Abstract
Background
Studies suggest that polymorphisms in the D4 dopamine receptor (DRD4) and opioid receptor, μ1 (OPRM1) genes are involved in differential response to the effects of alcohol and to alcohol cues. However, to date, the mechanisms that underlie these differences remain largely unknown.
Methods
Using functional magnetic resonance imaging, hemodynamic response in mesocorticolimbic structures after exposure to alcohol tastes was contrasted with a control taste and compared between DRD4 variable number of tandem repeats (VNTR) genotypes and OPRM1 A118G genotypes. Additionally, the effects of a priming dose of alcohol on this response were examined.
Results
The results indicated that DRD4 VNTR >7 repeat individuals (DRD4.L) had significantly greater response to alcohol cues in the orbitofrontal cortex, anterior cingulate gyrus, and striatum compared with individuals with <7 repeats (DRD4.S) prior to a priming dose of alcohol (p < 0.05), but not after a priming dose. In the OPRM1 comparisons, results showed that individuals with at least 1 copy of the OPRM1 + 118 G allele had greater hemodynamic response in mesocorticolimbic areas both before and after priming compared with those who were homozygous for the OPRM1 + 118 A allele. For the DRD4.L and OPRM1 + 118 G groups, brain response in the striatum was highly correlated with measures of alcohol use and behavior such that greater activity corresponded with greater frequency and quantity of alcohol use.
Conclusions
The DRD4 VNTR and OPRM1 A118G polymorphisms are associated with functional neural changes in mesocorticolimbic structures after exposure to alcohol cues. This provides evidence for the contributions of the DRD4 and OPRM1 genes in modulating neural activity in structures that are involved in the motivation to drink.
Keywords: DRD4, OPRM1, Alcohol, Craving, fMRI, Mesocorticolimbic
Although twin, family and adoption studies have established that alcohol dependence is influenced by genetic factors (Kendler et al., 1994; Rhee et al., 2003), the identification of specific genetic variations, and understanding how those variations contribute to the risk for alcohol dependence has proven to be difficult. This is in part due to the imprecision of behavioral phenotypes (e.g., a diagnosis of alcohol dependence) that are highly heterogeneous and, as a result, are likely influenced by a multitude of genes as well as a multitude of environmental factors. It is also driven by the lack of phenotypes that are directly tied to the neurophysiology of alcohol dependence. Clearly, the identification of genetic factors that influence the etiology of alcohol dependence will be aided by the adoption of approaches that have a stronger neurobiological component.
While cue-elicited subjective craving has proven useful as an endophenotype, the subjective report of craving is obviously distal to the neurobiological mechanisms that are invoked by exposure to alcohol cues (Kalivas and Volkow, 2005). fMRI measurement of hemodynamic activation of mesocorticolimbic areas represents a significant methodological improvement in research designed to elucidate the neurobiology of this endophenotype because it more accurately reflects the activation of the biological mechanisms that putatively mediate the development and expression of craving. Several recent studies have incorporated fMRI to examine the hemodynamic response of brain structures after exposure to alcohol cues. For example, visual alcohol stimuli were reported to induce significant activation of brain areas such as the fusiform gyrus, basal ganglia, and orbitofrontal gyrus, as compared to abstract control pictures (Wrase et al., 2002), while exposure to alcohol odors increased activation of the cerebellum and amygdala in alcohol dependent patients but not in controls (Schneider et al., 2001). In 2 recent studies that combined priming (sip of alcohol) with visual presentations (picture of alcoholic stimuli), it was reported that alcohol-related stimuli increased activation in the prefrontal cortex and anterior thalamus (George et al., 2001) and anterior limbic areas (Myrick et al., 2004). We have also recently reported that using real-time taste cues, activation in the mesocorticolimbic areas was greater during alcohol tastes compared to a novel control taste cue (i.e., litchi juice) and a resting baseline period. Furthermore, we found that activation in these substrates were correlated with state measures of alcohol craving [i.e., Alcohol Urge Questionnaire (AUQ)] as well as more stable measures of drinking behavior and problems [i.e., Alcohol Use Disorders Identification Test (AUDIT)] (Filbey et al., 2007).
In addition to the more recent neuroimaging work that implicates the dopaminergic (DAergic) system in craving, previous association studies also focused on genes that modulate functioning of the DAergic system. More specifically, both the D4 dopamine receptor (DRD4) and opioid receptor, μ1 (OPRM1) genes influence subjective craving to alcohol (Hutchison et al., 2002b; Ray and Hutchison, 2004, 2007). The DRD4 has a variable number of tandem repeats (VNTR) in exon 3 with common variants of 2, 4, and 7 repeats (Van Tol et al., 1992). Previous studies have suggested that the 7-repeat allele of the DRD4 VNTR is associated with greater craving for alcohol (Hutchison et al., 2002b), associated with the effects of medications designed to reduce craving for alcohol (Hutchison et al., 2003, 2006), subjective craving for tobacco after exposure to cues as well as activation of the prefrontal cortex (Hutchison et al., 2002a; McClernon et al., 2007), and craving after exposure to heroin cues (Shao et al., 2006). While there are studies that have not found similar associations (van den Wildenberg et al., 2007), it appears that, in general, the DRD4 VNTR plays a role in cue-elicited craving and this association is thought to be due to the role of the mesolimbic DAergic system in the rewarding effects of several substances, including alcohol and nicotine (Berridge, 2007).
The DAergic system also interacts with μ-opioid receptors, which are implicated in the reinforcing effects of alcohol (Gianoulakis, 2001). It has been suggested that DA release by ethanol is the consequence of increased opioidergic activity that inhibits GABAergic neurons, thereby, disinhibiting DAergic neurons (Erickson, 1996; Herz, 1997; Kreek, 1996). Behavioral genetic studies have examined variation in the gene coding for OPRM1, given the putative association between μ-opioid receptors and the reinforcing effects of several substances, including alcohol. One of the most widely studied polymorphisms of the OPRM1 gene is the +118A/G single nucleotide polymorphism (SNP) located in the +118 position in exon 1, which codes for the A to G substitution (rs1799971). Molecular studies of this polymorphism have initially suggested that the A to G substitution affects receptor activity for endogenous ligand β-endorphin leading to a gain in function, such that the G variant was thought to bind β-endorphin 3 times stronger than the A allele (Bond et al., 1998). In a more recent study of the functional significance of this SNP suggested that the G allele has deleterious effects on both mRNA and protein yield (Zhang et al., 2005). In short, although the specific nature and direction of the functional effects of this SNP are still unclear, the molecular literature suggests that this polymorphism is indeed functional.
While reports of an association between the A118G SNP and alcohol dependence have been inconsistent (Arias et al., 2006), previous studies using an endophenotype approach (i.e., focusing on subjective responses to alcohol and cue-induced craving) found that individuals with the G allele of the OPRM1 gene reported higher subjective feelings of intoxication, stimulation, sedation, and positive mood across rising levels of breath alcohol concentration (BAC), as compared with participants with the A allele (Ray et al., 2006). A study by van den Wildenberg et al. (2007) found that male carriers of the G allele of the OPRM1 gene reported higher levels of alcohol craving following cue-reactivity.
