Abstract
Background
Most of environmental mycobacteria have been previously demonstrated to resist free-living amoeba with subsequent increased virulence and resistance to antibiotics and biocides. The Mycobacterium avium complex (MAC) comprises of environmental organisms that inhabit a wide variety of ecological niches and exhibit a significant degree of genetic variability. We herein studied the intra-ameobal location of all members of the MAC as model organisms for environmental mycobacteria.
Results
Type strains for M. avium, Mycobacterium intracellulare, Mycobacterium chimaera, Mycobacterium colombiense, Mycobacterium arosiense, Mycobacterium marseillense, Mycobacterium timonense and Mycobacterium bouchedurhonense were co-cultivated with the free-living amoeba Acanthamoeba polyphaga strain Linc-AP1. Microscopic analyses demonstrated the engulfment and replication of mycobacteria into vacuoles of A. polyphaga trophozoites. Mycobacteria were further entrapped within amoebal cysts, and survived encystment as demonstrated by subculturing. Electron microscopy observations show that, three days after entrapment into A. polyphaga cysts, all MAC members typically resided within the exocyst.
Conclusions
Combined with published data, these observations indicate that mycobacteria are unique among amoeba-resistant bacteria, in residing within the exocyst.
Background
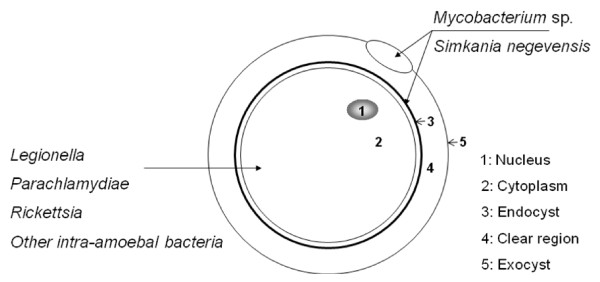
So-called amoeba-resistant bacteria are characterized by the ability to survive within free-living amoeba (FLA) trophozoites [1,2]. Some amoeba-resistant species have been further demonstrated to survive within the amoebal cyst which may act as a "Trojan horse" protecting the organisms from adverse environmental conditions [1]. The amoebal cyst is comprised of the nucleus and the cytoplasm embedded into three successive layers, i.e. the endocyst, the clear region and the outer exocyst. Despite the fact that specific location of amoeba-resistant bacteria into the amoebal cyst could modify the outcome of the organisms, precise location of intracystic organisms has not been systematically studied.
Most of environmental mycobacteria have been demonstrated to be amoeba-resistant organisms also residing into the amoebal cyst [3] (Table 1). The Mycobacterium avium complex (MAC) organisms have been used as model organisms for environmental mycobacteria, comprising of mycobacteria that are responsible for opportunistic infections and zoonoses [4-8]. M. avium and Mycobacterium intracellulare have been recovered from various sources, including fresh water [9-13] and hospital water supplies, in which FLA are frequently isolated [14-17]. Several experimental studies have further demonstrated M. avium-FLA interactions, including Acanthamoeba spp. [3,18-22] and Dictyostelium spp. [23-25]. M. avium and M. intracellulare have also been grown in the ciliated, unicellular protist Tetrahymena pyriformis [26]. It has been demonstrated that M. avium subsp. avium and M. avium subsp. paratuberculosis are able to survive within FLA [20-22], which results in their increased virulence [18,19] and protection against adverse situations including exposure to antibiotics [19]. The habitat of the recently described Mycobacterium chimaera (formerly sequevar MAC-A), isolated from respiratory tract specimens [27-29]; Mycobacterium colombiense (formerly sequevar MAC-X), isolated from the blood of an HIV-positive patient [30] and from enlarged lymph nodes in non-immunocompromised children [30-32];Mycobacterium arosiense isolated from bone lesions [33]; and Mycobacterium marseillense, Mycobacterium timonense and Mycobacterium bouchedurhonense isolated from respiratory tract specimens [34,35], remains however unknown. MAC species exhibit on-going evolutionary divergence as evidenced by the 97.9-98.71% ANI (Average Nucleotide Identity) between the genomes of M. avium subsp. paratuberculosis K10 (NC_000962) and M. avium strain 104 (NC_008595), the 3.7% 16S rRNA gene divergence between M. avium and M. timonense and between M. avium and M. chimaera, and the 7.2% rpoB gene sequence divergence between M. avium and M. colombiense [34].
Table 1.
