Abstract
Mechanisms that regulate the lifespan of CD4+CD8+ double positive (DP) thymocytes help shape the peripheral T cell repertoire. However, the molecular mechanisms that control DP thymocyte survival remain poorly understood. The Myb proto-oncogene encodes a transcription factor required during multiple stages of T cell development. We demonstrate that Myb mRNA expression is up-regulated in the small, pre-selection DP stage during T cell development. Using a conditional deletion mouse model, we demonstrate that Myb deficient DP thymocytes undergo premature apoptosis, resulting in a limited Tcrα repertoire biased towards 5’ Jα segment usage. Premature apoptosis occurs in the small pre-selection DP compartment in an αβTCR independent manner and is a consequence of decreased Bcl-xL expression. Forced Bcl-xL expression is able to rescue survival and re-introduction of c-Myb restores both Bcl-xL expression and the small pre-selection DP compartment. We further demonstrate that thymocytes become dependent on Bcl-xL for survival upon entering the quiescent, small pre-selection DP stage and c-Myb promotes transcription at the Bclx locus via a genetic pathway that is independent of the expression of TCF-1 or RORγt, two transcription factors that induce Bcl-xL expression in T cell development. Thus, Bcl-xL is a novel mediator of c-Myb activity during normal T cell development.
Introduction
The thymic cortex consists largely of quiescent DP thymocytes undergoing Vα-Jα recombination at the Tcrα locus. Production of a functional T cell receptor (TCR) α chain leads to replacement of the preTCR complex on DP thymocytes with surface expression of a mature αβTCR complex. Thymocytes that successfully assemble an αβTCR have three possible fates based on their reactivity towards self peptide-major histocompatibility complexes (MHC) (1). The vast majority of all DP thymocytes fail to produce an αβTCR with sufficient affinity towards the self peptide-MHC complex to become positively selected and undergo death by neglect. DP thymocytes that express an αβTCR with high affinity towards the self peptide-MHC complex are potentially auto-reactive and induced to die by negative selection. Less than 5% of all DP thymocytes produce an αβTCR with intermediate affinity towards self peptide-MHC complexes and receive the vital signal for positive selection. Positively selected DP thymocytes are granted continued survival and undergo commitment to either the CD4+ single positive (SP) or the CD8+ SP T cell lineage in a MHC dependent fashion (2).
T cell maturation relies on the balanced proliferation, survival and differentiation of thymocytes. During the DP stage, withdrawal from cell cycle coincides with the on set of Vα-Jα rearrangements at the Tcrα locus (3–5). The quiescent phenotype at this developmental stage has been attributed to the anti-proliferative function of the orphan nuclear receptor RORγt (6). DP thymocytes that fail to receive positive selection signals have a short intrinsic lifespan of 3–4 days before undergoing death by neglect (4, 5). To maximize the chance of assembling a productive αβTCR within this time frame, DP thymocytes are able to undergo multiple rounds of Vα-Jα rearrangements at the Tcrα locus, testing further distally 3’ located Jα segments (3, 7–9). This mechanism is known as receptor editing and is terminated by positive selection signals or death by neglect (10–12), ensuring only thymocytes that express a productive αβTCR will survive. Thus, the survival window of pre-selection DP thymocytes limits the progression of Tcrα rearrangements and thereby influences opportunities for positive selection and the generation of a diverse peripheral T cell repertoire (13). The lifespan of unsignaled DP thymocytes must therefore be precisely regulated to balance mechanisms that enforce death by neglect and those that enable sufficient receptor editing. Hypo-responsiveness to cytokine-mediated survival signals in pre-selection DP thymocytes has been reported to cause a decrease in Bcl-2 expression, glucose metabolism and cell volume, and increased sensitivity towards death by neglect (14–16). To counteract premature death by neglect, expression of the critical survival factor Bcl-xL, encoded by the proto-oncogene Bcl2l1, is greatly up-regulated in the DP stage of T cell development (17, 18). This efficient up-regulation has been attributed to the actions of RORγt and the HMG domain containing transcription factor TCF-1 (19, 20). Bcl-xL deficient DP thymocytes undergo premature apoptosis and display preferential use of proximal 5’ Jα segments (13, 18).
The Myb proto-oncogene encodes the highly conserved c-Myb transcription factor, which can function to activate or repress transcription in a context dependent manner (21, 22). The greatest amount of Myb expression is found in progenitor cells of each hematopoietic lineage and down-regulation of Myb expression is associated with differentiation (23–27). Myb−/− mutations are embryonic lethal and embryos die on day E15 due to severe anemia caused by failures of both erythroid and myeloid development (28). Evidence from several models point to the involvement of c-Myb in regulating the balance between survival, proliferation and differentiation that is required for normal hematopoiesis (29, 30). However, down stream mediators of c-Myb activities during hematopoiesis remain poorly understood.
Large amounts of Myb transcripts have been detected in the thymic cortex (26). We previously used conditional mutagenesis at the Myb locus to demonstrate that c-Myb is crucial at multiple stages of thymocyte differentiation, including transition from the DN to the DP stage, survival of pre-selection DP thymocytes and differentiation of CD4 SP thymocytes (31). To gain insight into the pro-survival role of c-Myb in DP thymocytes, the present study uses a Cd4-Cre expressing mouse strain to achieve efficient deletion at the Myb locus in DP thymocytes. We demonstrate that Myb deficient DP thymocytes undergo premature apoptosis due to decreased Bcl-xL expression. Premature apoptosis occurs in the small, pre-selection DP compartment and restricts the 3’ progression of Jα segment usage during Tcrα recombination. Forced Bcl-xL expression is able to rescue survival and re-introduction of c-Myb restores both Bcl-xL expression and the small pre-selection DP compartment. We demonstrate that c-Myb stimulates Bcl-xL expression at the level of transcription, through a genetic pathway independent of RORγt and TCF-1 expression. Finally, we demonstrate that survival is regulated differently in large and small pre-selection DP thymocytes, and that only the latter depend c-Myb and Bcl-xL for their survival.
Materials and methods
Mice
Mice were used at 4–6 weeks of age. Mybf/f and Mybf/w mice were previously described (31). Cd4-Cre (generous gift of Dr Christopher B. Wilson, University of Washington, Seattle, WA) and Bcl-2tg mice (generous gift of Dr Ellen V. Rothenberg, California Institute of Technology, Pasadena, CA) were previously described (32, 33). Tcrα−/− mice were purchased from Jackson Labs (Bar Harbor, ME). Tcf-1−/− and Rorγ−/− mice (generous gifts of Dr Zouming Sun, Beckman Research Institute of the City of Hope, Duarte, CA) were previously described (19, 34). These studies have been reviewed and approved by the Institutional Animal Care and Use Committee at the University of Virginia.
Flow cytometry and cell sorting
Single-cell suspensions of thymi from 4- to 6-week-old mice were prepared and stained with fluorochrome- or biotin-conjugated antibodies as described (31). Antibodies and reagents were purchased as follows: Caltag (Burlingame, CA), anti-CD4-APC (clone CT-CD4), and anti-CD8 APC-Alexa750 (clone 5H10); eBioscience (San Diego, CA), anti-CD90.2 PE (clone 53-2.1); Cell Signaling Technology (Beverly, MA), anti-Bcl-xL (clone 54H6); Jackson ImmunoResearch (West Grove, PA), F(ab’) Fragment Goat-anti-rabbit-IgG (H+L)-APC. Intracellular staining with anti-Bcl-xL was performed according to manufacturer’s recommendations. Apoptosis detection by flow cytometry was performed using the Annexin V-FITC or PE apoptosis detection kit (BD Biosciences) and 7-aminoactinomycin D (Molecular probes, Carlsbad, CA) according to manufacturer’s recommendations. Cells were analyzed on a FACSCalibur or Becton Dickinson FACSVantage SE Turbo Sorter with DIVA Option (BD Biosciences, San Jose, CA). Data was analyzed using FlowJo (Tree Star) software. Electronic cell sorting was done on a Becton Dickinson FACSVantage SE Turbo Sorter with DIVA Option.
qRT-PCR
Electronically sorted thymocytes were lysed and homogenized over Qiashredder columns (Qiagen, Valencia, CA). Total RNA was extracted using the RNeasy Mini kit (Qiagen) including on-column DNase digestion (RNase-free DNase Set: Qiagen) following the manufacturer’s recommendations. RNA was converted into cDNA using Superscript II First strand cDNA synthesis system with oligo(dT) primers (Invitrogen, Carlsbad, CA) following manufacturer’s recommendations.
qPCR was performed using Titanium Taq DNA polymerase (Clonetech, Mountain View, CA) and 1× SYBR Green (Invitrogen) in an Opticon DNA engine (MJ research; Waltham, MA). qPCR conditions are as follows, 3 minutes at 95°C, followed by 40 cycles of 95°C for 40s, 66°C for 20s, and 72°C for 30s. After a final extension step (72 C for 1min), melting curve analysis was executed. mRNA expression primers are as follows: c-Myb forward 5’ AACGAGCTGAAGGGACAGCA 3’, c-Myb reverse 5’ TGGCATGGTGTTCTCCCAAA 3’. Rorγ, Tcf1 and Bcl2l1 primers were previously described (35–37). The mRNA levels were normalized using Hprt-1 primers (forward: 5’ TGCCGAGGATTTGGAAAAAGTG 3’, reverse: 5’ CACAGAGGGCCACAATGTGATG 3’) as previously described (38).
mRNA expression microarray analysis
DP thymocytes from four Mybf/w Cd4-Cre Tcrα−/− and four Mybf/f Cd4-Cre Tcrα−/− mice were purified using MACS CD4 MicroBeads (Miltenyi Biotec, Auburn, CA) per the manufacturer’s protocol. Positive selection yielded populations of 91–99% purity as determined by flow cytometry. Total RNA (3–15 µg) was extracted as described above and assessed for its integrity by analysis on an Agilent Bioanalyzer. Generation of double stranded cDNA and biotin-labeled cRNA was synthesized per manufacturer’s instructions. Biotin-labeled cRNA from each mouse was individually hybridized to Mouse Genome 430 2.0 (Affymetrix, Santa Clara, CA) Gene Chip for 16 h, and scanned with the Affymetrix Gene-Array Scanner as previously described (39). c-RNA synthesis, hybridization and data collection were performed at the University of Virginia Gene Chip analysis Core Facility.
Pairwise analysis of control and mutant sample groups was performed using the GeneSifter microarray data analysis system (http://www.genesifter.net/) (VizX Labs LLC, Seattle, WA) to identify genes with statistically significant (p<0.01 Student’s t-test) >2 fold change in expression within the Gene Ontology group GO:0043067:regulation of programmed cell death (40).
