Abstract
The advance of novel technologies that will enable the detection of large sets of biomarker proteins, to greatly improve the sensitivity and specificity of an assay, represents a major objective in biomedical research. To demonstrate the power of mass spectrometry (MS) detection for large scale biomarker screening in cancer research, a simple, one-step approach for fast biomarker fingerprinting in complex cellular extracts is described. MCF-7 breast cancer cells were used as a model system. Fast proteomic profiling of whole cellular extracts was achieved on a linear trap quadrupole (LTQ) mass spectrometer by one of the following techniques: (a) data dependent liquid chromatography (LC)-MS/MS of un-labeled cell extracts, (b) data dependent LC-MS/MS with pulsed Q dissociation (PQD) detection of iTRAQ labeled samples, and (c) multiple reaction monitoring (MRM)-MS of low abundant proteins that could not be detected with data dependent MS/MS. The data dependent LC-MS/MS analysis of MCF-7 cells enabled the identification of 796 proteins (p<0.001) and the simultaneous detection of 156 previously reported putative cancer biomarkers. PQD detection of iTRAQ labeled cells resulted in the detection of 389 proteins and 64 putative biomarkers. MRM-MS analysis enabled the successful monitoring of a panel of low abundance proteins in one single experiment, highlighting the utility of this technique for targeted analysis in cancer investigations. These results demonstrate that MS-based technologies relying on a one-step separation protocol have the potential to revolutionize biomarker research and screening applications by enabling fast, sensitive and reliable detection of large panels of putative biomarkers. To further stimulate the exploration of proteins that have been previously reported in the literature to be differentially expressed in a variety of cancers, an extensive list of ~1,100 candidate biomarkers has been compiled and included in the manuscript.
Keywords: LC-MS/MS, MRM-MS, iTRAQ, Proteomics, Cancer, MCF-7
1. INTRODUCTION
Cancer is a life-threatening disease that is affecting humans worldwide. It is estimated that every year ~500,000 people will die from cancer in the US alone. With the early detection of the disease, the mortality rate is expected to be significantly reduced. Biomarker discovery/screening is an expanding field in proteomics aimed at the identification of proteins that change expression levels or post-translational modifications in response to the on-set of a disease. A variety of techniques have been explored for the identification and/or quantitation of cancer biomarkers including immunohistochemistry (IHC), Western blotting, enzyme-linked immunosorbent assays (ELISA), antibody/cell/tissue microarrays, protein-chips, two dimensional polyacrylamide gel electrophoresis (2D-PAGE), difference in-gel electrophoresis (DIGE), free flow electrophoresis and a broad range of MS-based approaches [(2D)-LC-MS/MS, matrix assisted laser desorption ionization (MALDI)-time-of-flight (TOF)-MS, isoelectric focusing (IEF)-LC/electrospray ionization (ESI)-TOFMS, IEF-LC/MALDI-TOF, MS imaging, surface enhanced laser desorption ionization (SELDI)-TOFMS and multiple reaction monitoring (MRM)-MS] [1-4]. Each of these technologies present advantages and disadvantages. For example, gel, array or chip-based methods enable high-throughput analysis, while immunoassay and MS-based methods enable sensitive detection. However, gel-based approaches present concerns related to reproducibility, sensitivity and dynamic range; microarray technology interfacing to mass spectrometry detection is not always straightforward; and, immuno-methods often suffer from low throughput and specificity.
To obviate many of the difficulties associated with existing technologies for biomarker detection, the development and implementation of novel, alternative techniques continues to remain of interest to many researchers. Recent improvements in the performance of mass spectrometry in terms of sensitivity, speed of analysis and capability to unambiguously identify protein components in a sample, have rapidly increased its applicability for biomarker discovery [5-13]. Several MS-based “omics” approaches, namely, proteomics, peptidomics, glycomics, phosphoproteomics, metabolomics and lipidomics have been reported [14]. Most importantly, a variety of quantitative MS strategies for protein differential expression profiling, including label-free and stable isotope labeling approaches, have been explored. Label free methods rely on measuring peak areas, intensities or spectral counts [15,16]. As the samples are processed independently, quantitation errors can be high. Therefore, label-based techniques such as metabolic, enzymatic or chemical labeling have been developed [17-27]. Of these, chemical labeling carries the advantage of being broadly applicable to samples of various origins. It is based on stable-isotope tagging of protein/peptide samples that upon MS analysis generate tag ratios that are proportional to abundance. A number of strategies including isotope coded affinity tags (ICAT) [24], mass coded abundance tags (MCAT) and isobaric tags for relative and absolute quantitation (iTRAQ) have been reported [25-27]. Interest in the latter approach has grown rapidly, as the method allows for the simultaneous quantitation of up to 4-8 different samples: peptides are labeled with isobaric tags at the N-terminus and the lysine side chains, and upon MS/MS fragmentation signature ions are generated that enable the comparison of protein expression levels in various samples.
Cancer biomarker research has been conducted by using a variety of specimens such as body fluids (i.e., serum, plasma, nipple aspirate fluid, urine, saliva), cell lines and tissues [6]. A major difficulty associated with these studies relates to the identification and quantitation of low abundance proteins (pg-ng/mL), particularly in plasma and serum specimens. Additional concerns relate to the lack of reproducibility, sensitivity and specificity of detection. Progress made in sample preparation, separation technology, mass spectrometry instrumentation, statistics and bioinformatics-tool development for proteomic studies has enabled the discovery of a large set of putative biomarkers for various cancers including bladder, breast, prostate, ovarian, colorectal, lung, endometrial, gastric, thyroid, esophageal, neuroendocrine, etc. [6-11,28,29]. In spite of these efforts, however, only very few biomarkers, namely, prostate specific antigen (PSA), carcinoembryonic antigen (CEA), cancer antigens 125 (CA 125), 153 (CA 15.3) and 19.9 (CA 19.9), alpha-fetoprotein (AFP), bladder tumor antigen, thyro-globulin, Her-2/neu, leptin, troponin I and B-type natriuretic peptide, have been approved by the FDA for clinical use. Many reviews and research papers have been published on this topic [2-14,28-41]. One of the most comprehensive reviews was conducted by Polanski and Anderson [41] who have compiled a list of 1261 proteins that were found to be differentially expressed in different types of human cancer, of which a sub-set of 260 most studied candidate biomarkers were listed in their manuscript.