Studies of interactions between MAC species and amoeba.
| Mycobacterium avium Species | Strains | Amoeba species | Survival in A. polyphaga | Reference | |
|---|---|---|---|---|---|
| Trophozoites | Cysts | ||||
| M. avium subsp. avium | M. avium 109 | A. castellanii | + | ? | [47] |
| M. avium subsp. avium | CIP104244T | A. polyphaga Linc-AP1 | + | + | [3] |
| M. intracellulare | CIP104243T | A. polyphaga Linc-AP1 | + | + | [3] |
| M. avium subsp. | |||||
| paratuberculosis | ? | A. castellanii CCAP1501 | + | ? | [22] |
| M. avium subsp. | |||||
| paratuberculosis | ? | A. castellanii CCAP1501 | + | + | [20] |
| M. avium subsp. avium | ? | D. discodium AX2 | + | ? | [24] |
| M. avium subsp. avium | ? | A. castellanii | + | ? | [48] |
| M. avium subsp. hominissuis | M. avium 104 | A. castellanii ATCC30234 | + | ? | [49] |
| M. avium | Serotype 4 | A. castellanii ATCC30872 | + | + | [21] |
| M. avium | ? | A. castellanii ATCC30234 | + | + | [18] |
| M. avium subsp. avium | ATCC 25291T | A. polyphaga Linc-AP1 | + | + | Present study |
| M. avium subsp. | |||||
| paratuberculosis | ATCC 19698T | - | + | + | - |
| M. avium subsp. hominissuis | IWGMT 49 | - | + | + | - |
| M. avium subsp. silvaticum | ATCC 49884T | - | + | + | - |
| M. intracellulare | ATCC 15985 | - | + | + | - |
| M. chimaera | DSM 446232T | - | + | + | - |
| M. colombiense | CIP 108962T | - | + | + | - |
| M. marseillense | CSUR P30T | - | + | + | - |
| M. timonense | CSUR P32T | - | + | + | - |
| M. bouchedurhonense | CSUR P34T | - | + | + | - |
| M. arosiense | DSM 45069T | - | + | + | - |
Using optic microscopy, electron microscopy and culturing methods, we herein used the MAC species as model organisms to study the location of environmental mycobacteria into the amoebal cyst and we further compared these observations with previously published data to find out that residing into the exocyst is a unique characteristic of environmental mycobacteria among amoeba-resistant organisms.
Results and Discussion
The 11 MAC strains (8 species) studied survived, but did not grow, after a 24-hour incubation in Page's modified Neff's Amoeba Saline (PAS) at 32°C. Microscopic examination of infected amoeba demonstrated that all MAC organisms were entrapped in A. polyphaga trophozoites and were visible in 3- to 5-μm large "Mycobacterium containing vacuoles" as early as 24 hours post-infection; 1 to 12 such vacuoles were observed per infected amoeba (Figure 1). The mean number of "Mycobacterium containing vacuoles" was not statistically different between the various MAC species. Electron microscopy observations revealed that, in the "Mycobacterium containing vacuoles" containing only one organism, there was a close apposition of the vacuole membrane all over the mycobacterial cell surface (Figure 2A, B), which was tightly apposed all over the organism cell wall, in contrast to organisms in vacuoles that contained several organisms as previously described in macrophages [36]. In this study, we did not resolved whether the presence of several mycobacteria within one vacuole resulted from the uptake of clumped mycobacteria, the replication of mycobacteria or the coalescence of several, single-organism vacuoles remains undetermined. In any case, our observations agree with previous studies that M. avium is initially entrapped in the vacuoles of amoebal trophozoites [18,23,24,21,22] and macrophages [36] (Table 1). In Dictyostelium, M. avium accumulated within vacuoles decorated with vacuolin, the Dictyostelium flotilin homologue, but it did not break the vacuole membrane, in contrast to Mycobacterium tuberculosis and Mycobacterium marinum. This result was linked to the absence of a particular region of difference (RD1), which in M. tuberculosis and M. marinum, encodes a type seven secretion system along with secreted effectors [23].
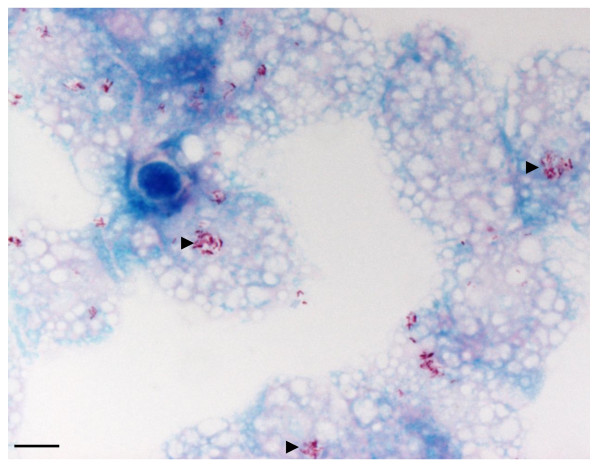
Figure 1.
