/036354659001800404. [DOI] [PubMed] [Google Scholar]
- 6.Sherman MF, Lieber L, Bonamo JR, et al. The long-term follow-up of primary anterior cruciate ligament repair. Defining a rationale for augmentation. Am J Sports Med. 1991;19:243–255. doi: 10.1177/036354659101900307. [DOI] [PubMed] [Google Scholar]
- 7.Casteleyn PP. Management of anterior cruciate ligament lesions: surgical fashion, personal whim or scientific evidence? Study of medium- and long-term results. Acta Orthop Belg. 1999;65:327–339. [PubMed] [Google Scholar]
- 8.Sandberg R, Balkfors B, Nilsson B, et al. Operative versus non-operative treatment of recent injuries to the ligaments of the knee. A. prospective randomized study. J Bone Joint Surg Am. 1987;69:1120–1126. [PubMed] [Google Scholar]
- 9.Galatz LM, Ball CM, Teefey SA, et al. The outcome and repair integrity of completely arthroscopically repaired large and massive rotator cuff tears. J Bone Joint Surg. 2004;86A:219–224. doi: 10.2106/00004623-200402000-00002. [DOI] [PubMed] [Google Scholar]
- 10.Petermann J, von Garrel T, Gotzen L. Non-operative treatment of acute medial collateral ligament lesions of the knee joint. Knee Surg Sports Traumatol Arthrosc. 1993;1:93–96. doi: 10.1007/BF01565459. [DOI] [PubMed] [Google Scholar]
- 11.Reider B, Sathy MR, Talkington J, et al. Treatment of isolated medial collateral ligament injuries in athletes with early functional rehabilitation. A. five-year follow-up study. Am J Sports Med. 1994;22:420–447. doi: 10.1177/036354659402200406. [DOI] [PubMed] [Google Scholar]
- 12.Lundberg M, Messner K. Long-term prognosis of isolated partial medial collateral ligament ruptures. A. ten-year clinical and radiographic evaluation of a prospectively observed group of patients. Am J Sports Med. 1996;24:160–163. doi: 10.1177/036354659602400207. [DOI] [PubMed] [Google Scholar]
- 13.Ross SM, Joshi R, Frank CB. Establishment and comparison of fibroblast cell lines from the medial collateral and anterior cruciate ligaments of the rabbit. In Vitro Cell Dev Biol. 1990;26:579–584. doi: 10.1007/BF02624206. [DOI] [PubMed] [Google Scholar]
- 14.Nagineni CN, Amiel D, Green MH, et al. Characterization of the intrinsic properties of the anterior cruciate and medial collateral ligament cells: an in vitro cell culture study. J Orthop Res. 1992;10:465–475. doi: 10.1002/jor.1100100402. [DOI] [PubMed] [Google Scholar]
- 15.Murphy PG, Hart DA. Influence of exogenous growth factors on the expression of plasminogen activators and plasminogen activator inhibitors by cells isolated from normal and healing rabbit ligaments. J Orthop Res. 1994;12:564–575. doi: 10.1002/jor.1100120413. [DOI] [PubMed] [Google Scholar]
- 16.Spindler KP, Clark SW, Nanney LB, et al. Expression of collagen and matrix metalloproteinases in ruptured human anterior cruciate ligament: an in situ hybridization study. J Orthop Res. 1996;14:857–861. doi: 10.1002/jor.1100140603. [DOI] [PubMed] [Google Scholar]
- 17.Pryce BA, Brent AE, Murchison ND, et al. Generation of transgenic tendon reporters, ScxGFP and Scx AP, using regulatory elements of the Scleraxis gene. Dev Dyn. 2007;236:1677–1682. doi: 10.1002/dvdy.21179. [DOI] [PubMed] [Google Scholar]
- 18.Schweitzer R, Chyung JH, Murtaugh LC, et al. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development. 2001;128:3855–3866. doi: 10.1242/dev.128.19.3855. [DOI] [PubMed] [Google Scholar]
- 19.Brent AE, Schweitzer R, Tabin CJ. A somitic compartment of tendon progenitors. Cell. 2003;113:235–248. doi: 10.1016/s0092-8674(03)00268-x. [DOI] [PubMed] [Google Scholar]
- 20.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 21.Corps AN, Robinson AHN, Movin T, et al. Increased expression of aggrecan and biglycan mRNA in Achilles tendinopathy. Rheumatology. 2005;45:291–294. doi: 10.1093/rheumatology/kei152. [DOI] [PubMed] [Google Scholar]
