Abstract
A C60+ cluster ion projectile is employed for sputter cleaning biological surfaces to reveal spatio-chemical information obscured by contamination overlayers. This protocol is used as a supplemental sample preparation method for time of flight secondary ion mass spectrometry (ToF-SIMS) imaging of frozen and freeze-dried biological materials. Following the removal of nanometers of material from the surface using sputter cleaning, a frozen-patterned cholesterol film and a freeze-dried tissue sample were analyzed using ToF-SIMS imaging. In both experiments, the chemical information was maintained after the sputter dose, due to the minimal chemical damage caused by C60+ bombardment. The damage to the surface produced by freeze-drying the tissue sample was found to have a greater effect on the loss of cholesterol signal than the sputter-induced damage. In addition to maintaining the chemical information, sputtering is not found to alter the spatial distribution of molecules on the surface. This approach removes artifacts that might obscure the surface chemistry of the sample and are common to many biological sample preparation schemes for ToF-SIMS imaging.
Imaging time of flight secondary ion mass spectrometry (ToF-SIMS) is an emerging bioanalytical tool that allows molecular-specific images of biological samples to be acquired with their native chemistry intact, at submicron resolution, and without employing labels. However, the preparation of biological samples remains a limiting step in applying the technique to answering specific biological questions [1–10]. Numerous methods have been developed to make biological samples more amiable to the vacuum environment required for ToF-SIMS, including freeze-drying with various rinses [2, 11, 12], metal imprinting [8], metal deposition [2], freeze etching [3], freeze fracture [4, 13], and vitrification [5]. These methods have proven to be useful but all have shortcomings, the most limiting being reproducibility.
Ultimately, a SIMS experiment should yield an information-rich image, composed of intense signals corresponding to the ions of interest. Sample preparation techniques might damage the surface of the sample to some degree. Optimization of the sample preparation can be problematic for tissues and cells because of inherent sample-to-sample variation and identical procedures can produce sample surfaces with varying degrees of damage. However, if the damage is confined to the surface of the sample, this interfering layer might be removed and the underlying, undamaged material analyzed.
Surface bombardment using cluster ion projectiles has the unique ability to remove molecular material with minimal damage accumulation to the underlying layers when compared to atomic projectiles [14, 15]. Also, these projectiles do not significantly redistribute material from one layer to the next. This approach has been successfully applied for several applications including molecular depth profiling [16–20] and 3-D imaging [12, 21, 22], sputtering has been used to clean inorganic materials for many years [23] but has not yet been used as a supplemental sample preparation method for SIMS imaging biological materials.
Here, we demonstrate that it is possible to use the unique properties of cluster ion projectiles to increase the reproducibility of current sample preparation methods for biological SIMS imaging. The surface specificity of SIMS makes it possible for important information to be obscured by a matrix overlayer or by a layer of damaged material at the surface. The approach presented here demonstrates that it is possible to uncover spatio-chemical information (the chemical identity of a compound as determined by the mass to charge ratio with respect to the area of the surface where the ion originated) from below the sample surface, thus overcoming some common artifacts caused by sample preparation.
In the presence of atmospheric water, frozen hydrated samples accumulate an ice layer on the top of the surface to be analyzed. Likewise, freeze-fractured samples can be completely covered in ice due to the unpredictability of the position in the fracture plane[24]. This ice might not damage the surface; however, it is a layer of material that masks the chemical information below the ice. Controlled removal of this layer could increase the reproducibility of these two sample preparation methods. To examine the usefulness of C60+ for removing a water layer from a sample without damaging the underlying chemical structure, a simulated biological sample was created by using physical vapor deposition (PVD). Briefly, an SEM finder grid was placed on a silicon substrate and cholesterol vapor was allowed to condense on the surface. After deposition, the grid was removed leaving behind a relief pattern of cholesterol with known spatial dimensions. The sample was then cooled to LN2 temperature for 1 h, which allowed water from the vacuum environment to condense on the surface forming an ice overlayer. After collecting a SIMS image, the ice layer was removed using a C60+ impact with a sputter dose of 6 × 1012 C60+/cm2. From the measured sputter yield of amorphous ice for a 40 keV C60+ (4000 H2O/ C60+) and the molecular density (3.075 × 1022 molecules/cm3) we calculate that ≤ 8 nm of ice is removed from the surface.
