Summary
Reversible protein phosphorylation is a signaling mechanism involved in all cellular processes. To create a systems view of the signaling apparatus in budding yeast, we generated an E-MAP (epistatic miniarray profile) comprised of 100,000 pair-wise, quantitative genetic interactions, including virtually all protein kinases and phosphatases and key cellular regulators. Quantitative genetic interaction mapping reveals factors working in compensatory pathways (negative genetic interactions; e.g. synthetic lethality) or those operating in linear pathways (positive genetic interactions; e.g. suppression). Within kinases, phosphatases, and their substrates, we found an enrichment of positive genetic interactions. To develop a global view of the signaling apparatus, we isolated “triplet genetic motifs” and assembled these into a higher-order map. The resulting network view provides new insights into signaling pathway regulation, and revealed a link between the cell cycle kinase, Cak1, the Fus3 MAP kinase, and a pathway that regulates chromatin integrity during transcription by RNA polymerase II.
Introduction
Phosphate-based signaling is critical to almost all major cellular processes and is ubiquitously present across archea, prokaryota and eukaryota (Kannan et al., 2007). Systems-wide studies in the post-genome era have provided unprecedented information about the activities of signaling proteins (Johnson and Hunter, 2005) and several thousand sites of protein phosphorylation have been mapped using mass spectrometry (Ficarro et al., 2002; Green and Pflum, 2007; Lee et al., 2006; Matsuoka et al., 2007; Olsen et al., 2006). The knowledge of kinase-substrate relationships has been expanded by both in vitro protein chip analysis (Ptacek et al., 2005) and large-scale genetic screens using a kinase over-expression strategy (Sopko et al., 2006). Bioinformatic efforts have focused on network-level analyses of phosphorylation, providing database resources for phosphorylation sites and signaling pathways (Diella et al., 2008; Lee et al., 2006; Zanzoni et al., 2007). In addition, the integration of context-dependent information (including protein interactions and cell-specific kinase expression) has helped to improve the specificity of phospho-consensus site assignments (Linding et al., 2007).
Despite these achievements, signaling networks have remained difficult to study. While phosphoproteomic datasets illuminate the magnitude and diversity of protein phosphorylation, the functional relevance for the majority of these phosphorylation sites remains unknown (Johnson and Hunter, 2005). Focused studies can elucidate the function of one specific kinase or one particular pathway, but often overlook important connections to components that do not directly participate in the pathway. In particular, two hallmarks of kinase signaling are the linear cascades of kinases important for signal amplification and the abundant cross-talk between these pathways in order to coordinate multiple cellular inputs/outputs.
Genetic interactions report on the extent to which the function of one gene depends on the presence of a second gene and can illuminate the functional organization of protein networks. Negative genetic interactions (synthetic sick/lethal interactions) describe cases where two mutations in combination cause a stronger growth defect than expected from the two single mutations. In contrast, positive genetic interactions correspond to cases where the double mutant is either no sicker (epistatic) or healthier (suppressive) than the sickest single mutant (Figure 1a). Negative genetic interactions are often found for proteins that work in compensatory pathways, while positive interactions can identify pairs of proteins that are in complex and/or function in the same pathway (Collins et al., 2007b; Kelley and Ideker, 2005; Roguev et al., 2008; Schuldiner et al., 2005; St. Onge et al., 2006). Two approaches, synthetic genetic array (SGA) technology (Tong et al., 2001) and diploid based synthetic lethality analysis on microarrays (dSLAM) (Pan et al., 2004), have been developed to identify synthetic sick/lethal (SSL) relationships on a genome-wide scale in S. cerevisiae. SSL interactions however, only represent a subset of the full epistatic interaction spectrum. Our interest in mapping the pathway architectures involved in phosphate-based signaling led us to use an SGA-based methodology that measures the entire range of epistatic relationships, termed epistatic mini array profile (or E-MAP) (Figure 1a) (Collins et al., 2007b; Collins et al., 2006; Roguev et al., 2008; Schuldiner et al., 2005; Schuldiner et al., 2006; Wilmes et al., 2008). The E-MAP method does not rely on the detection of individual phosphoproteins, but it rather provides the functional connections between signaling molecules, whether these are direct or indirect. In addition, genetic interactions are not limited to the detection of proteins in physical contact and are thus particularly useful in identifying more transient interactions, and those controlled by post-translational modification. Here we use our E-MAP approach to provide an unbiased, genetic overview of the functional connections within the signaling machinery.
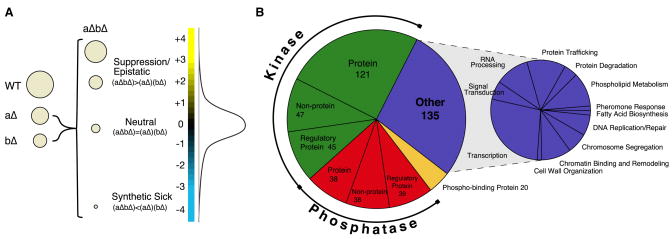
Figure 1. Epistasis analysis of the yeast signaling machinery.
a) The entire spectrum of genetic interactions. Genetic interactions range from negative (e.g. synthetic sick) to positive (e.g. suppression) where the growth rate of the double mutant is either less ((aΔbΔ) < (aΔ)(bΔ)) or greater ((aΔbΔ) > (aΔ)(bΔ)) than the product of the growth rates of the corresponding single mutants, respectively. As shown in the distribution curve of our data set, significantly negative (S ≤ −2.5) and positive (S ≥ 2.0) genetic interactions are rare. b) Composition of the signaling E-MAP. For a full list of genes analyzed in this study, see Supplementary Table 1.
Results and Discussion
Composition of the Signaling E-MAP
An E-MAP quantitatively records genetic interactions between pairs of mutations for a defined subset of genes. The data is generated by systematically constructing double mutant strains, measuring their growth rates, and converting this information to individual genetic interaction scores, both positive and negative (Collins et al., 2006; Schuldiner et al., 2006). To explore the signaling apparatus of S. cerevisiae, our genetic approach targeted almost all known protein kinases (121) and phosphatases (38) and their regulatory proteins (45 and 39, respectively), as well as non-protein kinases (47) and phosphatases (38) (Figure 1b, Supplementary Table 1). To include those components of the signaling machinery that are essential for viability, we employed the DAmP (Decreased abundance by mRNA perturbation) strategy to create 48 hypomorphic alleles. This method relies on the insertion of the antibiotic-resistance marker to disrupt the natural 3′UTR, which leads to destabilization of the mRNA transcripts, and, subsequently lowered protein levels (Schuldiner et al., 2005). In all, the signaling E-MAP contains 483 genes, including 135 genes that serve as reporters of a variety of major biological processes (Figure 1b, Supplementary Table 1). We comprehensively evaluated the pairwise genetic interactions of these 483 alleles, resulting in approximately 100,000 distinct, pair-wise genetic interaction measurements (Supplementary Figure 1, Supplementary Table 2). All data is available at http://interactome-cmp.ucsf.edu in an interactive and searchable format.