Western blotting
Electronically sorted DP thymocytes were lysed in (20 mM Tris, 7.4; 100 mM NaCl, 10 mM EDTA; 1 mM EGTA, 1% Triton X-100) (41) containing EDTA-free protease inhibitor cocktail (Roche, Indianapolis, IN). Fifteen milligrams of protein was fractionated on 10% SDS-polyacrylamide gels and transferred to Protran nitrocellulose transfer membranes (Whatman, Dassel, Gernmany). Membranes were blocked in PBS, 0.05% Tween-20 (PBS-T) with 5% nonfat dry milk for 1hr and then incubated with the appropriate primary antibody over night at 4°C. Membranes were washed three times in PBS-T and probed with anti-mouse-HRP, anti-rabbit-HRP or anti-armenian hamster-HRP conjugated antibody in PBS-T for 1 h at room temperature. After washing the membrane three times in PBS-T, the proteins were detected by enhanced chemiluminescence (Amersham, Piscataway, NJ). Western blotting primary antibodies: Millipore (Bedford, MA): anti-c-Myb (clone 1-1). Cell Signaling Technology (Beverly, MA): anti-Bcl-xL (clone 54H6) and anti-TCF1 (clone C63D9). Rockland (Gilbertsville, PA): anti-Mcl-1 (600-401-394). Anti-RORγ was a generous gift from Dr Dan R. Littman, NYU School of Medicine, NY (19).
In vitro survival assays
Freshly isolated total thymocytes were cultured in RPMI-1640 growth medium supplemented with 10% FBS, 100U/ml penicillin-streptomycin and 2 mM L-glutamine (Gibco, Carlsbad, CA) and 50 µM 2-mercaptoethanol (Sigma-Aldrich, St. Louis, MO) at 2×106 cells/ml with no treatment or 40 mM Z-VAD(OMe)-FMK (ICN Pharmaceuticals, Aurora, OH) for the indicated amount of time in a humidified chamber with an atmosphere of 5% CO2. To monitor the synchronized differentiation of large and small pre-selection DP thymocytes in vitro, total thymocytes were negatively selected over MACS CD4 MicroBeads (Miltenyi Biotec, Auburn, CA). Flow-through was >95% DN and ISP thymocytes as determined by flow cytometry and placed in culture for the indicated amounts of time.
Jα segment profiling
Total cellular RNA from electronically sorted DP thymocytes was subjected to reverse transcription, PCR and southern blotting as previously described (9, 13). PCR primers and southern blotting probes were as described (9, 42). End labeling of the probes with [α-32P]dATP (Perkin Elmer, Waltham, MA) was performed using T4 polynucleotide kinase (New England Biolabs, Ipswich, MA).
Chromatin Immunoprecipitation
Protein was crosslinked to chromatin by adding 1% formaldehyde to 107 total thymocytes/ml cold PBS for 10 minutes on ice. The reaction was stopped by adding 125 mM glycine for 5 minutes with rocking at room temperature. Cells were pelleted and washed once in cold PBS. Cell pellets were resuspended in 107 cells/ml cold cytoplasmic lysis buffer (10 mM Tris-HCl pH8, 85 mM KCl, 0.5% NP40, 1 mM PMSF and EDTA-free protease inhibitor cocktail, Roche, Indianapolis, IN) and incubated on ice for 10 minutes. Nuclei were resuspended at 107 cells/ml in ice cold sonication buffer (10 mM Tris-HCl pH8, 0.1 mM EDTA, 1% NP40, 1 mM PMSF, EDTA-free protease inhibitor cocktail) and sonicated using model W-375 cell disruptor (Ultrasonics Inc, Plainview, NY) to generate chromatin fragments 200–500 bp in size. Debris was cleared by centrifugation and sonication buffer was supplemented with 5% glycerol and 127 mM NaCl. Chromatin aliquots of 500 µl was pre-cleared using salmon sperm DNA/protein A agarose slurry (Millipore, Bedford, MA) for 1 hour and immunoprecipitated over night with either 3 µg of anti-RNA polymerase II CTD clone 4H8 (Abcam, Cambridge, MA) or mouse IgG2a,κ isotype control (BD Biosciences, San Jose, CA) with rotation at 4°C. Immune complexes were collected with 100 ml of salmon sperm DNA/protein A agarose slurry for 1 hour with rotation at 4°C. Beads were washed for 5 minutes with rotation at 4°C with low salt buffer (10 mM Tris-HCl pH8, 2 mM EDTA, 0.1% SDS, 1% NP40, 150 mM NaCl), high salt buffer (10 mM Tris-HCl pH8, 2 mM EDTA, 0.1% SDS, 1% NP40, 500 mM NaCl), LiCl buffer (10 mM Tris-HCl pH8, 1mM EDTA, 1% Deoxycholate, 1% NP40, 250 mM LiCl) and twice in TE. All wash buffers were supplemented with protease inhibitors and PMSF. Bound complexes were eluted off the beads in 500 µl elution (0.1 M, 1% SDS) buffer with rotation at room temperature for 30 minutes. Formaldehyde crosslinking was reversed in the presence of 200 mM NaCl at 65°C overnight. DNA was phenol/chloroform extracted following RNAse A and proteinase K treatment. RNA polymerase II localization at the thymic Bcl2l1 transcriptional start site was detected by qPCR. ChIP primer 1F 5’-TTGGACACCGACATCGAAAG-3’, 1R 5’-CGCGTGGAACGTTTATGGTT-3’ 2F 5’-ATTCCTCTGTCGCCTTCTGA-3’, 2R 5’-CCCCGGAAGGTCTTTTGTAT-3’.
Co-culture and transduction of thymocytes on OP9-DL1 stromal cells
Enriched DN thymocytes were allowed to differentiate on OP9-DL1 cells (generous gift from J.C. Zuniga-Pflucker, University of Toronto, Toronto, Canada) (43). 108 total thymocytes were depleted for DP thymocytes using MACS CD4 and CD8 MicroBeads (Miltenyi Biotec, Auburn, CA) per manufacturer’s protocol. Negatively selected thymocytes were cultured over night at 5 × 105/mL on 80% confluent OP9-DL1 mono-layer in flat-bottom 24-well culture plates with αMEM (Invitrogen Life Technologies, Carlsbad, CA) supplemented with 20% FBS, 100 U/ml penicillin-streptomycin and 2 mM L-glutamine (Invitrogen Life Technologies, Carlsbad, CA) and 5 ng/ml recombinant murine IL-7 (PeproTech, Rocky Hill, NJ). The following day, co-cultures are transduced by spinfection at 700 × g for 90 minutes using retroviral supernatants in the presence of 8 µg/ml polybrene (Sigma-Aldrich). pMIGR1 (gift from Dr Warren S. Pear, University of Pennsylvania, Philadelphia, PA) was previously described (44). The MIGR1-c-Myb vector was constructed by subcloning a HA-c-Myb cDNA construct into the BamHI/BglII site of MIGR1. The MIGR1-Bcl-xL vector was a kind gift from Dr. Thomas J. Braciale (University of Virginia, Charlottesville, VA). Retroviral supernatants were produced by transient CaPO4 co-transfection of 293T cells with the RetroMax packaging vector pCL-Eco (Imgenex, San Diego, CA) and the appropriate MIGR1 based expression vector. Seventy-two hrs post transduction co-cultures were harvested for flow cytometry or electronic cell sorting. For the detection of Bcl-xL protein expression in MIGR1-c-Myb transduced DP thymocytes, 40 µM of Z-VAD (ICN Pharmaceuticals, Aurora, OH) was added to co-cultures 48 hrs post transduction and 24 hrs prior to harvest.
Statistics
Differences between data sets were analyzed with two-tailed Student's t-test and a confidence level of 99% for the mRNA expression microarray and 95% for all other experiments.
Results
Myb is highly expressed in pre-selection DP thymocytes
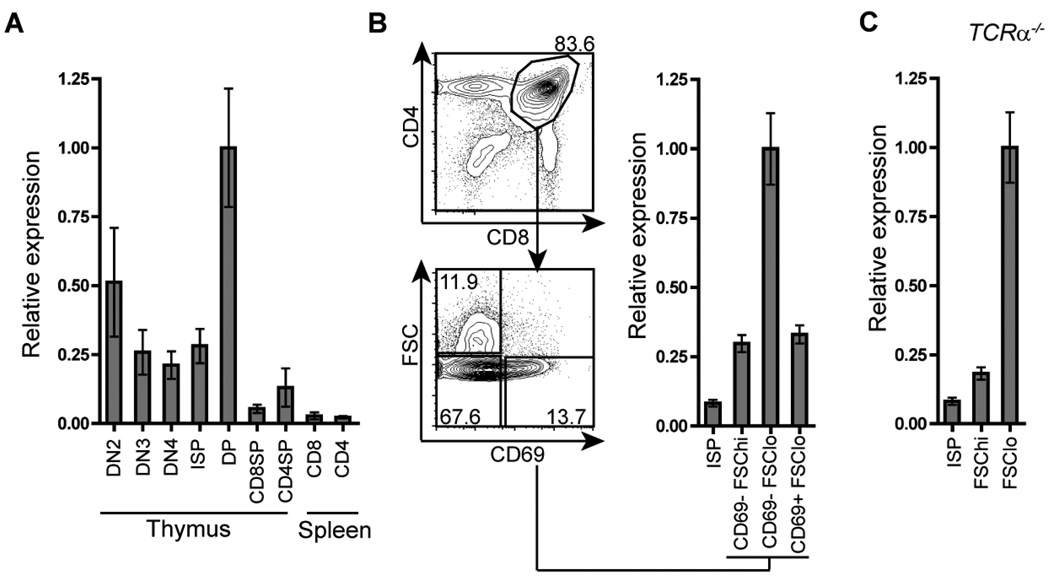
Myb mRNA is abundantly produced by DN and DP thymocytes compared to SP thymocytes and naïve CD4 and CD8 T cells (26, 31). To better resolve changes in Myb mRNA expression during αβT cell development, we performed quantitative reverse-transcription (qRT) PCR analysis on mRNA extracted from electronically sorted populations ranging from DN2 thymocytes to mature, naïve T cell subsets in the spleen (Fig. 1A). The greatest amount of Myb mRNA was detected in DN2 and DP thymocytes. Interestingly, the amount of Myb mRNA detected decreased in DN3, DN4 and ISP thymocytes compared to DN2 and DP thymocytes, suggesting that Myb mRNA expression decreases as developing thymocytes transit through the DN3, DN4 and ISP stages and then increases approximately 4-fold in the DP subset. Myb mRNA expression is greatly decreased in CD4 and CD8 SP thymocytes and naïve CD4 and CD8 T cells compared to DP thymocytes. The DP compartment was further sorted into CD69−FSChi, CD69−FSClo and CD69+FSClo subsets (45), referred to here as large pre-selection, small preselection and post-selection DP thymocytes respectively (Fig. 1B). The largest amount of Myb mRNA was detected in the small pre-selection DP subset (Fig. 1B). Moreover, this peak in expression was readily detectable in Tcrα−/− mice (Fig. 1C), demonstrating that increased Myb mRNA expression occurs in small pre-selection thymocytes in an αβTCR independent manner. Consistent with previous reports (26, 46, 47), a sharp decrease of Myb mRNA level was observed in the post-selection DP subset, suggesting that down-regulation of Myb mRNA expression may be a consequence of αβTCR mediated selection signals (Fig. 1B). In further support of this notion, mimicking positive selection by in vitro stimulation of Tcrα−/− DP thymocytes with phorbol 12-myristate 13-acetate (PMA) and Ca2+ ionophore A23187 (48) resulted in a rapid decrease in Myb mRNA level (Supplementary Fig. 1). These results demonstrate that Myb expression is dynamically regulated during T cell development. The abundance of Myb mRNA expression detected in the small pre-selection DP subset suggests that c-Myb may play a particularly important role at this stage during thymocyte development.
Figure 1. Expression of Myb mRNA during αβT cell development.