The A118G SNP of the OPRM1 gene has also received attention as a moderator of the effects of naltrexone, an opioid receptor antagonist, for the treatment of alcoholism. Specifically, in a double-blind placebo controlled study, we have found that this OPRM1 genotype moderated the effects of naltrexone on alcohol-induced “high” in the laboratory, such that carriers of the G allele reported greater naltrexone-induced blunting of alcohol “high” as compared to homozygotes for the A allele (Ray and Hutchison, 2007). This may explain the important clinical finding that carriers of the G-allele are more responsive to naltrexone for the treatment of alcohol dependence (Oslin et al., 2003), a medication that in turn is thought to reduce feelings of alcohol euphoria and stimulation (Drobes et al., 2004; McCaul et al., 2001; Swift et al., 1994). On the other hand, a study by McGeary et al. (2006) found that carriers of the G-allele reported higher urge to drink in a cue-reactivity paradigm while taking naltrexone, and the VA Cooperative Study has found no support for an association between this SNP and clinical response to naltrexone (Gelernter et al., 2007). Taken together, the literature on the A118G SNP of the OPRM1 gene suggests that this polymorphism may be associated with the acute effects of alcohol and possibly responses to naltrexone, even though studies testing the association between this polymorphism and the diagnostic phenotype of alcohol dependence have been largely inconclusive (Arias et al., 2006).
The present study is a secondary analysis of the possible genetic moderators of the mesocorticolimbic response to alcohol tastes we previously reported, although the current sample has increased significantly from n = 37 to n = 69 (Filbey et al., 2007; Hutchison et al., in press). Our aim is to extend the literature on the DRD4 VNTR and OPRM1 A118G SNP by testing whether a differential fMRI blood oxygenated level dependent (BOLD) response to alcohol taste-cues is associated with 2 candidate polymorphisms for alcohol dependence: the DRD4 VNTR and OPRM1 A118G SNP. In addition, the study was designed to test whether these associations might be different after a priming dose of alcohol. We previously found that neural response in incentive motivation structures of the brain is greater for alcohol taste cues compared with control taste cues. Based on this finding and our aforementioned work on the DRD4 VNTR and OPRM1 A118G SNP, we expected to find that these 2 genetic variants modulate these neural responses to alcohol cues mesocorticolimbic structures of the brain. We also expected that this response will be enhanced by alcohol priming.
Materials and Methods
Participants
Seventy-three heavy drinking, but otherwise healthy subjects took part in this fMRI study. Because we were interested in the effects of DRD4 VNTR and OPRM1, participants were classified according to their genotype. Twenty-three participants had DRD4 VNTR >7 repeats (and without the OPRM1 G allele) and were classified into the DRD4 long group (DRD4.L). Eleven participants had at least 1 copy of the OPRM1 G allele (and DRD4 VNTR <7 repeats) and were classified into the OPRM1 G allele group (OPRM1.G). Six subjects were carriers of both the DRD4 >7 repeat and OPRM1 G allele variants and were excluded from the analyses given that this sample was too small to examine additive and interactive (i.e., epistatic) genetic effects and could instead confound the analyses of main effects. Thirty-three participants had neither risk genotype (i.e., DRD4 VNTR <7 and OPRM1 AA genotype) and were classified into the control group. The observed allele frequencies were consistent with previous studies of primarily individuals of European Ancestry, which is approximately 20 to 30% frequency for the minor allele (e.g., Arias et al., 2006; Hutchison et al., 2002a, b; Ray and Hutchison, 2004, 2007). All participants were right-handed and did not have any present or past history of head injury. Participants signed written informed consents approved by the University of Colorado Human Research Committee. Demographic characteristics of the participants are summarized in Table 1.
Table 1.
Demographic Characteristics of the Participants
| DRD4. L (n = 21) | OPRM1.G (n = 11) | Controls (n = 31) | ||||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | |
| Age | 22.70 | 2.63 | 22.80 | 2.75 | 22.90 | 2.27 |
| Gender (M/F) | 14/7 | – | 5/6 | – | 20/11 | – |
| Education | 15.90 | 1.00 | 15.70 | 1.03 | 15.60 | 1.26 |
| No. of drinks/occasion | 5.29 | 2.59 | 5.80 | 2.78 | 4.72 | 2.17 |
| Largest no. of drinks/occasion | 12.20 | 4.32 | 14.90 | 7.68 | 13.10 | 6.09 |
| Times/month drank | 12.00 | 5.63 | 12.60 | 4.66 | 10.70 | 3.99 |
| No. of times drank >5 drinks/occasion | 6.79 | 5.54 | 6.60 | 5.64 | 6.77 | 4.93 |
The DRD4.L and OPRM1.G individuals did not differ significantly from the control group (individuals with the DRD4.S and OPRM1.A genotype).
Outcome Measures
Subjective Craving
The AUQ (Bohn, et al. 1995) was used to measure current craving for alcohol. This measure was collected immediately after the scanning session.
Drinking Behaviors
Quantity and frequency of alcohol use and problems related to alcohol use were measured by the AUDIT (Allen et al., 1997). This measure was collected prior to the scanning session.
Taste Cue Paradigm
We utilized an fMRI taste-cue paradigm previously reported to elicit BOLD response in mesocorticolimbic areas (Filbey et al., 2007) (see Fig. 1). Briefly, all taste stimuli were delivered to the participants via Teflon tubing using a computer-controlled delivery system. We used subject-relevant (i.e., alcohol most commonly consumed) alcohol stimuli and litchi juice as the control stimulus. We presented each subject with 4 echo planar imaging (EPI) runs consisting of pseudo-randomized 6 alcohol and 6 control trials to control for order effects. Two of the 4 runs were presented following consumption of an alcohol beverage (similar to the alcohol stimulus delivered during the scans) precalculated to result in BAC = 0.03 mg using a formula described by Watson (1998). The participants were given 10 minutes to consume the beverage and another 10 minutes for absorption prior to re-scanning. This is illustrated in Fig. 2. For this study, the order of priming dose was kept consistent across subjects such that the postpriming scans were always runs 3 and 4. While having a 2-session counterbalanced design would have been ideal, we felt that for this first study, it is more important to minimize having incomplete datasets by not requiring subjects to return for a second session.
Fig. 1.
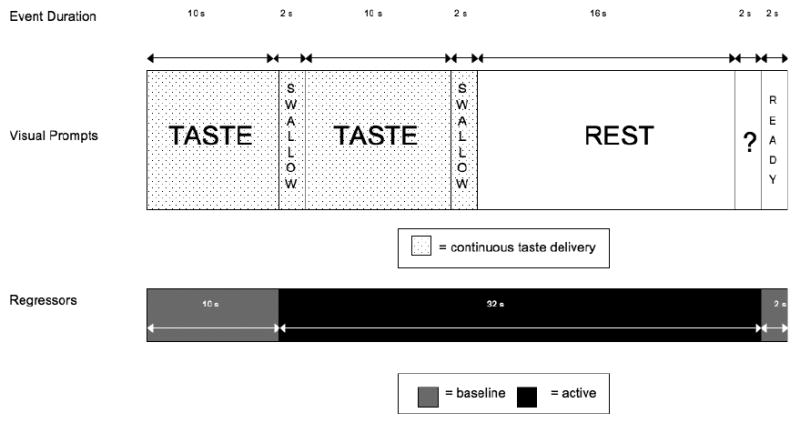
Schematic of a single taste cue trial. A single trial consisted of a continuous delivery of either alcohol or litchi juice presented for 24 seconds at the beginning of each trial (interspersed with 2 visual prompts to allow subjects to swallow). The total amount of liquid delivered during this period was 1 ml. The taste delivery was followed by a 16-second washout period. This was followed by an urge question presented on the screen for 2 seconds to which the subjects were asked to rate their current subjective urge to drink alcohol (“Please rate your urge to drink alcohol right now?”) using a scale of 1 (no urge at all) to 4 (very high urge) using a button box. Each trial ended with a 2-second “Ready” prompt screen. The active and baseline regressors are also indicated. Because tastes were not detected until mid-way through the 1 ml stimulus delivery (typically after the first swallow prompt) and took longer to dissipate, we modeled the active period to encompass the period between the initial swallow and the end of the washout period.