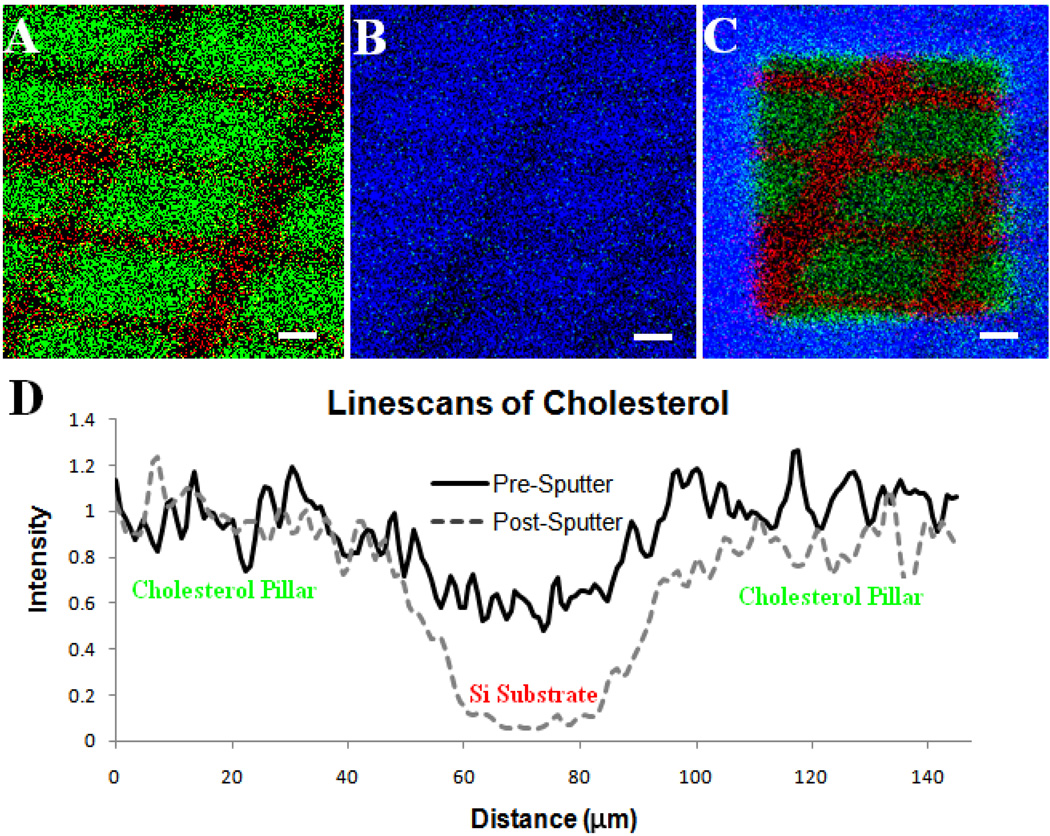
A SIMS image overlay of a patterned cholesterol film on a Si substrate is shown in Figure 1A prior to ice deposition. Here the patterned cholesterol islands are visible; Si is mapped in red and cholesterol is mapped in green. The second panel (Figure 1B) contains an image overlay following ice deposition; water is mapped in blue and cholesterol in green. The grid pattern has been covered by the ice and cannot be resolved. Following sputter removal, the grid pattern can once again be resolved and it is possible to detect molecular ion signals (Figure 1C). The intensity of line scans for cholesterol taken across one of the grid lines in the patterned film before water deposition and after water deposition and subsequent removal is shown in Figure 1D. These data demonstrate that there is minimal loss of lateral resolution following the removal of the ice over layer.
Figure 1.
(A.) Positive SIMS image of patterned cholesterol film on silicon created using physical vapor deposition, m/z 369 shown in green and m/z 28 in red. (B) The film was then cooled in vacuum (−196°C) and water was allowed to deposit, m/z 18 shown in blue and m/z 369 in green. The cholesterol layer is almost completely covered in water. (C.) After etching with a dose of 1013 C60+ ions/cm2 the patterned film beneath is revealed with maintained spatial integrity. (D.) Line scans across the film features reveal that the distribution of cholesterol on the surface is maintained when C60+ is used to remove the water overlayer versus when the surface is not cleaned with the C60+ nanotome. Scale bars are 25 µm.