Clusters of Mycobacterium colombiense (▶) in trophozoïtes of the free-living amoeba Acanthamoeba polyphaga Linc-AP1 (Ziehl Neelsen staining after a 24-hour incubation at 32°C). Scale bar = 10 μm.
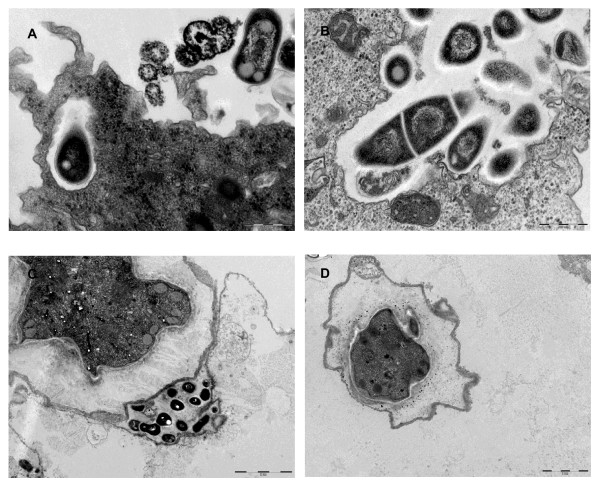
Figure 2.
Transmission electron-microscopy images of trophozoites and amoebal cysts infected by M. colombiense (A and B. Scale bar = 500 nm), M. avium, M. marseillense (C, D and E. Scale bar = 2 μm) Ec: Exocyst, Ed: Endocyst, Cr: Clear region, M: Mycobacterium, P: Phagosome.
Electron microscopy further disclosed that the 11 MAC strains under study were entrapped inside of the A. polyphaga cysts (Figure 2C, D). In all cases, the intracystic organisms were localized within the exocyst. In addition, M. marseillense could be observed in the clear region between the exocyst and the endocyst and in the inner side of the endocyst, and this was also the situation for M. intracellulare (Figures 2C, D) (Table 2). We further observed that a 36-hour exposure of the cysts to HCl did not affect the viability of the cysts, as new trophozoites emerged after 7-day incubation in peptone yeast extract-glucose (PYG) media at 32°C as determined by light microscopy. Sub-culturing such trophozoites on Middlebrook 7H10 agar yielded mycobacteria for all of the 8 MAC species (11 strains) under study after a 15-day incubation, whereas the cyst washing fluid remained sterile. Interestingly, we observed that these mycobacteria occupied a preferential location within the amoebal exocyst, where they were found in-between the two layers of the exocyst. Among the several Mycobacterium species reported to survive within amoebal cysts, such a particular feature has been previously illustrated only for M. avium in A. polyphaga cysts [21]; M. smegmatis [37]; M. abscessus, M. chelonae and M. septicum [3]; and M. xenopi [38]. Among intra-amoebal bacteria, location within the exocyst has also been reported for Simkania negevensis [39], despite the fact that S. negevensis organisms could also be observed within the cytoplasm of the cyst, depending on the strain under study [40]. Location within exocyst wall contrasts with the observation of Legionella pneumophila, which was found within the cytoplasm of pre-cysts and mature cysts of A. polyphaga [41] or non-entrapped within amoebal cysts [42]. Reviewing published data regarding amoebal-resistant bacterial species [1,2] found that 11/32 (34.37%) Mycobacterium species versus 1/28 (3.57%) non-mycobacterium amoebal-resistant bacterial species have been reported to survive within A. polyphaga exocyst (P = 0.003) (Figure 3). As both L. pneumophila and mycobacteria are pathogens, the intracystic location of organisms may not influence their virulence. The mechanisms and biological significance of this particular location remain to be studied. It has been established that A. polyphaga exocyst is composed of cellulose [43] and the authors have observed that mycobacteria encode one cellulose-binding protein and one or two cellulases which are indeed transcribed [44]. Cellulase encoded by mycobacteria may play a role in their unique exocyst location.
Table 2.