In an effort to gain a better control over cancer and disease management, the field of biomarker discovery continues to evolve. The development of novel methods that will enable sensitive screening for large panels of biomarkers, instead of just one, to greatly improve specificity, represents one of the greatest needs. A complementary search for proteins described in recent years as putative biomarkers has resulted in a collection of 1365 such proteins (~1100 unique), of which more than 800 are new additions to the list previously published by Polanski [41] (see Appendix A). To further facilitate the exploration and validation of these proteins as candidate cancer biomarkers, in this work, we are describing a quick and simple RPLC-ESI-MS/MS method that enabled the simultaneous identification of a panel of 156 putative markers (out of ~1100) in the MCF-7 breast cancer cells. We expect that the research presented in this manuscript will open new avenues for developing fast, specific and sensitive methods for unambiguous detection of biomarker panels, to increase our ability to diagnose disease at an early stage and identify relevant targets for cancer treatment.
2. EXPERIMENTAL
2.1. Reagents and materials
MCF-7 breast cancer cells, Eagle’s minimum essential medium-EMEM, fetal bovine serum-FBS, insulin, and trypsin/EDTA were from ATCC (Manassas, VA, USA). Phenol red-free DMEM (Dulbecco’s Modified Eagle’s Medium) was purchased from Invitrogen (Carlsbad, CA, USA). Trypsin (phenol red free) was obtained from SAFC Biosciences (Lenexa, Kansas, USA). Charcoal/dextran treated fetal calf serum was supplied by HyClone (Logan, UT, USA). Buffers and denaturing agents (TrisHCl, sodium chloride, formic acid, acetic acid, trifluoroacetic acid, dithiothreitol-DTT, urea), protease and phosphatase inhibitors (NaF, Na3VO4), β-estradiol (E2), tamoxifen (Tam), and L-glutamine were from Sigma (St. Louis, MO, USA). Ammonium bicarbonate was purchased from Aldrich (Milwaukee, WI, USA). RIPA lysis buffer was supplied by Upstate (Lake Placid, NY, USA). Sequencing-grade modified trypsin was from Promega Corp. (Madison, WI, USA). The iTRAQ reagent kit was from Applied Biosystems (Foster City, CA, USA). SPEC-PTC18 and SCX solid-phase extraction pipette tips were from Varian Inc. (Lake Forest, CA, USA). HPLC-grade solvents (methanol and acetonitrile) were supplied by Fisher Scientific (Fair Lawn, NJ, USA). Deionized water, used to prepare all aqueous solutions, was from a MilliQ ultrapure water system (Millipore, Bedford, MA, USA).
2.2. MCF-7 cell culture
MCF-7 cells were cultured in a humidified atmosphere of 5 % CO2, at 37 °C using one of the following conditions: (a) EMEM supplemented with FBS (10 %) and bovine insulin (10 μg/mL); (b) DMEM red-free supplemented with L-glutamine (4 mM), charcoal stripped fetal calf serum (10 %), insulin (1 μg/mL) and E2 (1 nM); and (c) DMEM red-free supplemented with L-glutamine (4 mM), charcoal stripped fetal calf serum (10 %), insulin (1 μg/mL), E2 (1 nM) and tamoxifen (Tam) (1 μM). After harvesting, cells were stored at -80 °C until used.
2.3. Cell lysis and protein extraction
Cells were processed according to a procedure described in detail elsewhere [42]. Briefly, cells were lysed (2 h at 4 °C with gentle rocking) with a solution prepared from 1 mL RIPA buffer (500 mM TrisHCl/pH 7.4, 10 mM EDTA, 1.5 M NaCl, 2.5 % deoxycholic acid, 10 % NP-40), phosphatase inhibitors (50 μL of 200 mM Na3VO4, and 100 μL of 100 mM NaF), 100 μL protease inhibitor cocktail (104 mM AEBSF, 4 mM bestatin, 2 mM leupeptin, 1.5 mM pepstatin A, 1.4 mM E-64, 0.08 mM aprotinin), and 8.75 mL iced-cold water. The extract was centrifuged at 13,000 rpm (Eppendorf centrifuge 5415D) and 4 °C for ~15 min. The supernatant containing the cell extract was removed, and the protein concentration was determined with the Bradford assay (595 nm) using a SmartSpec Plus spectrophotometer (Bio-Rad, Hercules, CA, USA). Typically, the extracts contained 1-4 mg/mL protein. Samples were stored at -80 °C until further treatment.
2.4. Protein digestion and C18/SCX clean-up
The protein extract was denatured (50 mM Tris, 8 M urea, 4.5 mM DTT) for 1 h at 60 °C, and digested with trypsin (substrate:enzyme ratio 50:1 w/w) for 24 h at 37 °C. The digestion process was quenched with glacial acetic acid. E2 cultured cell digests were cleaned-up with SPEC-PTC18 and SPEC-PTSCX solid-phase extraction pipette tips (Varian Inc., Lake Forest, CA, USA). For C18 clean-up, the sample was loaded on the cartridge in the tryptic digestion buffer quenched with glacial acetic acid, rinsed with CH3OH/H2O/TFA (5:95:0.1), and eluted with CH3CN/H2O/TFA (80:20:0.1). For SCX clean-up, the sample was brought to dryness, re-dissolved in CH3OH/CH3COOH 0.1 M (1:2), loaded on the cartridge, rinsed with CH3OH/CH3COOH 0.1 M (50:50), and eluted with CH3OH/NH4OH (98:2).
2.5. iTRAQ labeling of MCF-7 cells treated with estradiol and tamoxifen for protein differential expression analysis
E2 and Tam cultured cell extract digests were labeled with iTRAQ reagents after SPEC-PTC18 clean-up. SPEC-PTC18 clean-up prior to labeling was essential in order to avoid interferences from sample components that contain primary amino groups that may interfere with the labeling reaction (such as the Tris and ammonium bicarbonate buffers that were used during cell extract preparation). Protein digests (~100 μg) were dried to ~5 µL volumes, re-dissolved in 25 µL of iTRAQ dissolution buffer, and labeled with iTRAQ reagents (114, 115, 116, and 117) dissolved in 70 μL of ethanol. The manufacturer’s protocol was followed, with the exception that the labeling reaction was allowed to proceed for 2h, instead of 1h, at room temperature. E2 treated cells (A) were labeled with reagents 114 and 115, Tam treated cells (B) with reagents 116 and 117, and the labeled extracts were mixed in a ratio of (A):(A):(B):(B) of 1:1:1:1. The mixture was then cleaned-up with SPEC-PTSCX solid-phase extraction pipette tips to eliminate compounds that may interfere with MS analysis (see protocol described above).
