Abstract
Terms used to describe nervous system parts and their interconnections are rife with synonyms, partial correspondences, and even homonyms, making effective scientific communication unnecessarily difficult. To address this problem a new Topological Relations schema for the Relations module of BAMS (Brain Architecture Knowledge Management System) was created. It includes a representation of the qualitative spatial relations between nervous system parts defined in different neuroanatomical nomenclatures or atlases and is general enough to record data and metadata from the literature, regardless of description level or species. Based on this foundation a Projections Translations inference engine was developed for the BAMS interface that automatically translates neuroanatomical projection (axonal inputs and outputs) reports across nomenclatures from translated information. To make BAMS more useful to the neuroscience community three things were done. First, we implemented a simple schema for validation of the translated neuroanatomical projections. Second, more than 1,000 topological relations between brain gray matter regions for the rat were inserted, along with associated details. Finally, a case study was performed to enter all historical or legacy published information about terminology related to one relatively complex gray matter region of the rat. The bed nuclei of the stria terminalis (BST) were chosen and 21 different nomenclatures from 1923 to present were collated, along with 284 terms for parts (gray matter differentiations), 360 qualitative topological relations between parts, and more than 7,000 details about spatial relations between parts, all of which was annotated with appropriate metadata. This information was used to construct a graphical “knowledge map” of relations used in the literature to describe subdivisions of the rat BST.
Keywords: data mining, neuroinformatics, databases, neuroanatomical projections, neuroanatomy
Introduction
Effective science communication is based on an accurate and carefully defined vocabulary that is internally consistent and consistently applied (Bota and Swanson, 2008a). Unfortunately, the vocabulary used currently in neuroanatomy to describe the structural organization of the nervous system lacks the precision and rigor of many other scientific domains (Swanson, 2000). Terminologies for gray matter regions, fiber tracts, and other macroscopic parts like ventricles and surface features are not always properly defined and relationships between alternate or partially corresponding terms often are not yet established experimentally or are arbitrary and thus based on the force of authority rather than on data. This is a fundamental problem at the level of vocabulary that hinders the next level of analysis – determining structure–function relationships between the various parts of the nervous system. And to make matters worse, this fundamental problem is getting worse with an exponential explosion of terminologies at both the regional and individual neuron levels of nervous system organization (Bota and Swanson, 2008b).
The development of neuroanatomical ontologies (or structured vocabularies) for computer science applications is not possible without rigorous specification of relationships between concepts and terms (Larson and Martone, 2009), and the corresponding nervous system parts. Ideally this could be accomplished rigorously in a high-resolution 3D, resliceable computer graphics model for a particular species, and in a series of such models for various species. This would specify clearly, accurately, and visually the spatial relationships between different interpretations of boundaries and internal features. However, operational models like this do not yet exist, and until they do, relationships between parts defined in different nomenclatures can only be expressed qualitatively.
This paper describes an important extension of the Brain Architecture Knowledge Management System (BAMS)1 backend database that allows collation of topological spatial relations and associated metadata. The database schema is general enough to register any type of qualitative spatial comparison found in the literature, and can record neuroanatomical nomenclature relationships in any species. The relations and associated metadata can be accessed via the web accessible user interface, which now also includes an inference engine that translates records about neuroanatomical projections (physical axon pathways between gray matter regions) across nomenclatures. This latter feature is critical for understanding neural systems, circuits, or networks and is built upon the foundation laid by the Topological Relations schema.
These BAMS enhancements were developed in association with a systematic collation of neuroanatomical nomenclatures for one especially complex gray matter region in one species – the bed nuclei of the stria terminalis (BST) in the rat. The BST are a conspicuous group of gray matter nuclei in the cerebral nuclei or basal ganglia division of the cerebral hemisphere, and they play an important role in coordinating autonomic, neuroendocrine, and behavioral responses critical for survival of the individual and its species – like eating, drinking, reproduction, and defense (Dong and Swanson, 2003, 2004, 2006). The BST in the rat have been recognized and described as a stand-alone, recognizable unit for a relatively short time (de Groot, 1959), although parts were described considerably earlier (Johnston, 1923; Gurdjian, 1925). At present there is no consensus about a unitary parcellation scheme for the rat BST. Commonly used nomenclatures fall into two general classes: one, the older, emphasizes a fundamental medial–lateral division whereas the other emphasizes a fundamental anterior–posterior (or rostral–caudal) division. This dichotomy makes the translation of experimental results related to the BST difficult, although obviously a “knowledge map” of qualitative topological relations between BST parts defined in different nomenclatures is necessary for interpreting and integrating experimental data, and for the development of a commonly agreed nomenclature for this part of the rat rostral pallidum.
Our strategy of systematic collation of BST terms and associated nomenclature from the published literature was chosen for a number of reasons. First, the choice of one species avoids more complex issues associated with establishing homologous relationships between species (Bota and Arbib, 2004). Second, there is currently much more data about gray matter regions, fiber tracts, and interconnections in rat than any other vertebrate species (Bota and Swanson, 2007a). And third, the BST is complex enough that it probably supplies proof of principle for any similar problem in any species (Dong and Swanson, 2006).
Materials and Methods
Design of topological relations and associated tables
Database Schema
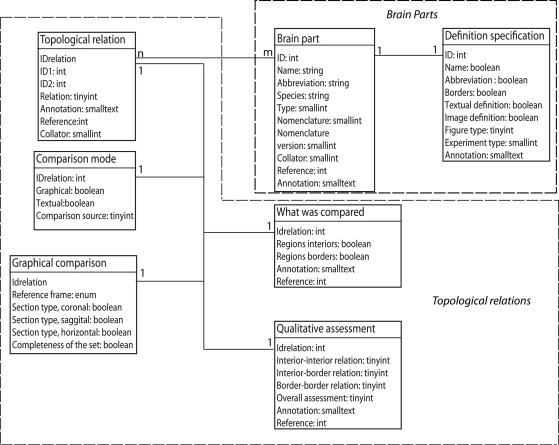
The database structure of BAMS includes five interrelated modules: Brain Parts, Relations, Cell Types, Connections, and Molecules. The Relations module backend extensions are implemented in MySQL and its entity-relationship (ER), which is shown in Figure 1. The scripts used to create web interfaces for all new BAMS extensions to be described are written in PHP.
Figure 1.
The ER structure of the new Topological Relations module of BAMS. The Brain Parts and the Topological Relations modules are shown in dotted boxes.
The Brain Parts module, which currently consists of two tables: Brain Parts and Definition Specification (see Table 1 for details), encodes for the definition of a nervous system part as found in a neuroanatomical nomenclature, and associated metadata (see below). Each nervous system part is defined in BAMS by a tuple that includes its name, abbreviation, species identified in, associated nomenclature, and part type (that is, gray matter, ventricle, or fiber tract). A nomenclature is defined as an internally consistent set of terms used by an author or group of authors to name and define various parts of the nervous system in a given species. A detailed description of this table is found in Bota et al. (2005).
Table 1.