Myb mRNA expression was analyzed by quantitative real-time PCR and normalized to the expression of Hprt-1 mRNA. The peak of relative Myb mRNA levels in each experiment is indicated as 1. Data are presented as mean +/− SEM, n=2, and are representative of two mice. (A) Relative Myb mRNA levels in DN2 (CD4−CD8−c-Kit+CD44+CD25+), DN3 (CD4−CD8−c-Kit−CD44−CD25+), DN4 (CD4−CD8−c-Kit−CD44−CD25−) thymocytes (66), ISP (TCRαloCD4−CD8+) and thymic and splenic CD4SP (TCRαhiCD4+CD8−) and CD8SP (TCRαhiCD8+CD4−) populations. (B) Relative Myb mRNA in ISP (TCRαloCD4−CD8+), large pre-selection DP (FSChiCD69−CD4+CD8+), small pre-selection DP (FSCloCD69−CD4+CD8+) and post-selection DP subsets (FSCloCD69+CD4+CD8+). (C) Relative Myb mRNA expression in Tcrα−/− ISP (CD4−CD8+), large pre-selection DP (CD4+CD8+FSChi) and small pre-selection DP (CD4+CD8+FSClo) subsets.
c-Myb is required for the survival of small pre-selection DP thymocytes
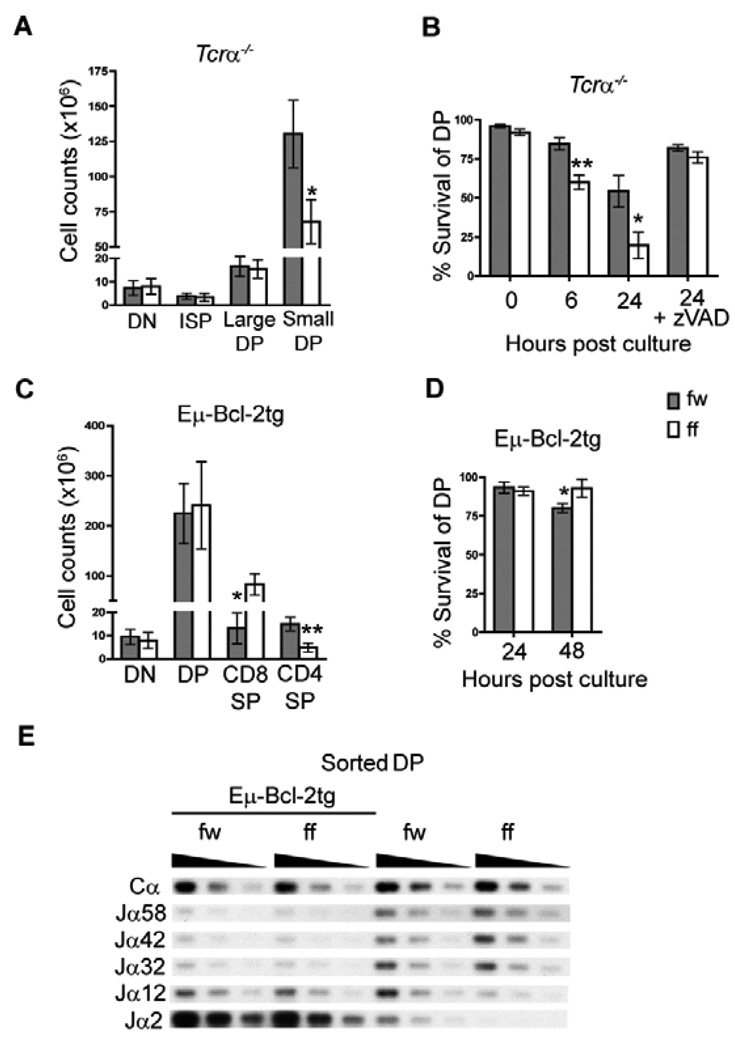
Apoptotic cell death in wildtype DP thymocytes occurs most commonly through αβTCR independent death by neglect, but also through αβTCR dependent negative selection. We have previously demonstrated that Lck-Cre mediated deletion in mice homozygous for the loxP targeted allele of Myb (Mybf/f) results in a Tcrα independent survival defect in DP thymocytes and impaired development of CD4SP thymocytes (31). We obtain the same phenotype with Cd4-Cre mediated deletion (Supplementary Fig. 2). However, Cd4-Cre mediated deletion of the Mybf allele is more efficient and we do not detect counter selection for the Mybf allele in CD4SP thymocytes as we did with Lck-Cre (31). Interestingly, we did not detect a difference in the number of large pre-selection DP thymocytes in thymi from Mybf/f Cd4-Cre Tcrα−/− mice compared to Mybf/w Cd4Cre Tcrα−/− controls, but we detected a severe reduction in the number of small pre-selection DP thymocytes (Fig. 2A). This finding in combination with abundant Myb mRNA expression suggests that c-Myb may be especially important for the survival of small preselection DP thymocytes. To determine if impaired survival of Mybf/f Cd4-Cre DP thymocytes is a result of apoptotic cell death we compared the ability of Mybf/w Cd4-Cre and Mybf/f Cd4-Cre DP thymocytes to survive in liquid culture in the presence or absence of the pan-caspase inhibitor Z-VAD. After 6 and 24 hrs in culture, decreased survival was detected in Mybf/f Cd4- Cre Tcrα−/− DP thymocytes compared to controls (Fig. 2B). Survival of Mybf/f Cd4-Cre DP thymocytes was restored to a level equivalent to controls after treatment with Z-VAD, suggesting that the decreased number of cells detected in c-Myb deficient DP thymocytes was due to apoptotic cell death. This result was further confirmed by increased TUNEL staining and caspase-3 activation in cultured DP thymocytes lacking c-Myb (Supplementary Fig. 3). Consistent with the notion that the requirement for c-Myb mediated survival is specific to the small pre-selection DP compartment, we observed a significant depletion of the small preselection DP compartment in Mybf/f Cd4-Cre compared to Mybf/w Cd4-Cre thymocytes 24 hrs post culture, which is prevented by the presence of Z-VAD (Fig. 2C). Thus, decreased survival in c-Myb deficient DP thymocytes is a consequence of increased apoptotic cell death that occurs in an αβTCR independent fashion in the small pre-selection DP thymocyte subset.
Figure 2. Mybf/f Cd4-Cre DP thymocytes undergo increased apoptotic cell death in a Tcrα independent fashion.
(A) The absolute number of 4-6-week-old Mybf/w (fw) Cd4-Cre Tcrα−/− and Mybf/f (ff) Cd4-Cre Tcrα−/− thymocyte subsets was calculated based on total thymic cellularity and the percentage of DN (CD4−CD8−), ISP (CD4−CD8+), large pre-selection DP (CD4+CD8+FSChi) and small pre-selection DP (CD4+CD8+FSClo) thymocytes. Data are presented as mean +/− SEM. n≥5, *p = 0.0068 (Student’s t-test). (B) Assessment of Tcrα−/− DP thymocyte survival after 6 and 24 hrs in culture. Where indicated, thymocytes were cultured in the presence of 40 µM Z-VAD. Cultures were stained for CD4, CD8, 7AAD, and Annexin 5. Percent survival of DP was defined as the percentage of 7AAD−Annexin 5− cells through a CD4+CD8+ gate. n≥5, *p = 0.011, **p = 0.0023. (C) Histograms display the forward scatter distribution of live DP thymocytes of the indicated genotype 24 hrs post culture in the presence or absence of Z-VAD through a 7AAD− Annexin 5− gate. Number above gate indicates the percentage of live DP thymocytes within a FSChi gate. (D) Absolute number of Bcl-2tg DN (CD4−CD8−), large DP (FSChiCD4+CD8+), small DP (FSCloCD4+CD8+) CD8SP (CD4−CD8+) and CD4SP (CD4+CD8−) thymocyte populations of 4-6-week-old Mybf/w Cd4-Cre and Mybf/f Cd4-Cre mice. Data are presented as mean +/− SEM. n=3, *p = 0.022, **p = 0.011. (E) Assessment of total Bcl-2tg DP thymocyte survival after 24 and 48 hrs in culture. n=3, *p = 0.030. (F) Histograms display the forward scatter distribution of live DP thymocytes of the indicated genotype 24 hrs post culture through a 7AAD− Annexin 5− gate. Number above gate indicates the percentage of live DP thymocytes within a FSChi gate. (G) Decreased 3’ Jα segment use in Mybf/f Cd4-Cre DP thymocytes due to impaired survival. cDNA was generated from sorted Mybf/w Cd4-Cre and Mybf/f Cd4-Cre DP thymocytes in the presence and absence of Bcl-2tg expression. PCR amplification was performed with serially (1/3) diluted cDNA samples using primers specific for Vα3 and Cα. PCR products were sequentially probed using oligonucleotide probes specific for the indicated Jα segments as previously described (9, 42). An internal Cα probe was used to normalize input cDNA. Data is representative of two independent experiments.
Apoptosis is an important component of normal T lymphopoiesis and can be triggered through the extrinsic or the intrinsic pathway (49). Spontaneous apoptosis of normal DP thymocytes in liquid culture is believed to mimic death by neglect and largely reflect the activity of the intrinsic apoptotic pathway in a cytochrome c/Apaf-1/caspase-9 apoptosome independent fashion (50). To better understand how the absence of c-Myb resulted in increased apoptosis of small pre-selection DP thymocytes we determined the pathway by which accelerated apoptosis takes place in Myb deficient pre-selection DP thymocytes. We cultured Mybf/f Cd4-Cre Tcrα−/− and Mybw/f Cd4-Cre Tcrα−/− thymocytes in the presence or absence of the caspase-8 specific inhibitor Z-IETD to examine a possible contribution by the extrinsic apoptotic pathway. While Z-IETD effectively inhibited apoptosis of anti-CD95 treated C57BL/6j DP thymocytes, it did not prevent premature apoptosis of Mybf/f Cd4-Cre Tcrα−/− DP thymocytes in vitro (Supplementary Fig. 4), arguing against a major role for the extrinsic apoptotic pathway in the survival defect caused by the lack of c-Myb. Thus, we turned our attention to members of the Bcl-2 family, which are key effectors of the intrinsic apoptotic pathway. The over-expression or abrogation of several Bcl-2 family members has been associated with dramatic effects on the lifespan of preselection DP thymocytes (17, 32, 35, 51, 52). To determine if forced expression of a pro-survival Bcl-2 family member could rescue survival, Mybf/f Cd4-Cre mice were crossed with Bcl-2tg expressing mice. The Bcl-2tg mice direct expression of the human Bcl-2 transgene to the T cell lineage (32), and fully restored both cellularity (Fig. 2D) and in vitro survival and maintenance of c-Myb deficient small DP thymocytes (Fig. 2E and F). These results indicate that c-Myb may counteract the intrinsic apoptotic pathway to promote the survival of preselection DP thymocytes. Interestingly, the Bcl-2tg did not restore but rather enhanced the reduced CD4SP:CD8SP ratio observed in Mybf/f Cd4-Cre thymocytes (Fig. 2D). Previous interpretation of the role for c-Myb in CD4SP lineage development (31, 53, 54) was confounded by the survival defect in c-Myb deficient pre-selection DP thymocytes, which limits opportunities for positive selection (13). Our observations in Mybf/f Cd4-Cre thymocytes carrying an Bcl-2tg demonstrate that c-Myb deficiency negatively affects CD4SP lineage representation independent of its ability to promote DP thymocyte survival.