2.6. RP-HPLC-ESI-MS/MS
A micro liquid chromatography instrument (Agilent Technologies, Palo Alto, CA, USA) coupled to an LTQ ion trap mass spectrometer (Thermo Electron Corp., San Jose, CA, USA), via an in-house developed on-column/no-split injection system [42], was used to perform the analysis. RPLC columns were prepared from 100 μm i.d. × 12 cm fused silica capillaries packed with 5 μm Zorbax SB-C18 particles (Agilent Technologies). A 1 cm long capillary (20 μm i.d. × 90 μm o.d.) inserted into the separation column served as a nanospray emitter. Mobile phase A was 0.01 % TFA in H2O:ACN (95:5 v/v), and mobile phase B was 0.01 % TFA in H2O:ACN (20:80 v/v). The volumetric flow rate in the separation column was ~160-180 nL/min, and the separation gradient was 3 h long from 0 % to 100 % eluent B. Sample volumes of 8 μL containing 1-2 μg/μL protein extract digest were injected for analysis.
A data-dependent acquisition method was developed to acquire the MS data [42]. The conditions were as follows: 1 MS scan (5 microscans averaged) followed by one zoom scan and one MS2 on the top 5 most intense peaks, ±5 m/z zoom scan width, dynamic exclusion enabled at repeat count 1 with a 30 s repeat duration, exclusion list size of 200, 60 s exclusion duration, and ±1.5 m/z exclusion mass width. Collision induced dissociation (CID) parameters were set at the following values: isolation width 3 m/z, normalized collision energy 35 %, activation Q at 0.25, and activation time 30 ms. For the analysis of iTRAQ labeled peptides in the E2/Tam treated MCF-7 cell extracts, the pulse Q dissociation (PQD) mode was used, and the parameters were set as follows: isolation width at 3 m/z, normalized collision energy at 35 %, activation Q at 0.7, and activation time (T) at 0.1 ms. The Bioworks 3.3 software (Thermo Electron Corp., San Jose, CA, USA) and a Homo sapiens SwissProt protein database were used for protein identification. To enable the search, two missed tryptic cleavage sites per peptide were allowed, with a 2 amu peptide tolerance and a 1 amu fragment ion tolerance. Only fully tryptic fragments were considered for peptide matching with the database entries. For identification of iTRAQ labeled peptides, five chemical modifications at the N terminal peptide and lysine groups were allowed, and all peptides were assigned to unique references. Other modifications were not included in the search. Carbamylation on lysine residues was observed (<4 %), however, due to low quality tandem mass spectra displayed by carbamylated peptides, to reduce the false positive identification rates, such modifications were not allowed in the final database search. Data filtering parameters were as follows: (a) Xcorr = 1.9 for z = 1, Xcorr = 2.2 for z = 2, and Xcorr = 3.8 for z ≥ 3, (b) only different peptides were enabled for each protein, and (c) only top matching proteins with p<0.001 were considered in the analysis. The p-value represents the probability of a random match, and was calculated by the Bioworks software based on the parameters that characterize the quality of a tandem mass spectrum. MRM data acquisition included the development of 3 h long LC-MS/MS runs with 6 segments (~20 min long) and 6-9 scan events/segment. Alternatively, some ions were monitored for the duration of the entire LC-MS/MS experiment. MRM was performed using the same CID settings as described for data dependent analysis. The mass window that was monitored around the product ion was m/z=±1.5.
SwissProt accession numbers for the list of identified and filtered proteins were uploaded on the Gene Ontology/AmiGO website (http://amigo.geneontology.org/cgi-bin/amigo/go.cgi), queried by the “Database ID” search field in the “Genes or proteins” search category, and filtered by the “Biological process” or “Cellular component” “Ontology” filter. The “Gene Products” filters were set to “All.” Gene product associations were displayed for the Homo sapiens species, and the output results were processed in Excel by manually searching for different categories of proteins.
3. RESULTS AND DISCUSSION
A previous study in our lab, that involved the analysis of MCF-7 cells by 2D-SCX-LC-MS/MS, enabled the identification of ~1,859 proteins (p<0.001) of which ~200 were involved in cancer-relevant cellular processes [42]. The work involved the preliminary generation of strong cation exchange sample sub-fractions followed by individual LC-MS/MS analysis of each fraction. The purpose of the present research was to evaluate whether a simple, one-step LC-MS/MS approach, involving the analysis of un-separated cell extracts, could enable the detection of a large number (or panel) of proteins that could be used for biomarker fingerprinting applications.
To demonstrate the power of LC-MS/MS for rapid proteomic profiling, three of the most relevant data acquisition/processing strategies were investigated for the analysis of MCF-7 breast cancer cellular extracts. These strategies included data dependent LC-MS/MS analysis of unlabeled extracts for biomarker screening applications, data dependent LC-PQD-MS/MS of iTRAQ labeled samples for differential expression profiling, and MRM-MS for targeted analysis of low abundant proteins. The results were compared to the ones generated by the more tedious, time-consuming, large-scale 2D-SCX-LC-MS/MS experiment [42]. Data-dependent LC-MS/MS analysis techniques aim at generating a comprehensive proteome profile of complex cellular extracts. They rely on selecting the most intense ions that co-elute during LC-MS analysis for MS fragmentation and structure elucidation. By properly selecting the timing of various MS events to coincide with the LC elution time-window of peptides, sequentially lower intensity ions are selected for fragmentation (see experimental section). Eventually, this “smart” sample analysis strategy enables the detection of a very large number of peptides in complex samples, being, thus, very useful for large-scale screening applications. The method is applicable to both qualitative and quantitative investigations. However, peptides belonging to low abundant proteins, such as the case of many biomarkers, are often overlooked by data dependent detection approaches. Specialized techniques, such as targeted MRM detection, have been developed for the analysis of such proteins.