Details of the tables of the Topological Relations module, implemented in BAMS and shown in Figure 1.
| Table name | Field name | Allowed values | Comment |
|---|---|---|---|
| Definition specification | The table in included in Brain Parts module. It stores metadata about the mode of brain parts definitions. | ||
| ID | integer | Brain part unique identifier (see Figure 1). | |
| Name | boolean | Specifies whether name of the brain part is explicitly written in the collated reference. | |
| Abbreviation | boolean | Specifies whether the abbreviation of the brain part is explicitly written in the collated reference. | |
| Textual definition | boolean | Specifies whether the brain part was defined textually in the collated reference. | |
| Image definition | boolean | Specifies whether the brain part is shown in a graphical format in the collated reference. | |
| Figure type | integer | Specifies the type of figure that used to define the brain part. It is related to a table that stores different types of graphical format (BrainMetaFig, not shown in Figure 1). Examples include “drawing” or “image”. A textual annotation is also associated to each entry in BrainMetaFig. | |
| Experiment type | integer | Encodes for the experimental procedure used for tissue staining and mapping. It is related to a table that encodes for different types of staining procedures (not shown in Figure 1). | |
| Annotation | text | Allows association of textual comments; can be collated from the associated reference, or a comment inserted by collator. | |
| Topological relation | Encodes for topological relations between two brain parts. | ||
| IDrelation | integer | Unique identifier for inserted qualitative relations | |
| ID1,;ID2 | integers | Brain parts unique numerical identifiers. | |
| Relation | integers | Identifier of one of the eight qualitative spatial relations (see Text for details). | |
| Annotation | text | Allows association of textual comments to the inserted relation. | |
| Reference | integer | Identifier of the associated reference. Related to the Reference table. | |
| Collator | integer | Collator numerical identifier. | |
| Comparison mode | Encodes for basic metadata related to the method of comparison used in the associated reference, or employed by collator. | ||
| IDrelation | integer | Unique identifier for inserted qualitative relations. Allows 1:1 relationship with table Topological Relationships. | |
| Graphical | boolean | Encodes for the explicit graphical (i.e. drawing or image) comparison of related regions in the associated reference. | |
| Textual | boolean | Encodes for the explicit textual comparison of related regions in the associated reference. | |
| Comparison source | boolean | Encodes for the source type that was used for comparison. See Text for details. | |
| Graphical comparison | Encodes for mapping metadata, associated with the comparison of pairs of brain parts defined in different nomenclatures. | ||
| IDrelation | integer | Unique identifier for inserted qualitative relations. Allows 1:1 relationship with table Topological Relationships. | |
| Reference frame | enumeration | Encodes for the mapping method, collated from the associated reference, or employed by collator. See Text and Table 2 for details. | |
| Section type (coronal, saggital, transversal) | boolean | Encodes for cutting plane used for both related brain parts. The three planes are most used in neuroanatomy. We used three Boolean fields instead of a single one, because some references include more than one plane for brain parts comparison. | |
| Set completeness | boolean | Encodes for completeness of the sets of sections used for comparison in the associated reference. | |
| What was compared | Encodes for metadata associated to the relations of topological components of compared brain parts | ||
| IDrelation | integer | Unique identifier for inserted qualitative relations. Allows 1:1 relationship with table Topological Relationships. | |
| Regions interiors (borders) | boolean | Encode for explicit comparison, either collated reference or made by collator, of regions and borders of the brain parts pair. | |
| Annotation | text | Allows association of textual comments; can be collated from the associated reference, or a comment inserted by collator. | |
| Qualitative assessment | Encodes for metadata qualitative details associated with the explicit comparison of interiors and borders of the brain parts pair | ||
| IDrelation | integer | Unique identifier for inserted qualitative relations. Allows 1:1 relationship with table Topological Relationships. | |
| Interior–interior; interior–border; border–border relation | integer | Qualitative assessments of the precision of comparison of interiors and borders of brain parts pair. Can be either collated from the associated reference, or inferred by the collator. | |
| Overall assessment | integer | Qualitative assessment of the precision of comparison performed by authors in the associated reference, or inferred by collator. | |
| Annotation | text | Allows association of textual comments; can be collated from the associated reference, or a comment inserted by collator. |
The new Topological Relations table in the Relations module encodes for qualitative spatial relations between nervous system parts in different nomenclatures, seen as distinct and unitary objects. These relations can be inserted directly by collators from associated references, or they can be results of the general topological inference engine described in detail in Bota et al. (2005). A topological relation between two nervous system parts is modeled in BAMS as an instance of a spatial relation with the constraint that the two related parts should be defined in different nomenclatures. Its attributes, as shown in Figure 1, are inherited from the Brain Parts table. However, collator name, the associated annotation, and the reference can differ from those recorded with the nervous system parts.
The Comparison Mode table encodes for the type of comparison that was specifically made. A qualitative spatial comparison can be made graphically or textually, or both, or neither way. These combinations are encoded in the pair of Boolean variables graphical and textual. The last possibility (neither) is usually associated when collators establish a spatial relation between two nervous system parts solely on their own expertise or when the relation is the result of a previous inference performed in BAMS. The variable comparison source records the source (reference) that was used to collate the associated spatial relation. Its allowed values are, “one of the original references”, “a different reference”, “the collator based on information found in one of the original references”, “the collator based on information found in a different reference”, and “collator's inference”. These five values cover all possibilities that can be found in recording or interpreting a topological spatial relation from the literature.
The Graphical Comparison table records metadata associated with the images or maps that were explicitly used in the process of qualitative comparison (see also Table 1 for details), and found in the associated references. The variable reference frame refers to the method of comparison employed in the associated reference or by the collator. Its allowed values are listed in Table 2. An example of graphical comparison onto a standard reference frame (atlas) is shown in Swanson and Petrovich (1998).
Table 2.
The set of possible values of the variable reference frame, associated with the mode of graphical comparison of two nervous system parts defined in different nomenclatures.
| Inserted value | Graphical comparison mode |
|---|---|
| 1 | comparison was made onto a standard frame of reference (atlas) |
| 2 | Comparison was made onto one of the compared atlases |
| 3 | Comparison was made onto one of the compared sets of images |
| 4 | It is unclear from the collated information how the graphical comparison was performed |
| 5 | Not applicable |
| 6 | No explicit graphical comparison was made |
The actual process of qualitative (topological) comparison of two nervous system parts is encoded in two tables, What was Compared and Qualitative Assessment, shown in Figure 1. Both tables have a 1:1 relationship with Topological Relations because each qualitative relation entered in BAMS has a unique set of comparisons. This approach is not original; it was adapted from the methodology used by Felleman and van Essen (1991) to evaluate the precision of definitions for various areas of the primate (macaque) cerebral cortex (see also Table 1 for details). However, this extension of BAMS is the first effort to explicitly represent the procedure involved in directly comparing nervous system parts. The table What was Compared encodes for those topological parts (interior and border) that were explicitly compared: an author may compare explicitly the interiors of two nervous system parts, or their borders, or both, or none of them. The explicit comparison of only the interiors of a pair of nervous system parts defined in different nomenclatures is specific to those parts drawn or mapped without borders. This is the case when authors are either unsure of the exact position of borders, or when borders are not simple lines but instead intermediary zones with a nonzero width. Whenever two adjacent parts are separated by an intermediary zone, the process of drawing a zero-width border becomes difficult if not misleading. The Qualitative Assessment table is directly adapted from Felleman and van Essen (1991) and encodes for the precision of comparisons reported by neuroanatomists. The allowed values for all assessments are simply, “high”, “low”, or “unknown”. Both tables include annotation fields that can be used to associate text relevant to the topological comparison.
The Qualitative Assessment and What was Compared tables (see Table 1 for details) include an additional field for references that is filled whenever explicit topological comparisons were performed in publications different from the publication being collated, or when comparisons were made solely by the collators (reference value “1”).