Fig. 2.
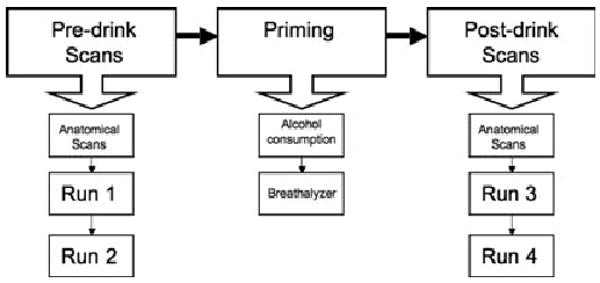
Schematic of the fMRI protocol. During the fMRI session, subjects underwent 2 sets of scanning. The first set of scans (“predrink scans”) were without alcohol priming and consisted of anatomical scans and a counterbalanced order of 2 fMRI runs. These scans were followed by a break from scanning (“priming”), where subjects were taken out of the magnet and allowed to stretch while consuming a priming dose of alcohol within 10 minutes followed by another 10 minutes to allow absorption. Subjects were then breathalyzed and their BACs recorded. They were immediately placed back into the scanner for the second set of scans (“postdrink scans”), which was identical to the first set.
Data Acquisition
The functional EPI images were acquired on a GE 3T scanner (Milwaukee, WI). Because the orbitofrontal cortex (OFC) is involved in the craving/reward system and can suffer from severe signal dropout caused by susceptibility effects, we used a volume-selective z-shim EPI technique to acquire the functional images (Du et al., 2007). Other parameters of the EPI data acquisition were as follows: repetition time (TR) = 2000 ms, echo time (TE) = 26 ms, flip angle = 77°, field of view (FOV) = 22 cm, matrix size = 64 × 64, slice thickness = 4 mm without inter-slice gap, and collection time = 9:11 (includes 11 TRs of stabilization scans that were excluded from the analyses). Because the effective TR was 1000 ms in the z-shim slices, a lower flip angle of 62° was used to maximize the image signal intensity in these slices.
For a 2-stage registration of the EPI images, high-resolution T1-weighted FLAIR part-head images (29 axial slices of part head, matrix = 256 × 192, collection time = 2:59) were acquired using the same slice angles, thickness, and gap as the EPI images. Another high-resolution full-head 3D structural image was collected in the coronal plane using an inversion-recovery spoiled gradient recalled sequence (TI = 500 ms, flip angle = 10°, slice thickness = 1.4 mm, 256 × 256 matrix, 220 × 220 mm FOV, bandwidth = 15.6kHz, 124 slices, collection time = 9:12).
During data acquisition, head restraints were placed using a foam pillow. To mark the right side of the brain the right forehead was marked using a vitamin E capsule. Visual instructions was presented using a goggle system (Resonance Technology, Inc., Northridge, CA) and responses to the urge questions were recorded using 2 fiber-optics response pads with 2 response buttons each placed in each hand. Gustatory stimuli were delivered using an Infinity Controller controlled by a presentation computer running E-Prime.
DRD4 Assay
The DRD4 exon III 48 bp VNTR was assayed by a modification (Anchordoquy et al., 2003) of the method of Sander et al. (1997) using the primer sequences given in Lichter et al. (1993). After amplification, an aliquot of PCR product was analyzed with an ABI PRISM® 3100 Genetic Analyzer using protocols supplied by the company. Additional details of the method can be found in Anchordoquy et al. (2003) and at http://ibgwww.colorado.edu/genotyping_lab/dopamine_d4_receptor.html.
Allele sizes were scored by 2 investigators independently, and inconsistencies were reviewed and rerun when necessary. For analysis individuals with at least 1 allele ≥7 repeats were classified as having the long DRD4 L genotype. Those with fewer than 7 repeats were classified as having the short DRD4 S genotype. A rationale for this classification has been suggested by Wang et al., (2004) who reported that the 7-repeat allele arose as the result of a relatively recent mutational event.
OPRM1 Assay
An ABI PRISM 7500 instrument was used to conduct 5′-nuclease (TaqMan) assays of the OPRM1 SNP using assays commercially available from Applied Biosystems. This method relies on allele-specific hybridization of oligonucleotide probes (Livak, 1999).
Data Analysis
fMRI Data Preprocessing
The first 7 volumes of all EPI runs were discarded to allow the MR signal to reach steady state. Motion correction was carried out using FMRIB's Software Library (FSL), http://www.fmrib.ox.ac.uk/fsl) Motion Correction using FMRIB's Linear Image Registration Tool (McFLIRT) Version 5.0, and indicated that all of the participants had minimal head movement of <1 mm within a run. Four subjects (1 DRD4.L, 2 controls, and 1 with both DRD4.L and OPRM1.G genotype) did not complete their scans and are excluded from all of the analyses.
fMRI data analyses were carried out using FMRI Expert Analysis Tool (FEAT) Version 5.63, part of FSL using the following prestatistics processing: nonbrain tissue/skull removal using Brain Extraction Tool (BET), spatial smoothing using a Gaussian kernel of FWHM 5 mm, mean-based intensity normalization of all volumes by the same factor and highpass temporal filtering (Gaussian-weighted least-squares straight line fitting, with sigma = 50.0 s). Time-series statistical analysis was carried out using FMRIB's Improved Linear Model (FILM) with local autocorrelation correction.
Explanatory variables (i.e., taste and baseline periods for alcohol and control trials separately) were created by convolving the stimulus timing files with a double gamma hemodynamic response function in FEAT. A multiple linear regression analysis was performed to estimate the hemodynamic parameters for the different explanatory variables and a corresponding t-statistic indicates the significance of the activation of the stimulus. Contrast maps were created by contrasting alcohol taste vs. control taste conditions. Statistical maps were then registered to the Montreal Neurological Institute (MNI) template with a 2-step process. First, EPI images were registered to the part-head high resolution T1-weighted image acquired in the same plane as the EPI images. The part-head anatomical image was then registered to the high resolution full-head image, which was subsequently registered to the 152 brain average MNI template. These registration steps were performed using FLIRT.
To be consistent with our previously reported findings (Filbey et al., 2007), we utilized anatomically defined a priori region of interest (ROI) masks of structures within the reward-craving pathway of the brain frequently reported in the craving literature, namely the striatum, the ventral tegmental area/midbrain (VTA/midbrain), OFC, and the medial prefrontal cortex (MPFC) (Kalivas and Volkow, 2005; Volkow et al., 2007). The VTA/midbrain mask was created using MRIcro software and the Talairach and Tournoux brain atlas was used as a guide for defining anatomical landmarks. The striatum, MPFC, and OFC masks were obtained from the Nielsen and Hansen's volume of interest online database (Nielsen and Hansen, 2002).
Between-Group Analyses
To examine the genetic moderators of response to alcohol cues and priming, we used a 2 × 2 × 2 (genotype × cue × priming) study design. After the preprocessing steps, higher-level analysis was carried out using FMRIB's Local Analysis of Mixed Effects (FLAME). Z (Gaussianised T/F) statistic images were thresholded using GRF-theory-based maximum height thresholding with a significance threshold of 1-tailed p < 0.05. Peak loci of activation were obtained using MRI3dX (version 5.5; http://www.aston.ac.uk/lhs/staff/singhkd/mri3dX) and anatomical localization was confirmed by the Talairach Daemon Database and verified by the Talairach and Tournoux brain atlas.