3.1. Large-scale data-dependent analysis
In our study, the one-step LC-MS/MS approach involving data dependent analysis of un-labeled and iTRAQ labeled MCF-7 cellular extracts resulted in the identification of 796 and 389 proteins, respectively. The estrogen positive MCF-7 cells were cultured under relevant conditions that involved the presence of E2 at concentrations commensurate with the physiological levels of this hormone, i.e., 1 nM. In addition, for testing the technology for iTRAQ labeled samples, the cells were cultured in the presence of E2 (10 pM) and Tam (1 μM)-a non-steroidal drug prescribed in breast cancer therapy. The concentration of E2 was chosen at such low levels to not counteract the Tam effect, while the concentration of Tam was chosen at the typical clinical treatment levels. Table 1 summarizes the overall findings. To increase the total number of identified proteins, six LC-MS/MS replicates of each sample were performed. The data were filtered by selecting only proteins that had p<0.001 and that were matched by peptides that passed the Xcorr vs. charge state filter set at 1.9, 2.2 and 3.8, respectively. The data dependent LC-MS/MS analysis of whole cellular extracts enabled the detection of 796 proteins, i.e., ~40 % of the SCX pre-fractionated samples. The number of unique peptide matches per protein dropped proportionally, but on the average was maintained at ~2.5 peptides/protein. PQD detection of iTRAQ labeled samples typically resulted in the identification of only ~50-60 % of the un-labeled proteins that could have been identified with conventional collision induced dissociation (CID)-MS, most probably due to the lower fragmentation efficiency of the PQD vs. the CID process. PQD is a novel MS detection approach that was recently implemented on linear ion trap instruments for precursor ion activation and dissociation [43]. The technique allows for the trapping of low m/z ions, and is particularly useful for the analysis of peptides that are labeled with isobaric tags that upon MS/MS fragmentation break down from the parent peptide and produce low m/z signature ions for quantitative evaluations.
Table 1.
Protein identification in MCF-7 cell extracts by various sample processing and data acquisition strategies. Conditions: (A) MCF-7 cells were cultured in insulin/EMEM. Cellular extracts were pre-fractionated by SCX into 16 sample sub-fractions and analyzed separately by LC-MS/MS. The data reflect combined results from all SCX-LC-MS/MS runs (total sample amount injected in 16 fractions was ~42 μg) [42]. (B) MCF-7 cells were cultured in E2/DMEM. Whole cellular extract were analyzed by LC-MS/MS. The data reflect combined results from six runs (~16 μg sample/LC injection). (C) MCF-7 cells were cultured in E2/Tam/DMEM and labeled with iTRAQ reagents. Whole cellular extract were analyzed by LC-MS/MS. The data reflect combined results from five runs (~16 μg sample/LC injection). MS data filtering was accomplished by selecting only proteins with p<0.001 that were matched by peptides that passed the Xcorr vs. charge state filter set at 1.9, 2.2 and 3.8, respectively. Specific conditions are provided in the experimental section.
| Method of analysis | # Proteins (P<0.001)/ | # Peptides unique (P<0.001) | % False positive (Peptides/Proteins All unique) | |
|---|---|---|---|---|
| A | SCX-LC-MS/MS data dependent analysis (Insulin culture) | 1,859 | 6,597 | 0.3/2.4 |
| B | LC-MS/MS data dependent analysis (E2 culture) | 796 | 1,906 | 1.7/3.1 |
| C | LC-MS/MS iTRAQ data dependent analysis (E2/Tam culture) | 389 | 829 | 2.3/4.1 |
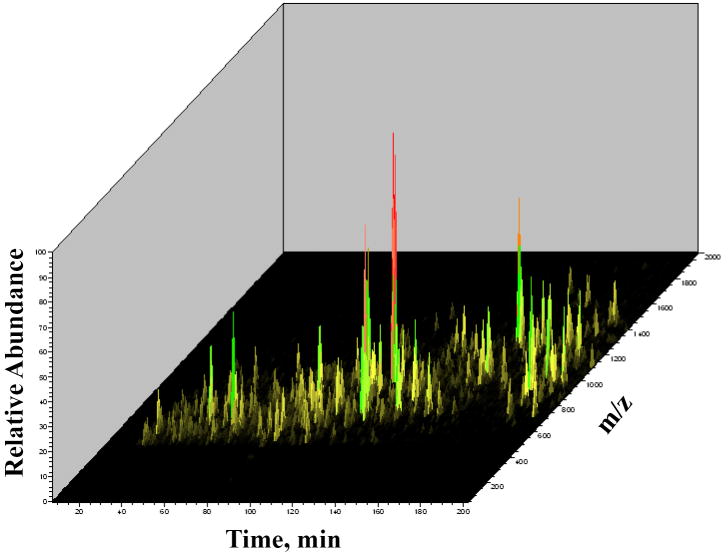
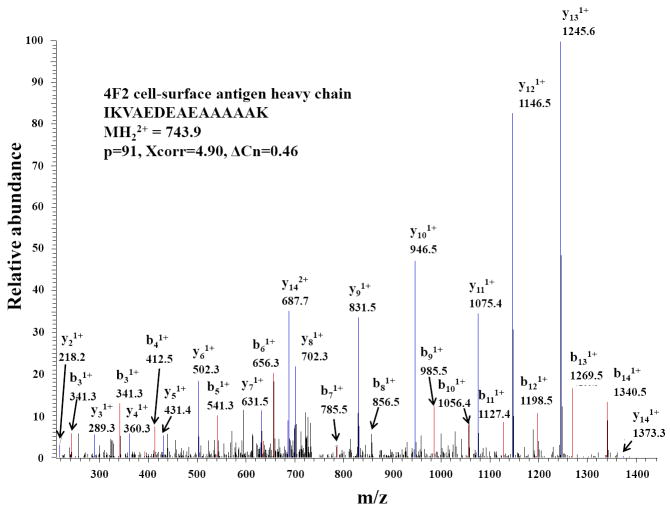
Overall, relevant to this research was the fact that the one-step LC-MS/MS analysis strategy enabled the identification of a large set, i.e., hundreds, of proteins. Important to note, that the rate of false positive identifications, while somewhat higher than in the SCX fractionated samples (as expected for a smaller data set), was preserved at values <5 %, as commonly required for the evaluation of large proteomic datasets. The rate of false positive identifications was calculated by reversing the protein sequences in the human protein database that was downloaded from SwissProt, and searching the raw MS/MS data against a forward-reversed database. Figure 1 represents a 2D view of a typical LC-MS/MS experiment performed on whole MCF-7 extracts. A uniform distribution of sample components, with no major peptide overlaps or strong peaks (typically belonging to detergents) that could compromise the quality of the analysis and ultimately the interpretation of results, is observed. A tandem mass spectrum of a representative peptide (IKVAEDEAEAAAAAK) belonging to 4F2 cell-surface antigen heavy chain protein (CD98 antigen), a putative biomarker, is provided in Figure 2. The most relevant b and y ions are marked in the spectrum. Full y-ion series and good scores (Xcorr, ΔCn and p-value) confirm the amino acid sequence of the peptide.