- 22.Halper J, Griffin A, Hu W, et al. In vitro culture decreases the expression of TGFβ, Hsp47 and type I procollagen and increases the expression of CTGF in avian tendon explants. J Musculoskelet Neuronal Interact. 2005;5:53–63. [PubMed] [Google Scholar]
- 23.DiCesare P, Hauser N, Lehman D, et al. Cartilage oligomeric matrix protein (COMP) is an abundant component of tendon. FEBS Lett. 1994;354:237–240. doi: 10.1016/0014-5793(94)01134-6. [DOI] [PubMed] [Google Scholar]
- 24.Oldberg A, Antonsson P, Lindblom K, et al. A collagen-binding 59-kd protein (fibromodulin) is structurally related to the small interstitial proteoglycans PG-S1 and PG-S2 (decorin) EMBO J. 1989;8–9:2601–2604. doi: 10.1002/j.1460-2075.1989.tb08399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hauser N, Paulsson M, Kale AA, et al. Tendon extracellular matrix contains pentameric thrombospondin-4 (TSP-4) FEBS Lett. 1995;368:307–310. doi: 10.1016/0014-5793(95)00675-y. [DOI] [PubMed] [Google Scholar]
- 26.Anderson DM, Arredondo J, Hahn K, et al. Mohawk is a novel homeobox gene expressed in the developing mouse embryo. Dev Dyn. 2006;235:792–801. doi: 10.1002/dvdy.20671. [DOI] [PubMed] [Google Scholar]
- 27.Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morphol. 1951;88:1. [PubMed] [Google Scholar]
- 28.Wakahara T, Kusu N, Yamauchi H, et al. fibin, a novel secreted lateral plate mesoderm signal, is essential for pectoral fin bud initiation in zebrafish. Dev Biol. 2007;303:527–535. doi: 10.1016/j.ydbio.2006.11.041. [DOI] [PubMed] [Google Scholar]
- 29.Brandau O, Meindl A, Fassler R, et al. A novel gene, Tendin, is strongly expressed in tendons and ligaments and shows high homology with Chondromodulin-I. Dev Dyn. 2001;221:72–80. doi: 10.1002/dvdy.1126. [DOI] [PubMed] [Google Scholar]
- 30.Shukunami C, Oshima Y, Hiraki Y. Molecular cloning of tenomodulin, a novel Chondromodulin-I related gene. Biochem Biophys Res Commun. 2001;280:1323–1327. doi: 10.1006/bbrc.2001.4271. [DOI] [PubMed] [Google Scholar]
- 31.Koyama E, Shibukawa Y, Nagayama M, et al. A distinct cohort of progenitor cells participates in synovial joint and articular cartilage formation during mouse limb skeletogenesis. Dev Biol. 2008;316:62–73. doi: 10.1016/j.ydbio.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagai Y, Miyata K, Sun GP, et al. Aldosterone stimulates collagen gene expression and synthesis via activation of ERK1/2 in rat renal fibroblasts. Hypertension. 2005;46:1039–1045. doi: 10.1161/01.HYP.0000174593.88899.68. [DOI] [PubMed] [Google Scholar]
- 33.Hayashi M, Ninomiya Y, Parsons J, et al. Differential localization of mRNAs of collagen types I and II in chick fibroblasts, chondrocytes, and corneal cells by in situ hybridization using cDNA probes. J Cell Biol. 1986;102:2302–2309. doi: 10.1083/jcb.102.6.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fong KD, Trindade MC, Wang Z, et al. Microarray analysis of mechanical shear effects on flexor tendon cells. Plast Reconstr Surg. 2005;116:1393–1404. doi: 10.1097/01.prs.0000182345.86453.4f. [DOI] [PubMed] [Google Scholar]
- 35.Wu H, Du J, Zheng Q. Expression ofMMP-1 in cartilage and synovium of experimentally induced rabbit ACLT traumatic osteoarthritis: immunohistochemical study. Rheumatol J. 2008 Jul 3; doi: 10.1007/s00296-008-0636-2. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 36.Elliott SF, Coon CI, Hays E, et al. Bcl-3 is an interleukin-1-responsive gene in chondrocytes and synovial fibroblasts that activates transcription of the matrix metalloproteinase 1 gene. Arthritis Rheum. 2002;46:3230–3239. doi: 10.1002/art.10675. [DOI] [PubMed] [Google Scholar]
- 37.Jeong JG, Kim JM, Cho H, et al. Effects of IL-1beta on gene expression in human rheumatoid synovial fibroblasts. Biochem Biophys Res Commun. 2004;324:3–7. doi: 10.1016/j.bbrc.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 38.Zhang HG, Hyde K, Page GP, et al. Novel tumor necrosis factor alpha-regulated genes in rheumatoid arthritis. Arthritis Rheum. 2004;50:420–431. doi: 10.1002/art.20037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.