The module that encodes for the definition of a nervous system part – Brain Parts, which is central to the whole ER organization of BAMS – was also augmented with a second table that further specifies how a nervous system part was defined. The Definition Specification table shown in Figure 1 includes metadata that explicitly encode the mode of definition and description for the nervous system part in an associated reference. The fields, “name”, “abbreviation”, “borders”, and “textual/image definition” specify whether each of these variables have been stated explicitly in the associated reference. A nervous system part can be defined only as a drawing, or as a name without a graphical representation, or as an abbreviation with no reference to a complete name or graphical representation. Ideally, the definition of a nervous system part is in textual format with a graphical representation as supporting information (high quality version of Figure 9)2, but either may be absent in particular references. To account for these situations, we considered two Boolean variables, textual definition and image definition, respectively. The field, “Figure type” encodes for the type of graphical representation used in the associated text, and allows the values, “drawing”, “image”, “drawing/image”, and “none”. This field is associated with an additional table (not shown in Figure 1) that allows association of collator annotations. The variable, “Experiment type” encodes for the experimental procedure used for tissue staining and mapping. The present schema allows simple string values like “Nissl stain” or “rapid Golgi stain” and it can be extended easily in the future to allow detailed description of the experimental procedure employed.
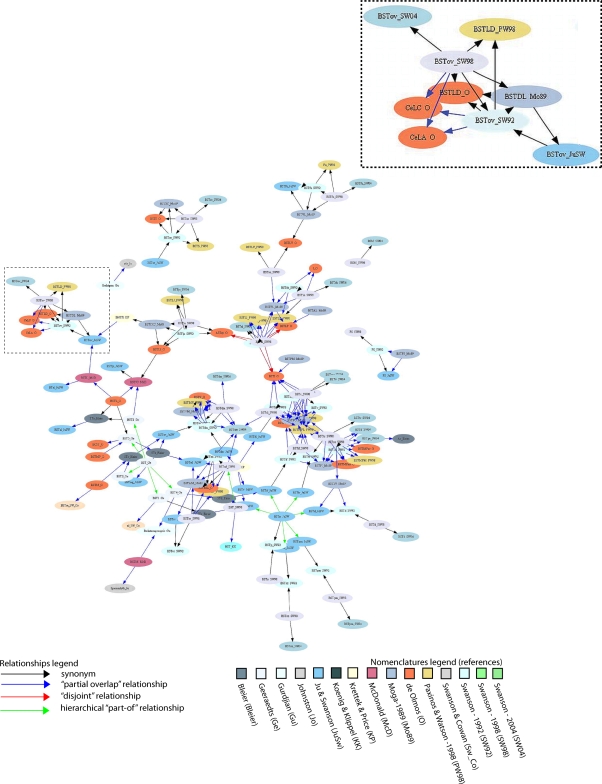
Figure 9.
Graphical display of BST-related terms defined in 15 nomenclatures, and their semantic relations (“the knowledge map”). The graph was obtained using GraphViz's Neato tool (Gansner and North, 1999; Bota and Swanson, 2007b). A higher resolution version of this Figure can be found at the URL: http://brancusi.usc.edu/bkms/bst-relations.png. The inset of the Figure shows the semantical grouping of terms BSTov (Swanson 92/98/04; Ju/Swanson), BSTDL defined in Moga-89 nomenclature, BSTLD in Paxinos/Watson-98, and two related amygdalar nuclei defined in the deOlmos-85 nomenclature.
Web interface
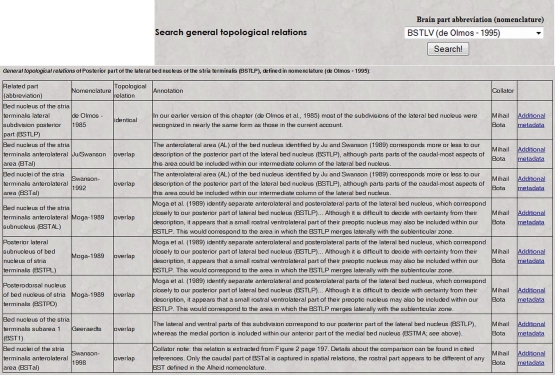
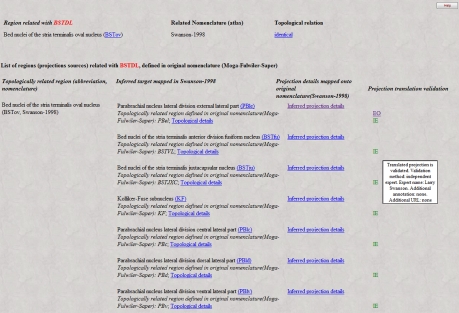
As with previous BAMS extensions, a web interface was developed so that users can view metadata and details about topological relations that are inserted in the backend database. A simple select tool (Figure 2, inset) was added to the Brain Parts web interface (Bota et al., 2005)3,4, and it can be used to search general topological relations of a nervous system part of interest, along with associated details. This search tool includes only those nervous system parts where at least one topological relation is recorded with one or more parts defined in one or more other nomenclatures. A page returned by such a search is shown in Figure 2. It includes the related nervous system parts, nomenclatures where they were identified, topological relations with the searched nervous system part, and the associated annotations. The table shown in Figure 2 includes links called “Additional metadata” that allow users to view the metadata described in the preceding section. An example of a qualitative spatial comparison that can be accessed by users is shown in Figure 3. For easier access to metadata, the page is divided into three categories: general metadata (collated reference and associated annotation), metadata associated with the mode of comparison, and qualitative assessment of the spatial comparison.
Figure 2.
The result of a search in BAMS for topological relations by regions defined in different neuroanatomical nomenclatures (inset; it can be accessed directly at the URL: http://brancusi. usc.edu/bkms/brain/search_bname_con.php). The result of the search will include the related regions, their nomenclatures, and associated annotated and metadata. The link “Additional metadata” will lead to details related to the comparison method, shown in Figure 3 (see Text for details).
Figure 3.
Users can further access details of the comparison process shown in Figure 2, and of the precision of comparison that are collated from the associated reference. See Text for details.
The topological comparison data and metadata can also be accessed from the main page that is returned when searching BAMS for a nervous system part (Bota et al., 2005)4. The topological relations and associated metadata also can be accessed by hitting the “Details” button, shown in the upper part of the page. The returned page includes details about the definition of the nervous system part if they are recorded in BAMS. Thus, this page includes the fields for the Definition Specification table, as shown in Figure 4.
Figure 4.
Metadata stored in the “Definition specification” table, which can be associated with a nervous system region recorded in BAMS are listed together with its definition. This example refers to the part BST2, defined by Geeraedts et al. (1990).
Results
Translation of projection reports across nomenclatures
Having established the topological relationships between different neuroanatomical nomenclatures for the same and for different nervous system gray matter regions, it is now possible to take the next major step, that is, the automatic translation of reported axonal connections (projections or pathways) between different gray matter regions – which constitutes the organization of nervous system macroconnectivity (Bota et al., 2003). For this, we used the extensions of the BAMS Relations module described above, along with a simplified algorithm for inferring topological relations (Egenhofer and Franzosa, 1991; Bota and Arbib, 2004), to implement a Projections Translations inference engine that automatically translates reports of neuroanatomical connections in the Connections module across nomenclatures. To reduce processing time the Projections Translations inference engine takes into account only qualitative relationships that yield unequivocal results (i.e., a single possible answer; Egenhofer and Franzosa, 1991; Bota and Arbib, 2004).
Whenever a nervous system gray matter region defined in one nomenclature is topologically related in BAMS with at least one gray matter region defined in another nomenclature, and the related region is associated with a projection report(s), then the inference engine first evaluates the topological relations of the experimental pathway tracer injection sites and labeled cell bodies, pathways, and/or terminal fields (seen as compact 2D objects) with the source and target regions.
This second new algorithm implemented for the BAMS web interface translates injection site and resulting labeled pathway information from the original nomenclature to the reference nomenclature. The algorithm works under the assumption that compared regions are convex 2D objects, and the allowed topological relations are a subset of U = {meet, disjoint, overlap, covers, contains, iscovered, iscontained, identical} (Egenhofer and Franzosa, 1991).