Abundance of mycobacteria in A. polyphaga strain Linc-AP1 and their preferential location in amoebal cyst wall.
| MAC species | No. of vacuoles that contain mycobacteria | Location in amoebal cyst wall |
|---|---|---|
| M. timonense | 1.3 ± 0.5 vacuoles | Exocyst |
| M. bouchedurhonense | 2.1 ± 1.7 vacuoles | Exocyst |
| M. marseillense | 2.4 ± 1.4 vacuoles | Exocyst, clear region, cytoplasm |
| M. avium (M. avium subsp. avium) | 2.6 ± 2.2 vacuoles | Exocyst |
| M. chimaera | 3.6 ± 2.6 vacuoles | Exocyst, cytoplasm |
| M. intracellulare | 4.6 ± 4.8 vacuoles | Exocyst, Endocyst |
| M. colombiense | 5.7 ± 6.2 vacuoles | Exocyst, cytoplasm |
| M. arosiense | 9.4 ± 15.2 vacuoles | Exocyst |
Figure 3.
Preferential localisation of Mycobacterium sp. and other amoeba-resistant bacterial organisms in amoebal cyst.
Moreover, we observed that all MAC species can survive within such A. polyphaga cyst. This occurrence did not merely result from the potential contamination of the amoeba by extra-amoebal mycobacteria, since we destroyed any MAC organism left on the surface of cysts by incubating the cysts in HCl, a method previously demonstrated to kill remaining trophozoites, immature cysts and extra-amoebal M. avium [21]. We checked the efficacy of this process by incubating the rinsing buffer on Middlebrook and found no growth of mycobacteria, which indicated that the HCl had indeed destroyed any extracystic MAC organisms. The fact that all of the MAC species survived in the exocyst may be relevant to the persistence of these organisms in the environment despite adverse conditions. Non-tuberculous mycobacteria, including M. avium, have been shown to persist up to 26 months in drinking water systems despite filtration and ozonation [45]. Also, M. intracellulare and other non-tuberculous mycobacteria have been shown to be protected against 15 mg/liter of free-chlorine for 24 hours by entrapment within A. polyphaga cysts [3]. Therefore, free-living amoeba cysts may be a "Trojan horse" for MAC organisms and protect them from adverse environmental conditions, including high concentrations of chlorine, as previously reported for other environmental mycobacteria.
Conclusion
The data presented herein on MAC species illustrate that survival within the amoebal exocyst is a significant feature of environmental mycobacteria. This particular location, preserving mycobacteria from adverse environment, nevertheless allow them to rapidly escape from the amoebal cyst. The mechanisms for such unique location remain to be established in environmental mycobacteria.
Methods
Mycobacterium strains
M. avium subsp. avium ATCC 25291T, M. chimaera DSM 446232T, M. colombiense CIP 108962T, M. arosiense DSM45069T [33], M. marseillense CSURP30T, M. timonense CSURP32T and M. bouchedurhonense CSURP34T [35] reference strains that were previously identified by 16S rRNA and rpoB gene sequencing [34] were subcultured on Middlebrook 7H10 agar (Becton Dickinson, Le Pont de Claix, France) for 7 days at 30°C under a 5% CO2 atmosphere. Cells were washed in 1.5 ml phosphate buffered saline (PBS), pH 7.3, by centrifugation at 8,600 g, and the inoculum was adjusted to 106 bacteria/ml in PBS.
Infection of amoeba
The A. polyphaga strain Linc-AP1 was obtained from T. J. Rowbotham, Public Health Laboratory, Leeds, United Kingdom and cultured at 28°C for 3 days in 150 cm3 culture flasks (Corning, New York USA) that contained 30 ml PYG broth [46]. Amoebal cells were harvested by centrifugation at 500 g for 10 min. The pellet was suspended twice in PAS to obtain 5 × 105 cells/ml. One milliliter of this suspension was dropped into each well of a 12-well microplate (Corning) and incubated at 33°C for 7 days. The microplate, prepared as described above, was used for culturing the mycobacteria. Each well of the microplate was inoculated with a final concentration of 106 mycobacteria/ml (MOI = 10). The inoculum was sonicated for 5 min at 234 watts (BRANSON 2210; Branson Ultrasonics Corporation, Danbury, CT, USA) in order to limit mycobacteria cell clumping. The microplate was centrifuged at 1,000 g for 30 min and incubated at 33°C under a humidified, 5% CO2 atmosphere. This microplate was examined daily for 15 days for cytopathic effects and the presence of intra-amoebal organisms by shaking, cytocentrifugation at 200 g for 10 min and Ziehl-Neelsen staining.