Correlation Analyses
To determine the relationship between the BOLD response and behavior related to alcohol use, Pearson correlations were performed between the self-reported alcohol behavior and craving measures (i.e., AUDIT, AUQ, and urge ratings) and ROI maximum percent signal change values using SPSS Statistical Software vs. 11 (http://www.spss.com). The maximum percent signal change per contrast for each ROIs were calculated using Featquery (part of FEAT).
Results
Subjective Measures
The t-tests revealed no significant difference between the groups in any of the subjective measures of alcohol use and alcohol-related problems as measured by the AUDIT and the AUQ. However, the OPRM1.G individuals did differ from the controls in their difference score between post- minus prepriming in scanner litchi urge rating such that OPRM1.G individuals appeared to have a greater difference between their postpriming and prepriming urge rating for litchi cues (the negative difference score in the OPRM1.G individuals, but not in the controls, indicates greater urge ratings to litchi juice prior to the priming dose of alcohol) (Table 2).
Table 2.
Results of Subjective Measures
| DRD4. L (n = 22) | OPRM1.G (n = 11) | Controls (n = 31) | ||||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | |
| Total AUDIT score | 12.80 | 4.95 | 12.11 | 4.70 | 11.10 | 4.90 |
| Baseline AUQ | 18.30 | 17.20 | 15.90 | 8.97 | 14.60 | 7.06 |
| Postscan AUQ | 19.70 | 5.84 | 15.30 | 8.33 | 17.60 | 7.65 |
| Postscan AUQ minus baseline AUQ | 2.19 | 5.62 | −0.36 | 10.90 | 2.72 | 5.00 |
| In-scanner alcohol urge rating (before priming) | 1.60 | 0.51 | 1.50 | 0.63 | 1.63 | 0.66 |
| In-scanner litchi urge rating (before priming) | 1.40 | 0.53 | 1.57 | 0.90 | 1.46 | 0.52 |
| In-scanner alcohol craving (after priming) | 2.02 | 0.79 | 1.54 | 0.63 | 1.97 | 6.81 |
| In-scanner litchi craving (after priming) | 1.91 | 0.64 | 1.51 | 0.743 | 1.81 | 0.65 |
| Post- minus prepriming in-scanner litchi urge rating | 0.53 | 0.58 | −0.06* | 0.53 | 0.33 | 0.39 |
| Post- minus prepriming in-scanner alcohol urge rating | 0.42 | 0.71 | 0.04 | 0.59 | 0.29 | 0.39 |
Mean values and standard deviations of subjective measures for each group are summarized in this table. T-tests revealed a significant difference between OPRM1.G individuals and controls (individuals with DRD4.S and OPRM1.A genotype) in the difference score between postscan minus prescan in scanner litchi urge ratings (p < 0.05). The groups did not differ in any of the other subjective measures.
t = −2.457, p = 0.02.
BOLD Response
Our imaging analyses indicated that individuals with ≥7 DRD4 VNTR had greater BOLD response compared with those with <7 repeat alleles in all of the ROIs but only before the priming dose of alcohol (p < 0.05). After the priming dose of alcohol, the DRD4.L group did not have significantly greater activation in the ROIs compared with the control group. The control group, however, showed greater activation in all of the ROIs (see Table 3 and Fig. 3).
Table 3.
Greater Areas of Activity in DRD4.L Group Compared With the Control Group
| Region | BA | Peak Z-score | TLRC |
|---|---|---|---|
| DRD4.L > Controls | |||
| Prepriming | |||
| L OFC* | 47 | 3.17 | −42, 43, −5 |
| L subcallosal gyrus | 25 | 3.10 | −10, 19, −15 |
| L IFG | 47 | 2.92 | −36, 29, −15 |
| R putamen | – | 2.79 | 24, 11, −7 |
| L OFC* | 47 | 2.54 | −14, 23, −15 |
| R insula | 13 | 2.41 | 28, 17, −7 |
| R OFC | 11 | 2.26 | 40, 45, −13 |
| R OFC | 11 | 2.21 | 12, 25, −15 |
| R VTA/midbrain | – | 1.90 | 10, −17, −15 |
| Postpriming | |||
| None | |||
| Controls > DRD4.L | |||
| Prepriming | |||
| R medial frontal gyrus | 10 | 3.65 | 20, 43, −1 |
| L ACG | 32 | 3.38 | 16, 33, 7 |
| R caudate | – | 3.05 | 22, 21, 5 |
| R ACG | 32 | 2.90 | 16, 41, −1 |
| L caudate | – | 2.66 | −18, 27, 1 |
| R IFG | 11 | 1.78 | 26, 31, −25 |
| Postpriming | |||
| R IFG | 47 | 3.93 | 23, 31, −11 |
| L OFC | 11 | 3.64 | −24, 35, −13 |
| L caudate | – | 3.58 | 22 27 −7 |
| R globus pallidus | – | 3.57 | 20, −13, −5 |
| L thalamus | – | 3.35 | −10, −3, −13 |
| R VTA/midbrain | – | 3.30 | 14, −9, −9 |
| R putamen | – | 3.21 | −10, 5, −9 |
| R ACG | 32 | 2.99 | 18, 33, −11 |
| L VTA/midbrain | – | 2.87 | −6, −19, −9 |
| L putamen | – | 2.80 | −24, 9, −7 |
| R thalamus | – | 2.70 | 18, −5, 13 |
L, left; R, right; OFC, orbitofrontal cortex; IFG, inferior frontal gyrus; VTA, ventral tegmental area; ACG, anterior cingulate gyrus.
Without priming, the DRD4.L subjects had greater BOLD response in all of the ROIs compared to the controls (p < 0.05). None of these structures was greater in the DRD4.L subjects after priming. Maximum loci of activation are listed for each substrate as anatomical labels, Brodmann area (BA) corresponding peak z-score, and Talairach (TLRC) co-ordinates corresponding of greater BOLD response in the risk allele group compared with the nonrisk allele group.
Foci of peak activation (i.e., L OFC) that overlap with OPRM1.G individuals.
Fig. 3.