The process of inference of projections across nomenclatures is as follows:
(1) For region A in nomenclature X, and A is not associated with any connectivity reports in BAMS.
(2) Construct the set of regions {Bk}k = 1,n that are in topological relations {identical, is covered, is contained} with A and identified in nomenclature Yk = 1,n and Yk≠X.
(3) Construct the set of regions {B′k}k = 1,n′ that are in topological relations {covers, contains} with A and identified in nomenclature Yk = 1,n and Yk≠X.
(4) For any Bk construct the set of regions {Cl} that are associated in BAMS with projections reports from Bk and Cl are identified in nomenclature Y k.
(5) For any B′k construct the set of regions {C′l} that are associated in BAMS with projections reports from B′k and C′l are identified in nomenclature Ykwith the constraints that both injection sites and terminal fields cover the whole extent of B′k and C′l, respectively.
(6) For any Cl construct the set {A2} that are identified in the original nomenclature X and are in topological relations {identical, covers, contains} with Cl.
(7) For any C′l construct the set {A′2} that are identified in the original nomenclature X and are in topological relations {iscovered, iscontained} with C′l.
The “negative projections”, i.e. those projections that have been shown to be absent are assumed to be projections with qualitative strength “does not exist” (Bota et al., 2005), and the location of the terminal field, or of injection site, are identical with the target region, or projecting region, respectively.
Thus, the inference engine first constructs the set of nervous system gray matter regions where the translated injection site and transported label information may be located according to the translation algorithm presented in detail in Bota and Arbib (2004). This set of regions is then reduced to those yielding unequivocal results from the Topological Relations inference engine. Finally, these steps are repeated for all nervous system gray matter regions that are associated with neuroanatomical (axonal) projections, and the results are topologically related back to the reference gray matter regions and nomenclature. In other words, the Projections Translations inference engine in the Relations module mines tables in the Connections module for projection records mapped onto different nomenclatures and associates them with the reference gray matter regions.
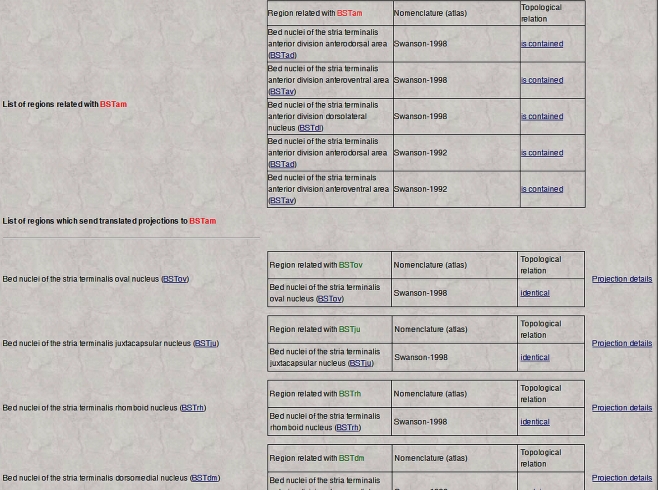
Thus, whenever a gray matter region is topologically related with regions defined in different nomenclatures and associated with projection reports, the main BAMS page (which contains summary information) will include two more links named “Afferent projections (inputs) translated from other nomenclatures” or “Efferent projections (outputs) translated from other nomenclatures”. Clicking on either link will return a page that looks like the one shown in Figure 5.
Figure 5.
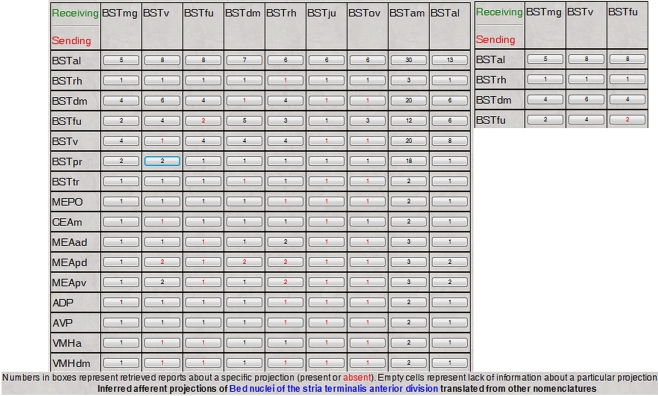
An example of neuroanatomical Projections Translations inference engine output. The screenshot lists the inferred projections of BSTrh defined in rat Swanson (2004) nomenclature. The topologically-based Projections Translations inference engine first lists all nervous system regions related with the region of interest and defined in different nomenclatures. The second table is made of those regions that receive neuroanatomical projections recorded in their original nomenclatures and translated into the one of interest (Swanson, 2004), links to details of the translated projections, and of the topological relations between original and translated regions. See Text for details.
The output of the Topological Relations inference engine first lists all gray matter regions defined in other nomenclatures that are spatially related to the reference region (region of interest). The second table shown in Figure 5 contains three columns: Topologically related regions (those gray matter regions inferred to send or receive projections from the gray matter regions listed in the first table), Inferred projections sources or targets in the new nomenclature, and Projection details mapped onto the original nomenclature (experimental results that were mapped onto the original atlas or nomenclature and recorded in BAMS). Additionally, each inferred source or target is listed with the topological related region identified in the original nomenclature. From the example shown in Figure 5, the set of regions found as targets of the BSTrh (defined in the Swanson, 2004 rat brain nomenclature; BST, bed nuclei of the stria terminalis) includes the anterior amygdaloid area (AAA), nucleus accumbens (ACB), and Accessory facial nucleus (ACVII) – all defined in the Swanson (1998) nomenclature. These regions are therefore included in the distinct set of targets, respectively, of the BSTrh (Figure 5). The supporting evidence for this inferred set is displayed on the right side of Figure 5 and includes a link to the original projections, that is, the projections that have been translated into the new or reference nomenclature. Additionally each topological relation between the original and translated targets in the second column includes a link called “Topological relation” that retrieves details that include the related regions and nomenclatures, the topological relation, associated annotations as collated from the original references, collators’ names and reference details.
The Topological Relations inference engine was also implemented to reconstruct projection patterns of nervous system gray matter regions from topological and hodological information associated with their subregions. The process of reconstructing these projection patterns (i.e. answering to the general query “What is the projection pattern of region made of several subregions?”) was described in detail elsewhere (Bota et al., 2005). This approach is combined with a general form of the Topological Relations inference engine that translates projection patterns for each of the subregions included in the region of interest. Thus, the algorithm described above runs for each subregion included in the region of interest.
An example of the output of this inference process is shown in Figure 6. Details about each projection in the matrix (Figure 6) can be accessed by clicking the corresponding buttons, with the results displayed in tabular format. Like the interface designed to infer projection patterns that was described in detail earlier (Bota et al., 2005), the interface shown in Figure 6 includes the option of transforming the matrix of reports into a color-coded matrix showing the maximal strength of each projection.
Figure 6.
The reconstructed and translated projections matrix of the anterior division of the rat BST, as defined in Swanson (2004) nomenclature. The inset shows a magnified portion of the matrix.
Finally, the Topological Relations inference engine was added to the interfaces that construct user-defined projection matrices (Bota et al., 2005; also see BAMS User Manual)5. For any empty cell of a user-defined matrix (for example, there is no information in the system associated with that particular projection), the inference engine translates projection information into the nomenclature that was used to create the matrix. In other words, the inference engine seeks to complete the user-defined matrices with data mapped onto different nomenclatures.