Superposition of Literature-Derived Network with Signaling E-MAP
To evaluate the genetic interaction data, we first examined how the data recapitulated known phosphorylation/dephosphorylation events. To this end, we manually curated from the literature the majority of well-characterized kinase- and phosphatase-substrate relationships (654 and 141, respectively) (Figure 2a, Supplementary Table 3). Of these 795 pair-wise connections, our E-MAP contained data for 252 (32%) of them (Supplementary Table 4). We analyzed the genetic interaction patterns for different subsets of the manually curated set of known phosphorylation events: kinases and their substrates, phosphatases and their substrates, two kinases that share the same substrate, two phosphatases that share the same substrate, and kinase-phosphatase pairs that target a common substrate (Figure 2b; Supplementary Tables 4, 5). As a metric to describe the trends within these different subsets, we employed a ratio of highly positive (S ≥ 2.0) to negative (S ≤ −2.5) genetic interactions (P to N ratio). Importantly, the relative ratios obtained for the different subsets were independent of the thresholds used to define the positive and negative genetic interactions (see Methods). The E-MAP data revealed a significant enrichment of positive genetic interactions for known kinase- and phosphatase-substrate pairs. While the E-MAP as a whole displayed a P to N ratio of 0.53, this ratio was almost inverted for characterized kinase- and phosphatase-substrate relationships (P to N ratio: 1.78; p= 1.8 × 10−3) (Figure 2b, Supplementary Tables 3, 4, 5). For example, we detected positive genetic interactions between the CAK1 kinase and two of its substrates, CTK1 and SMK1 (Figure 2a(1)) (Ostapenko and Solomon, 2005; Schaber et al., 2002). These trends might be rationalized as follows: if the phosphorylated form of a substrate is important for cell growth, then either mutation of the substrate, or loss of its phosphorylation through mutation of the cognate kinase, will result in impaired cell growth. Once the kinase has been deleted (and the level of substrate phosphorylation has decreased), additional deletion of the substrate will result in a less severe growth defect than expected from the two single mutations, thus giving rise to a positive genetic interaction. Similar logic can be applied to phosphatase-substrate relationships that display positive genetic interactions (e.g. SIT4-GCN2 (Cherkasova and Hinnebusch, 2003) or SIT4-SAP155 (Luke et al., 1996), Figure 2a(2)) if the dephosphorylated form of a substrate is important for cell growth. Conversely, pairs of kinases (or phosphatases) that act on the same substrate can display negative interactions amongst themselves, if they are partially or completely redundant. Indeed, we observed an enrichment of negative genetic interactions in these cases (P to N ratio: 0.21; p= 2.5 × 10−3) (Figure 2b, Supplementary Table 5), including a negative interaction between the CLA4 and STE20 kinases, both of which phosphorylate Myo3 (Wu et al., 1997), and between PPH21 and PPH22, duplicated PP2A phosphatase family members known to act redundantly on a number of different substrates (Figure 2a (3, 4)) (Oficjalska-Pham et al., 2006).
Figure 2. Comparison of the literature-derived signaling network to the genetic interaction data.
a) A network diagram of characterized phosphorylation and dephosphorylation events between kinases and their substrates (green arrows) and phosphatases and their substrates (red arrows), manually curated from the literature (see Supplementary Table 2). Below this are specific examples of kinase-substrate (1) and phosphatase-substrate relationships (2), and cases where two kinases (3), two phosphatases (4) or one kinase and one phosphatase (5) target one or more substrates. Blue and yellow edges correspond to negative and positive genetic interactions, respectively. The thickness of the edge correlates with the strength of the genetic interaction. b) Ratios of highly positive (S ≥ 2.0) to negative (S ≤ −2.5) genetic interactions (P to N ratio). Above dashed line: The P to N ratios of previously published datasets (early secretory pathway (ESP)(Schuldiner et al., 2005) and chromosome function(Collins et al., 2007b)), the signaling E-MAP, only protein kinases and protein phosphatases (K vs. K or P vs. P), and protein kinases versus protein phosphatases (K vs. P) are presented. Below dashed line: The P to N ratios for known kinase-substrate and phosphatase-substrate pairs (K → S, P → S) (out of these 252 relationships that we genetically tested, we observed significant genetic interactions (positive and negative) for 25 of them (10%)), as well as kinase-kinase, phosphatase-phosphatase (K,K → S; P,P → S) and kinase-phosphatase pairs (K,P → S) that share one or more substrates are shown. To obtain these ratios and subsequent p-values, we also included regulatory subunits known to direct kinase/phosphatase activity towards specific substrates (see Methods and Supplementary Table 3).
The most striking P to N ratio was observed for kinase-phosphatase pairs that target a common substrate. These pairs of counterbalancing enzymes proved to be much more likely to display positive genetic interactions (P to N ratio: 5.5; p=2.8 × 10−3) (Figure 2b, Supplementary Table 5). For example, a positive genetic interaction was observed between deletions of the SNF1 kinase and the phosphatase SIT4, both of which regulate the phosphorylation status of the Gln3 transcription factor (Figure 2a (5)) (Bertram et al., 2002; Tate et al., 2006; Wang et al., 2003). In these cases, if optimal cell growth requires a substrate to maintain a particular level of phosphorylation, then mutation of its phosphatase (or kinase) will perturb the steady state phosphorylation levels, resulting in a growth phenotype. By additionally deleting the counterbalancing kinase (or phosphatase), wild-type phosphorylation levels may be restored, and the growth defect may be suppressed. We therefore contend that this quantitative genetic platform can be used to identify kinase-substrate pairs, phosphatase-substrate pairs, and kinase-pairs, phosphatase-pairs and kinase-phosphatase pairs that share a common substrate (see Supp. Tables 2 and 6 for complete list of genetic interactions).
While we found a significant enrichment of positive interactions within the set of kinase-substrate, phosphatase-substrate and kinase-phosphatase pairs that share a common substrate, many of the literature-curated relationships were not captured by our genetic analysis (Supplementary Table 5). There are several reasons why we would not expect a perfect correlation between these two datasets. First, many signaling pathways are only fully activated in response to a particular stimulus (i.e. high salt/Hog1). Since our genetic interaction screen was carried out under standard growth conditions, most pathways are only operating at basal levels. As a result, their perturbation may not yield a significant growth phenotype. Generating E-MAP data under a number of different stresses would help detect many of these stimulus-specific genetic interactions. Second, there are many duplicated copies of kinases or phosphatases that exist in the yeast genome (e.g. MKK1 and MKK2) and this redundancy can mask potential genetic interactions. In these cases, triple mutants could provide richer interaction profiles and help identify additional connections. Despite these limitations, we detected a significant number of genetic interactions that correlated with previously described network connections, illustrating the predictive power of this dataset.
To investigate the characteristics of genetic interactions among components of the signaling apparatus, we analyzed the P to N ratio among and between the protein kinases and phosphatases. Compared to previous data sets, including an E-MAP of chromosome function (Collins et al., 2007b) which had a P to N ratio of 0.49, we found that there was a large enrichment of positive genetic interactions within the set of protein kinases and protein phosphatases (P to N ratio: 0.78; p=1.0 × 10−3) and between kinases and phosphatases (P to N ratio: 0.87; p=5.1 × 10−3) (Figure 2b, Supplementary Figure 2, Supplementary Table 5). The trend towards positive genetic interactions could stem from the fact that kinases and phosphatases often work together in linear pathways.