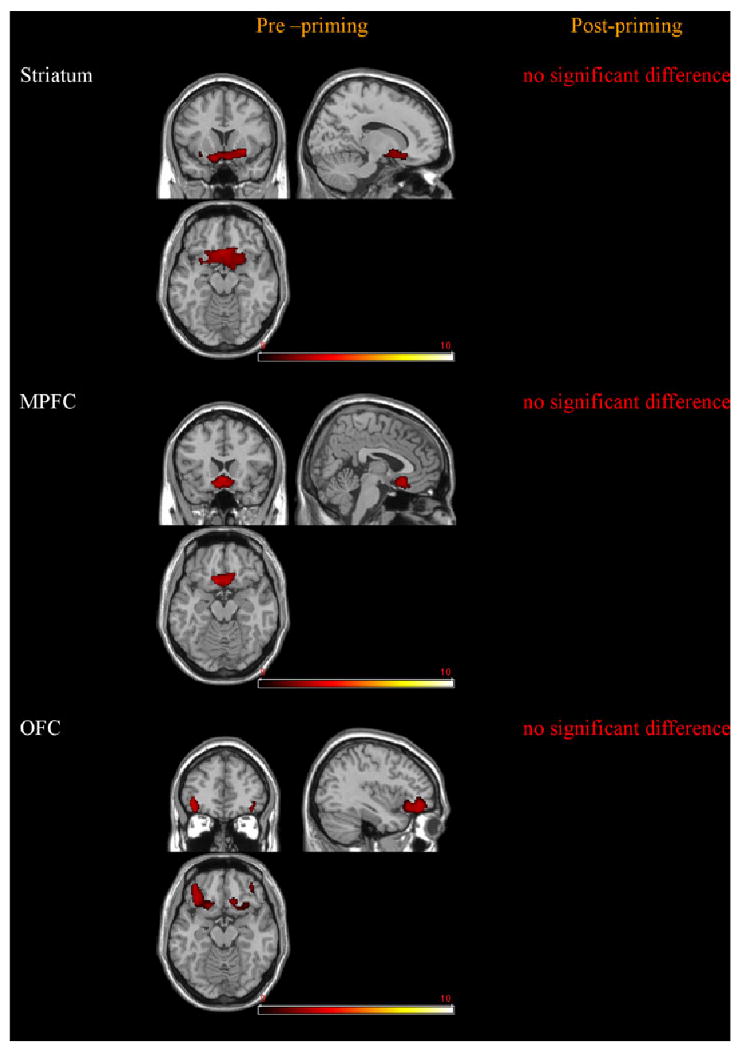
Greater activity in the DRD4.L subjects compared with control subjects. The DRD4.L subjects had significantly greater BOLD response compared with the controls in response to alcohol taste cues in the all areas before priming. After priming, the DRD4.L subjects did not have increased response in the ROIs compared with the controls (p < 0.05). The right side of the image reflects right-hemispheric activation. The colorscale represents z-scores.
The OPRM1 comparisons showed that the OPRM1.G group had significantly greater activation compared with the control group both before and after the priming dose. Before the priming dose of alcohol, greater activity in the OPRM1.G group was found in the OFC, VMPFC, and striatum (see Table 3), (p < 0.05). After priming, greater activity in the OPRM1.G group remained in these ROIs (see Table 4 and Fig. 4).
Table 4.
Greater Areas of Activity in OPRM1.G Group Compared With the Control Group
| Region | BA | Peak Z-score | TLRC |
|---|---|---|---|
| OPRM1.G > Controls | |||
| Prepriming | |||
| L ACG | 25 | 2.44 | −1, 23, −9 |
| L OFC* | 11 | 2.41 | −26, 35, −13 |
| L VS/NAc | – | 2.27 | −30, 9, −11 |
| L OFC* | 11 | 2.25 | −40, 37, −13 |
| R IFG | 47 | 2.24 | 30, 21, −13 |
| R VS/NAc | – | 2.17 | 28, 7, −13 |
| R IFG | 47 | 2.15 | 28, 19, −13 |
| R ACG | 32 | 1.99 | 10, 29, −11 |
| Postpriming | |||
| L IFG | 47 | 2.36 | −28, 33, −7 |
| L globus pallidus | – | 2.13 | −18, 1, −5 |
| L ACG | 24 | 2.10 | −12, 27, −3 |
| R OFC | 11 | 2.08 | 15, 55, −17 |
| L ACG | 25 | 2.04 | −2, 11, −5 |
| L OFC | 11 | 2.01 | −16, 53, −17 |
| R claustrum | – | 1.88 | 30, 11, −7 |
| R putamen | – | 1.87 | 18, 15, −7 |
| R putamen | – | 1.82 | 20, 17, −7 |
| R IFG | 47 | 1.81 | 34, 23, −9 |
| R medial frontal gyrus | 11 | 1.72 | 4, 47, −11 |
| Controls > OPRM1.G | |||
| Prepriming | |||
| R middle frontal gyrus | 10 | 3.83 | 30, 51, −1 |
| R ACG | 32 | 3.03 | 16, 39, 1 |
| R globus pallidus | – | 2.47 | 18, −5, 5 |
| R caudate | – | 2.46 | 22, 19, 7 |
| L thalamus | – | 2.04 | −18, −9, 9 |
| R IFG | 47 | 1.94 | 16, 15, −23 |
| L middle frontal gyrus | 10 | 1.86 | −22, 51, −1 |
| R IFG | 11 | 1.80 | 26, 31, −25 |
| L VTA/midbrain | – | 1.65 | −15, −16, −13 |
| Postpriming | |||
| R IFG | 47 | 3.00 | 18, 13, −23 |
| L IFG | 47 | 2.22 | −16, 15, −23 |
| L IFG | 11 | 2.13 | −24, 29, −25 |
| R medial frontal gyrus | 9 | 1.89 | 6, 51, 23 |
| L medial frontal gyrus | 10 | 1.83 | −18, 49, −3 |
| L medial frontal gyrus | 10 | 1.70 | −12, 47, 1 |
| L medial frontal gyrus | 10 | 1.70 | −6, 55, 15 |
| R IFG | 47 | 1.67 | 26, 25, −23 |
L, left; R, right; ACG, anterior cingulate gyrus; OFC, orbitofrontal cortex; VS/NAc, ventral striatum/nucleus accumbens; IFG, inferior frontal gyrus; VTA, ventral tegmental area.
The OPRM1.G subjects had greater BOLD response in all of the ROIs (except the VTA) before and after priming compared with the controls. Maximum loci of activation are listed for each substrate as anatomical labels, corresponding Brodmann area (BA), peak z-score and Talairach (TLRC) co-ordinates of greater BOLD response in the risk allele group compared with the nonrisk allele group (p < 0.05).
Foci of activation (i.e., L OFC) that overlap with DRD4.L individuals.
Fig. 4.
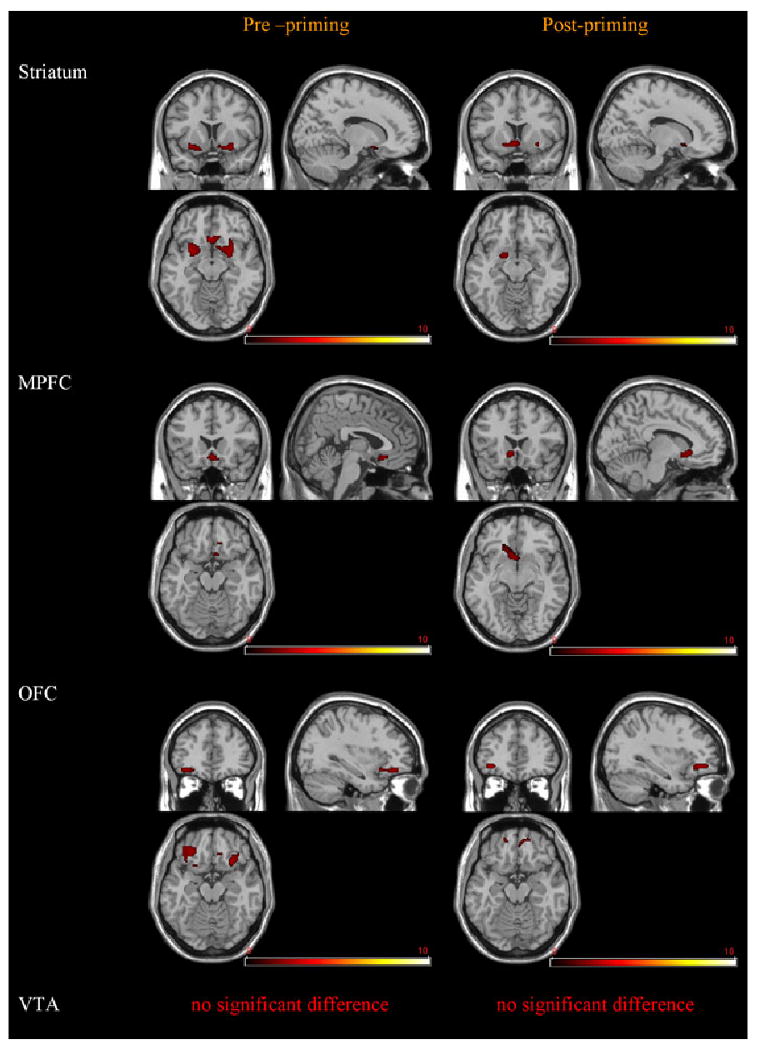
Greater activity in the OPRM.G subjects compared with controls. The OPRM1.G subjects had significantly greater BOLD response compared with the controls in response to alcohol taste cues in the ventral striatum, ventromedial PFC, and OFC before the priming dose of alcohol (p < 0.05). After the priming dose, striatal, ventromedial PFC, and OFC areas had greater response in the OPRM1.G group compared with the control group (p < 0.05). The right side of the image reflects left-hemispheric activation. The colorscale represents z-scores.