The inference algorithm described above is not complete in terms of the set of topological relations used. However, it is sound because all used three relations: {identical, covers, contains} ensure that at least a part of the injection site and the terminal field, respectively, are mapped onto the reference nomenclature from the original ones. Potential false positive and negative results may occur from the relative disadvantages of the employed staining technique and from the collation process itself. In order to reduce the latter, the collators associate annotations to each of the inserted relations.
Validation of Translated Projections Data
The problem of validation of results of inferences performed by neuroinformatics knowledge management systems is crucial for correctness and completeness of the implemented algorithms. Since the data collated in BAMS refer to results of experiments and measurements, the validation of any inference result should be made in terms of results of independent experiments recorded in the system, or evaluations of field experts. We implemented here a simple schema for validation of the translated projections across different nomenclatures.
The validation process is as follows: let X and Y be two neuroanatomical nomenclatures (atlases), both recorded in BAMS. The projections inference engine described above translates a projection of the general semantic form “region A projects to region B”, originally mapped onto nomenclature X to another projection mapped onto Y, with the general semantic form “region A′ projects to region B′”. We implemented in BAMS a simple schema that validates translated projections at the general semantic level A projects to B. This schema allows multiple validation records to be associated with a translated neuroanatomical projection.
The basic principle of the schema is that the validation process can be performed both automatically and manually. First, the validation inference engine searches the database for any projections reports mapped onto nomenclature X and which will either validate or invalidate the sentence “region A′ projects to region B′”. Whenever the validation engine will find projections reports in BAMS’ database that either validate or invalidate the statement above, a code “EO” (validation through an experiment performed independently, and mapped on the original nomenclature X; see Figure 9) is associated with any translated projection report shown in Figure 5. Users can click on this code and the result of this action will include the details of the projections inserted in BAMS and mapped onto nomenclature X.
The second step of the validation process includes association of any statement made by human agents with regard of the validity of the translation process. Human agents are defined here as the authors of the references associated with experiments that validate or contradict the projections translation inferences, experts in the field that are registered in BAMS, and BAMS’ collators and curators. In order to allow human agents to associate validation statements to the translated projections, we implemented a simple database schema with three tables, from which the table Translation_validation, shown in Table 3, is the most important one. The validation database schema implements four types of statements supporting or contradicting the translated neuroanatomical projections inserted by human agents, and shown in Table 4. Each of these statement types can be associated with free text annotations and URL's pointing to papers that have not yet been included in BAMS (see Figure 9 for an example).
Table 3.
Details of the Translation_validation table, implemented in BAMS as part of the validation schema.
| Table name | Field name | Allowed values | Comment |
|---|---|---|---|
| Translation_validation | Id_projecting_region | integer | Unique numerical identifier for the projecting brain region. |
| Id_target_region | integer | Unique numerical identifier for the target brain region. | |
| Id_expert | smallinteger | Unique identifier of the human agent that performed the validation, and recorded in BAMS. | |
| Validation_method | boolean | Encodes for validation or invalidation of a translated projection, performed by a human agent. | |
| Validation_type | smallinteger | Unique identifier for the types of validation that can be performed by human agents. | |
| Annotation | text | Additional text inserted by the human agent, associated with the validation action. | |
| URL | varchar(255) | Additional string that contains an URL address to a published reference that is not recorded yet in BAMS. |
Table 4.
Human agents types and associated codes implemented in BAMS as part of the validation schema. See Text for details.
| Numerical ID | Type of validation | Code |
|---|---|---|
| 1 | Validation performed by one of authors of the collated reference(s) | AA |
| 2 | Validation performed by an independent expert, who is not one of the authors of the collated reference(s) | IE |
| 3 | Validation performed by a user registered in BAMS | RE |
| 4 | Validation performed by one of BAMS’ collators | EC |
Thus the validation schema implemented in BAMS allows accumulation of supporting or contradicting evidence, which can be extracted automatically from the backend database, or inserted by experts.
An example of translated projections validation is shown in Figure 8. The projections of the dorsolateral bed nucleus of the stria terminalis (BSTDL), defined in the Moga–Fulwiler–Saper nomenclature (the above nomenclature Y) are inferred by the translation inference engine from the pattern of projections of the oval nucleus of BST (BSTov), defined in the Swanson (1998) nomenclature (the above nomenclature X). The Moga–Fulwiler–Saper is an extension of Moga-1989 nomenclature (Moga et al., 1989; see also Table 5) with the parabrachial nucleus parcellation scheme defined in Fulwiler and Saper (1984). For each translated projection, a set of validation codes is associated (the legend of codes and colors is not shown in Figure 8, but it is implemented in the web interface).
Figure 8.
Example of validation of the process of neuroanatomical projections translation. A portion of the projections pattern of BSTDL (Moga–Fulwiler–Saper nomenclature), which is the result of translation of projections of BTov (Swanson, 1998 nomenclature) is validated both automatically (code EO) by projections reports mapped onto the original nomenclature (Moga–Fulwiler–Saper), and manually, by experts (code IE). See Text for details.
Table 5.
The rat neuroanatomical nomenclatures inserted in BAMS used to define BST nuclei and related regions.
| Nomenclature | Number of BST-related regions inserted in BAMS | Reference |
|---|---|---|
| Bayer | 3 | Bayer (1987) |
| Bleier | 7 | Bleier et al. (1979) |
| Craigie | 3 | Craigie (1925) |
| de Groot | 1 | de Groot (1959) |
| de Olmos and Ingram | 5 | de Olmos and Ingram (1972) |
| de Olmos 1985 | 64 | de Olmos et al. (1985) |
| Alheid et al., 1995 | 18 | Alheid et al. (1995) |
| Geeraedts | 8 | Geeraedts et al. (1990) |
| Gurdjian | 2 | Gurdjian (1925) |
| Johnston | 2 | Johnston (1923) |
| Ju and Swanson | 30 | Ju and Swanson (1989) |
| König and Klippel | 1 | König and Klippel (1963) |
| Krettek and Price | 30 | Krettek and Price (1978) |
| McDonald | 4 | McDonald (1983) |
| Moga | 19 | Moga et al. (1989) |
| Paxinos and Watson 1986 | 16 | Paxinos and Watson (1986) |
| Pellegrino | 1 | Pellegrino et al. (1979) |
| Swanson – 1992 | 21 | Swanson (1992) |
| Swanson – 1998 | 24 | Swanson (1998) |
| Swanson – 2004 | 19 | Swanson (2004) |
| Swanson and Cowan | 6 | Swanson and Cowan (1979) |
The first translated projection is associated with two validation codes, EO and IE. The first code refers to neuroanatomical projection reports already existent in BAMS that validate the projection from BSTDL to PBle, and which are automatically associated with the translated projection. Thus, the projection from BSTDL to PBle, inferred by the translation inference engine from a projection mapped on another nomenclature, is automatically validated by the results of an independent tract-tracing experiment mapped onto the original nomenclature (Moga–Fulwiler–Saper). Clicking on the link associated with the EO code will retrieve the details of the projection, as described elsewhere (Bota et al., 2005). The second code, IE, colored in green, which encodes for supporting evidence, is the validation manually inserted by an independent expert. The inset shown in Figure 8 provides the details of the validation: the name of the expert, associated annotation, and URL to a published reference.
Each translated projection can be associated with any number of validation statements, which will be displayed as shown in Figure 8.
Collation and Curation of Inserted Data
As with previous BAMS extensions, we inserted here data and metadata mostly from the primary literature (published, reviewed research articles). However, relations between nervous system parts are also discussed in neuroscience atlases, review articles, and books. Hence, we also inserted information from these types of references. In addition, attention was focused on avoiding as much as possible the insertion of topological relations and associated details based only on the collator's expert knowledge. Details about spatial comparisons include multiple annotation fields, and all of them were used to support the recorded data and metadata.