Signaling E-MAP Reveals Factors Involved in Histone Htz1 Deposition and Acetylation
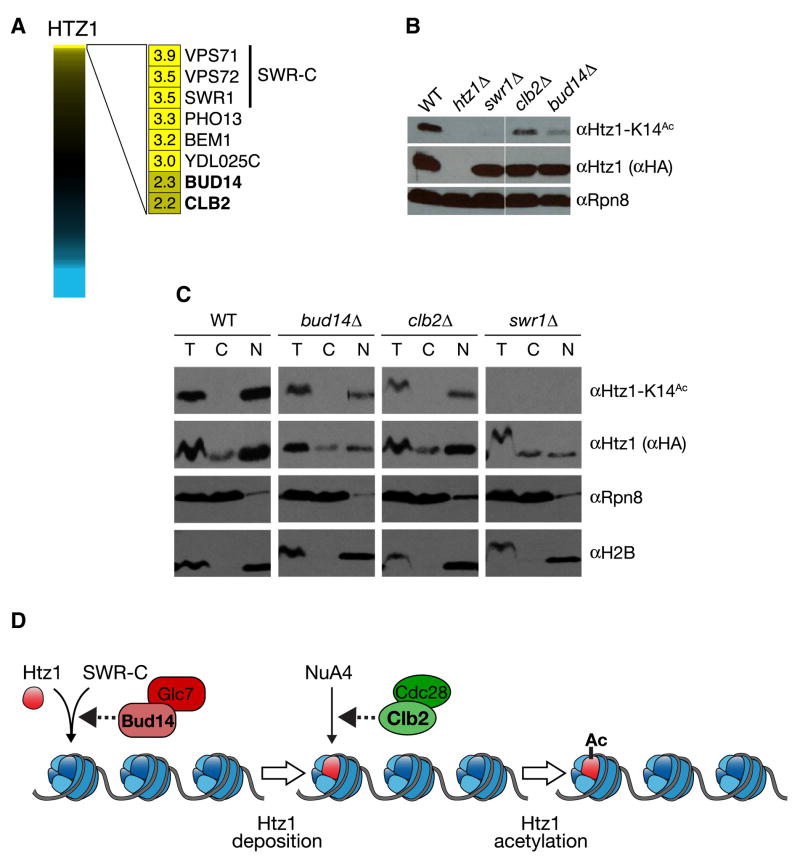
To test the predictive power of the E-MAP data, we examined the positive genetic interactions we identified with the histone H2A variant, Htz1. Htz1 is incorporated into chromatin by the SWR-C chromatin-remodeling complex (Korber and Horz, 2004) and is subsequently acetylated on its amino-terminus by the histone acetyltransferase, NuA4 (Babiarz et al., 2006; Keogh et al., 2006b; Millar et al., 2006). Consistent with this, HTZ1 displayed positive genetic interactions with SWR1 (+3.9), the catalytic subunit of SWR-C, as well as with VPS71 (+3.5) and VPS72 (+3.5), two other components of the complex (Figure 3a). Significant positive interactions were also observed with other factors not previously known to function with Htz1. These included Bud14, a regulatory subunit for the phosphatase Glc7 (Cullen and Sprague, 2002) and Clb2, a B-type cyclin that regulates Cdc28 (CDK) activity (Cullen and Sprague, 2002; Rua et al., 2001) (Figure 3a). Deletion of either BUD14 or CLB2 resulted in a marked decrease in Htz1 lysine 14 acetylation (Htz1-K14Ac; Figure 3b), indicating that these proteins impinge on the abundance of a chromatin-associated form of this histone variant (Keogh et al., 2006b). Fractionation experiments determined that deletion of BUD14 resulted in almost a 3-fold decrease in nuclear Htz1, which correlated with the decrease (~4 fold) in the K14-acetylated form of the histone (Figure 3c). The Bud14-associated phosphates Glc7 has been shown to impinge on chromatin structure by dephosphorylating serine 10 of histone H3, a mark linked to chromosome transmission (Hsu et al., 2000). Therefore, Glc7/Bud14 may regulate Htz1 incorporation by SWR-C into chromatin, either directly or indirectly, potentially through histone H3 (Figure 3d).
Figure 3. Identification of factors involved in histone Htz1 depostion and acetylation.
a) Full spectrum of HTZ1 genetic interactions. Genes with the strongest positive genetic interactions are highlighted. b) clb2Δ or bud14Δ profoundly reduce the total pool of Htz1-K14Ac. Whole cell extracts were isolated from isogenic strains containing Htz1 with a C-terminal HA3-tag, resolved by SDS-PAGE and immunoblotted as indicated. Rpn8 was used as a loading control. c) bud14Δ reduces both chromatin-associated Htz1 and Htz1-K14Ac, while clb2Δ specifically reduces the latter pool. Isogenic strains containing Htz1.HA3 were separated into T(otal), C(ytoplasmic) or N(uclear) fractions and immunoblotted with the indicated antibodies. Enrichment of the proteasome component Rpn8 and the chromatin component H2B in appropriate fractions (soluble cytoplasm and insoluble nucleus) demonstrate efficient separation. Using the H2B levels as a loading control, Htz1-K14Ac was decreased 3.7- and 3.0-fold in bud14Δ and clb2Δ respectively. By comparison, nuclear Htz1 was decreased 2.7-fold in bud14Δ but comparable to WT in clb2Δ. d) Schematic illustration of Htz1 deposition by SWR-C and acetylation by NuA4. The potential points of action of Bud14 and Clb2 in this pathway are indicated.
Conversely, the effect of clb2Δ was primarily seen at the level of Htz1 acetylation. Here the levels of nuclear Htz1 were unperturbed by deletion of CLB2, but Htz1-K14Ac was decreased 3 fold (Figure 3c). This implies that Clb2 exerts a regulatory function on NuA4, the enzyme complex that adds this acetylation mark (Figure 3d). An independent study determined that Yng2, a component of NuA4 required for its histone acetyl transferase activity (Krogan et al., 2004), is a target of Clb2-depenedent Cdc28 phosphorylation on two consensus CDK sites (T185 and S188) (L. Holt, J. Villen, S. Gygi and D. Morgan, unpublished data). This connection is also in agreement with previous reports that suggested Htz1 function is cell-cycle regulated (Dhillon et al., 2006). Although further work will be required to work out the mechanistic details of Htz1 regulation, these data confirm the predictive power of individual positive genetic interactions from the E-MAP.