Decreased lifespan of DP thymocytes in vivo results in the predominant usage of 5’ proximal Tcrα Jα segments due to limited 3’ progression of rearrangements along the Jα locus (13). To confirm our in vitro survival data, we compared Jα segment usage in DP thymocytes from Mybf/w Cd4-Cre and Mybf/f Cd4-Cre mice, with or without Bcl-2tg expression. cDNA generated from sorted DP thymocytes was subjected to a PCR-based assay (9), in which products amplified by Vα3 and Cα segment specific primers were sequentially probed for a selection of Jα segments ranging from 5’ (proximal) to 3’ (distal) in location (9, 42). A probe against the Cα segment demonstrates equal loading. Comparison of Jα segment profiles revealed a severe decrease in the usage of the distally located Jα12 and Jα2 gene segments by Mybf/f Cd4-Cre DP thymocytes compared to controls (Fig. 2E). In contrast, both Mybf/w Cd4-Cre and Mybf/f Cd4-Cre DP thymocytes displayed preferential usage of the distal Jα2 segment in the presence of Bcl-2tg expression, consistent with prolonged survival independent of c-Myb. Thus, c-Myb deficient DP thymocytes do not progress to distal Jα segment usage as a direct consequence of premature apoptosis occurring in vivo.
Decreased Bcl-xL expression in Myb deficient DP thymocytes
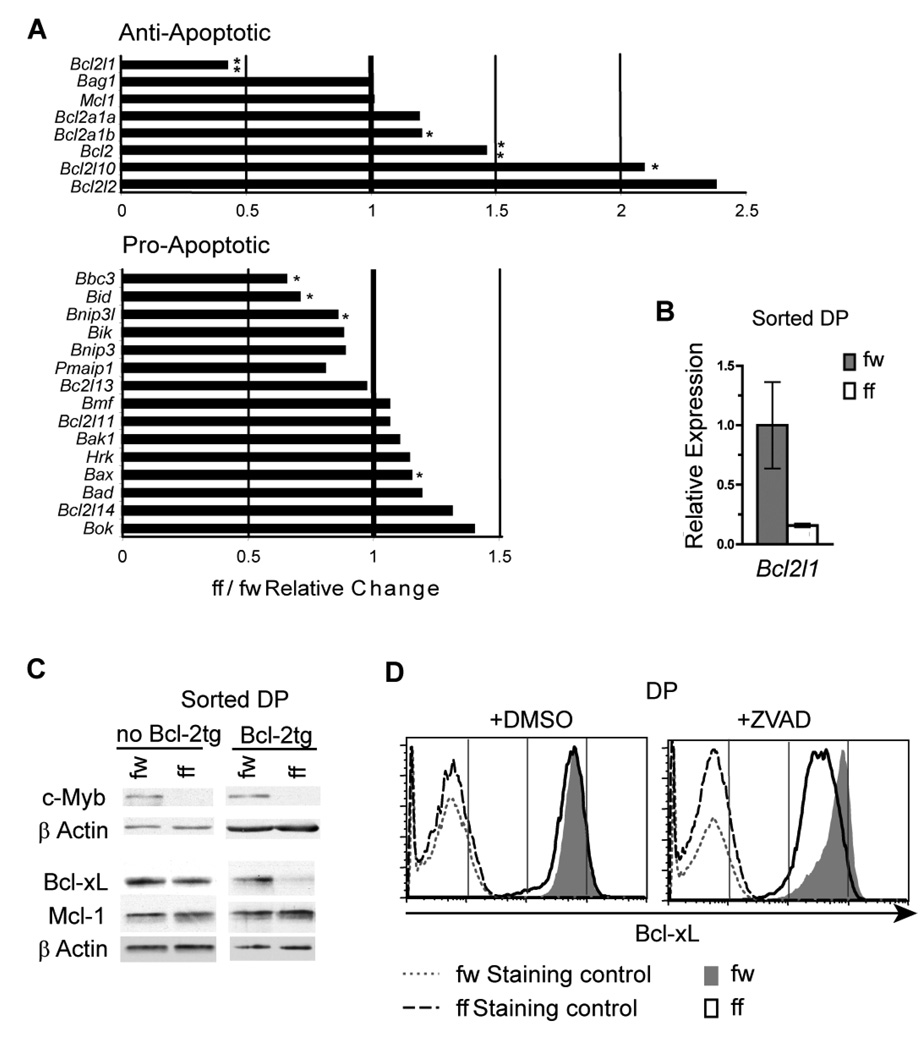
To identify candidate c-Myb target genes that mediate protection from apoptosis through the intrinsic pathway, we performed mRNA expression microarray analysis to compare gene expression profiles in DP thymocytes purified over magnetic beads from four Mybf/w Cd4-Cre Tcrα−/− and four Mybf/f Cd4-Cre Tcrα−/− DP mice. All eight samples were individually hybridized to Mouse Genome 430 2.0 Gene Chips and the resulting dataset was subjected to several selection criteria. We identified genes that displayed a >2 fold differential expression within the gene ontology group Regulation of programmed cell death (GO:0043067) (40) (Supplementary Table 1). This analysis revealed a statistically significant 2.4 fold (p<0.01) decrease in Bcl2l1 transcript levels. In addition, while no pro-apoptotic members were clearly up-regulated, Bcl2l1 was the only anti-apoptotic family member significantly down-regulated in Mybf/f Cd4-Cre Tcrα−/− DP thymocytes (Fig. 3A). The expression of Bcl-xL, like that of c-Myb, is specifically up-regulated in DP thymocytes (17, 18), making Bcl2l1 an attractive candidate down-stream target of c-Myb. The decrease in Bcl2l1 mRNA level identified in the microarray experiment was validated in sorted small Mybf/w Cd4-Cre Tcrα−/− and Mybf/f Cd4-Cre Tcrα−/− DP thymocytes by qRT-PCR (Fig. 3B).
Figure 3. Decreased Bcl-xL expression in Mybf/f Cd4-Cre Tcrα−/− DP thymocytes.
(A) Changes in mRNA expression of Bcl-2 family members in c-Myb deficient pre-selection DP thymocytes detected by mRNA expression microarray analysis. mRNA expression microarray analysis was performed comparing DP thymocytes from Mybf/w Cd4-Cre Tcrα−/− and Mybf/f Cd4-Cre Tcrα−/− mice. Bar graphs represent mean of relative mutant:control mRNA expression of anti- and pro-apoptotic Bcl-2 family members in DP thymocytes. Genes of the Bcl-2 super family were manually selected from the complete GO:0043067:regulation of programmed cell death ontology list (40). n=4, *p < 0.05, **p < 0.01 (Student’s t-test). (B) Bcl2l1 mRNA expression was analyzed by quantitative real-time PCR in electronically sorted Mybf/w Cd4-Cre Tcrα−/− and Mybf/f Cd4-Cre Tcrα−/− FSClo DP thymocytes and normalized to Hprt-1 mRNA expression. Results are representative of 3 separate experiments. n=2. (C) Western blot of whole cell lysates from electronically sorted Mybf/w Cd4-Cre Tcrα−/− and Mybf/f Cd4-Cre Tcrα−/− DP thymocytes with and without Eµ-Bcl-2tg expression probed for c-Myb, Bcl-xL, Mcl-1 and β-actin. Results are representative of 3 separate experiments. (D) Intracellular staining for Bcl-xL, CD4 and CD8 in Mybf/w Cd4-Cre Tcrα−/− and Mybf/f Cd4-Cre Tcrα−/− DP thymocytes 24 hrs post culture with either 2 µl/ml DMSO or 40 µM Z-VAD. All antibodies except the anti-Bcl-xL primary antibody were used in the staining controls. Results are representative of 3 separate experiments.
Surprisingly, we did not detect decreased expression of Bcl-xL protein by western blotting (Fig. 3C), despite undetectable levels of residual c-Myb protein in sorted Mybf/f Cd4-Cre Tcrα−/− DP thymocytes. However, since the protein half-life of Bcl-xL greatly exceeds that of c-Myb (55–57) we reasoned c-Myb deficient thymocytes that lack expression of Bcl-xL might die rapidly leaving, mainly cells that maintain sufficient expression of Bcl-xL to survive. To address this possibility, Bcl-xL protein expression was examined in Mybf/w Cd4-Cre Tcrα−/− and Mybf/f Cd4-Cre Tcrα−/− DP thymocytes that either carried aBcl-2tg or were treated with Z-VAD in liquid culture. Strikingly, both approaches revealed a marked decrease in Bcl-xL protein in c-Myb deficient compared to c-Myb sufficient DP thymocytes (Fig. 3C and 3D), suggesting that impaired survival masked the reduction in Bcl-xL protein in freshly isolated pre-selection DP thymocytes lacking c-Myb. We also compared Mcl-1 protein expression in Mybf/f Cd4-Cre Tcrα−/− and Mybf/f Cd4-Cre Tcrα−/− DP thymocytes with or without Bcl-2tg expression. Mcl-1 is another pro-survival member of the Bcl-2 family recently reported to play a role in the survival of DP thymocytes (58). Consistent with our mRNA expression microarray result, we did not detect a decrease in the amount of Mcl-1 protein in Mybf/f Cd4-Cre Tcrα−/− DP thymocytes with or without Bcl-2tg expression (Fig. 3C). These results demonstrate that the reduced amount of Bcl-xL protein in Mybf/f Cd4-Cre Tcrα−/− DP thymocytes carrying an Bcl-2tg is protein specific among Bcl-2 family members.
Small but not large pre-selection DP thymocytes are dependent on Bcl-xL for survival
It has been well established that Bcl-xL is a crucial survival factor in DP thymocytes (17, 18). Our observation that Bcl-xL expression is decreased in c-Myb deficient pre-selection DP thymocytes and that the number of small but not large DP thymocytes is reduced in Mybf/f Cd4-Cre Tcrα−/− compared to Mybf/w Cd4-Cre Tcrα−/− littermates (Fig. 2A) collectively suggest that c-Myb and Bcl-xL may be important for maintaining the survival of small pre-selection DP thymocytes specifically. To better characterize the requirement for the pro-survival function of Bcl-xL as pre-selection DP thymocytes differentiate from large to small, we monitored Bcl-xL protein expression and cell survival as Mybf/w Cd4-Cre Tcrα−/− and Mybf/f Cd4-Cre Tcrα−/− DP thymocytes spontaneously differentiate from the large to the small pre-selection DP stage in vitro. Adapting a previously described assay (61), thymocytes were depleted of the DP population on magnetic beads and placed in liquid culture, allowing DN and ISP thymocytes that have received preTCR signals in vivo to differentiate into the large and small pre-selection DP stage followed by death by neglect in a synchronous manner. After 20 and 36 hrs, in vitro differentiated DP thymocytes from both Mybf/f Cd4-Cre Tcrα−/− and Mybf/w Cd4-Cre Tcrα−/− mice consisted predominantly of large and small pre-selection DP thymocytes respectively (Fig. 4A). At 20 hrs post culture, reduced intracellular Bcl-xL protein was readily detectable without Z-VAD treatment in Mybf/f Cd4-Cre Tcrα−/− DP thymocytes compared to Mybf/w Cd4-Cre Tcrα−/− DP thymocytes (Fig. 4B) while no significant difference was detected in their ability to survive (Fig. 4C). This result demonstrates that Bcl-xL protein expression is significantly reduced in Mybf/f Cd4-Cre Tcrα−/− large DP thymocytes without negatively impacting their survival and is consistent with a model where c-Myb and Bcl-xL are not required for the survival of large pre-selection DP thymocytes. This is in contrast to 36 hrs post culture, when we observed no difference in Bcl-xL protein expression (Fig. 4B) but significantly impaired survival in Mybf/f Cd4-Cre Tcrα−/− DP thymocytes (Fig. 4C), indicative of a survival defect in DP thymocytes harboring reduced Bcl-xL protein. This phenotype highly resembles our observations in total thymocytes without Z-VAD treatment, likely because the in vivo DP compartment is predominantly composed of small pre-selection DP thymocytes at any given time regardless of the sufficient Myb expression (Fig. 2C). Collectively, these results support a model where thymocyte survival becomes dependent on Bcl-xL at a point near the end of the proliferative large pre-selection DP stage or as developing thymocytes enter the quiescent small pre-selection stage.