To further investigate the value of employing C60+ in sample preparation, axial slices from a nine-day-old mouse embryo were studied. This model system was chosen because results from a previous study carried out by Ding et. al. revealed increased cholesterol concentration in specific regions of the embryo [25]. The samples were freeze-dried for this imaging experiment without the use of any matrix or cyro-protectant; tissue slices were excised and either rinsed in 18 MΩ water or incubated in a 100 µM solution of soluble cholesterol for 30 min prior to rinsing in 18 MΩ water. After rinsing, the tissue samples were plunge frozen in LN2 and introduced into the analysis chamber of the mass spectrometer at LN2 temperature. The sample temperature was raised to −80° C, allowing the ice to sublime into the vacuum (1 × 10−8 torr) of the analysis chamber. Examination of the non-incubated tissue yielded insufficient cholesterol signal to establish differential distribution in the tissue slice (both sputtered and non-sputtered samples, data not shown).
SIMS images of the cholesterol-incubated tissue were taken directly after freeze-drying. In these images the cholesterol signal was weak and did not localize to any specific area of the tissue (Figure 2B). The C60+ probe was used to sputter-clean the surface to expose the presumably undisturbed bulk of the tissue. The tissue was sputtered with a dose of 1 × 1013 C60+/cm2, which removed approximately 50 nm of tissue from the surface. The thickness of the amount removed was calculated using an estimated sputter yield of 500 molecules/ C60+ and an estimated molecular density of 1 × 1021 molecules/cm3. These values were taken from experiments performed using the disaccharide trehalose [16]. The amount of material removed to uncover cholesterol localization is relatively small compared to the thickness of the tissue slice (~ 300 µm). The subsequent image (Figure 2C) shows increased cholesterol signal as well as localization to the genital ridge of the tissue in agreement with previously published results [25]. These results suggest that the damage created during preparation of the tissue samples without fixation, might only be confined to the shallow depths (tens of nanometers) below the surface.
Figure 2.
(A.) Scanning ion image (SIM) of an axial slice from a 9 day old mouse embryo. The gut and the genital ridge are identified by white arrows. The images in (B) and (C) are SIMS images of cholesterol (m/z 366–370) from the tissue in the SIM image. The image in (B) was taken prior to a sputter dose of 1 × 1013 C60+/cm2 and the image in (C) was taken after nanotome sputtering. Scale bars are 100 µm.
The changes in signal observed after removal of the surface of the tissue are shown in Table 1. After normalizing to total signal and image pixels, the cholesterol signal increased by 110 % and the phosphocholine (PC) signal decreased by 40 %. An increase in cholesterol signal following sputtering is an unexpected result, as it has been shown that depth profiling of cholesterol leads to an 1.2 to 4.5 fold drop in signal before a steady state is reached [21, 22, 26]. It is important toemphasize that these experiments were carried out at cold temperatures (T ≤ −80 °C), because it has been shown that there is significant migration of cholesterol in tissue at temperatures above 0 °C in vacuum conditions [22, 27]. The observed increase suggests the sample prep method affects the surface distribution of molecules in the pre-sputtered sample to a significant degree. The decrease in PC is expected; however, the magnitude is less than expected based on depth profiling of a pure dipalmitoylphosphatidylcholine (DPPC) film in which signal decreases by approximately 3.5 times less than the initial value [18]. These changes suggest that there are more intact cholesterol and DPPC molecules available for ionization after sputtering than before due to the removal of sample damage.
Table 1.
Normalized ion signal following a sputter dose of 1 × 1013 C60+/cm2
| PC (m/z 184)* | Cholesterol (m/z 366–370)** | |
|---|---|---|
| Tissue Slice 1 | 70.4 | 202.6 |
| Tissue Slice 2 | 52.1 | 207.8 |
| Tissue Slice 3 | 58.4 | 221.4 |
| Average | 60 ± 9 | 211 ± 10 |
For comparisons of normalized signal before and after sputtering
p < 0.001
p = 0.01 (Students t-test).