Correlations Between Subjective Measures and BOLD Response
Pearson correlations revealed that in the DRD4.L subjects, peak activity in the striatum and medial PFC before priming was marginally correlated with the total AUDIT score (r = 0.43, p = 0.06; r = 0.38, p = 0.10, respectively). The total AUDIT score was not correlated with peak activity in any of the ROIs after the priming dose in the DRD4.L group. There was a significant positive correlation between postscan AUQ and peak activity in the OFC after the priming dose (r = 0.52, p = 0.02) and a trend-level significance for correlations between in-scanner urge ratings for alcohol and peak activity in the medial PFC prior to the priming dose of alcohol (r = 0.46, p = 0.08).
OPRM1.G individuals also showed significantly correlated peak activity in the striatum and AUDIT total before priming (r = 0.71, p = 0.05). After the priming dose, a significant negative correlation with the AUDIT was found in the medial PFC (r = −0.82, p = 0.01) as well as a marginally significant negative correlation with the OFC (r = −0.65, p = 0.08). No significant correlations were found between the AUQ and in-scanner urge ratings, and peak activity in the ROIs in the OPRM1.G individuals.
In the control group, only 1 significant correlation was found and it was between peak activity in the medial PFC and AUDIT total before priming (r = 0.36, p = 0.05). There were no significant correlations between the AUQ and in-scanner urge ratings, and peak activity in ROIs in the controls.
Discussion
We previously reported that substrates within the mesocorticolimbic pathway have increased neural response to alcohol taste cues compared to an appetitive control taste. In the present study, we add that this response is modulated by 2 functional polymorphisms that regulate receptors within the mesocorticolimbic pathway. Specifically, our present findings suggest that (1) the DRD4 VNTR and the OPRM1 A118G SNP were associated with increased cue-elicited activation of mesocorticolimbic structures, and, that (2) this activation is extended by a priming dose of alcohol in some structures but not others in the OPRM1.G individuals, and (3) activity in the striatum correlated with drinking behavior (i.e., AUDIT score) among individuals with the G allele of the OPRM1 A118G SNP and the DRD4.L VNTR, whereas controls showed significant correlations with activity in MPFC. Differences in alcohol use and alcohol-related problems, indicated by AUDIT scores, were observed in a sample that was, on average, well above the cut-off score for harmful alcohol use [i.e., 8 or more, cf Allen et al. (1997) and Saunders et al. (1993)]. The positive correlations in the striatum within the DRD4.L and OPRM1 G group suggest that alcohol-related problems may be the consequence of a heightened biological response to alcohol cues. However, a direct assessment of this relationship must be performed to ascertain the exact nature of this association. Overall, the present findings suggest that the DRD4 VNTR and OPRM1 SNP may influence the development of incentive salience.
The findings of an association between the A118G SNP of the OPRM1 gene and ROIs examined in this study suggest that carriers of the G-allele had significantly greater activation in all of the ROIs compared to homozygotes for the A-allele. These findings extend the previous work suggesting that carriers of the G allele are more sensitive to the rewarding effects of alcohol (Ray and Hutchison, 2004, 2007). This provides support for a pharmacogenetic mechanism whereby carriers of the G allele appear to experience the acute effects of alcohol differently. More specifically, if opioid receptors in the carriers of the G allele have greater affinity for opioids released during ingestion of alcohol, and this binding acts to disinhibit DAergic cell bodies, greater activity in the DAergic system would be expected in G allele carriers compared with A allele carriers.
These findings should be interpreted in the context of the clinical literature suggesting that carriers of the G allele are more responsive to naltrexone, an opiate blocker thought to act selectively for μ-opioid receptors, as a pharmacotherapy for alcoholism (Oslin et al., 2003; Ray and Hutchison, 2007). Specifically, the present results suggest that differential activation of this circuitry in the brain, as a function of OPRM1 genotype, may be underlying some of the differential behavioral responses to alcohol previously reported (Ray and Hutchison, 2004). Future pharmacogenetic studies using fMRI technology may be useful in elucidating the specific brain mechanisms by which genotype may predict differential responses to a pharmacotherapy such as naltrexone. In summary, while the literature on the A118G SNP of the OPRM1 gene suggests that this polymorphism may be functional at the cellular (Bond et al., 1998; Zhang et al., 2005), behavioral (Ray and Hutchison, 2004, 2007), and clinical levels (Oslin et al., 2003), the present study suggests that it may also be functional at the neuronal level.
The DRD4 VNTR genotype has also been shown to predict differential responses to alcohol cues and alcohol craving (Hutchison et al., 2002a,b; MacKillop et al., 2007), yet little is known about the neural correlates associated with this genotype. Unfortunately, the current sample size does not allow us to test for additive genetic effects, given that only 5 participants had the risk alleles for both genotypes (i.e., G allele of the OPRM1 gene and 7 repeats of the DRD4 gene). The fact that the DRD4.L repeat group showed greater activity in the prepriming stage of the experiment may have been due to the novelty of the cues, whereas the diminished effect for the DRD4.L vs. DRD4.S contrast in the postpriming dose scans may have been due to habituation (both at the behavioral and neural level). Thus, the effects of priming are potentially confounded by differential time and fatigue for the postpriming runs because as mentioned in the Materials and Methods sections, priming was not presented in a counterbalanced order across subjects (i.e., postpriming scans were always runs 3 and 4). Overall, while the functional significance of the DRD4 VNTR remains largely unknown, our findings provide further evidence for variation of the polymorphism and the possibility that the DRD4 7-repeat variant is associated with less functional dopamine transmission.
In conclusion, the present study applied an empirically driven approach to the examination of the neural correlates and functional significance of 2 a priori candidate genes for alcohol use disorders. Results revealed significant differences in activation of several brain regions of interest, with most ROIs focusing on structures involved in the reward pathway of alcohol, thereby providing support for the role of these genetic variants in neural responses to alcohol cues. A unique feature of this study is the use of fMRI to examine manipulation of the endophenotypes of interest through the administration of alcohol cues and a priming dose of alcohol. The results observed at the fMRI level represent differential neural responses to these manipulations and increase our confidence in the genetic findings. These results await replication and should be interpreted in light of the study's limitations. Ultimately, additional studies combining pharmacogenetic and imaging techniques are needed to effectively translate these biological and behavioral findings into more effective and targeted treatments for alcohol use disorders.