Encystment and excystment of infected amoeba
In 25 cm3 culture flasks (Corning), 10 ml of amoeba that had been infected for 48 hours were rinsed once with encystment buffer adapted from [21] (0.1 M KCl, 0.02 M Tris, 8 mM MgSO4, 0.4 mM CaCl2, 1 mM NaHCO3). After centrifugation at 500 g, the pellet was resuspended in 10 ml of fresh encystment buffer and incubated for 3 days at 32°C. The excystment of the cysts was examined by light microscopy. Amoebal cysts were pelleted by centrifugation at 1,000 g for 10 min and treated with 3% (vol/vol) HCl as previously described [21]. Treated cysts were then washed three times with PAS buffer. Half of the sample was processed for electron microscopy (see above), and the other part was incubated for 7 days in PYG medium at 33°C. Intra-amoebal mycobacteria were released by lysing the monolayer with 1 ml of 0.5% sodium dodecyl sulfate, followed by two successive passages through a 27-gauge needle [3]. The presence of viable mycobacteria was documented by detecting colonies on Middlebrook 7H10 agar inoculated with 200 μl of the cell lysate and incubated at 30°C for 15 days. The identities of the mycobacteria were confirmed by Ziehl-Neelsen staining and partial rpoB gene sequencing using primers Myco-F (5'-GGCAAGGTCACCCCGAAGGG-3') and Myco-R (5'-AGCGGCTGCTGGGTGATCATC-3') [34]. All experiments were repeated three times.
Electron microscopy
Non-ingested mycobacteria were eliminated by rinsing the amoebal monolayer twice with sterile PBS. The amoeba monolayer that was previously infected by MAC species was then fixed in 2% glutaraldehyde and 0.1 M cacodylate buffer overnight. After this first fixation, the bacteria were fixed in 2% glutaraldehyde and 0.33% acroleine in a 0.07 M cacodylate buffer for 1 hour. After washing in 0.2 M cacodylate buffer, the bacteria were post-fixed in 1% osmium bioxide in 0.1 M potassium ferrycyanure for 1 hour and dehydrated in an ascending series of ethanol concentrations, and after 100% ethanol, the dehydration was finished in propylene oxide, and the samples were embedded in an Epon 812 resin. Sections (70 nm) were stained with 5% uranyl acetate and lead citrate before examination with a transmission electron microscope (Philips Morgagni 268D, Eindhoven, the Netherlands). For the determination of mycobacterial abundance, we made observations on a total of 30 A. polyphaga trophozoites for each of the 8 MAC species. In order to determine the total number of mycobacteria per trophozoite, we recorded the total number of vacuoles with one Mycobacterium organism and the total number of vacuoles with > 1 Mycobacterium organism. We also made observations on a total of 30 A. polyphaga organisms for each of the 8 MAC species in order to determine their intracystic location, which was considered as intracystic when apposed to the cyst wall and reaching into the cyst wall (between the endo- and the exocyst). These observations were performed in triplicate.
Statistical tests
Comparison among amoeba-resistant bacterial species [2] as for their survival within exocyst was done using the χ2 test and corrected by Mantel Haenszel method. Comparaisons of mean ± standard deviation of the number of infected vacuoles were done using the ANOVA test. A P value < 0.05 was considered to be significant.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
IBS performed the experiments, he interpreted data and wrote the manuscript.
MD designed the experiment, he provided support, interpreted data and wrote the manuscript. Both authors have read and approved the final version of the manuscript.
Contributor Information
Iskandar Ben Salah, Email: bensalahiskandar@gmail.com.
Michel Drancourt, Email: michel.drancourt@univmed.fr.
Acknowledgements
The authors acknowledge Bernard Campagna for his help with the electron microscopy observations.