Figure 4. Small but not large pre-selection DP thymocytes are acutely sensitive to reduced intracellular Bcl-xL.
Total Mybf/w Cd4-Cre Tcrα−/− and Mybf/f Cd4-Cre Tcrα−/− thymocytes were depleted of DP thymocytes using MACS CD4 MicroBeads. Negatively selected cells were subsequently cultured for 20 or 36 hrs and analyzed by flow cytometry. Results are representative of three separate experiments. (A) Histogram overlays present the change in FSC of Mybf/w Cd4-Cre Tcrα−/− and Mybf/f Cd4-Cre Tcrα−/− DP thymocytes from 20 to 36 hrs post culture. (B) Intracellular staining for Bcl-xL measured by flow cytometry at 20hrs and 36 hrs post culture through a live DP thymocyte gate. (C) Assessment of DP thymocyte survival after 20 hrs and 36 hrs in culture by staining for CD4, CD8, 7AAD and Annexin 5. Results are presented as mean +/− SEM. n≥3, *p < 0.005 (Student’s t-test).
Exogenous c-Myb restores the expression of Bcl-xL in c-Myb deficient pre-selection DP thymocytes
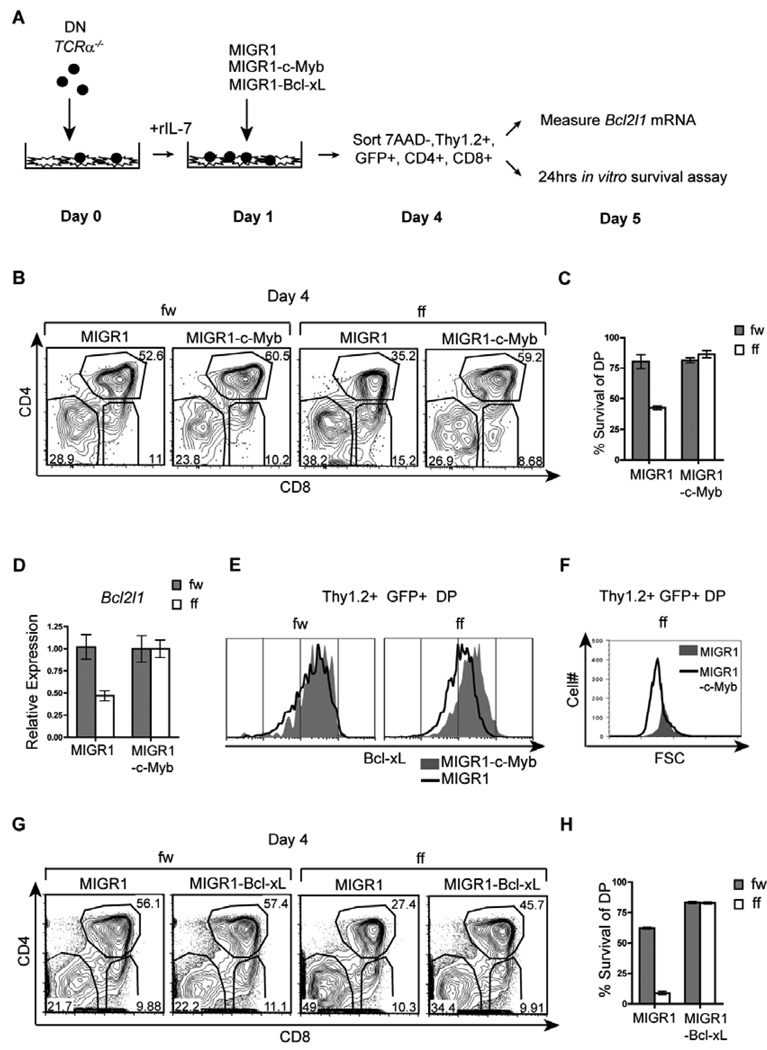
To confirm that Bcl-xL is a down-stream effector of c-Myb mediated survival in DP thymocytes, we determined if an exogenous source of c-Myb could restore Bcl-xL expression and cell survival in Mybf/f Cd4-Cre Tcrα−/− DP thymocytes. We utilized an in vitro system in which thymocytes from Mybf/w Cd4-Cre Tcrα−/− and Mybf/f Cd4-Cre Tcrα−/− mice were depleted of the DP population on magnetic beads and placed in co-culture with OP9-Delta-like 1 (DL1) stromal cells (43). Thymocytes were subsequently transduced with a c-Myb cDNA containing retrovirus (MIGR1-c-Myb) and allowed to differentiate into DP thymocytes in co-culture. Transduced live DP thymocytes (7AAD−, Thy1.2+, GFP+, CD4+CD8+) were electronically sorted 72hrs post-infection and subsequently either analyzed for Bcl2l1 mRNA expression or cultured for an additional 24 hrs to assess survival (Fig. 5A). A decreased percentage of MIGR1 transduced Mybf/f Cd4-Cre Tcrα−/− DP thymocytes was observed on day 4 of co-culture compared to MIGR1 transduced control DP thymocytes (Fig. 5B), consistent with impaired survival in pre-selection DP thymocytes lacking c-Myb. Further, decreased survival upon additional culture of MIGR1 transduced Mybf/f Cd4-Cre Tcrα−/− DP thymocytes confirmed that the defect is intrinsic to the DP compartment and not a consequence of impaired DN to DP transition (Fig. 5C). By the same criteria, MIGR1-c-Myb transduced Mybf/f Cd4-Cre Tcrα−/− thymocytes displayed complete restoration of survival. Importantly, qRT-PCR revealed restored expression of Bcl2l1 mRNA and protein in sorted MIGR1-c-Myb transduced Mybf/f Cd4-Cre Tcrα−/− DP thymocytes compared to MIGR1-c-Myb transduced control DP thymocytes (Fig. 5D and 5H), suggesting that c-Myb regulates Bcl2l1 expression in pre-selection DP thymocytes. In addition, we repeatedly observed increased accumulation of Mybf/f Cd4-Cre Tcrα−/− FSClo DP thymocytes when transduced with MIGR1-c-Myb (Fig. 5F). This observation is consistent with an absolute requirement for c-Myb and Bcl-xL mediated survival by small but not large pre-selection DP thymocytes. Finally, we demonstrate that MIGR1-Bcl-xL transduced thymocytes also restore Mybf/f Cd4-Cre Tcrα−/− DP thymocyte survival (Fig. 5G and H). Taken together, these findings demonstrate that Bcl-xL acts as a physiological effector of c-Myb function in small pre-selection DP thymocytes to prolong a critical survival window.
Figure 5. An exogenous source of c-Myb restores Bcl-xL expression and survival in Mybf/f Cd4-Cre Tcrα−/− DP thymocytes.
(A) Experimental plan to rescue survival of DP thymocytes by retroviral transduction in OP9-DL1 stromal cell cultures. (B) Day 4 co-cultures transduced with MIGR1-c-Myb or MIGR1 were analyzed by flow cytometry for CD4 and CD8 surface expression through a Thy1.2+7AAD−GFP+ gate. Results are representative of 4 separate experiments. (C) DP thymocytes transduced with MIGR1-c-Myb or MIGR1 were electronically sorted on day 4 of co-culture and cultured for an additional 24hrs to assess spontaneous apoptosis as measured by Annexin 5 and 7AAD staining. Results are presented as mean +/− SEM, n=2, and are representative of 4 separate experiments. (D) Transduced DP thymocytes from 4B were electronically sorted and Bcl2l1 mRNA expression was analyzed by quantitative real-time PCR. Data is normalized to Hprt-1 mRNA expression. Results are presented as mean +/− SEM, n=2, and are representative of 3 separate experiments. (E) Intracellular staining for Bcl-xL of MIGR1-c-Myb or MIGR1 transduced Mybf/w Cd4-Cre Tcrα−/− and Mybf/f Cd4-Cre Tcrα−/− DP thymocytes. To retain Mybf/f Cd4-Cre Tcrα−/− DP thymocytes with reduced Bcl-xL protein, 40 µM Z-VAD was added to the co-cultures 48 hrs prior to intracellular staining on day 5. Results are representative of 3 separate experiments. (F) Forward light scatter profiles of MIGR1-c-Myb or MIGR1 transduced Mybf/f DP thymocytes on day 4 of co-culture measured by flow cytometry. Results are representative of 3 separate experiments. (G) Day 4 co-cultures transduced with either MIGR1-Bcl-xL or MIGR1 were analyzed for CD4 and CD8 expression by flow cytometry through a Thy1.2+7AAD−GFP+ gate. Results are representative of 3 separate experiments. (H) DP thymocytes transduced with either MIGR1-Bcl-xL or MIGR1 were sorted on day 4 of co-culture and cultured for an additional 24hrs to assess spontaneous apoptosis as measured by Annexin 5 and 7AAD staining. Results are presented as mean +/− SEM, n=2, and are representative of 3 separate experiments.