Mapping Genetic Interactions onto Characterized Signaling Pathways
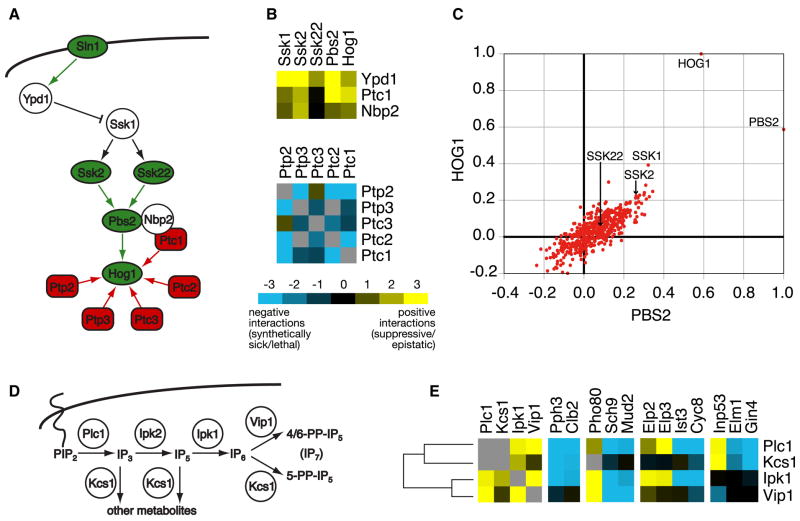
We next explored how the genetic interactions mapped within signaling cascades, involving multiple kinases and/or phosphatases. The HOG (High Osmolarity Glycerol) pathway is a well-characterized MAP kinase cascade that controls the response to osmotic shock (Figure 4a) (Hohmann, 2002). The Sln1 branch of the pathway is activated by the Sln1 osmosensor, which leads to activation of two MAP kinase kinase kinases (MAPKKKs), Ssk2 and Ssk22. These two kinases then phosphorylate a dedicated MAP kinase kinase (MAPKK), Pbs2, which in turn dually phosphorylates and activates the MAP kinase (MAPK) Hog1. Under iso-osmotic conditions, the pathway is down-regulated by the Ypd1 phosphotransferase, and Ptc1, a phosphatase that, together with its scaffolding protein Nbp2, controls Hog1 dephosphorylation (Figure 4a). Mutations of these negative regulators (YPD1, PTC1 and NBP2) primarily displayed positive genetic interactions with the activating proteins in the pathway (Figure 4b), presumably suppressive relationships that counterbalance the levels of phosphorylation in the pathway. In contrast, the genes coding for protein phosphatases that control Hog1 basal and maximal activity (PTC1, PTC2, PTC3, PTP2, PTP3) show largely negative genetic interactions among themselves (Figure 4b), suggesting a certain degree of redundancy.
Figure 4. Mapping genetic interaction data onto known signaling pathways.
a) General schematic of the Sln1 branch of the HOG pathway. Protein kinases and phosphatases are denoted as green and red, respectively, while green and red arrows correspond to phosphorylation and dephosphorylation actions, respectively. b) The negative pathway regulators YPD1, PTC1, and NBP2, show strong positive genetic interactions with the pathway activators SSK1, SSK2, SSK22, PBS2, and HOG1. In contrast, genes coding for the protein phosphatases (PTC1, PTC2, PTC3, PTP2, and PTP3) acting on HOG1 show primarily negative genetic interactions among themselves. c) Scatter plot of the correlation coefficients of pbs2Δ and hog1Δ with all genetic profiles in the signaling E-MAP. d) Schematic illustration of the inositol polyphosphate pathway. e) A subset of genetic interactions for PLC1, KCS1, IPK1, and VIP1. In b) and e), negative and positive genetic interactions are indicated by blue or yellow squares, respectively. Gray squares represent genetic interactions not tested.
In addition to the direct genetic interaction score between gene pairs, each gene possesses a genetic interaction profile which describes its interactions with all other genes in the E-MAP (see http://interactome-cmp.ucsf.edu (2008)). The genetic interaction profile provides a high resolution phenotype, and functionally related genes often have similar interaction profiles. For example, hog1Δ and pbs2Δ showed the most highly correlated profiles among the HOG pathway genes, indicating they are the most functionally related pair of proteins in this pathway (Figure 4c). Also correlated, but to a lesser extent, are ssk1Δ and ssk2Δ, which we believe is due to the fact that Pbs2 can also be activated by an alternative MAPKKK, Ste11 (O’Rourke and Herskowitz, 2004). Interestingly, ssk2Δ is more highly correlated with hog1Δ and pbs2Δ when compared to ssk22Δ (Figure 3c), which suggests that Ssk2 is the predominant MAPKKK for pathway activation under the growth conditions of our assay. Consistent with that the interactions of ssk2Δ with genes encoding the negative pathway regulators YPD1, PTC1, and NBP2 are much more positive than those seen with ssk22Δ (Figure 4b). Overall, the information encapsulated within both the individual pair-wise scores and the correlation coefficients between the genetic interaction profiles can be used to resolve pathway architectures more powerfully than any single observation.
The signaling E-MAP also contains the majority of small molecule kinases and phosphatases (Figure 1b, Supplementary Table 1), including several enzymes involved in inositol polyphosphate metabolism. Inositol phosphate signaling is initiated by the phospholipase Plc1, resulting in the formation of IP3, which is converted to IP5 by Ipk2, then to IP6 by Ipk1 (Figure 4d) (Alcazar-Roman and Wente, 2008). IP6 can subsequently become diphosphorylated by Kcs1 or Vip1 to respectively yield the IP7 metabolites, 5-PP-IP5 or 4/6-PP-IP5 (Figure 4d) (Mulugu et al., 2007; Saiardi et al., 2004). Genetic analysis of these enzymes (except for ipk2Δ, which had mating and sporulation defects) revealed that all members of the pathway displayed positive genetic interactions with each other, except for VIP1 and KCS1 (Figure 4e). This is consistent with a linear pathway for the synthesis of IP6, which then branches to yield the different IP7 metabolites (Figure 4d) (Alcazar-Roman and Wente, 2008). Although the overall genetic interaction profiles for these four enzymes are similar, there are almost no examples where all four enzymes display the same interactions with a given mutant. For example, all components except VIP1 displayed strong negative interactions with the cyclin CLB2 and PPH3, a phosphatase linked to cell cycle progression (Keogh et al., 2006a). Since Plc1, Ipk1 and Kcs1 – but not Vip1 – are involved in the synthesis of the 5-PP-IP5 metabolite, these data may imply a functional connection of the 5-PP-IP5 metabolite with CLB2 and PPH3. Furthermore, only PLC1 and IPK1 showed positive genetic interactions with ELP2 and ELP3, two proteins linked to transcriptional elongation and tRNA function (Svejstrup, 2007), which suggests that PLC1 and IPK1, or the metabolites they synthesize, impinge on those processes (Figure 4e). Our results concur with recent evidence that specific inositide metabolites are involved in distinct cellular functions (Alcazar-Roman and Wente, 2008) and thus give rise to a wide range of genetic interaction patterns.
Overall, the inositol polyphosphate and MAP kinase cascades are two examples where the signaling E-MAP supports previously described network connections, and provides new functional insight into these well-characterized pathways.