Figure 1.
2D-view data dependent LC-MS/MS analysis of an MCF-7 cell extract (generated with the Xcalibur 2.2 software from Thermo Electron Corp). Conditions: MCF-7 cells were cultured in E2/DMEM; Sample injection volumes were 8 μL, and the protein concentration in the sample subjected to LC-MS/MS analysis was ~1-2 μg/μL; The LC separation column was a 100 μm i.d. × 12 cm fused silica capillary packed with 5 μm Zorbax SB-C18 particles; The composition of mobile phase A was H2O:CH3CN (95:5 v/v) and of mobile phase B was H2O:CH3CN (20:80 v/v), each containing 0.01 % CF3COOH; The separation gradient, running from 0 to 100 % B, was 3 h long; The volumetric flow rate through the LC separation column was ~170-180 nL/min.
Figure 2.
Tandem mass spectrum of a representative peptide belonging to putative biomarker protein 4F2 cell-surface antigen heavy chain (CD98 antigen). Conditions are provided in the experimental section; Specific LC-MS/MS conditions are provided in Figure 1.
3.2. Reproducibility
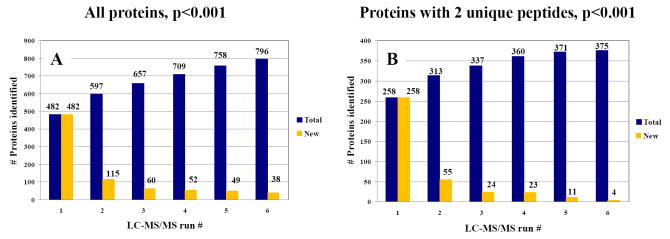
Large-scale proteomic studies that target biomarker discovery and screening rely on the capability to identify the same set of proteins over multiple LC/MS experiments. In this work, on the average, ~430 proteins were identified in each of the LC-MS/MS runs. Proper sample cleanup and an adequately chosen LC gradient were essential to enable the identification of a large number of proteins (see experimental section and prior work where the analysis of a cellular extract without cleanup and with a short LC gradient resulted in the identification of only 95 proteins [42]). The total number of proteins identified in six LC-MS/MS runs was 796 and 375, if all proteins (Figure 3A), or only proteins matched by 2 or more unique peptides were considered in the analysis (Figure 3B), respectively. In estimating the reproducibility of our data, we considered only the condition of proteins with p<0.001. For unambiguous protein identification, most often, 2 or more unique peptide matches/protein are required. The number of identified proteins increased with every single LC-MS/MS analysis (e.g, from 258 in the 1st run, to 313, 337, 360, 371 and 375 in the 2nd, 3rd, 4th, 5th and 6th runs, respectively, in Figure 3B), however, the rate of increase dropped from the first to the last replicate run, and it leveled off after five-six LC-MS/MS runs. As expected, the rate of increase was smaller for proteins matched by 2 or more unique peptides. Five-to-six replicate analyses seemed to be enough to evaluate reproducibility and reach saturation in terms of identified proteins. The typical protein overlap between two consecutive runs was >60 %, while between three runs was >50 %, regardless of using CID or PQD tandem MS detection. As a result of small changes in peptide LC retention times (<2-3 %), and peptide elution with different background ions, different peptides are selected for fragmentation during data dependent MS. Thus, from one chromatographic run to another, the total number of identified peptides, as well as the nature of peptide species and proteins that are identified, can change. However, for proteins matched by ≥2 unique peptides, the reproducibility of protein identifications between consecutive runs was higher, i.e., >90 %. We must note, though, that if a protein is not identified in every single run, does not mean that its identification is incorrect. Most likely, the protein is in lower abundance in the mix than the other proteins, and its matching peptide is not picked for tandem MS in every single run. Thus, the protein is missed. As a good tandem mass spectrum is sufficient for reliable peptide identification, proteins matched by such peptides should not be discarded. Having, however, two peptide matches per protein instead of just one, increases the likelihood of detection during data dependent analysis and helps with the identification of protein isoforms.
Figure 3.
Reproducibility of protein identifications in six replicate LC-MS/MS analyses of MCF-7 cell extracts (DMEM/E2 cultures). (A) All proteins with p<0.001 that were matched by peptides that passed the Xcorr vs. charge state filter (1.9, 2.2, 3.8) were considered in the analysis. Total unique proteins identified in six runs was 796; (B) Only proteins with p<0.001 that were matched by at least two unique peptides that passed the Xcorr vs. charge state filter (1.9, 2.2, 3.8) were considered in the analysis. Total unique proteins identified in six runs was 375. Other conditions were the same as provided in Figure 1.