Acknowledgments
This study was funded by an NIAAA RO1 grant to Dr. Hutchison.
Contributor Information
Francesca M. Filbey, Department of Psychology, University of Colorado at Boulder, Boulder, Colorado
Lara Ray, Department of Psychology, University of Colorado at Boulder, Boulder, Colorado, Brown University Center for Alcohol and Addiction Studies, Providence, Rhode Island.
Andrew Smolen, Institute for Behavioral Genetics, University of Colorado at Boulder, Boulder, Colorado.
Eric D. Claus, Department of Psychology, University of Colorado at Boulder, Boulder, Colorado
Amy Audette, Department of Psychology, University of Colorado at Boulder, Boulder, Colorado.
Kent E. Hutchison, Department of Psychology, University of Colorado at Boulder, Boulder, Colorado
References
- Allen JP, Litten RZ, Fertig JB, Babor T. A review of research on the Alcohol Use Disorders Identification Test (AUDIT) Alcohol Clin Exp Res. 1997;21:613–619. [PubMed] [Google Scholar]
- Anchordoquy HC, McGeary C, Liu L, Krauter KS, Smolen A. Genotyping of three candidate genes following whole genome preamplification of DNA collected from buccal cells. Behav Genet. 2003;33:73–78. doi: 10.1023/a:1021007701808. [DOI] [PubMed] [Google Scholar]
- Arias A, Feinn R, Kranzler HR. Association of an Asn40Asp (A118G) polymorphism in the mu-opioid receptor gene with substance dependence: a meta-analysis. Drug Alcohol Depend. 2006;83:262–268. doi: 10.1016/j.drugalcdep.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Berridge KC. The debate over dopamine's role in reward: the case for incentive salience. Psychopharmacology (Berl) 2007;191:391–431. doi: 10.1007/s00213-006-0578-x. [DOI] [PubMed] [Google Scholar]
- Bohn MJ, Krahn DD, Staehler BA. Development and initial validation of a measure of drinking urges in abstinent alcoholics. Alcohol Clin Exp Res. 1995;19:600–606. doi: 10.1111/j.1530-0277.1995.tb01554.x. [DOI] [PubMed] [Google Scholar]
- Bond C, LaForge KS, Tian M, Melia D, Zhang S, Borg L, Gong J, Schluger J, Strong JA, Leal SM, Tischfield JA, Kreek MJ, Yu L. Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci U S A. 1998;95:9608–9613. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobes DJ, Anton RF, Thomas SE, Voronin K. Effects of naltrexone and nalmefene on subjective response to alcohol among non-treatment-seeking alcoholics and social drinkers. Alcohol Clin Exp Res. 2004;28:1362–1370. doi: 10.1097/01.alc.0000139704.88862.01. [DOI] [PubMed] [Google Scholar]
- Du YP, Dalwani M, Wylie K, Claus E, Tregellas JR. Reducing susceptibility artifacts in fMRI using volume-selective z-shim compensation. Magn Reson Med. 2007;57:396–404. doi: 10.1002/mrm.21150. [DOI] [PubMed] [Google Scholar]
- Erickson CK. Review of neurotransmitters and their role in alcoholism treatment. Alcohol Alcohol. 1996;31(Suppl. 1):5–11. [PubMed] [Google Scholar]
- Filbey FM, Claus E, Audette AR, Niculescu M, Banich MT, Tanabe J, Du YP, Hutchison KE. Exposure to the taste of alcohol elicits activation of the mesocorticolimbic neurocircuitry. Neuropsychopharmacology. 2008;33:1391–1401. doi: 10.1038/sj.npp.1301513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Gueorguieva R, Kranzler HR, Zhang H, Cramer J, Rosenheck R, Krystal JH. Opioid receptor gene (OPRM1, OPRK1, and OPRD1) variants and response to naltrexone treatment for alcohol dependence: results from the VA Cooperative Study. Alcohol Clin Exp Res. 2007;31:555–563. doi: 10.1111/j.1530-0277.2007.00339.x. [DOI] [PubMed] [Google Scholar]
- George MS, Anton RF, Bloomer C, Teneback C, Drobes DJ, Lorberbaum JP, Nahas Z, Vincent DJ. Activation of prefrontal cortex and anterior thalamus in alcoholic subjects on exposure to alcohol-specific cues. Arch Gen Psychiatry. 2001;58:345–352. doi: 10.1001/archpsyc.58.4.345. [DOI] [PubMed] [Google Scholar]
- Gianoulakis C. Influence of the endogenous opioid system on high alcohol consumption and genetic predisposition to alcoholism. J Psychiatry Neurosci. 2001;26:304–318. [PMC free article] [PubMed] [Google Scholar]
- Herz A. Endogenous opioid systems and alcohol addiction. Psychopharmacology (Berl) 1997;129:99–111. doi: 10.1007/s002130050169. [DOI] [PubMed] [Google Scholar]
- Hutchison K, Haughey H, Niculescu M, Schacht J, Kaiser A, Stitzel J, Horton WJ, Filbey F. The incentive salience of alcohol: Translating the effects of the genetic variant in CNR1. Arch Gen Psychiatry. 2008 doi: 10.1001/archpsyc.65.7.841. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison KE, LaChance H, Niaura R, Bryan A, Smolen A. The DRD4 VNTR polymorphism influences reactivity to smoking cues. J Abnorm Psychol. 2002a;111:134–143. doi: 10.1037//0021-843x.111.1.134. [DOI] [PubMed] [Google Scholar]
- Hutchison KE, McGeary J, Smolen A, Bryan A, Swift RM. The DRD4 VNTR polymorphism moderates craving after alcohol consumption. Health Psychol. 2002b;21:139–146. [PubMed] [Google Scholar]
- Hutchison KE, Ray L, Sandman E, Rutter MC, Peters A, Davidson D, Swift R. The effect of olanzapine on craving and alcohol consumption. Neuropsychopharmacology. 2006;31:1310–1317. doi: 10.1038/sj.npp.1300917. [DOI] [PubMed] [Google Scholar]
- Hutchison KE, Wooden A, Swift RM, Smolen A, McGeary J, Adler L, Paris L. Olanzapine reduces craving for alcohol: a DRD4 VNTR polymorphism by pharmacotherapy interaction. Neuropsychopharmacology. 2003;28:1882–1888. doi: 10.1038/sj.npp.1300264. [DOI] [PubMed] [Google Scholar]
- Kalivas PW, Volkow ND. The neural basis of addiction: a pathology of motivation and choice. Am J Psychiatry. 2005;162:1403–1413. doi: 10.1176/appi.ajp.162.8.1403. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Heath AC, Kessler RC, Eaves LJ. A twin-family study of alcoholism in women. Am J Psychiatry. 1994;151:707–715. doi: 10.1176/ajp.151.5.707. [DOI] [PubMed] [Google Scholar]
- Kreek MJ. Cocaine, dopamine and the endogenous opioid system. J Addict Dis. 1996;15:73–96. doi: 10.1300/J069v15n04_05. [DOI] [PubMed] [Google Scholar]
- Lichter JB, Barr CL, Kenedy JL, Van Tol HHM, Kidd KK, Livak KJ. A hypervariable segment in the human dopamine receptor D4 (DRD4) Human Mol Genet. 1993;2:767–773. doi: 10.1093/hmg/2.6.767. [DOI] [PubMed] [Google Scholar]