c-Myb controls Bcl2l1 transcription by a genetically distinct pathway that is independent of TCF-1 and RORγt expression
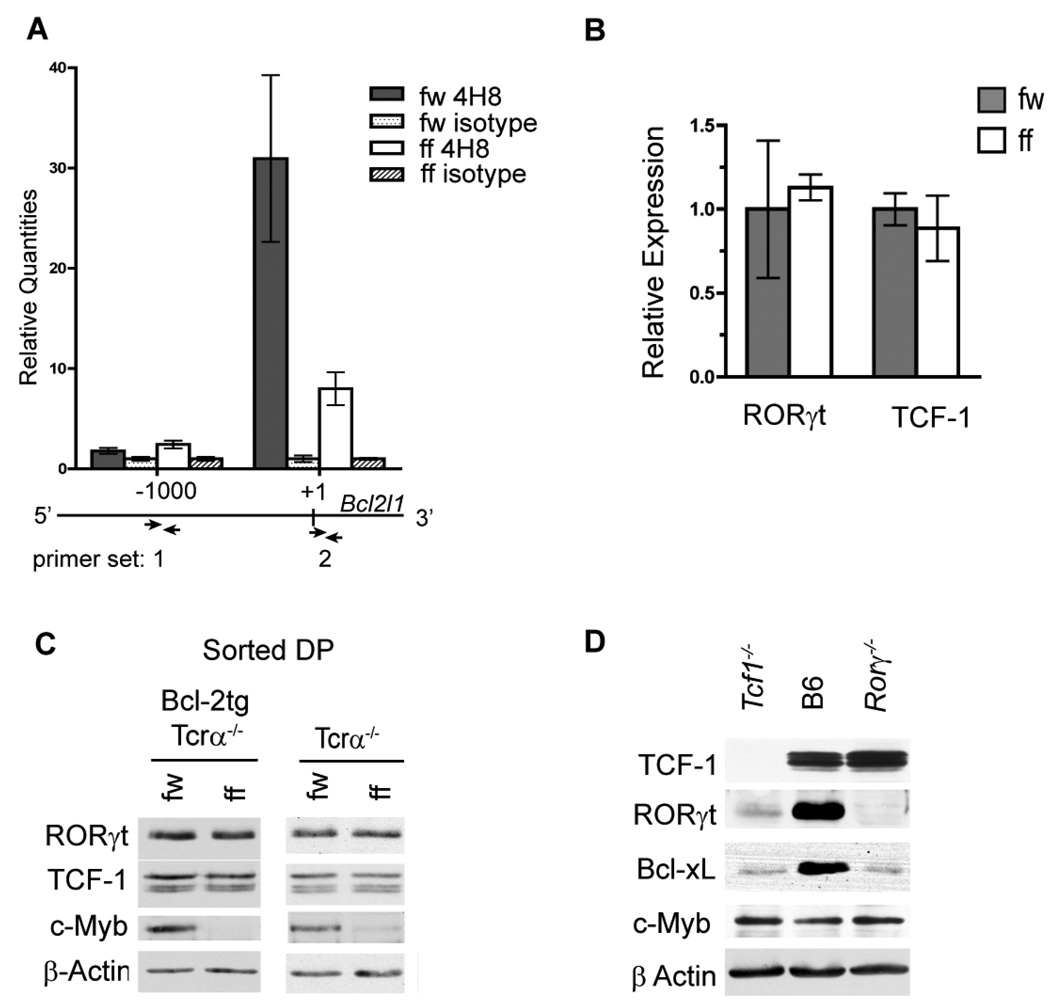
To determine if the decrease in Bcl-xL expression in c-Myb deficient DP thymocytes is a consequence of reduced transcription at the Bcl2l1 locus we measured RNA polymerase II localization at the previously identified transcriptional start site (primer set 2) (59) by chromatin immunoprecipitation (ChIP). Primers that amplify a site approximately 1kb up-stream of the transcriptional start site (primer set 1) were used as negative control. qPCR of DNA precipitated with anti-RNA polymerase II revealed a 31-fold enrichment of over isotype control at the transcriptional start site in total Mybf/w Cd4-Cre Tcrα−/− thymocytes compared to an 8-fold enrichment in Mybf/f Cd4-cre Tcrα−/− thymocytes (Fig. 6A right). No enrichment was detected in either sample using the negative control primer set 1 (Fig. 6A left), demonstrating that c-Myb regulates Bcl-xL expression in pre-selection DP thymocytes by promoting transcription at the Bcl2l1 locus. DNA sequence analysis identified three potential c-Myb binding sites in the Bcl2l1 promoter region that are conserved between humans and mice. However, anti-c-Myb ChIP in Mybf/w Cd4-Cre Tcrα−/− thymocytes did not reveal enrichment of c-Myb localization at either of the three potential binding sites compared to Mybf/f Cd4-Cre Tcrα−/− thymocytes (Supplementary Fig. 5). The efficacy of the anti-c-Myb ChIP was verified by a statistically significant enrichment at a previously identified c-Myb binding site within the Cd53 promoter (60) of Mybf/w Cd4-Cre Tcrα−/− thymocytes. These results suggest that c-Myb regulates Bcl2l1 expression at the level of transcription, but likely not through direct binding to the three conserved c-Myb binding sites in the Bcl2l1 promoter.
Figure 6. c-Myb promotes transcription at the Bcl2l1 locus independent of TCF-1 and RORγt expression.
(A) RNA polymerase II ChIP in Mybf/w Cd4-Cre Tcrα−/− and Mybf/f Cd4-Cre Tcrα−/− total thymocytes. Precipitated genomic DNA was amplified using primer sets specific to either the transcription initiation site (+1) or a negative control site approximately 1kb upstream (−1000bp) by quantitative real-time PCR. Results are normalized to IgG control samples and presented as mean +/− SEM, n=3. Data are representative of 2 independent experiments. (B) Rorγ and Tcf1 mRNA levels in sorted Mybf/w Cd4-Cre Tcrα−/− and Mybf/f Cd4-Cre Tcrα−/− FSClo DP thymocytes measured by quantitative real-time PCR and normalized to Hprt-1 mRNA expression. Data are presented as mean +/− SEM, n=2, and representative of 2 separate experiments. (C) Western blot of whole cell lysates from sorted Mybf/w Cd4-Cre Tcrα−/− and Mybf/f Cd4-Cre Tcrα−/− FSClo DP thymocytes with and without of Eµ-Bcl-2tg expression probed for TCF-1, RORγt, c-Myb and β-actin. Results are representative of 3 separate experiments. (D) Western blot of whole cell lysates from Rorγ−/−, C57BL/6j, and Tcf1−/− total thymocytes probed for TCF-1, RORγt, c-Myb, Bcl-xL and β-actin. Results are representative of 2 separate experiments.
Deficiency of TCF-1 or RORγt results in the premature apoptosis of DP thymocytes due to decreased Bcl-xL expression (19, 20). To determine if the absence of c-Myb results in altered expression of TCF-1 or RORγt, we compared their mRNA and protein expression in sorted Mybf/w Cd4-Cre Tcrα−/− and Mybf/f Cd4-Cre Tcrα−/− DP thymocytes. To reveal potential changes in protein expression that might be masked by impaired survival we also measured the amount of TCF-1 and RORγt in Mybf/w Cd4-Cre Tcrα−/− and Mybf/f Cd4-Cre Tcrα−/− DP thymocytes that carry the Bcl-2tg. mRNA expression microarray and qRT-PCR did not reveal statistically significant changes in Tcf1 or Rorγ mRNA levels (Data not shown and Fig. 6B). Moreover, western blot analysis using electronically sorted small DP thymocytes did not reveal differences in protein expression of TCF-1 or RORγt between Mybf/w Cd4-Cre Tcrα−/− and Mybf/f Cd4-Cre Tcrα−/− with or without the EM-Bcl-2 transgene (Fig. 6C). Thus, decreased expression of Bcl-xL in c-Myb deficient DP thymocytes is not the result of impaired TCF-1 or RORγt expression.
To determine if TCF-1 or RORγt promote Bcl-xL expression indirectly through modulating c-Myb expression, we compared c-Myb protein expression in Tcf1−/− and Rorγ−/− to wildtype total thymocytes (19, 34). We also detected no difference in the amount of c-Myb contained in Tcf1−/−, Rorγ−/− and B6 thymocytes by western blot, suggesting that TCF-1 and RORγt do not control c-Myb expression during T cell development. Interestingly, we observed an apparent decrease in the amount of RORγt protein in Tcf1−/− thymocytes but no change in the amount of TCF-1 protein in Rorγ−/− thymocytes (Fig. 6D), suggesting that TCF-1 may indirectly stimulate Bcl-xL expression by up-regulating RORγt expression during T cell development. Taken together, our results demonstrate that c-Myb promotes transcription at the Bcl2l1 locus in DP thymocytes via a genetically distinct pathway, independent of RORγt and TCF-1 expression.
Discussion
Apoptosis is an essential process underlying normal lymphocyte development and dysregulated control of apoptosis can lead to autoimmune disease or immunodeficiency. The intrinsic lifespan of pre-selection DP thymocytes regulates the composition and diversity of the Tcrα repertoire and is therefore critical to normal T cell development (13). Thymocytes enter the DP stage as large proliferating cells and initiation of Tcrα recombination coincides with the transition of large pre-selection DP thymocytes into a small, quiescent subset that is prone to death by neglect (10, 15, 62). We previously reported that c-Myb is required for the survival of DP thymocytes in an αβTCR independent fashion (31). We now demonstrate that c-Myb expression is up-regulated in small, pre-selection DP thymocytes where it promotes survival by controlling the expression of Bcl-xL. Thymocytes have developed a mechanism to efficiently increase Bcl-xL expression in the pre-selection DP compartment to promote survival and increase the opportunity for assembling an αβTCR (13, 17, 18). Our observation that c-Myb deficient DP thymocytes exhibit a Tcrα repertoire that is skewed toward the use of 5’ located Jα segments demonstrates that c-Myb plays a crucial role in determining the window of time that is available for developing thymocytes to produce a diverse Tcrα repertoire.
Spontaneous apoptosis of DP thymocytes in liquid culture mimics death by neglect and is thought to be controlled by the balanced functions of Bcl-xL and the pro-apoptotic Bcl-2 family members Bax, Bak and Bim in a cytochrome c/Apaf-1/caspase-9 apoptosome independent fashion (13, 50–52). Our mRNA expression microarray experiments identified a unique decrease in Bcl2l1 mRNA expression among the anti-apoptotic Bcl-2 family members but no obvious changes in the expression of pro-apoptotic Bcl-2 family members in c-Myb deficient DP thymocytes. This finding, combined with our ability to rescue survival in c-Myb deficient DP thymocytes with exogenously supplied Bcl-xL, is consistent with previous work that described a non-redundant role for Bcl-xL in DP thymocyte survival (18, 19). A recent report examined the synergy of Bcl-xL and Mcl-1 in promoting DP thymcyte survival implying functional redundancy in vivo (58). To clarify this point, we carefully analyzed Mcl-1 expression and detected no difference in the level of Mcl-1 mRNA or protein in c-Myb deficient DP thymocytes. Thus, our results demonstrate that reduced expression of Bcl-xL alone is sufficient to have a significant impact on pre-selection DP thymocyte survival.
Few effectors of c-Myb activity have been identified and tested in physiologically relevant models to date. We demonstrate that the efficient up-regulation of Bcl-xL expression in pre-selection DP thymocytes is controlled at least in part at the level of transcription by c-Myb as RNA polymerase II localization at the Bcl2l1 transcription start site was decreased in c-Myb deficient thymocytes. Consistent with a direct role for c-Myb in promoting Bcl2l1 transcription, we identified three consensus c-Myb binding sites in the mouse Bcl2l1 promoter (59) that are conserved between mice and humans. However, we were unable to directly localize c-Myb to these sites by ChIP assay. Thus, c-Myb may promote Bcl-xL transcription through an indirect mechanism, although it remains possible that c-Myb may tether to the Bcl2l1 promoter through protein-protein interactions independently of its DNA binding domain, or interact with as yet unidentified regulatory regions that control Bcl2l1 transcription. Irrespective of the precise mode of regulation, we demonstrate that c-Myb controls transcription at the Bcl2l1 locus in small preselection DP thymocytes and that Bcl-xL is a down-stream effector of c-Myb activity during normal T cell development. The implications of this finding may reach beyond normal T cell development to the association of duplication events at the Myb locus in over eight percent of human T-ALL and attendant defects in the control of survival, proliferation and differentiation in these tumors (63, 64).