Complete details have been inserted here for 1,020 topological relations between gray matter regions defined in 21 different rat brain nomenclatures, along with associated metadata, mostly from collated references (including the BST data considered with next). This volume of data makes BAMS one the most complete neuroanatomy database in terms of qualitative comparisons of nervous system parts in different parcellation schemes (see also BrainInfo)6.
As described in detail elsewhere (Bota and Swanson, 2007b, 2008b), BAMS has six collators and two curators. Neuroanatomical terms, their definitions, and associated metadata are manually checked for mistakes, clarity, and consistency. As with other modules of BAMS, each data entry is paired with a free text annotation that allows users and authors of original references to check for completeness. Whenever possible, we check our entries with the authors of the collated references. We welcome new collators and feedback from the web users and the authors of the inserted references.
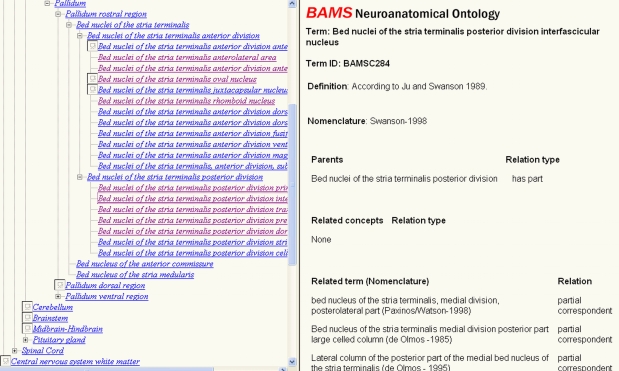
Based on work done here, we have updated the BAMS Ontology (Bota and Swanson, 2008b)7 with terms defined in other rat brain nomenclatures and their relations with the concepts of the ontology. The allowed semantical relations in the present version of the ontology are: identical term (when both the topological relation between designated brain regions and used terms are identical), synonym, and partial correspondence (which summarizes topological relations “overlap”, “part of”, and “includes”). An example of the updated web interface of the BAMS Ontology is shown in Figure 7. We have also updated the XML version of the Ontology, which can be downloaded and transformed in other formats (for example, OWL) by users. BAMS Ontology contains so far 1,881 rat neuroanatomical concepts and related terms, and more than 2,200 relations.
Figure 7.
The updated BAMS Ontology (http://retzius. usc.edu/bkms/bams-ontology.html) includes neuroanatomical terms defined in different rat nervous system nomenclatures, and their relations with concepts. The ontology can be browsed online, and downloaded as an XML file, found at the same URL.
Case Study: The Rat Bed Nuclei of the Stria Terminalis (BST)
The first step in establishing relations between different parts of the rat BST was to insert the relevant terms and nomenclatures found in the literature. An attempt was made to insert comprehensively all terms and nomenclatures relevant to the rat BST in the literature. The list of nomenclatures and number of terms associated with the rat BST are provided in Table 5. Overall, 284 rat BST-related terms defined in 21 different rat brain nomenclatures were inserted.
The second step was to insert relations between terms defined in the different nomenclatures, as collated from the literature. The qualitative relationships explicitly stated in the literature were primarily inserted, with collator interpretation kept to a minimum. A total of 360 qualitative topological relations were inserted, and annotated with all metadata described above. Overall, more than 7,000 details about spatial relations between rat BST nuclei defined in various nomenclatures were inserted into the backend database of BAMS. This information is the result of the most comprehensive effort to collate relationships between rat BST parts defined in various parcellation schemes and published up to now.
The third and last step was to represent graphically the set of rat BST topological relations collated in BAMS. The approach is similar to the one described for the rat retina in Bota and Swanson (2007b), and the result is shown in Figure 8.
Not all terms related to rat BST parts have been captured in Figure 9 because some terms are not associated with topological relations in BAMS. In addition, Figure 9 was simplified by merging the nomenclatures of de Olmos et al. (1985) and Alheid et al. (1995), which reduced the number of nodes and relationships. These two nomenclatures are similar, and displaying both of them unnecessarily complicates the graph in Figure 9. Another simplification involved merging the topological relations, “includes”, “is included”, “contains”, “is contained”, and “overlap” in a single “partial overlap” relationship. As a result, only four relations between gray matter regions were used, instead of nine (eight topological and one hierarchical “part-of”). To “compress” the graph several hierarchical “part-of” relationships between parts defined in the same nomenclature were used. And finally, the number of “disjoint” relations was minimized because these relations do not provide additional information to this type of graph.
The graph shown in Figure 9 includes 146 nodes (that is, names of BST parts, or of neighboring parts, that are defined in 15 rat nomenclatures) and 271 relations. Overall, the graph reflects relations inserted in BAMS and thus the completeness and precision of comparisons reported in the literature. As expected, the majority of relations (158/271) shown in Figure 9 are of the type “partial overlap” because five possible topological relations are included and the majority of nomenclatures follow either a medial–lateral or an anterior–posterior scheme. The graph is not fully connected, and includes several unconnected “islands” of related names. This is the case for the BST defined by Gurdjian (acronym: Bednterm_Gu; Gurdjian, 1925) and that defined by Johnston (acronym: stb_Jo; Johnston, 1923). These two nuclei are interrelated, but neither is connected with any subsequent nomenclature. The graph includes several clusters of names (terms) that mainly consist of parts in the Swanson parcellation schemes (Swanson, 1992, 1998, 2004) or the Paxinos/de Olmos nomenclatures (de Olmos, 1985; Paxinos and Watson, 1986, 1998; Alheid et al., 1995). This means that the majority of relations published in the literature refer to these two groups of nomenclatures.
Based on relatively comprehensive, internally consistent ontogenetic/developmental (Bayer, 1987), cytoarchitectural (Ju and Swanson, 1989), chemoarchitectural (Ju et al., 1989), and connectional/hodological (Dong et al., 2001; Dong and Swanson, 2003, 2004, 2006) criteria, the anterior–posterior division of the BST was chosen as the standard or reference scheme. Therefore, the Swanson nomenclatures are considered variations on a standard or reference nomenclature for the rat BST. If the BST parts defined in the Swanson nomenclatures are followed in Figure 9, it can be seen that each of the clusters is composed of parts from either the anterior or the posterior division, and of terms related to them but defined in other nomenclatures. Thus, several parts are either separated (BSTd, BSTpm, BSTsz; lower right side of Figure 9) or form clusters (BSTov, BSTse, BSTfu, BSTju). The farthest right concentration of terms consists of the BSTif, BSTpr, and BSTtr – all belonging to the posterior division of the BST. To the left of this cluster the BSTmg, BSTv, and BSTdl (anterior division of the BST) form another cluster. Finally, the center of the graph is occupied predominantly by the terms BSTal, BSTad, BSTam, and BSTdm – all of which belong the anterior division of the BST. In other words, the anterior–posterior divisions of the Swanson nomenclatures are roughly preserved in the graph shown in Figure 9.
This graph can also be used to discover new relations between previously unrelated parts. For example, the BSTl nucleus defined in the Geraaedts nomenclature (acronym: BSTl_Ge; Geeraedts et al., 1990) is considered to be identical with the STa nucleus defined in the Bleier nomenclature (acronym: STa; Bleier et al., 1979), which in turn overlaps with the principal nucleus of the BST as defined by Ju and Swanson (acronym: BSTpr; Ju and Swanson, 1989). Therefore, BSTl also overlaps with BSTpr. In summary, the set of qualitative topological relations inserted in BAMS, and shown in graphical format, can be used to infer new relations either manually or using automated algorithms. However, many of these relations are equivocal (Egenhofer and Franzosa, 1991; Bota et al., 2005) or even contradictory, necessitating collator expertise.