- Livak K. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- Mackillop J, Menges DP, McGeary JE, Lisman SA. Effects of craving and DRD4 VNTR genotype on the relative value of alcohol: an initial human laboratory study. Behav Brain Funct. 2007;3:11. doi: 10.1186/1744-9081-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaul ME, Wand GS, Stauffer R, Lee SM, Rohde CA. Naltrexone dampens ethanol-induced cardiovascular and hypothalamic-pituitary-adrenal axis activation. Neuropsychopharmacology. 2001;25:537–547. doi: 10.1016/S0893-133X(01)00241-X. [DOI] [PubMed] [Google Scholar]
- McClernon FJ, Hutchison KE, Rose JE, Kozink RV. DRD4 VNTR polymorphism is associated with transient fMRI-BOLD responses to smoking cues. Psychopharmacology (Berl) 2007;194:433–441. doi: 10.1007/s00213-007-0860-6. [DOI] [PubMed] [Google Scholar]
- McGeary JE, Monti PM, Rohsenow DJ, Tidey J, Swift R, Miranda R., Jr Genetic moderators of naltrexone's effects on alcohol cue reactivity. Alcohol Clin Exp Res. 2006;30:1288–1296. doi: 10.1111/j.1530-0277.2006.00156.x. [DOI] [PubMed] [Google Scholar]
- Myrick H, Anton RF, Li X, Henderson S, Drobes D, Voronin K, George MS. Differential brain activity in alcoholics and social drinkers to alcohol cues: relationship to craving. Neuropsychopharmacology. 2004;29:393–402. doi: 10.1038/sj.npp.1300295. [DOI] [PubMed] [Google Scholar]
- Nielsen F, Hansen L. Automatic Anatomical Labeling of Talairach Coodinates and Generation of Volumes of Interest via the BrainMap Database. Paper presented at the 8th International Conference on Functional Mapping of the Brain; Sendai, Japan. 2002. [Google Scholar]
- Oslin DW, Berrettini W, Kranzler HR, Pettinati H, Gelernter J, Volpicelli JR, O'Brien CP. A functional polymorphism of the mu-opioid receptor gene is associated with naltrexone response in alcohol-dependent patients. Neuropsychopharmacology. 2003;28:1546–1552. doi: 10.1038/sj.npp.1300219. [DOI] [PubMed] [Google Scholar]
- Ray LA, Hutchison KE. A polymorphism of the mu-opioid receptor gene (OPRM1) and sensitivity to the effects of alcohol in humans. Alcohol Clin Exp Res. 2004;28:1789–1795. doi: 10.1097/01.alc.0000148114.34000.b9. [DOI] [PubMed] [Google Scholar]
- Ray LA, Hutchison KE. Effects of naltrexone on alcohol sensitivity and genetic moderators of medication response: a double-blind placebo-controlled study. Arch Gen Psychiatry. 2007;64:1069–1077. doi: 10.1001/archpsyc.64.9.1069. [DOI] [PubMed] [Google Scholar]
- Ray R, Jepson C, Patterson F, Strasser A, Rukstalis M, Perkins K, Lynch KG, O'Malley S, Berrettini WH, Lerman C. Association of OPRM1 A118G variant with the relative reinforcing value of nicotine. Psychopharmacology (Berl) 2006;188:355–363. doi: 10.1007/s00213-006-0504-2. [DOI] [PubMed] [Google Scholar]
- Rhee SH, Hewitt JK, Young SE, Corley RP, Crowley TJ, Stallings MC. Genetic and environmental influences on substance initiation, use, and problem use in adolescents. Arch Gen Psychiatry. 2003;60:1256–1264. doi: 10.1001/archpsyc.60.12.1256. [DOI] [PubMed] [Google Scholar]
- Sander T, Harms H, Dufeu P, Kuhn S, Rommelspacher H, Schmidt LG. Dopamine D4 receptor exon III alleles and variation of novelty seeking in alcoholics. Am J Med Genet. 1997;74:483–487. doi: 10.1002/(sici)1096-8628(19970919)74:5<483::aid-ajmg5>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Saunders JB, Aasland OG, Babor TF, de la Fuente JR, Grant M. Development of the Alcohol Use Disorders Identification Test (AUDIT): WHO Collaborative Project on early detection of persons with harmful alcohol consumption —II. Addiction. 1993;88:791–804. doi: 10.1111/j.1360-0443.1993.tb02093.x. [DOI] [PubMed] [Google Scholar]
- Schneider F, Habel U, Wagner M, Franke P, Salloum JB, Shah NJ, Toni I, Sulzbach C, Honig K, Maier W, Gaebel W, Zilles K. Subcortical correlates of craving in recently abstinent alcoholic patients. Am J Psychiatry. 2001;158:1075–1083. doi: 10.1176/appi.ajp.158.7.1075. [DOI] [PubMed] [Google Scholar]
- Shao C, Li Y, Jiang K, Zhang D, Xu Y, Lin L, Wang Q, Zhao M, Jin L. Dopamine D4 receptor polymorphism modulates cue-elicited heroin craving in Chinese. Psychopharmacology (Berl) 2006;186:185–190. doi: 10.1007/s00213-006-0375-6. [DOI] [PubMed] [Google Scholar]
- Swift RM, Whelihan W, Kuznetsov O, Buongiorno G, Hsuing H. Naltrexone-induced alterations in human ethanol intoxication. Am J Psychiatry. 1994;151:1463–1467. doi: 10.1176/ajp.151.10.1463. [DOI] [PubMed] [Google Scholar]
- Van Tol HH, Wu CM, Guan HC, Ohara K, Bunzow JR, Civelli O, Kennedy J, Seeman P, HB N, Jovanovic V. Multiple dopamine D4 receptor variants in the human population. Nature. 1992;358:149–152. doi: 10.1038/358149a0. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Fowler JS, Wang GJ, Swanson JM, Telang F. Dopamine in drug abuse and addiction: results of imaging studies and treatment implications. Arch Neurol. 2007;64:1575–1579. doi: 10.1001/archneur.64.11.1575. [DOI] [PubMed] [Google Scholar]
- Watson P. Total body water and blood alcohol levels: updating the fundamentals. In: Crow K, Batt R, editors. Human Metabolism of Alcohol. CRC Press; Florida: 1998. pp. 41–56. [Google Scholar]
- van den Wildenberg E, Janssen RG, Hutchison KE, van Breukelen GJ, Wiers RW. Polymorphisms of the dopamine D4 receptor gene (DRD4 VNTR) and cannabinoid CB1 receptor gene (CNR1) are not strongly related to cue-reactivity after alcohol exposure. Addict Biol. 2007;12:210–220. doi: 10.1111/j.1369-1600.2007.00064.x. [DOI] [PubMed] [Google Scholar]
- Wang E, Ding YC, Flodman P, Kidd JR, Kidd KK, Grady L, Ryder OA, Spence MA, Swanson JM, Moyzis RK. The genetic architecture of selection at the human dopamine receptor D4 (DRD4) gene locus. Am J Human Genet. 2004;74:931–944. doi: 10.1086/420854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrase J, Grusser SM, Klein S, Diener C, Hermann D, Flor H, Mann K, Braus DF, Heinz A. Development of alcohol-associated cues and cue-induced brain activation in alcoholics. Eur Psychiatry. 2002;17:287–291. doi: 10.1016/s0924-9338(02)00676-4. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang D, Johnson AD, Papp AC, Sadee W. Allelic expression imbalance of human mu opioid receptor (OPRM1) caused by variant A118G. J Biol Chem. 2005;280:32618–32624. doi: 10.1074/jbc.M504942200. [DOI] [PubMed] [Google Scholar]