References
- Greub G, Raoult D. Microorganisms resistant to free-living amoebae. In Microbiol Rev. 2004;17:413–433. doi: 10.1128/CMR.17.2.413-433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas V, McDonnell G, Denyer SP, Maillard JY. Free-living amoeba and their intracellular pathogenic microorganisms: risks for water quality. FEMS Microbiol Rev. in press . [DOI] [PubMed]
- Adekambi T, Ben Salah S, Khlif M, Raoult D, Drancourt M. Survival of environmental mycobacteria in Acanthamoeba polyphaga. Appl Environ Microbiol. 2006;2:5974–5981. doi: 10.1128/AEM.03075-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortoli E, Cichero P, Piersimoni C, Simonetti MT, Gesu G, Nista D. Use of BACTEC MGIT 960 for recovery of mycobacteria from clinical specimens: multicenter study. J Clin Microbiol. 1999;37:3578–3582. doi: 10.1128/jcm.37.11.3578-3582.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turenne CY, Wallace R Jr, Behr MA. Mycobacterium avium in the postgenomic era. Clin Microbiol Rev. 2007;20:205–229. doi: 10.1128/CMR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajko DM, Chin DP, Gonzalez PC, Nassos PS, Hopewell PC, Reingold AL, Horsburgh CR Jr, Yakrus MA, Ostroff SM, Hadley WK. Mycobacterium avium complex in water, food, and soil samples collected from the environment of HIV-infected individuals. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;9:176–182. [PubMed] [Google Scholar]
- Karakousis PC, Moore RD, Chaisson RE. Mycobacterium avium complex in patients with HIV infection in the era of highly active antiretroviral therapy. Lancet Infect Dis. 2004;4:557–565. doi: 10.1016/S1473-3099(04)01130-2. [DOI] [PubMed] [Google Scholar]
- Lauzi S, Pasotto D, Amadori M, Archetti IL, Poli G, Bonizzi L. Evaluation of the specificity of the gamma-interferon test in Italian bovine tuberculosis-free herds. Vet J. 2000;160:17–24. doi: 10.1053/tvjl.1999.0444. [DOI] [PubMed] [Google Scholar]
- Falkinham JO, Norton CD, LeChevallier MW. Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other mycobacteria in drinking water distribution systems. Appl Environ Microbiol. 2001;67:1225–1231. doi: 10.1128/AEM.67.3.1225-1231.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkinham JO, Iseman MD, de Haas P, van Soolingen D. Mycobacterium avium in a shower linked to pulmonary disease. J Water Health. 2008;6:209–213. doi: 10.2166/wh.2008.032. [DOI] [PubMed] [Google Scholar]
- Hilborn ED, Yakrus MA, Covert TC, Harris SI, Donnelly SF, Schmitt MT, Toney S, Bailey SA, Stelma GN Jr. Molecular comparison of Mycobacterium avium isolates from clinical and environmental sources. Appl Environ Microbiol. 2008;74:4966–4968. doi: 10.1128/AEM.02900-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Dantec C, Duguet JP, Montiel A, Dumoutier N, Dubrou S, Vincent V. Occurrence of mycobacteria in water treatment lines and in water distribution systems. Appl Environ Microbiol. 2002;68:5318–5325. doi: 10.1128/AEM.68.11.5318-5325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos R, Oliveira F, Fernandes J, Goncalves S, Macieira F, Cadete M. Detection and identification of mycobacteria in the Lisbon water distribution system. Water Sci Technol. 2005;52:177–180. [PubMed] [Google Scholar]
- Aronson T, Holtzman A, Glover N, Boian M, Froman S, Berlin OG, Hill H, Stelma G Jr. Comparison of large restriction fragments of Mycobacterium avium isolates recovered from AIDS and non-AIDS patients with those of isolates from potable water. J Clin Microbiol. 1999;37:1008–1012. doi: 10.1128/jcm.37.4.1008-1012.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Moulin GC, Stottmeier KD, Pelletier PA, Tsang AY, Hedley-Whyte J. Concentration of Mycobacterium avium by hospital hot water systems. JAMA. 1988;260:1599–1601. doi: 10.1001/jama.260.11.1599. [DOI] [PubMed] [Google Scholar]
- Goslee S, Wolinsky E. Water as a source of potentially pathogenic mycobacteria. Am Rev Respir Dis. 1976;113:287–292. doi: 10.1164/arrd.1976.113.3.287. [DOI] [PubMed] [Google Scholar]