3.3. Biological relevance of identified proteins
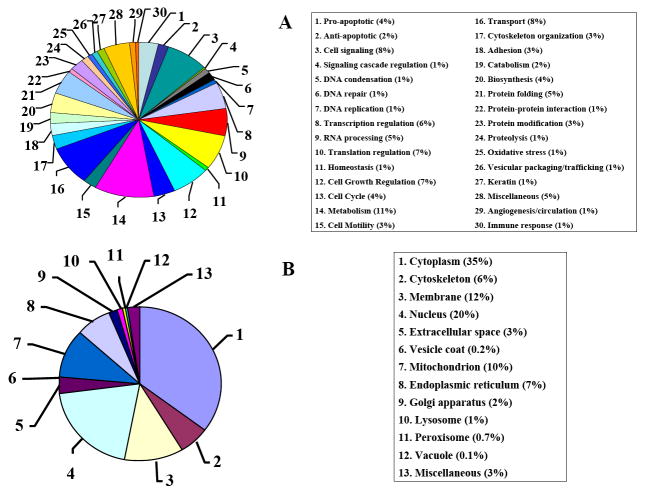
The 796 proteins that were identified in this study were categorized, based on biological process and cellular location, using the Gene Ontology identification tool (Figure 4). As displayed in Figure 4A, the major biological processes associated with the identified proteins including metabolism (88), transport (64), cell growth/regulation (56), translation/regulation (56) and cell signaling (64). Most importantly, we identified ~200 proteins with a role in essential biological processes related to cancer: 54 in apoptosis, 14 in DNA repair, 65 in cell growth regulation, 39 in cell cycle regulation and 24 in cell adhesion. As seen in Figure 4B, the majority of the identified proteins were localized in the cytoplasm (278), the nucleus (160) and the cell membrane (96). The capability to assign a biological process and cellular location for most of the identified proteins is of great importance in biomarker screening. Several proteins that have been recognized previously as putative cancer biomarkers have been also identified in this study (see next section of the manuscript). Known biomarker proteins such as cathepsin D, an aspartyl endoproteinase [44], have known cellular location (i.e., lysosome) and well defined biological function (i.e., proteolysis). In contrast, annexin A3, a protein that belongs to a structurally-related family of calcium- and phospholipid-binding proteins, has recently been proposed as a putative biomarker for lung cancer adenocarcinoma. Although dysregulation of several annexins has been linked to cancer, and multiple reports provide vast information about these proteins, information about annexin A3 is limited [45]. GO assigns the cytoplasm as the cellular location for this protein and its biological function is determined to be in cell signaling. Both assignments for annexin A3 are broad in nature, but are a step forward toward the identification of specific functions and roles for this protein in cancer.
Figure 4.
Categorization of identified proteins in MCF-7 cell extracts (DMEM/E2 cultures) by data dependent LC-MS/MS analysis. (A) Biological process; (B) Cellular location. Conditions were the same as provided in Figure 1. The pie charts were created with MS multiconsensus results generated from six LC-MS/MS runs.
3.4. Biomarker fingerprinting
Prior to performing our work, we conducted a literature search for putative biomarkers that were reported in research papers and reviews published within the recent few years. This search resulted in a list of ~1100 proteins including ~800 additional entries to the ~260 proteins previously published by Polanski. In Appendix A we list all but the proteins that were unique to the Polanski list [41]. The table provides the full protein name, the SwissProt annotation number (if reported), the types of cancer the protein has been correlated to, the sample source in which the protein was identified, the up/down regulation status of the protein in the sample, the method(s) used for the analysis of the protein, and the reference from where the information was compiled. Essential to note that many of these recently reported proteins were identified/quantified not by conventional, but novel, MS-based technologies. The proteins are listed by their name in alphabetical order, those with names starting with a number being listed first.
In the present work, the combined results of the six replicate data dependent LC-ESI-MS/MS analyses of E2 treated cells, that comprised 796 proteins (p<0.001), were compared with the list of ~1100 proteins gathered from the literature. A total of 156 putative biomarkers (from the list of ~1100) were identified in the E2 cultured MCF-7 cells (see Appendix B). Provided in the table are the SwissProt annotation number, the probability of a random match (p-value, as reported by the Bioworks software), the protein sequence coverage, the molecular weight (MW) of the protein, and the unique peptide hits for each protein. Additional information such as the type of cancer in which the protein has been identified, its up/down regulation status, its function, the methods of analysis that were previously used to identify the protein, and the corresponding references are reported. The p-value is reported as -10xlg(p). The function/biological process involvement for each protein was extracted from the ExPASy Proteomics Server website (http://www.expasy.ch/). Most of these biomarker proteins have been identified in multiple types of cancer, and 54 have been correlated with breast cancer. As reproducibility is an important aspect to take into consideration, the number of LC-MS/MS runs (out of 6) in which any given protein was identified is also provided in the table. The second data dependent experiment performed in our laboratory, that involved the analysis of iTRAQ labeled E2 and Tam treated MCF-7 cells (five LC-MS/MS runs), enabled the detection of 389 proteins (p<0.001) and 16 up/down regulated proteins with >2 fold change in expression levels. These results and the statistical corroboration of the differentially expressed proteins under E2 and Tam culture conditions are discussed in detail in a previous manuscript [46]. Relevant for this study, however, was the fact that similar to the data dependent profiling protocol, the iTRAQ experiment enabled the detection of 64 additional proteins that regardless of no change in expression level during the E2/Tam treatment, were found to be up- or down-regulated in a variety of biological experiments that targeted the discovery of cancer biomarkers. Thus, the iTRAQ method could have been used for differential expression analysis in any of these reported studies. These proteins were marked with an “*” in Appendix B.
During a biological experiment that is devised to study a particular disease, proteins will change expression levels not only as a result of the intended scope, because these proteins have relevance to the disease, but also as a result of numerous cellular processes that are inherently induced, as a side effect, by the biological treatment. To avoid the enumeration of such proteins, the candidate markers enlisted in Appendix B were searched for their biological relevance and were all found to be involved in processes with key relevance to cancer (i.e., cell proliferation, apoptosis, metastasis, tumorigenesis, DNA replication/transcription/repair, RNA processing and metabolism, etc., - function and biological process assignment was performed by GO). The ability to identify 156 candidate biomarker proteins that have been shown to change expression level by different methods, in various cell lines and cancer types, and in various biological studies, supports the idea that the one-step LC-MS/MS approach could be used effectively for a broad range of comprehensive cancer biomarker fingerprinting applications (we note that a method needs to be able of detecting/identifying first a protein, prior to quantifying it). The list that we put forward includes known markers such as cathepsin D, 14-3-3-sigma, CD54 and CD98 antigens, TCTP, profilin, stathmin, calreticulin precursor, serpinH1 precursor, as well as classes of proteins such as heat shock proteins, keratins, annexins, calcium binding proteins and peroxiredoxins. For example, peroxiredoxin-6 (P30041) is a cytosolic protein that is expressed in response to oxidative stress, and it has been shown that its up-regulation increases the proliferation and metastatic potential of breast cancer cells [47]; SerpinH1 (P50454) precursor is up-regulated in metastatic carcinomas, and it could be used for tumor prognosis [48]; TCTP (P13693, translationally controlled tumor protein) is involved in Ca2+ binding and transport, cell growth, apoptosis and allergic response [18]; 4F2hc cell-surface antigen heavy chain (P08195, CD98 antigen) is a protein that participates in normal/neoplastic cell growth, is involved in sodium-independent high affinity transport of large neutral amino acids, and has been reported to be up-regulated in tumor cells [49]; and, Cathepsin D precursor (P07339), an aspartic-type endopeptidase active in intracellular protein breakdown, has been found up-regulated in colorectal and breast cancers and was associated with cancer metastasis [41,44,50].