Analysis of “Triplet Genetic Motifs” (TGMs)
With the goal to extract new pathway connections, we further analyzed the genetic interaction data by extracting triplet motifs. In these “triplet genetic motifs” (or TGMs), sets of three genes were mutually connected to each other by positive (S ≥ 2.0) or negative (S ≤ −2.5) genetic interactions. Triplet motifs are the simplest motifs, apart from binary pairings, and we propose that gene triplets displaying genetic interactions are very likely to be functionally connected. There are four distinct types of TGMs: all three genetic interactions are positive (Type I); two positive, one negative (Type II); one positive, two negative (Type III); and all three negative (Type IV) (Figure 5a). Using only the genes coding for protein kinases, we generated each type of TGM (Supplementary Table 7) and assembled a network diagram where the motifs were connected if they shared one or two nodes (Figure 5b). We posit that TGMs comprised of three positive genetic interactions (Type I) will be enriched for factors functioning in the same pathway whereas all negative TGMs (Type IV) would potentially correspond to genes functioning in three distinct, redundant pathways (Figure 5a). Type II (++−) and Type III (+−−) TGMs represent intermediate cases. For example, a Type II TGM was observed for PTK2, HAL5, and SAT4, three kinases that are all involved in modulating the major electrogenic transporters of yeast (Pma1, Trk1, Trk2) (Supplementary Figure 3) (Goossens et al., 2000; Mulet et al., 1999). Ptk2 phosphorylates and activates the H+-ATPase Pma1, thereby increasing the electrical membrane potential, while the Trk1,2 K+ transport system is positively regulated by Hal5 and Sat4 kinases, and is a major consumer of electrical potential. Together, these two systems determine the steady-state membrane potential and thereby regulate secondary active transport systems. Due to their partial redundancy, HAL5 and SAT4 showed a negative genetic interaction, however, both genes displayed positive genetic interactions with PTK2, likely a suppressive relationship between the generator and the consumers of the membrane potential. Interestingly, the PTK2-HAL5 edge displayed negative genetic interactions with PBS2 (resulting in a Type III TGM) (Supplementary Figure 3), which illustrates how cells with a compromised ability to control ion homeostasis are unable to tolerate an additional mutation in a separate/parallel pathway that is important for responding to salt stress.
Figure 5. Triplet genetic motifs (TGMs) from the signaling E-MAP.
a) A schematic of the four types of triplet genetic motifs: all three positive (Type I); two positive, one negative (Type II); one positive, two negative (Type III); and all three negative (Type IV). b) A network diagram of all TGMs involving three kinases using highly negative (S ≤ −2.5) and positive (S ≥ 2.0) genetic interactions. The motifs were connected if they shared one or two nodes; blue and yellow edges correspond to negative and positive genetic interactions, respectively. The thickness of the edges is correlated with the strength of the genetic interaction. TGMs highlighted in grey are discussed in the text. c) Comparison of the ratio of the four types of TGMs that contain one or more kinase or phosphatase. The ratios are obtained by normalizing the number of genetic triplet motifs within each set (i.e. Types II, III, and IV) to the number of TGMs that are all positive (Type I). For a complete list of all TGMs, see Supplementary Table 5.
The three kinases most frequently found in TGMs were CLA4, which has been functionally linked to cytokinesis (12 TGMs); BUB1, which is essential for proper spindle checkpoint function (13 TGMs); and the AMP-activated kinase SNF1, required for glucose-repressed transcription and thermotolerance (14 TGMs) (Figure 5b). These three kinases were also the most genetically promiscuous when we extended this analysis to the other genes on the signaling E-MAP (Supplementary Figure 4), suggesting they occupy central signaling nodes in budding yeast.
Consistent with the pattern observed with the pair-wise genetic interactions (Figure 2b, Supplementary Figure 2), we found an overall enrichment of positive genetic interactions among and between kinases and phosphatases participating in the TGMs. When the TGMs were restricted to contain only kinases and phosphatases, we found an approximately equal number of the four types of motifs (Figure 5c). As non-kinase/phosphatase genes were introduced, the proportion of TGMs with positive genetic interactions decreased (Figure 5c).
Cak1 and Fus3 Kinases Function in the Set2/Rpd3C(S) Pathway
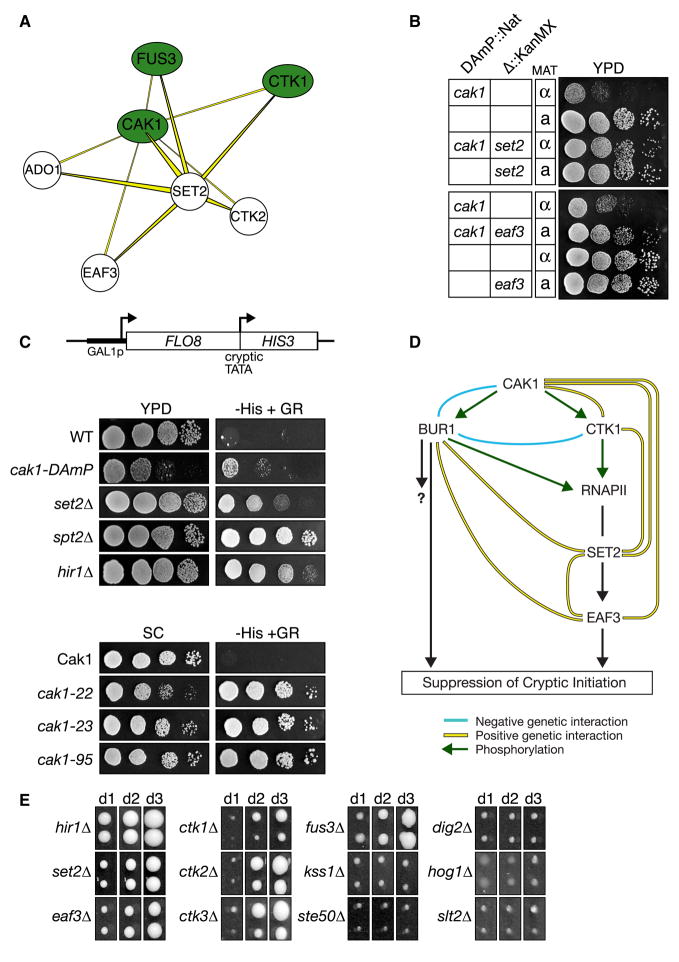
To uncover previously uncharacterized signaling pathways, we next focused on the Type I (+++) TGM, where only one protein was required to be either a kinase or phosphatase. We assembled a Type I TGM list, restricted to factors that were not physically associated (purification enrichment scores < 0.2 (Collins et al., 2007a)), since we wanted to enrich for pathways rather than protein complexes. Taking the 47 most significant Type I TGMs, as defined by the product of their three individual genetic interaction scores, we assembled a network diagram (Supplementary Figure 5) to identify regions containing high-density, positive genetic interactions. Several such clusters were discovered, including the SET2-CAK1 pair, which showed positive genetic interactions with CTK1, CTK2, EAF3, ADO1 and FUS3 (Figure 6a, Supplementary Figure 5). Several of these proteins have previously been demonstrated to act together in a pathway that is required for efficient transcriptional elongation by RNA polymerase II (RNAPII). In this cascade, the kinase Ctk1 (with its regulatory proteins, Ctk2 and Ctk3), phosphorylates Rpb1, the largest subunit of RNAPII, on its C-terminal domain (Cho et al., 2001). This modification recruits the methyltransferase Set2 to methylate Lys36 of histone H3 during transcriptional elongation (Hampsey and Reinberg, 2003). This mark, in turn, recruits Eaf3, a component of the Rpd3C(S) histone deacetylation complex, which then deacetylates histones H3 and H4 in the coding region of genes, resulting in chromatin compaction, an event that ultimately suppresses spurious cryptic initiation (Carrozza et al., 2005; Joshi and Struhl, 2005; Keogh et al., 2005).