- von Reyn CF, Waddell RD, Eaton T, Arbeit RD, Maslow JN, Barber TW, Brindle RJ, Gilks CF, Lumio J, Lahdevirta J. Isolation of Mycobacterium avium complex from water in the United States, Finland, Zaire, and Kenya. J Clin Microbiol. 1993;31:3227–3230. doi: 10.1128/jcm.31.12.3227-3230.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo JD, Falkow S, Tompkins LS, Bermudez LE. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect Immun. 1997;65:3759–3767. doi: 10.1128/iai.65.9.3759-3767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miltner EC, Bermudez LE. Mycobacterium avium grown in Acanthamoeba castellanii is protected from the effects of antimicrobials. Antimicrob Agents Chemother. 2000;44:1990–1994. doi: 10.1128/AAC.44.7.1990-1994.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mura M, Bull TJ, Evans H, Sidi-Boumedine K, McMinn L, Rhodes G, Pickup R, Hermon-Taylor J. Replication and long-term persistence of bovine and human strains of Mycobacterium avium subsp. paratuberculosis within Acanthamoeba polyphaga. Appl Environ Microbiol. 2006;72:854–859. doi: 10.1128/AEM.72.1.854-859.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert M, Birkness K, White E, Fields B, Quinn F. Mycobacterium avium bacilli grow saprozoically in coculture with Acanthamoeba polyphaga and survive within cyst walls. Appl Environ Microbiol. 1998;64:2256–2261. doi: 10.1128/aem.64.6.2256-2261.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whan L, Grant IR, Rowe MT. Interaction between Mycobacterium avium subsp. paratuberculosis and environmental protozoa. BMC Microbiol. 2006;6:63. doi: 10.1186/1471-2180-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn M, Rohde KH, Russell DG, Soldati T. Infection by tubercular mycobacteria is spread by nonlytic ejection from their amoeba hosts. Science. 2009;323:1729–1733. doi: 10.1126/science.1169381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skriwan C, Fajardo M, Hagele S, Horn M, Wagner M, Michel R, Krohne G, Schleicher M, Hacker J, Steinert M. Various bacterial pathogens and symbionts infect the amoeba Dictyostelium discoideum. Int J Med Microbiol. 2002;291:615–624. doi: 10.1078/1438-4221-00177. [DOI] [PubMed] [Google Scholar]
- Unal C, Steinert M. Dictyostelium discoideum as a model to study host-pathogen interactions. Methods Mol Biol. 2006;346:507–515. doi: 10.1385/1-59745-144-4:507. [DOI] [PubMed] [Google Scholar]
- Strahl ED, Gillaspy GE, Falkinham JO III. Fluorescent acid-fast microscopy for measuring phagocytosis of Mycobacterium avium. Mycobacterium intracellulare, and Mycobacterium scrofulaceum by Tetrahymena pyriformis and their intracellular growth. Appl Environ Microbiol. 2001;67:4432–4439. doi: 10.1128/AEM.67.10.4432-4439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bills ND, Hinrichs SH, Aden TA, Wickert RS, Iwen PC. Molecular identification of Mycobacterium chimaera as a cause of infection in a patient with chronic obstructive pulmonary disease. Diagn Microbiol Infect Dis. 2009;63:292–295. doi: 10.1016/j.diagmicrobio.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Schweickert B, Goldenberg O, Richter E, Gobel UB, Petrich A, Buchholz P, Moter A. Occurrence and clinical relevance of Mycobacterium chimaera sp. nov., Germany. Emerg Infect Dis. 2008;14:1443–1446. doi: 10.3201/eid1409.071032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortoli E, Rindi L, Garcia MJ, Chiaradonna P, Dei R, Garzelli C, Kroppenstedt RM, Lari N, Mattei R, Mariottini A, Mazzarelli G, Murcia MI, Nanetti A, Piccoli P, Scarparo C. Proposal to elevate the genetic variant MAC-A, included in the Mycobacterium avium complex, to species rank as Mycobacterium chimaera sp. nov. Int J Syst Evol Microbiol. 2004;54:1277–1285. doi: 10.1099/ijs.0.02777-0. [DOI] [PubMed] [Google Scholar]
- Murcia MI, Tortoli E, Menendez MC, Palenque E, Garcia MJ. Mycobacterium colombiense sp. nov., a novel member of the Mycobacterium avium complex and description of MAC-X as a new ITS genetic variant. Int J Syst Evol Microbiol. 2006;56:2049–2054. doi: 10.1099/ijs.0.64190-0. [DOI] [PubMed] [Google Scholar]
- Esparcia O, Navarro F, Quer M, Coll P. Lymphadenopathy caused by Mycobacterium colombiense. J Clin Microbiol. 2008;46:1885–1887. doi: 10.1128/JCM.01441-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuorenmaa K, Ben Salah I, Barlogis V, Chambost H, Drancourt M. Mycobacterium colombiense and pseudotuberculous lymphadenopathy. Emerg Infect Dis. 2009;15:619–620. doi: 10.3201/eid1504.081436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang D, Herlin T, Stegger M, Andersen AB, Torkko P, Tortoli E, Thomsen VO. Mycobacterium arosiense sp. nov., a slowly growing, scotochromogenic species causing osteomyelitis in an immunocompromised child. Int J Syst Evol Microbiol. 2008;58:2398–2402. doi: 10.1099/ijs.0.65503-0. [DOI] [PubMed] [Google Scholar]