We have used a C60 ion beam as a nanotome to remove nanometers of interfering surface material from biologically relevant samples without significant damage to the underlying surface. The information uncovered would otherwise be unobservable with a surface technique such as ToF-SIMS imaging. Using known and estimated sputter yields and molecular densities, it is possible to remove material in a controlled and quantifiable manner. This method of controlled etching might increase the number of analyzable biological samples for ToF-SIMS imaging. By removing the uppermost area of the surface, where much of the sample-preparation-induced damage occurs, reproducibility between samples might be significantly improved.
Acknowledgement
This work was supported by grants from the National Institutes of Health (2R01EB002016-16 N.W./A.G.E and GM069338, N.W). A.G.E. is supported as a Marie Curie Chair from the European Union 6th Framework.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Chandra S, Morrison GH. Sample Preparation of Animal-Tissues and Cell-Cultures for Secondary Ion Mass-Spectrometry (Sims) Microscopy. Biol. Cell. 1992;74:31–42. doi: 10.1016/0248-4900(92)90006-m. [DOI] [PubMed] [Google Scholar]
- 2.Altelaar AF, Klinkert I, Jalink K, de Lange RP, Heeren RMA, Piersma SR. Gold-enhanced biomolecular surface imaging of cells and tissue by SIMS and MALDI mass spectrometry. Anal. Chem. 2006;78:734–742. doi: 10.1021/ac0513111. [DOI] [PubMed] [Google Scholar]
- 3.Piehowski PD, Kurczy ME, Willingham D, Parry S, Heien ML, Winograd N, Ewing AG. Freeze-etching and vapor matrix deposition for ToF-SIMS imaging of single cells. Langmuir. 2008;24:7906–7911. doi: 10.1021/la800292e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quong JN, Knize MG, Kulp KS, Wu KJ. Molecule-specific imaging analysis of carcinogens in breast cancer cells using time-of-flight secondary ion mass spectrometry. Appl. Surf. Sci. 2004;231-2:424–427. [Google Scholar]
- 5.Parry S, Winograd N. High-resolution TOF-SIMS imaging of eukaryotic cells preserved in a trehalose matrix. Anal. Chem. 2005;77:7950–7957. doi: 10.1021/ac051263k. [DOI] [PubMed] [Google Scholar]
- 6.Cannon DM, Pacholski ML, Winograd N, Ewing AG. Molecule specific imaging of freeze-fractured, frozen-hydrated model membrane systems using mass spectrometry. J. Am. Chem. Soc. 2000;122:603–610. [Google Scholar]
- 7.Roddy TP, Cannon DM, Meserole CA, Winograd N, Ewing AG. Imaging of freeze-fractured cells with in situ fluorescence and time-of-flight secondary ion mass spectrometry. Anal. Chem. 2002;74:4011–4019. doi: 10.1021/ac0255734. [DOI] [PubMed] [Google Scholar]
- 8.Sjovall P, Lausmaa J, Nygren H, Carlsson L, Malmberg P. Imaging of membrane lipids in single cells by imprint-imaging time-of-flight secondary ion mass spectrometry. Anal. Chem. 2003;75:3429–3434. doi: 10.1021/ac0207675. [DOI] [PubMed] [Google Scholar]
- 9.Kurczy ME, Piehowski PD, Parry S, Jiang J, Chen G, Winograd N. Which is more important in bioimaging SIMS experiments: the sample preparation or the nature of the projectile? Appl. Surf. Sci. 2008;255:1298–1304. doi: 10.1016/j.apsusc.2008.05.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunelle A, Laprevote O. Recent advances in biological tissue Imaging with time-of-flight secondary ion mass spectrometry: Polyatomic ion sources, sample preparation, and applications. Curr. Pharm. Des. 2007;13:3335–3343. doi: 10.2174/138161207782360618. [DOI] [PubMed] [Google Scholar]
- 11.Sjovall P, Lausmaa J, Johansson B. Mass spectrometric imaging of lipids in brain tissue. Anal. Chem. 2004;76:4271–4278. doi: 10.1021/ac049389p. [DOI] [PubMed] [Google Scholar]