These results are remarkable in demonstrating the effectiveness of the data-dependent LC-MS/MS strategy for the detection of biomarker protein panels, and clearly support the potential applicability of the method for biomarker fingerprinting in complex cellular extracts. The advantages of using LC-MS/MS, as opposed to other more conventional techniques, are numerous. For instance, mass spectrometry offers sensitive detection, throughput, resolving power, as well as capability for analyte structure elucidation [2,4,6,9]. Most importantly, LC-MS/MS offers the potential for simultaneous screening for a large numbers of biomakers, to significantly outperform the sensitivity and specificity of assays that rely on a single marker. The larger the panel of biomarkers that is used to assess sensitivity and specificity, the more reliable the outcome of an assay is expected to be (we note that sensitivity refers to the probability of a test being positive in individuals that have the disease, while specificity refers to the probability of a test being negative in individuals that do not have the disease). Alternative, more conventional methods for biomarker research such as immunohistochemistry, 2D-PAGE, DIGE, Western blot, ELISA, and microarray based techniques involve complex and lengthy protocols, and, so far, did not offer yet the desirable sensitivity, specificity and reproducibility. The LC-MS/MS protocol that we have presented in this manuscript enables fingerprinting of more than 70 biomarkers in any single run. About ~72 % of the 156 putative biomarkers were identified within at least 4 runs. A total of 74 proteins were consistently identified in all 6 runs, with an additional 26 proteins identified in 5 LC-MS/MS runs. As a result, we believe that this method could bring much value in terms of sensitivity and specificity to launching MS-based screening in the clinical setting.
3.5. MRM-MS targeted analysis
A proteomic platform for cancer biomarker discovery can be regarded as a 3-step process. First, the proteomic platform should enable the identification of low-abundance proteins that could be potential markers of the disease. Next, the platform should facilitate the quantitation of putative biomarker candidates, i.e., of the proteins that were found to display different expression levels between cancerous and normal cell states. Ultimately, the platform should enable the validation of the biomarker candidates. Data-dependent MS/MS profiling strategies, as also demonstrated in this work, can be successfully utilized for protein identification and quantitation. For the detection of low abundance proteins and accurate quantitation studies, MRM-MS analysis has gained widespread popularity. Immuno-based methods such as Western blotting, and/or MRM-MS, are frequently used for the validation of data dependent MS methods. MRM-MS-based techniques are targeted screening approaches that strategically complement data-dependent analysis [51-55]. We note that while MRM-MS analysis is applicable mainly to the identification and quantitation of peptides with known MS/MS fragmentation pattern, the method offers significant improvements in detection sensitivity, reproducibility and throughput. An MRM experiment is performed by monitoring through MS only pre-selected pairs of precursor peptide ions and their characteristic product ions (i.e., selected MS transitions). The technique is highly specific, as interference from other peptide species with the same LC elution time, same m/z, and same fragment ions, is highly unlikely. Most importantly, the technique is applicable to the analysis of both, cell cultures and primary tissues. If the samples are spiked with isotope-labeled peptide standards with the same amino acid sequence as the target peptides, MRM analysis can be successfully applied not only for targeted identification, but also for absolute and relative quantitation of low abundant components in complex samples.
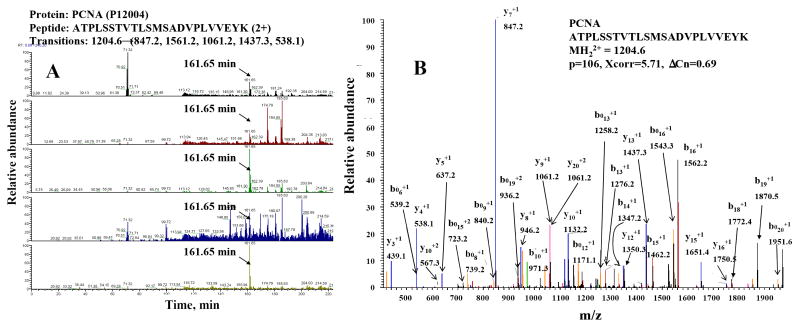
The applicability of MRM-MS for the detection of biomarkers in such complex peptide mixtures was investigated for sets of 5-50 proteins, by setting up transitions for 2-4 peptides per protein. Most of these proteins were searched by peptides that typically could not be identified by data dependent analysis of un-fractionated, whole cellular extracts, which is an indicator of these proteins being present in much lower abundance in the cell than other proteins that could be identified by data dependent MS. MRM-MS transitions for such peptides were selected based on the peptide fragmentation patterns that were generated in the SCX pre-fractionation experiment that enabled the detection of 1859 proteins (see Table 1). We note, however, that while it is preferable to have experimental peptide fragmentation data, machine learning algorithms can be developed to enable the theoretical prediction of such fragmentation patterns. The outcome of an MRM-MS experiment is provided for PCNA (proliferating cell nuclear antigen), which is a protein with a role in the regulation of cell cycle progression and DNA replication that is frequently used as a marker associated with the proliferative capacity of cancer cells. Extracted ion chromatograms (EIC) for selected transitions of a PCNA peptide (ATPLSSTVTLSMSADVPLVVEYK) are provided in Figure 5A. The strategy that we implemented for MRM analysis involved monitoring the top five most intense transitions for a peptide with known fragmentation pattern. The more transitions are monitored for a specific peptide, the narrower the m/z window for EIC generation and the narrower the time window for monitoring the MRM transition during the LC elution of the peptide of interest, the more sensitive and specific the analysis can be. If the precise elution time of a peptide is not known, transitions can be monitored for the entire length of LC analysis, as shown in Figure 5A. If using such conditions, the specificity of analysis can be ensured by using five or more transitions for each peptide. In the case of the PCNA peptide, all transitions displayed the presence of the fragment ion at the LC elution time of the parent peptide (161.65 min), confirming its presence in the extract. The tandem mass spectrum for this peptide, as generated by the SCX-LC-MS/MS experiment, illustrates the most intense fragment ions that were used for MRM identification (Figure 5B). Our typical detection limits in complex cellular extracts, when using data dependent analysis, were ~4 fmol injected on the LC column. As PCNA was not detectable in the whole extract without SCX pre-fractionation, we estimate that its concentration in the extract analyzed by data dependent MS and MRM-MS was below 0.5 nM (corresponding to 4 fmol/8 μL injection).