- Ben Salah I, Adekambi T, Raoult D, Drancourt M. rpoB sequence-based identification of Mycobacterium avium complex species. Microbiology. 2008;154:3715–3723. doi: 10.1099/mic.0.2008/020164-0. [DOI] [PubMed] [Google Scholar]
- Ben Salah I, Cayrou C, Raoult D, Drancourt M. Mycobacterium marsilliense sp. nov., Mycobacterium timonense sp. nov., and Mycobacterium bouchedurhonense sp. nov., members of the Mycobacterium avium complex. Int J Syst Evol Microbiol. 2009;59:2803–2808. doi: 10.1099/ijs.0.010637-0. [DOI] [PubMed] [Google Scholar]
- de Chastellier C. The many niches and strategies used by pathogenic mycobacteria for survival within host macrophages. Immunobiology. 2009;214:526–542. doi: 10.1016/j.imbio.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Sharbati-Tehrani S, Stephan J, Holland G, Appel B, Niederweis M, Lewin A. Porins limit the intracellular persistence of Mycobacterium smegmatis. Microbiology. 2005;151:2403–2410. doi: 10.1099/mic.0.27969-0. [DOI] [PubMed] [Google Scholar]
- Drancourt M, Adekambi T, Raoult D. Interactions between Mycobacterium xenopi, amoeba and human cells. J Hosp Infect. 2007;65:138–142. doi: 10.1016/j.jhin.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Kahane S, Dvoskin B, Mathias M, Friedman MG. Infection of Acanthamoeba polyphaga with Simkania negevensis and S. negevensis survival within amoebal cysts. Appl Environ Microbiol. 2001;67:4789–4795. doi: 10.1128/AEM.67.10.4789-4795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsaro D, Greub G. Pathogenic potential of novel Chlamydiae and diagnostic approaches to infections due to these obligate intracellular bacteria. Clin Microbiol Rev. 2006;19:283–297. doi: 10.1128/CMR.19.2.283-297.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilvington S, Price J. Survival of Legionella pneumophila within cysts of Acanthamoeba polyphaga following chlorine exposure. J Appl Bacteriol. 1990;68:519–525. doi: 10.1111/j.1365-2672.1990.tb02904.x. [DOI] [PubMed] [Google Scholar]
- Garcia MT, Jones S, Pelaz C, Millar RD, Abu KY. Acanthamoeba polyphaga resuscitates viable non-culturable Legionella pneumophila after disinfection. Environ Microbiol. 2007;9:1267–1277. doi: 10.1111/j.1462-2920.2007.01245.x. [DOI] [PubMed] [Google Scholar]
- Ben Sallah I, Ghigo E, Drancourt M. Free-living amoeba, a training field for macrophage resistance of mycobacteria. Clin Microbiol Infect. 2009;15:894–905. doi: 10.1111/j.1469-0691.2009.03011.x. [DOI] [PubMed] [Google Scholar]
- Mba Medie F, Ben Salah I, Drancourt M, Henrissat B. Paradoxal conservation of a set of three cellulose-targeting genes in Mycobacterium tuberculosis complex organisms. Microbiology. in press . [DOI] [PubMed]
- Hilborn ED, Covert TC, Yakrus MA, Harris SI, Donnelly SF, Rice EW, Toney S, Bailey SA, Stelma GN Jr. Persistence of nontuberculous mycobacteria in a drinking water system after addition of filtration treatment. Appl Environ Microbiol. 2006;72:5864–5869. doi: 10.1128/AEM.00759-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greub G, La Scola B, Raoult D. Amoebae-resisting bacteria isolated from human nasal swabs by amoebal coculture. Emerg Infect Dis. 2004;10:470–477. doi: 10.3201/eid1003.020792. [DOI] [PubMed] [Google Scholar]
- Danelishvili L, Wu M, Stang B, Harriff M, Cirillo SL, Cirillo JD, Bildfell R, Arbogast B, Bermudez LE. Identification of Mycobacterium avium pathogenicity island important for macrophage and amoeba infection. Proc Natl Acad Sci USA. 2007;104:11038–11043. doi: 10.1073/pnas.0610746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna-Prasad BNGSK. Preliminary report on engulfment and retention of mycobacteria by trophozoites of axenically grown Acanthamoeba castellanii Douglas 1930. Curr Sci. 1978;45:245–247. [Google Scholar]
- Tenant R, Bermudez LE. Mycobacterium avium genes upregulated upon infection of Acanthamoeba castellanii demonstrate a common response to the intracellular environment. Curr Microbiol. 2006;52:128–133. doi: 10.1007/s00284-005-0218-4. [DOI] [PubMed] [Google Scholar]