With this study, we have made clear that c-Myb, RORγt and TCF-1 are all necessary components in the transcriptional network responsible for the critical up-regulation of Bcl-xL expression in the pre-selection DP compartment. The expression of these three transcription factors is up-regulated in DP thymocytes. The induction of both RORγt and TCF-1 expression is known to require signaling through the preTCR (36, 65) as well as cessation of IL-7 receptor signaling during T cell development (61). Our finding that RORγt protein expression is diminished in thymocytes that lack TCF-1 supports a model where TCF-1 indirectly promotes Bcl-xL expression by regulating RORγt expression (Fig. 7). Signals that mediate the up-regulation of c-Myb during the DP stage remain elusive. However, western blot analysis demonstrates that c-Myb protein expression does not control nor is itself controlled by RORγt or TCF-1 protein expression. Thus, our data demonstrate that c-Myb is a component of a distinct genetic pathway that promotes transcription at the Bcl2l1 locus in DP thymocytes independent of TCF-1 and RORγt expression.
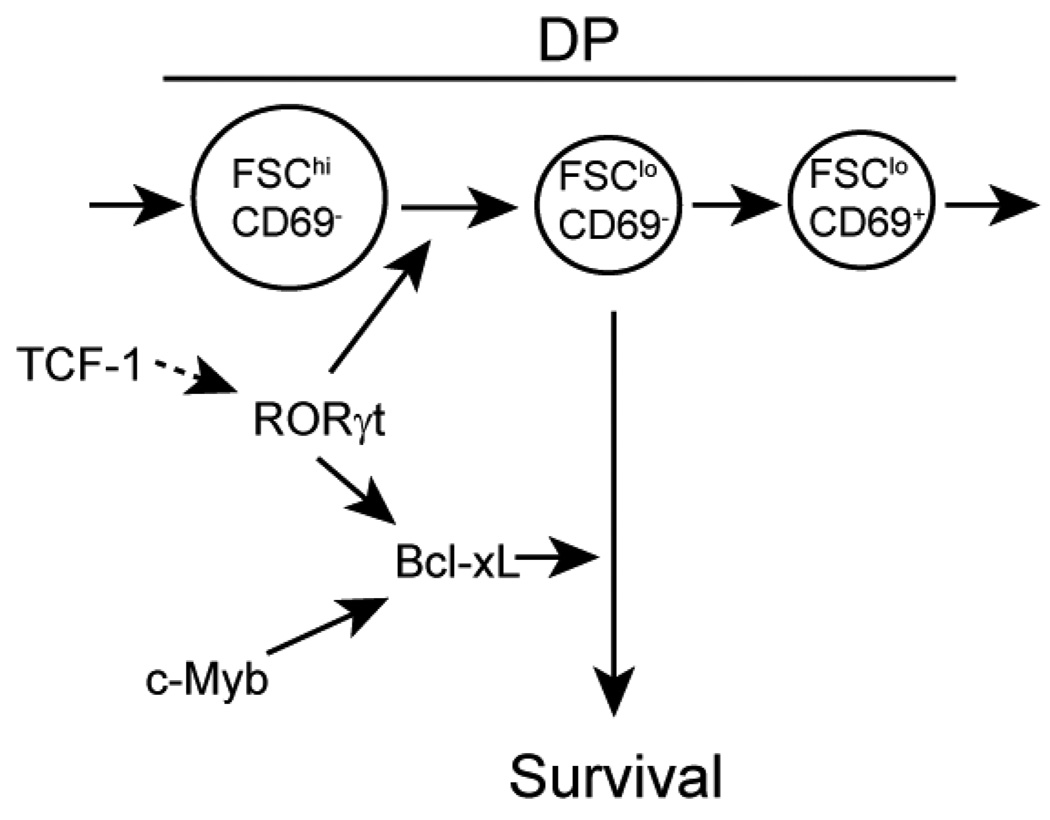
Figure 7. A model depicting the regulatory network that controls Bcl-xL expression in preselection DP thymocytes.
Bcl-xL is necessary for the survival of small pre-selection DP thymocytes during αβT cell development. c-Myb, RORγt and TCF-1 are all required to efficiently up-regulate Bcl-xL expression in DP thymocytes. RORγt also promotes the large to small pre-selection DP transition. Increased c-Myb expression in small pre-selection DP thymocytes promotes the expression of Bcl-xL by a genetic pathway independent of RORγt and TCF-1. Expression of RORγt appears to be TCF-1 dependent.
Our studies have identified a previously unrecognized difference in the requirement for Bcl-xL in the maintenance of the small but not the large pre-selection DP thymocyte compartment. c-Myb deficient small pre-selection DP thymocytes are rapidly eliminated from the thymus upon loss of Bcl-xL protein expression while we detected no significant decrease in the cellularity or survival of large preselection DP thymocytes despite the loss of Bcl-xL in the absence of c-Myb. In addition, the continued survival of large pre-selection DP thymocytes after the loss of Bcl-xL protein offers a likely explanation to why reduced Bcl-xL protein is readily detectable in freshly isolated Rorγ−/− and Tcf1−/− thymocytes but not Mybf/f Cd4-Cre Tcrα−/− thymocytes. Rorγ−/− DP thymocytes are blocked in the large pre-selection stage due to an inability to withdraw from cell cycle (6, 19). The Tcf1−/− DP compartment is also mainly composed of large pre-selection DP thymocytes (data not shown), possibly due to insufficient amounts of RORγt. In contrast, the Mybf/f Cd4-Cre DP compartment consists of mainly small pre-selection DP thymocytes. Since only the small pre-selection DP thymocytes appear to undergo increased apoptosis after the loss of Bcl-xL expression, only those cells remain that produce sufficient amounts of Bcl-xL to survive. The developmental block observed at the large pre-selection stage in Rorγ−/− but not Mybf/f Cd4-Cre mice suggests that RORγt is required for both transition to as well as the survival of the small pre-selection DP compartment while c-Myb is mainly important for the latter.
Supplementary Material
Acknowledgements
We thank Dr. David W. Mullins for his help with the GeneSifter software and Ms. Joanne Lannigan and Mr. Michael Solga of the University of Virginia Flow Cytometry Core Facility for their help and advice. The authors are indebted to Dr. Alfred Singer for advice and discussion. We thank Drs Ulrike M. Lorenz, Kodi S. Ravichandran and Jeremy A. Daniel for comments on the manuscript.
Footnotes
This work was supported in part by grant CA85842 from the National institutes of Health (to TPB).
References
- 1.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu. Rev. Immunol. 2003;21:139. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 2.Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat. Rev. Immunol. 2008;8:788. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petrie HT, Livak F, Schatz DG, Strasser A, Crispe IN, Shortman K. Multiple rearrangements in T cell receptor alpha chain genes maximize the production of useful thymocytes. J. Exp. Med. 1993;178:615. doi: 10.1084/jem.178.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egerton M, Shortman K, Scollay R. The kinetics of immature murine thymocyte development in vivo. Int. Immunol. 1990;2:501. doi: 10.1093/intimm/2.6.501. [DOI] [PubMed] [Google Scholar]
- 5.Penit C, Lucas B, Vasseur F. Cell expansion and growth arrest phases during the transition from precursor (CD4−8−) to immature (CD4+8+) thymocytes in normal and genetically modified mice. J. Immunol. 1995;154:5103. [PubMed] [Google Scholar]
- 6.Xi H, Schwartz R, Engel I, Murre C, Kersh GJ. Interplay between RORgammat, Egr3, and E proteins controls proliferation in response to pre-TCR signals. Immunity. 2006;24:813. doi: 10.1016/j.immuni.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 7.Wang F, Huang CY, Kanagawa O. Rapid deletion of rearranged T cell antigen receptor (TCR) Valpha-Jalpha segment by secondary rearrangement in the thymus: role of continuous rearrangement of TCR alpha chain gene and positive selection in the T cell repertoire formation. Proc. Natl. Acad. Sci. USA. 1998;95:11834. doi: 10.1073/pnas.95.20.11834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang C, Kanagawa O. Ordered and coordinated rearrangement of the TCR alpha locus: role of secondary rearrangement in thymic selection. J. Immunol. 2001;166:2597. doi: 10.4049/jimmunol.166.4.2597. [DOI] [PubMed] [Google Scholar]
- 9.Villey I, Caillol D, Selz F, Ferrier P, de Villartay JP. Defect in rearrangement of the most 5' TCR-J alpha following targeted deletion of T early alpha (TEA): implications for TCR alpha locus accessibility. Immunity. 1996;5:331. doi: 10.1016/s1074-7613(00)80259-9. [DOI] [PubMed] [Google Scholar]
- 10.Petrie HT, Livak F, Burtrum D, Mazel S. T cell receptor gene recombination patterns and mechanisms: cell death, rescue, and T cell production. J. Exp. Med. 1995;182:121. doi: 10.1084/jem.182.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turka LA, Schatz DG, Oettinger MA, Chun JJ, Gorka C, Lee K, McCormack WT, Thompson CB. Thymocyte expression of RAG-1 and RAG-2: termination by T cell receptor cross-linking. Science. 1991;253:778. doi: 10.1126/science.1831564. [DOI] [PubMed] [Google Scholar]
- 12.Brandle D, Muller C, Rulicke T, Hengartner H, Pircher H. Engagement of the T-cell receptor during positive selection in the thymus down-regulates RAG-1 expression. Proc. Natl. Acad. Sci. USA. 1992;89:9529. doi: 10.1073/pnas.89.20.9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo J, Hawwari A, Li H, Sun Z, Mahanta SK, Littman DR, Krangel MS, He YW. Regulation of the TCRalpha repertoire by the survival window of CD4(+)CD8(+) thymocytes. Nat. Immunol. 2002;3:469. doi: 10.1038/ni791. [DOI] [PubMed] [Google Scholar]
- 14.Gratiot-Deans J, Merino R, Nunez G, Turka LA. Bcl-2 expression during T-cell development: early loss and late return occur at specific stages of commitment to differentiation and survival. Proc. Natl. Acad. Sci. USA. 1994;91:10685. doi: 10.1073/pnas.91.22.10685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu Q, Park JH, Doan LL, Erman B, Feigenbaum L, Singer A. Cytokine signal transduction is suppressed in preselection double-positive thymocytes and restored by positive selection. J. Exp. Med. 2006;203:165. doi: 10.1084/jem.20051836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rathmell JC, Farkash EA, Gao W, Thompson CB. IL-7 enhances the survival and maintains the size of naive T cells. J. Immunol. 2001;167:6869. doi: 10.4049/jimmunol.167.12.6869. [DOI] [PubMed] [Google Scholar]
- 17.Grillot DA, Merino R, Nunez G. Bcl-XL displays restricted distribution during T cell development and inhibits multiple forms of apoptosis but not clonal deletion in transgenic mice. J. Exp. Med. 1995;182:1973. doi: 10.1084/jem.182.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma A, Pena JC, Chang B, Margosian E, Davidson L, Alt FW, Thompson CB. Bclx regulates the survival of double-positive thymocytes. Proc Natl Acad Sci U S A. 1995;92:4763. doi: 10.1073/pnas.92.11.4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani A, Brenner-Morton S, Mebius RE, Littman DR. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 2000;288:2369. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- 20.Ioannidis V, Beermann F, Clevers H, Held W. The beta-catenin--TCF-1 pathway ensures CD4(+)CD8(+) thymocyte survival. Nat. Immunol. 2001;2:691. doi: 10.1038/90623. [DOI] [PubMed] [Google Scholar]
- 21.Nomura T, Tanikawa J, Akimaru H, Kanei-Ishii C, Ichikawa-Iwata E, Khan MM, Ito H, Ishii S. Oncogenic activation of c-Myb correlates with a loss of negative regulation by TIF1beta and Ski. J. Biol. Chem. 2004;279:16715. doi: 10.1074/jbc.M313069200. [DOI] [PubMed] [Google Scholar]
- 22.Dai P, Akimaru H, Tanaka Y, Hou DX, Yasukawa T, Kanei-Ishii C, Takahashi T, Ishii S. CBP as a transcriptional coactivator of c-Myb. Genes. Dev. 1996;10:528. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 23.Westin EH, Gallo RC, Arya SK, Eva A, Souza LM, Baluda MA, Aaronson SA, Wong-Staal F. Differential expression of the amv gene in human hematopoietic cells. Proc. Natl. Acad. Sci. USA. 1982;79:2194. doi: 10.1073/pnas.79.7.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bender TP, Kuehl WM. Differential expression of the c-myb proto-oncogene marks the pre-B cell/B cell junction in murine B lymphoid tumors. J. Immunol. 1987;139:3822. [PubMed] [Google Scholar]