Discussion
Solutions to the problem of multiple different, partly overlapping neuroanatomical terminologies for the same species are crucial for understanding structure–function relationships within and between the many parts of the nervous system. Thus, any robust neuroanatomical ontology must include synonyms and partial correspondences for its core concepts. In the absence of 3D computer graphics structural models that can be used effectively as templates, the available comparative information remains confined to qualitative topological relations that are reported in the literature or if necessary inferred by expert collators.
Efforts to relate nervous system parts defined in different parcellation schemes or nomenclatures are not unique to BAMS. Other neuroinformatics systems, including BrainInfo5, include terms used in several rat and mouse nomenclatures, along with URL pointers to a set of standard terms initially defined in macaque monkey or human nomenclatures. The Foundational Model of Anatomy (FMA)8 includes a set of standard or reference terms for human brain parts, as well as presumed synonyms for them. Efforts to match 3D renderings of the macaque monkey brain with partial ontologies have also been made (Bezgin et al., 2009). The Neuroinformatics Information Framework (NIF)9 combines several available ontologies and vocabularies (Bug et al., 2008). Its files are deposited in the OBO Foundry in OWL format and can be used for automatic inference (Bug et al., 2008).
The schema implemented in BAMS fully documents topological relations collated from the literature, and thus is general enough to be adopted as a standard for curation of qualitative spatial relations between nervous system parts defined in different neuroanatomical nomenclatures, regardless of species. The user interface we designed allows users to browse and inspect details of topological relations registered in the system.
Based on the Topological Relations and associated tables it was then possible to implement a simplified version of a translation topological inference engine that automatically reconstructs neuroanatomical macroprojection patterns across nomenclatures. This new Projections Translations feature of BAMS, which links the Relations and Connections modules, enhances several existing interfaces. The inference engine constructs the input projection (afferents) and output projection (efferents) profiles of nervous system gray matter regions from data mapped onto different nomenclatures, infers a global projection pattern for a region based on translated connection data for all of its reported subregions, and seeks to complete user-defined connection matrices. This inference engine is helpful to users because it integrates online information collated and mapped on different neuroanatomical nomenclatures and atlases. To our knowledge, BAMS is the first neuroinformatics system to include an online data mining tool for answering the question, “What are the projection patterns of nervous system gray matter region A?”, and is not limited by the nomenclature or atlas entered by the user. Moreover, each translated neuroanatomical projection can be associated with statements that confirm or contradict the inference. The validation process is either automatic, i.e., independent experimental results mapped onto the original nomenclature are mined from the BAMS’ database, or can be inserted manually by experts. To our knowledge, BAMS is also the first neuroinformatics system that implemented a simple schema for validation of inferences.
The Relations module of BAMS was also populated here with enough data and metadata to become useful to the neuroscience community. This information can be used to create “knowledge maps” of terms that are reported in the literature to be related. BAMS is the first system to include a complete set of terms used for the rat BST, stretching from 1923 to now.
We are aware of the limitations of the approach described in this paper. Ideally, one should represent 3D relationships between brain parts defined in different atlases, but this depends on the advent of 3D sliceable models. We agree that topological relationships between non-convex geometric objects (Papadimitriou et al., 1999) would describe better relations between brain parts. However, our approach is a minimal one, since it aims to capture as much information from the literature as possible, where the relationships between parts may be expressed in a single sentence, without any graphical support. The implementation of an algorithm for non-convex objects will first involve the transformation of the BAMS database into a spatial one, and second will prevent us recording information from many references. Moreover, the comparison will always have to be made by pairs of original slices, a task which is virtually impossible to perform for older nomenclatures that are still used by neuroscientists. However, we implemented a general topological algorithm (Bota et al., 2005), which takes into account the relationships per pairs of Atlas Levels. Moreover, the present extension of the module includes several annotation fields, which can be used by collators to describe the spatial relations between compared parts.
There are several obvious ways the work reported here will be extended next. First, the collated nomenclatures and related terms will be incorporated into NIF and aligned with ontologies already in that system. Second, the “Experiment type” variable included in the ER schema (Figure 1; Table 1) will be extended to a representation complex enough to capture the most important aspects of current and future nervous system preparation methods. Third, the automated translation inference engine will be extended to the other neural characters that can be collated in BAMS (for example, molecules), and it will be refined to include the equivocal results of topological inferences. Fourth, the inserted topological relations will be tested using automated algorithms. And fifth, more nervous system terms and nomenclatures, and the relations between them, will be inserted for the rat, and this effort will be extended to the mouse and human.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
3Direct link: http://brancusi.usc.edu/bkms/brain/search_bname_con.php
4Online user manual: http://brancusi.usc.edu/bkms/user_manual.pdf
References
- Alheid G. F., de Olmos J. S., Beltramino C. A. (1995). ‘Amygdala and extended amygdala,’ in The Rat Nervous System. 2nd Edn, ed. Paxinos G. (Amsterdam: Academic Press; ), 495–578 [Google Scholar]
- Bayer S. A. (1987). Neurogenetic and morphogenetic heretogeneity in the bed nucleus of the stria terminalis. J. Comp. Neurol. 265, 47–64 10.1002/cne.902650105 [DOI] [PubMed] [Google Scholar]
- Bezgin G., Reid A. T., Schubert D., Kötter R. (2009). Matching spatial with ontological brain regions using Java tools for visualization, database access, and integrated data analysis. Neuroinformatics 7, 7–22 10.1007/s12021-008-9039-5 [DOI] [PubMed] [Google Scholar]
- Bleier R., Cohn P., Siggelkow I. R. (1979). ‘A cytoarchitectonic atlas of the hypothalamus and hypothalamic third ventricle of the rat,’ in Handbook of the Hypothalamus, eds. Morgane P. J., Panksepp J. (New York: Marcel Dekker; ). pp. 137–220 [Google Scholar]
- Bota M., Arbib M. A. (2004). Integrating databases and expert systems for the analysis of brain structures, connections, and homologies. Neuroinformatics 2, 20–59 10.1385/NI:2:1:019 [DOI] [PubMed] [Google Scholar]