- 12.Fletcher JS, Lockyer NP, Vaidyanathan S, Vickerman JC. TOF-SIMS 3D Biomolecular Imaging of Xenopus laevis Oocytes Using Buckminsterfullerene (C60) Primary Ions. Anal. Chem. 2007;79:2199–2206. doi: 10.1021/ac061370u. [DOI] [PubMed] [Google Scholar]
- 13.Ostrowski SG, Van Bell CT, Winograd N, Ewing AG. Mass spectrometric imaging of highly curved membranes during Tetrahymena mating. Science. 2004;305:71–73. doi: 10.1126/science.1099791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng LL, Wucher A, Winograd N. Chemically alternating Langmuir-Blodgett thin films as a model for molecular depth profiling by mass spectrometry. J. Am. Soc. Mass Spectr. 2008;19:96–102. doi: 10.1016/j.jasms.2007.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng J, Wucher A, Winograd N. Molecular depth profiling with cluster ion beams. J. Phys. Chem B. 2006;110:8329–8336. doi: 10.1021/jp0573341. [DOI] [PubMed] [Google Scholar]
- 16.Cheng J, Winograd N. Depth profiling of peptide films with TOF-SIMS and a C-60 probe. Anal. Chem. 2005;77:3651–3659. doi: 10.1021/ac048131w. [DOI] [PubMed] [Google Scholar]
- 17.Wagner MS. Molecular depth profiling of multilayer polymer films using time-of-flight secondary ion mass spectrometry. Anal. Chem. 2005;77:911–922. doi: 10.1021/ac048945c. [DOI] [PubMed] [Google Scholar]
- 18.Kozole J, Szakal C, Kurczy M, Winograd N. Model multilayer structures for three-dimensional cell imaging. Appl. Surf. Sci. 2006;252:6789–6792. [Google Scholar]
- 19.Conlan XA, Lockyer NP, Vickerman JC. Is proton cationization promoted by polyatomic primary ion bombardment during time-of-flight secondary ion mass spectrometry analysis of frozen aqueous solutions? Rapid Commun. Mass Spectrom. 2006;20:1327–1334. doi: 10.1002/rcm.2446. [DOI] [PubMed] [Google Scholar]
- 20.Kurczy ME, Kozole J, Parry SA, Winograd N, Ewing AG. Relative Quantification of Cellular Sections with Molecular Depth Profiling ToF-SIMS Imaging. Appl. Surf. Sci. 2008;255:1068–1070. doi: 10.1016/j.apsusc.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones EA, Lockyer NP, Vickerman JC. Mass spectral analysis and imaging of tissue by ToF-SIMS - The role of buckminsterfullerene, C-60(+), primary ions. Int. J. Mass Spectrom. 2007;260:146–157. [Google Scholar]
- 22.Jones EA, Lockyer NP, Vickerman JC. Depth profiling brain tissue sections with a 40 keV C-60(+) primary ion beam. Anal. Chem. 2008;80:2125–2132. doi: 10.1021/ac702127q. [DOI] [PubMed] [Google Scholar]
- 23.Farnsworth HE, Schlier RE, George TH, Burger RM. Application of the Ion Bombardment Cleaning Method to Titanium, Germanium, Silicon, and Nickel as Determined by Low-Energy Electron Diffraction. J. Appl. Phys. 1958;29:1150–1161. [Google Scholar]
- 24.Roddy TP, Cannon DM, Ostrowski SG, Winograd N, Ewing AG. Identification of cellular sections with imaging mass spectrometry following freeze fracture. Anal. Chem. 2002;74:4020–4026. doi: 10.1021/ac025574w. [DOI] [PubMed] [Google Scholar]
- 25.Ding JX, Jiang D, Kurczy M, Nalepka J, Dudley B, Merkel EI, Porter FD, Ewing AG, Winograd N, Burgess J, Molyneaux K. Inhibition of HMG CoA reductase reveals an unexpected role for cholesterol during PGC migration in the mouse. Bmc Dev. Biol. 2008;8:14. doi: 10.1186/1471-213X-8-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozole J, Wucher A, Winograd M. Energy deposition during molecular depth profiling experiments with cluster ion beams. Anal. Chem. 2008;80:5293–5301. doi: 10.1021/ac8002962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sjovall P, Johansson B, Lausmaa J. Localization of lipids in freeze-dried mouse brain sections by imaging TOF-SIMS. Appl. Surf. Sci. 2006;252:6966–6974. [Google Scholar]