- 25.Duprey SP, Boettiger D. Developmental regulation of c-myb in normal myeloid progenitor cells. Proc. Natl. Acad. Sci. USA. 1985;82:6937. doi: 10.1073/pnas.82.20.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ess KC, Witte DP, Bascomb CP, Aronow BJ. Diverse developing mouse lineages exhibit high-level c-Myb expression in immature cells and loss of expression upon differentiation. Oncogene. 1999;18:1103. doi: 10.1038/sj.onc.1202387. [DOI] [PubMed] [Google Scholar]
- 27.Gonda TJ, Sheiness DK, Bishop JM. Transcripts from the cellular homologs of retroviral oncogenes: distribution among chicken tissues. Mol Cell Biol. 1982;2:617. doi: 10.1128/mcb.2.6.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, Pietryga DW, Scott WJ, Jr, Potter SS. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 29.Oh IH, Reddy EP. The myb gene family in cell growth, differentiation and apoptosis. Oncogene. 1999;18:3017. doi: 10.1038/sj.onc.1202839. [DOI] [PubMed] [Google Scholar]
- 30.Greig KT, Carotta S, Nutt SL. Critical roles for c-Myb in hematopoietic progenitor cells. Semin. Immunol. 2008;20:247. doi: 10.1016/j.smim.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 31.Bender TP, Kremer CS, Kraus M, Buch T, Rajewsky K. Critical functions for c-Myb at three checkpoints during thymocyte development. Nat. Immunol. 2004;5:721. doi: 10.1038/ni1085. [DOI] [PubMed] [Google Scholar]
- 32.Strasser A, Harris AW, Cory S. bcl-2 transgene inhibits T cell death and perturbs thymic self-censorship. Cell. 1991;67:889. doi: 10.1016/0092-8674(91)90362-3. [DOI] [PubMed] [Google Scholar]
- 33.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, Wilson CB. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 34.Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, van de Wetering M, Oosterwegel M, Wilson A, MacDonald HR, Clevers H. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374:70. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- 35.Verschelde C, Michonneau D, Trescol-Biemont MC, Berberich I, Schimpl A, Bonnefoy-Berard N. Overexpression of the antiapoptotic protein A1 promotes the survival of double positive thymocytes awaiting positive selection. Cell. Death. Differ. 2006;13:1213. doi: 10.1038/sj.cdd.4401814. [DOI] [PubMed] [Google Scholar]
- 36.Goux D, Coudert JD, Maurice D, Scarpellino L, Jeannet G, Piccolo S, Weston K, Huelsken J, Held W. Cooperating pre-T-cell receptor and TCF-1-dependent signals ensure thymocyte survival. Blood. 2005;106:1726. doi: 10.1182/blood-2005-01-0337. [DOI] [PubMed] [Google Scholar]
- 37.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 38.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic. Acids. Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallagher P, Bao Y, Serrano SM, Laing GD, Theakston RD, Gutierrez JM, Escalante T, Zigrino P, Moura-da-Silva AM, Nischt R, Mauch C, Moskaluk C, Fox JW. Role of the snake venom toxin jararhagin in proinflammatory pathogenesis: in vitro and in vivo gene expression analysis of the effects of the toxin. Arch. Biochem. Biophys. 2005;441:1. doi: 10.1016/j.abb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 40.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. Embo. J. 2003;22:4478. doi: 10.1093/emboj/cdg434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riegert P, Gilfillan S. A conserved sequence block in the murine and human TCR J alpha region: assessment of regulatory function in vivo. J. Immunol. 1999;162:3471. [PubMed] [Google Scholar]
- 43.Schmitt TM, Zuniga-Pflucker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 44.Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC, Pendergast AM, Bronson R, Aster JC, Scott ML, Baltimore D. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780. [PubMed] [Google Scholar]
- 45.Testi R, D'Ambrosio D, De Maria R, Santoni A. The CD69 receptor: a multipurpose cell-surface trigger for hematopoietic cells. Immunol. Today. 1994;15:479. doi: 10.1016/0167-5699(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 46.Mick VE, Starr TK, McCaughtry TM, McNeil LK, Hogquist KA. The regulated expression of a diverse set of genes during thymocyte positive selection in vivo. J. Immunol. 2004;173:5434. doi: 10.4049/jimmunol.173.9.5434. [DOI] [PubMed] [Google Scholar]
- 47.Hoffmann R, Bruno L, Seidl T, Rolink A, Melchers F. Rules for gene usage inferred from a comparison of large-scale gene expression profiles of T and B lymphocyte development. J. Immunol. 2003;170:1339. doi: 10.4049/jimmunol.170.3.1339. [DOI] [PubMed] [Google Scholar]
- 48.Takahama Y, Nakauchi H. Phorbol ester and calcium ionophore can replace TCR signals that induce positive selection of CD4 T cells. J. Immunol. 1996;157:1508. [PubMed] [Google Scholar]
- 49.Opferman JT. Apoptosis in the development of the immune system. Cell. Death. Differ. 2008;15:234. doi: 10.1038/sj.cdd.4402182. [DOI] [PubMed] [Google Scholar]
- 50.Marsden VS, O'Connor L, O'Reilly LA, Silke J, Metcalf D, Ekert PG, Huang DC, Cecconi F, Kuida K, Tomaselli KJ, Roy S, Nicholson DW, Vaux DL, Bouillet P, Adams JM, Strasser A. Apoptosis initiated by Bcl-2-regulated caspase activation independently of the cytochrome c/Apaf-1/caspase-9 apoptosome. Nature. 2002;419:634. doi: 10.1038/nature01101. [DOI] [PubMed] [Google Scholar]
- 51.Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, Adams JM, Strasser A. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 1999;286:1735. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- 52.Rathmell JC, Lindsten T, Zong WX, Cinalli RM, Thompson CB. Deficiency in Bak and Bax perturbs thymic selection and lymphoid homeostasis. Nat. Immunol. 2002;3:932. doi: 10.1038/ni834. [DOI] [PubMed] [Google Scholar]
- 53.Maurice D, Hooper J, Lang G, Weston K. c-Myb regulates lineage choice in developing thymocytes via its target gene Gata3. Embo. J. 2007;26:3629. doi: 10.1038/sj.emboj.7601801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lieu YK, Kumar A, Pajerowski AG, Rogers TJ, Reddy EP. Requirement of c-myb in T cell development and in mature T cell function. Proc. Natl. Acad. Sci. USA. 2004;101:14853. doi: 10.1073/pnas.0405338101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mason KD, Carpinelli MR, Fletcher JI, Collinge JE, Hilton AA, Ellis S, Kelly PN, Ekert PG, Metcalf D, Roberts AW, Huang DC, Kile BT. Programmed anuclear cell death delimits platelet life span. Cell. 2007;128:1173. doi: 10.1016/j.cell.2007.01.037. [DOI] [PubMed] [Google Scholar]
- 56.Bies J, Wolff L. Oncogenic activation of c-Myb by carboxyl-terminal truncation leads to decreased proteolysis by the ubiquitin-26S proteasome pathway. Oncogene. 1997;14:203. doi: 10.1038/sj.onc.1200828. [DOI] [PubMed] [Google Scholar]
- 57.Klempnauer KH, Bonifer C, Sippel AE. Identification and characterization of the protein encoded by the human c-myb proto-oncogene. Embo. J. 1986;5:1903. doi: 10.1002/j.1460-2075.1986.tb04443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dzhagalov I, Dunkle A, He YW. The anti-apoptotic Bcl-2 family member Mcl-1 promotes T lymphocyte survival at multiple stages. J. Immunol. 2008;181:521. doi: 10.4049/jimmunol.181.1.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grillot DA, Gonzalez-Garcia M, Ekhterae D, Duan L, Inohara N, Ohta S, Seldin MF, Nunez G. Genomic organization, promoter region analysis, and chromosome localization of the mouse bcl-x gene. J. Immunol. 1997;158:4750. [PubMed] [Google Scholar]
- 60.Lang G, White JR, Argent-Katwala MJ, Allinson CG, Weston K. Myb proteins regulate the expression of diverse target genes. Oncogene. 2005;24:1375. doi: 10.1038/sj.onc.1208301. [DOI] [PubMed] [Google Scholar]
- 61.Yu Q, Erman B, Park JH, Feigenbaum L, Singer A. IL-7 receptor signals inhibit expression of transcription factors TCF-1, LEF-1, and RORgammat: impact on thymocyte development. J. Exp. Med. 2004;200:797. doi: 10.1084/jem.20032183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rathmell JC, Vander Heiden MG, Harris MH, Frauwirth KA, Thompson CB. In the absence of extrinsic signals, nutrient utilization by lymphocytes is insufficient to maintain either cell size or viability. Mol. Cell. 2000;6:683. doi: 10.1016/s1097-2765(00)00066-6. [DOI] [PubMed] [Google Scholar]
- 63.Clappier E, Cuccuini W, Kalota A, Crinquette A, Cayuela JM, Dik WA, Langerak AW, Montpellier B, Nadel B, Walrafen P, Delattre O, Aurias A, Leblanc T, Dombret H, Gewirtz AM, Baruchel A, Sigaux F, Soulier J. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood. 2007;110:1251. doi: 10.1182/blood-2006-12-064683. [DOI] [PubMed] [Google Scholar]
- 64.Lahortiga I, De Keersmaecker K, Van Vlierberghe P, Graux C, Cauwelier B, Lambert F, Mentens N, Beverloo HB, Pieters R, Speleman F, Odero MD, Bauters M, Froyen G, Marynen P, Vandenberghe P, Wlodarska I, Meijerink JP, Cools J. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat. Genet. 2007;39:593. doi: 10.1038/ng2025. [DOI] [PubMed] [Google Scholar]
- 65.Xi H, Kersh GJ. Sustained early growth response gene 3 expression inhibits the survival of CD4/CD8 double-positive thymocytes. J. Immunol. 2004;173:340. doi: 10.4049/jimmunol.173.1.340. [DOI] [PubMed] [Google Scholar]
- 66.Godfrey DI, Kennedy J, Suda T, Zlotnik A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8- triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J. Immunol. 1993;150:4244. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.