Figure 6. The Cak1 kinase functions in the Ctk1/Set2/Rpd3C(S) pathway that suppresses cryptic initiation by RNA polymerase II.
a) Subset of Type I TGMs that share the Cak1-Set2 positive genetic interaction. For a complete picture of all Type I TGMs see Supplementary Figure 4. b) Tetrad analysis and serial 10-fold dilution spot testing demonstrates that the slow growth of the cak1-DAmP allele is strongly suppressed by set2Δ or eaf3Δ. c) CAK1 suppresses cryptic transcription initiation. The GAL1-FLO8-HIS3 reporter (Nourani et al., 2006) is described in Methods. The HIS3 gene product is only produced when transcription aberrantly initiates from a cryptic promoter within FLO8, as when chromatin structure in the transcribing gene is disrupted (Carrozza et al., 2005; Joshi and Struhl, 2005; Keogh et al., 2005). His3 expression was monitored by 10-fold dilution spotting of the indicated strains onto synthetic complete (SC) medium +/− histidine with galactose/raffinose (2%/1% respectively) as the carbon source. The lower panels employed a series of cak1-temperature sensitive alleles (Espinoza et al., 1998) at their semi-permissive temperature (SC, 32°C, 48hrs; -HIS, 32°C, 96hrs). d) Possible schematic of this pathway controlling intergenic chromatin fidelity. e) The MAP kinase Fus3 regulates cryptic initiation. All strains contain a GAL1Sp-FLO8::HIS reporter, similar to that in Figure 6c (see Methods). Reporter strains were pinned in duplicate onto SC-HIS with galactose/raffinose as the carbon source, incubated at 30°C and photographed on days indicated. Panels are standardized to facilitate cross-comparison of reporter expression in each strain/time-point.
Since CAK1 displayed positive genetic interactions with multiple factors implicated in this pathway, we predicted that it would be an integral component of the RNAPII transcriptional elongation control process. The positive genetic interactions of CAK1 with SET2 and EAF3 were confirmed by tetrad analysis and spot testing, which showed that eaf3Δ and set2Δ suppressed the slow growth phenotype observed in a cak1-DAmP background (Figure 6b). In contrast, the positive genetic interaction between CAK1 and CTK1 corresponded to an epistatic interaction since the double mutant was no sicker than either of the two single mutants (Supplementary Figure 6b). To validate the genetic interaction observed between CAK1 and CTK1, we expression-profiled both mutants using DNA microarrays. This revealed a very significant overlap in mRNA expression of genes affected in either mutant, implying that these two kinases function together in vivo (Supplementary Figure 6c, Supplementary Table 8). Furthermore, out of a large collection of kinase mutant expression-profiles, the ctk1Δ profile was most similar to that of the cak1-DAmP strain (S. van Wageningen and F. Holstege, unpublished data).
Finally, the cak1-DAmP strain, as well as other previously characterized CAK1 mutants (cak1-22, cak1-23, cak1-95) (Espinoza et al., 1998), showed a significant increase of transcription initiation from a cryptic internal TATA site within FLO8 (Figure 6c). A similar phenotype has previously been observed with set2Δ and deletions of the chromatin assembly factors SPT2 and HIR1 (Figure 6c) (Carrozza et al., 2005; Kaplan et al., 2003; Nourani et al., 2006). In agreement with these data, Ostapenko and Solomon reported that Cak1 phosphorylates Ctk1 in vivo (Ostapenko and Solomon, 2005), and we found that deletion of CTK1 (or CTK2 and CTK3) also results in a cryptic initiation defect (Figure 6e) (Cheung et al., 2008; Youdell et al., 2008).
Interestingly Bur1, a kinase known to suppress spurious transcription initiation, is another well characterized Cak1 substrate (Yao and Prelich, 2002). We previously showed that deletion of SET2 or EAF3 suppressed the lethality associated with bur1Δ (Keogh et al., 2005), suggesting cross-talk between the two pathways (Figure 6d). The negative genetic interaction between CAK1 and BUR1 (Supplementary Figure 6d) suggests Bur1 is involved in another, parallel pathway. Overall these data demonstrate that Cak1 acts as a key regulator controlling two kinases cascades involved in regulating intergenic chromatin integrity (Figure 6d).
Both CAK1 and SET2 also displayed positive genetic interactions with FUS3, a MAP kinase involved in the mating pathway (Figure 6a). Further testing revealed that fus3Δ gave rise to a cryptic initiation defect as well (Figure 6e). Other factors known to work with FUS3 in the mating pathway (KSS1, STE50, DIG2) (Chen and Thorner, 2007) or other MAP kinases (HOG1 and SLT2) did not display positive genetic interactions with CAK1 and SET2, and accordingly did not have the same transcriptional defect (Figure 6e). Work by Young and Colleagues demonstrated that Fus3 localizes to actively transcribed genes by chromatin immunoprecipitation (Pokholok et al., 2006). Taken together with the genetic interaction data, these findings imply that Fus3 may phosphorylate one or more chromatin factors involved in suppressing cryptic initiation. Further work will be required to reveal the mechanistic details of this intriguing connection between the mating pathway and transcriptional chromatin integrity
Perspective
In summary, we have systematically and quantitatively mapped the pair-wise genetic interactions within the yeast signaling machinery. The entire genetic interaction matrix of 100,000 pair-wise interactions is available in a web-based searchable format, as illustrated in Supplementary Figure 7, at http://interactome-cmp.ucsf.edu. We have analyzed the quantitative genetic interaction data using triplet genetic motifs (TGMs) which we show can be a powerful approach for globally studying signaling pathways, and uncovering mechanistic details of how specific proteins function within these cascades. Our data set, when integrated with other genome-wide data sets, will help to understand and model the phosphate-based signaling behaviour of yeast cells. Furthermore, this signaling E-MAP also provides a launching platform for other analyses. While the current screen was carried out on rich media, many signaling pathways depend on specific stimuli. Condition-dependent screens will, therefore, reveal dynamic changes in the signaling apparatus. We also plan to compare the genetic interaction profiles of complete kinase gene deletions with those of catalytically dead mutants and analog-sensitive kinase alleles (Bishop et al., 2000). Such analyses should allow us to separate the catalytic activities from the scaffolding functions of these proteins. Comparison of the genetic architecture of the signaling networks between different yeast species (Roguev et al., 2008) and higher organisms (Bakal et al., 2008) will be instructive in understanding how regulation via reversible phosphorylation is conserved and how it has evolved. Finally, epistasis analyses have significant potential, not only for functionally connecting proteins into pathways and complexes, but also for identifying combinations of genes, or genes operating at critical nodes at the intersection of signaling pathways, that can serve as appropriate therapeutic targets.
Methods
Strains were constructed and E-MAP experiments were performed as previously described (Collins et al., 2007b; Collins et al., 2006; Schuldiner et al., 2005).