- Bota M., Dong H.-W., Swanson L. W. (2003). From gene networks to brain networks. Nat. Neurosci. 6, 795–799 10.1038/nn1096 [DOI] [PubMed] [Google Scholar]
- Bota M., Dong H.-W., Swanson L. W. (2005). Brain architecture management system. Neuroinformatics 3, 15–48 10.1385/NI:3:1:015 [DOI] [PubMed] [Google Scholar]
- Bota M., Swanson L. W. (2007a). Online workbenches for neural network connections. J. Comp. Neurol. 500, 807–814 10.1002/cne.21209 [DOI] [PubMed] [Google Scholar]
- Bota M., Swanson L. W. (2007b). The neuron classification problem. Brain Res. Rev 56, 76–88 10.1016/j.brainresrev.2007.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bota M., Swanson L. W. (2008a). 1st INCF Workshop on Neuroanatomical Nomenclature and Taxonomy. Nature Precedings Available at http://dx.doi.org/10.1038/npre.2008.1780.1
- Bota M., Swanson L. W. (2008b). BAMS neuroanatomical ontology: design and implementation. Front. Neuroinform 2, 2. 10.3389/neuro.11.002.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bug W. J, Ascoli G. A., Grethe J. S., Gupta A., Fennema-Notestine C., Laird A. R., Larson S. D., Rubin D., Shepherd G. M., Turner J. A., Martone M. E. (2008). The NIFSTD and BIRNLex vocabularies: building comprehensive ontologies for neuroscience. Neuroinformatics 6, 175–194 10.1007/s12021-008-9032-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie E. H. (1925). An Introduction to the Finer Anatomy of the Central Nervous System Based Upon That of the Albino Rat Philadelphia: Blakiston [Google Scholar]
- de Groot J. (1959). The rat hypothalamus in stereotaxic coordinate. J. Comp. Neurol. 113, 389–400 10.1002/cne.901130304 [DOI] [PubMed] [Google Scholar]
- de Olmos J., Alheid G. F., Beltramino C. A. (1985). ‘Amygdala,’ in The Rat Nervous System. Vol. I. Forebrain and Midbrain, ed. Paxinos G. (Sydney: Academic Press; ), 223–334 [Google Scholar]
- de Olmos J., Ingram W. R. (1972). The projection field of the stria terminalis in the rat brain. An experimental study. J. Comp. Neurol. 146, 303–336 10.1002/cne.901460303 [DOI] [PubMed] [Google Scholar]
- Dong H.-W., Petrovich G. D., Swanson L. W. (2001). Topography of projections from amygdala to bed nuclei of the stria terminalis. Brain Res. Brain Res. Rev. 38, 192–246 10.1016/S0165-0173(01)00079-0 [DOI] [PubMed] [Google Scholar]
- Dong H.-W., Swanson L. W. (2003). Projections from the rhomboid nucleus of the bed nuclei of the stria terminalis: implications for cerebral hemisphere regulation of ingestive behaviors. J. Comp. Neurol. 46, 434–472 10.1002/cne.10758 [DOI] [PubMed] [Google Scholar]
- Dong H.-W., Swanson L. W. (2004). Projections from bed nuclei of the stria terminalis, posterior division: implications for cerebral hemisphere regulation of defensive and reproductive behaviors. J. Comp. Neurol. 471, 396–433 10.1002/cne.20002 [DOI] [PubMed] [Google Scholar]
- Dong H.-W., Swanson L. W. (2006). Projections from bed nuclei of the stria terminalis, anteromedial area: cerebral hemisphere integration of neuroendocrine, autonomic, and behavioral aspects of energy balance. J. Comp. Neurol. 494, 142–178 10.1002/cne.20788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egenhofer M., Franzosa R. (1991). Point-set topological spatial relations. Int. J. Geogr. Inf. Syst. 5, 161–174 10.1080/02693799108927841 [DOI] [Google Scholar]
- Felleman D. J., van Essen D. C. (1991). Distributed hierarchical processing in the primate cerebral cortex. Cereb. Cortex 1, 1–47 10.1093/cercor/1.1.1-a [DOI] [PubMed] [Google Scholar]
- Fulwiler C. E., Saper C. B. (1984). Subnuclear organization of the efferent connections of the parabrachial nucleus in the rat. Brain Res. 319, 219–259 [DOI] [PubMed] [Google Scholar]
- Gansner E. R., North S. C. (1999). An open graph visualization system and its applications to software engineering. Softw. Pract. Exp. 30, 1–5 [Google Scholar]
- Geeraedts L. M. G., Nieuwenhuys R., Veening J. G. (1990). Medial forebrain bundle of the rat: III Cytoarchitecture of the rostral (telencephalic) part of the medial forebrain bundle bed nucleus. J. Comp. Neurol. 294, 507–536 10.1002/cne.902940403 [DOI] [PubMed] [Google Scholar]
- Gurdjian E. S. (1925). Olfactory connections in the albino rat, with special reference to the stria medullaris and anterior commissure. J. Comp. Neurol. 38, 127–163 10.1002/cne.900380202 [DOI] [Google Scholar]
- Johnston J. B. (1923). Further contributions to the study of the evolution of the forebrain. J. Comp. Neurol. 35, 337–380 10.1002/cne.900350502 [DOI] [Google Scholar]
- Ju G., Swanson L. W. (1989). Studies on the cellular architecture of the bed nuclei of the stria terminalis in the rat: I. Cytoarchitecture. J. Comp. Neurol. 280, 587–602 10.1002/cne.902800409 [DOI] [PubMed] [Google Scholar]
- Ju G., Swanson L. W., Simerly R. B. (1989). Studies on the cellular architecture of the bed nuclei of the stria terminalis in the rat: II. Chemoarchitecture. J. Comp. Neurol. 280, 603–621 10.1002/cne.902800410 [DOI] [PubMed] [Google Scholar]
- König J. F. R., Klippel R. A. (1963). The Rat Brain: A Stereotaxic Atlas of the Forebrain and Lower Parts of the Brain Stem. Baltimore, Wilkins & Wilkins [Google Scholar]
- Krettek J. E., Price J. L. (1978). A description of the amygdaloid complex in the rat and cat with observations on intra-amygdaloid axonal connections. J. Comp. Neurol. 178, 255–280 10.1002/cne.901780205 [DOI] [PubMed] [Google Scholar]
- Larson S. D., Martone M. (2009). Ontologies for neuroscience: what are they and what are they good for? Front. Neurosci. 3, 60–67 10.3389/neuro.01.007.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald A. J. (1983). Neurons of the bed nucleus of the stria terminalis: a Golgi study in the rat. Brain Res. Bull. 10, 111–120 10.1016/0361-9230(83)90082-5 [DOI] [PubMed] [Google Scholar]
- Moga M. M., Saper C. B., Gray T. S. (1989). Bed nucleus of the stria terminalis: cytoarchitecture, immunohistochemistry, and projections to the parabrachial nucleus in the rat. J. Comp. Neurol. 283, 283–332 10.1002/cne.902830302 [DOI] [PubMed] [Google Scholar]
- Papadimitriou C. H., Suciu D., Vianu V. (1999). Topological queries in spatial databases. J. Comp. Sys. Sci. 58, 29–53 10.1006/jcss.1998.1597 [DOI] [Google Scholar]
- Paxinos G., Watson C. (1986). The Rat Brain in Stereotaxic Coordinates. San Diego, Academic Press; 3099697 [Google Scholar]
- Paxinos G., Watson C. (1998). The Rat Brain in Stereotaxic Coordinates, 2nd Edn.San Diego, Academic Press [Google Scholar]
- Pellegrino J. S., Pellegrino A. S., Cushman A. J. (1979). A Stereotaxic Atlas of the Rat Brain. New York: Plenum Press; 393066 [Google Scholar]
- Swanson L. W. (1992). Brain Maps: Structure of the Rat Brain. Amsterdam, Elsevier [Google Scholar]
- Swanson L. W. (1998). Brain Maps: Structure of the Rat Brain, 2nd Edn.Amsterdam, Elsevier [Google Scholar]
- Swanson L. W. (2000). What is the brain? Trends Neurosci 23, 519–527 10.1016/S0166-2236(00)01639-8 [DOI] [PubMed] [Google Scholar]
- Swanson L. W. (2004). Brain Maps: Structure of the Rat Brain. A Laboratory Guide with Printed and Electronic Templates for Data, Models and Schematics, 3rd Edn.Amsterdam, Elsevier [Google Scholar]
- Swanson L. W., Cowan W. M. (1979). The connections of the septal region in the rat. Brain Res. 379, 399–403 10.1016/0006-8993(86)90799-7 [DOI] [PubMed] [Google Scholar]
- Swanson L. W., Petrovich G. D. (1998). What is the amygdala? Trends Neurosci 21, 323–331 10.1016/S0166-2236(98)01265-X [DOI] [PubMed] [Google Scholar]