To highlight strong genetic interactions, we introduced a cutoff of S ≤ −2.5 for negative interactions and S ≥ 2.0 for positive interactions. Positive to negative (P to N) ratios were computed for the subsets depicted in Figure 2a. The sets consisting of kinase and phosphatase pairs that act on the same substrates were compared with a subset of the signaling E-MAP that contains only kinases, phosphatases and regulatory subunits. Two-tailed P-values were computed using Fisher’s exact test. The remaining sets in Figure 2a were compared with the chromosome function E-MAP. Here, two-tailed P-values were computed using chi-square with Yates’ correction. Importantly, the relative ratios obtained were independent of the thresholds used to define the positive and negative genetic interactions (data not shown). Kinases, phosphatases, kinase-substrate pairs and phosphatase-substrate pairs are listed in Supplementary Tables 3,4,5
The preparation of yeast whole cell extracts by TCA (Figure 3b) and cell fractionation experiments (Figure 3c) were performed essentially as described (Keogh et al., 2006b). The GAL1-FLO8-HIS3 reporter (Figure 6c) consists of: (i) the FLO8 promoter replaced by that of galactose-regulated GAL1/10, and (ii) the HIS3 gene inserted out of frame into FLO8 (flo8Δ (+1729–02505)::HIS3 (+1–663)) such that HIS3 product is only translated when transcription initiates from a cryptic promoter within FLO8(Nourani et al., 2006). In the almost identical GAL1Sp-FLO8-HIS reporter (Figure 6e), GAL1Sp is an attenuated version of the GAL1 promoter, missing 1.5 copies of the 5′ GAL4-UAS (Mumberg et al., 1994). Reporter strains (Supplementary Table 9) were created by direct transformation, or mating followed by tetrad dissection. HIS3 expression was monitored by spotting the appropriate strains onto synthetic complete (SC) medium +/− histidine with either glucose (2%) or galactose/raffinose (2%/1% respectively) as the carbon source.
Gene expression profiling was carried out as previously described (van de Peppel et al., 2005). P-values for the overlap of the Venn diagrams were obtained using the hypergeometric test with a population size of 6389 genes.
Supplementary Material
Acknowledgments
The authors thank A. Roguev, C. Kaplan, C. J. Ingles, P. Aguilar, T. Walther, J. H. Morris and members of the Krogan, Shokat and Keogh labs for advice and comments. We also thank W. A. Lim for discussion on MAP kinase cascades. We are grateful to A. Chan, E. Cheng, Y. Nijati, and Li Jieying for technical assistance. We thank F. Winston and S. Buratowski for providing yeast strains and B. Strahl, S. Hahn, and D. Morgan for sharing unpublished data. The work was supported by an Ernst Schering Postdoctoral Fellowship (D. F.), the Biophysics Graduate Group at UCSF (H. B.), the National Science Foundation (G. C. and D. K.), the New York Speakers Fund for Biomedical Research (M-C. K.), the Howard Hughes Medical Institute (K. M. S.) and the NIH (N. J. K. and K. M. S.), and from Sandler Family Funding (N. J. K.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcazar-Roman AR, Wente SR. Inositol polyphosphates: a new frontier for regulating gene expression. Chromosoma. 2008;117:1–13. doi: 10.1007/s00412-007-0126-4. [DOI] [PubMed] [Google Scholar]
- Babiarz JE, Halley JE, Rine J. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 2006;20:700–710. doi: 10.1101/gad.1386306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakal C, Linding R, Llense F, Heffern E, Martin-Blanco E, Pawson T, Perrimon N. Phosphorylation Networks Regulating JNK Activity in Diverse Genetic Backgrounds. Science. 2008;322:453–456. doi: 10.1126/science.1158739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram PG, Choi JH, Carvalho J, Chan TF, Ai W, Zheng XFS. Convergence of TOR-nitrogen and Snf1-glucose signaling pathways onto Gln3. Mol Cell Biol. 2002;22:1246–1252. doi: 10.1128/MCB.22.4.1246-1252.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Chen RE, Thorner J. Function and regulation in MAPK signaling pathways: Lessons learned from the yeast Saccharomyces cerevisiae. Biochim Biophys Acta, Mol Cell Res. 2007;1773:1311–1340. doi: 10.1016/j.bbamcr.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasova VA, Hinnebusch AG. Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Genes Dev. 2003;17:859–872. doi: 10.1101/gad.1069003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung V, Chua G, Batada N, Landry CR, Michnick SW, Hughes TR, Winston F. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 2008;6:2550–2562. doi: 10.1371/journal.pbio.0060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 2001;15:3319–3329. doi: 10.1101/gad.935901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FCP, Weissman JS, Krogan NJ. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007a;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007b;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Collins SR, Schuldiner M, Krogan NJ, Weissman JS. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. GenomeBiology. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Sprague GF., Jr The Glc7p-interacting protein Bud14p attenuates polarized growth, pheromone response, and filamentous growth in Saccharomyces cerevisiae. Eukaryotic Cell. 2002;1:884–894. doi: 10.1128/EC.1.6.884-894.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon N, Oki M, Szyjka SJ, Aparicio OM, Kamakaka RT. H2A.Z functions to regulate progression through the cell cycle. Mol Cell Biol. 2006;26:489–501. doi: 10.1128/MCB.26.2.489-501.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diella F, Gould CM, Chica C, Via A, Gibson TJ. Phospho..ELM: a database of phosphorylation sites-update 2008. Nucleic Acids Res. 2008;36:D240–D244. doi: 10.1093/nar/gkm772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza FH, Farrell A, Nourse JL, Chamberlin HM, Gileadi O, Morgan DO. Cak1 is required for Kin28 phosphorylation and activation in vivo. Mol Cell Biol. 1998;18:6365–6373. doi: 10.1128/mcb.18.11.6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- Goossens A, De la Fuente N, Forment J, Serrano R, Portillo F. Regulation of yeast H+-ATPase by protein kinases belonging to a family dedicated to activation of plasma membrane transporters. Mol Cell Biol. 2000;20:7654–7661. doi: 10.1128/mcb.20.20.7654-7661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KD, Pflum MKH. Kinase-Catalyzed Biotinylation for Phosphoprotein Detection. J Am Chem Soc. 2007;129:10–11. doi: 10.1021/ja066828o. [DOI] [PubMed] [Google Scholar]
- Hampsey M, Reinberg D. Tails of intrigue: Phosphorylation of RNA polymerase II mediates histone methylation. Cell. 2003;113:429–432. doi: 10.1016/s0092-8674(03)00360-x. [DOI] [PubMed] [Google Scholar]
- Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JY, Sun ZW, Li X, Reuben M, Tatchell K, Bishop DK, Grushcow JM, Brame CJ, Caldwell JA, Hunt DF, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- Johnson SA, Hunter T. Kinomics: methods for deciphering the kinome. Nat Methods. 2005;2:17–25. doi: 10.1038/nmeth731. [DOI] [PubMed] [Google Scholar]
- Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Kannan N, Taylor SS, Zhai Y, Venter JC, Manning G. Structural and functional diversity of the microbial kinome. PLoS Biol. 2007;5:467–478. doi: 10.1371/journal.pbio.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- Kelley R, Ideker T. Systematic interpretation of genetic interactions using protein networks. Nat Biotechnol. 2005;23:561–566. doi: 10.1038/nbt1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MC, Kim JA, Downey M, Fillingham J, Chowdhury D, Harrison JC, Onishi M, Datta N, Galicia S, Emili A, et al. A phosphatase complex that dephosphorylates gamma H2AX regulates DNA damage checkpoint recovery. Nature. 2006a;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional Set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Mennella TA, Sawa C, Berthelet S, Krogan NJ, Wolek A, Podolny V, Carpenter LR, Greenblatt JF, Baetz K, Buratowski S. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006b;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber P, Horz W. SWRred not shaken; mixing the histones. Cell. 2004;117:5–7. doi: 10.1016/s0092-8674(04)00296-x. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Baetz K, Keogh MC, Datta N, Sawa C, Kwok TCY, Thompson NJ, Davey MG, Pootoolal J, Hughes TR, et al. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc Natl Acad Sci. 2004;101:13513–13518. doi: 10.1073/pnas.0405753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TY, Huang HD, Hung JH, Huang HY, Yang YS, Wang TH. dbPTM: an information repository of protein post-translational modification. Nucleic Acids Res. 2006;34:D622–D627. doi: 10.1093/nar/gkj083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linding R, Jensen LJ, Ostheimer GJ, van Vugt MATM, Jorgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke MM, Della Seta F, Di Como CJ, Sugimoto H, Kobayashi R, Arndt KT. The SAP, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol Cell Biol. 1996;16:2744–2755. doi: 10.1128/mcb.16.6.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR Substrate Analysis Reveals Extensive Protein Networks Responsive to DNA Damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Millar CB, Xu F, Zhang K, Grunstein M. Acetylation of H2AZ lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 2006;20:711–722. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulet JM, Leube MP, Kron SJ, Rios G, Fink GR, Serrano R. A novel mechanism of ion homeostasis and salt tolerance in yeast: the Hal4 and Hal5 protein kinases modulate the Trk1-Trk2 potassium transporter. Mol Cell Biol. 1999;19:3328–3337. doi: 10.1128/mcb.19.5.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulugu S, Bai W, Fridy PC, Bastidas RJ, Otto JC, Dollins DE, Haystead TA, Ribeiro AA, York JD. A Conserved Family of Enzymes That Phosphorylate Inositol Hexakisphosphate. Science. 2007;316:106–109. doi: 10.1126/science.1139099. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Mueller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourani A, Robert F, Winston F. Evidence that Spt2/Sin1, an HMG-like factor, plays roles in transcription elongation, chromatin structure, and genome stability in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:1496–1509. doi: 10.1128/MCB.26.4.1496-1509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke SM, Herskowitz I. Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol Biol Cell. 2004;15:532–542. doi: 10.1091/mbc.E03-07-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oficjalska-Pham D, Harismendy O, Smagowicz WJ, Gonzalez de Peredo A, Boguta M, Sentenac A, Lefebvre O. General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1. Mol Cell. 2006;22:623–632. doi: 10.1016/j.molcel.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Ostapenko D, Solomon MJ. Phosphorylation by Cak1 regulates the C-terminal domain kinase Ctk1 in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:3906–3913. doi: 10.1128/MCB.25.10.3906-3913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Yuan DS, Xiang D, Wang X, Sookhai-Mahadeo S, Bader JS, Hieter P, Spencer F, Boeke JD. A robust toolkit for functional profiling of the yeast genome. Mol Cell. 2004;16:487–496. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated Signal Transduction Kinases Frequently Occupy Target Genes. Science. 2006;313:533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- Roguev A, Bandyopadhyay S, Zofall M, Zhang K, Fischer T, Collins SR, Qu H, Shales M, Park HO, Hayles J, et al. Conservation and Rewiring of Functional Modules Revealed by an Epistasis Map in Fission Yeast. Science. 2008a;322:405–410. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rua D, Tobe BT, Kron SJ. Cell cycle control of yeast filamentous growth. Curr Opin Microbiol. 2001;4:720–727. doi: 10.1016/s1369-5274(01)00274-0. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Bhandari R, Resnick AC, Snowman AM, Snyder SH. Phosphorylation of proteins by inositol pyrophosphates. Science. 2004;306:2101–2105. doi: 10.1126/science.1103344. [DOI] [PubMed] [Google Scholar]
- Schaber M, Lindgren A, Schindler K, Bungard D, Kaldis P, Winter E. CAK1 promotes meiosis and spore formation in Saccharomyces cerevisiae in a CDC28-independent fashion. Mol Cell Biol. 2002;22:57–68. doi: 10.1128/MCB.22.1.57-68.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Weissman JS, Krogan N. Quantitative genetic analysis in Saccharomyces cerevisiae using epistatic miniarray profiles (E-MAPs) and its application to chromatin functions. Methods. 2006;40:344–352. doi: 10.1016/j.ymeth.2006.07.034. [DOI] [PubMed] [Google Scholar]
- Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell. 2006;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- St Onge RP, Mani R, Oh J, Proctor M, Fung E, Davis RW, Nislow C, Roth FP, Giaever G. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat Genet. 2006;39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svejstrup JQ. Elongator complex: how many roles does it play? Curr Opin Cell Biol. 2007;19:331–336. doi: 10.1016/j.ceb.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Tate JJ, Feller A, Dubois E, Cooper TG. Saccharomyces cerevisiae Sit4 Phosphatase Is Active Irrespective of the Nitrogen Source Provided, and Gln3 Phosphorylation Levels Become Nitrogen Source-responsive in a sit4-deleted Strain. J Biol Chem. 2006;281:37980–37992. doi: 10.1074/jbc.M606973200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AHY, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CWV, Bussey H, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- van de Peppel J, Kettelarij N, van Bakel H, Kockelkorn TTJP, van Leenen D, Holstege FCP. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol Cell. 2005;19:511–522. doi: 10.1016/j.molcel.2005.06.033. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang X, Jiang Y. Interaction with Tap42 is required for the essential function of Sit4 and type 2A phosphatases. Mol Biol Cell. 2003;14:4342–4351. doi: 10.1091/mbc.E03-02-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, Collins SR, Whitworth GB, Kress TL, Weissman JS, et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Lytvyn V, Thomas DY, Leberer E. The phosphorylation site for Ste20p-like protein kinases is essential for the function of myosin-I in yeast. J Biol Chem. 1997;272:30623–30626. doi: 10.1074/jbc.272.49.30623. [DOI] [PubMed] [Google Scholar]
- Yao S, Prelich G. Activation of the Bur1-Bur2 cyclin-dependent kinase complex by Cak1. Mol Cell Biol. 2002;22:6750–6758. doi: 10.1128/MCB.22.19.6750-6758.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youdell ML, Kizer KO, Kisseleva-Romanova E, Fuchs SM, Duro E, Strahl BD, Mellor J. Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol Cell Biol. 2008;28:4915–4926. doi: 10.1128/MCB.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanzoni A, Ausiello G, Via A, Gherardini PF, Helmer-Citterich M. Phospho3D: a database of three-dimensional structures of protein phosphorylation sites. Nucleic Acids Res. 2007;35:D229–D231. doi: 10.1093/nar/gkl922. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.