Figure 5.
MRM-MS based detection of a putative biomarker peptide belonging to PCNA (ATPLSSTVTLSMSADVPLVVEYK) that is not identifiable in the MCF-7 cell extract by data dependent analysis. Conditions: Whole MCF-7 cellular tryptic digests were analyzed by a 3 h long LC-MS/MS gradient; Specific LC-MS/MS conditions are provided in Figure 1; MRM/CID conditions are provided in the experimental section. (A) EICs that reflect five characteristic transitions at the elution time of the peptide; (B) Tandem mass spectrum of the peptide (as generated by SCX-LC-MS/MS).
4. CONCLUSIONS
Data-dependent RPLC-ESI-MS/MS proteomic strategies have been investigated for biomarker fingerprinting in cancerous cells, and demonstrated for the simultaneous detection of up to ~800 (p<0.001) proteins and 156 putative biomarkers. The reproducibility of biomarker identification within three consecutive LC-MS/MS runs was better than 72 %. These technologies are applicable to both qualitative and quantitative screening and/or discovery applications. Targeted MRM-MS methods can successfully complement data dependent approaches to increase throughput and detect low abundance proteins in complex cellular extracts. As the panel of 156 putative biomarkers was collected from literature data reflecting various experimental conditions, biological treatments, cell lines and methods/detection systems (all reporting the identification of only a few markers), the ability to simultaneously identify such a set of proteins by using a simple LC-MS/MS protocol will enable researchers to devise fast, but comprehensive experiments with utility for a broad variety of cancer-relevant studies. In comparison to conventional methods for biomarker screening, the LC-MS/MS technologies reported in this manuscript exhibit a number of advantages in terms of sensitivity, specificity, speed of analysis and cost, and could open new venues for the unambiguous detection, differential expression analysis and validation of panels of biomarkers. Overall, the results presented in this manuscript demonstrate the impact that LC-MS/MS technology will have in early disease detection, therapy and cancer research.
Supplementary Material
Acknowledgments
This project was partially supported by Award Number R21CA126669-01A1 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Footnotes
AUTHORS’ CONTRIBUTIONS Data acquisition and processing: JMA, MP, XY, DS; GO charts: DS and MP; literature search: DR; table assembly: everyone. IML conceived the study, coordinated the work and drafted the final version of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jenny M. Armenta, Email: jmab06@vbi.vt.edu.
Milagros Perez, Email: miperez@vbi.vt.edu.
Xu Yang, Email: yangx@vbi.vt.edu.
Danielle Shapiro, Email: shapirod@vt.edu.
Debby Reed, Email: dgreed@vbi.vt.edu.
Leepika Tuli, Email: ltuli@vbi.vt.edu.
Carla V. Finkielstein, Email: finkielc@vt.edu.
Iulia M. Lazar, Email: lazar@vbi.vt.edu.
References
- 1.Smith L, Lind MJ, Welham KJ, Cawkwell L. Cancer. 2006;107:232. doi: 10.1002/cncr.22000. [DOI] [PubMed] [Google Scholar]
- 2.Kumar S, Mohan A, Guleria R. Biomarkers. 2006;11:385. doi: 10.1080/13547500600775011. [DOI] [PubMed] [Google Scholar]
- 3.He Q-Y, Chiu J-F. J Cell Biochem. 2003;89:868. doi: 10.1002/jcb.10576. [DOI] [PubMed] [Google Scholar]
- 4.Srinivas PR, Verma M, Zhao Y, Srivastava S. Clin Chem. 2002;48:1160. [PubMed] [Google Scholar]
- 5.Vlahou A, Fountoulakis M. J Chromatogr B. 2005;814:11. doi: 10.1016/j.jchromb.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 6.Lu M, Faull KF, Whitelegge JP, He J, Shen D, Saxton RE, Chang HR. Biomark Insights. 2007;2:347. [PMC free article] [PubMed] [Google Scholar]
- 7.Simpson RJ, Bernhard OK, Greening DW, Moritz RL. Curr Opin Chem Biol. 2008;12:72. doi: 10.1016/j.cbpa.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 8.McDonald WH, Yates JR. Dis Markers. 2002;18:99. doi: 10.1155/2002/505397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo T, Lee CS, Wang W, DeVoe DL, Balgley BM. Electrophoresis. 2006;27:3523. doi: 10.1002/elps.200600094. [DOI] [PubMed] [Google Scholar]
- 10.Smith JC, Lambert J-P, Elisma F, Figeys D. Anal Chem. 2007;79:4325. doi: 10.1021/ac070741j. [DOI] [PubMed] [Google Scholar]
- 11.Duncan MW, Hunsucker SW. Exp Biol Med. 2005;230:808. doi: 10.1177/153537020523001105. [DOI] [PubMed] [Google Scholar]
- 12.Sawyers CL. Nature. 2008;452:548. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 13.Shau H, Chandler GS, Whitelegge JP, Gornbein JA, Faull KF, Chang HR. Brief Funct Genomic Proteomic. 2003;2:147. doi: 10.1093/bfgp/2.2.147. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Wei D, Yap Y, Li L, Guo S, Chen F. Mass Spectrom Rev. 2007;26:403. doi: 10.1002/mas.20132. [DOI] [PubMed] [Google Scholar]
- 15.Liu HB, Sadygov RG, Yates JR. Anal Chem. 2004;76:4193. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 16.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Mol Cell Proteomics. 2005;4:1487. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Julka S, Regnier F. J Proteome Res. 2004;3:350. doi: 10.1021/pr0340734. [DOI] [PubMed] [Google Scholar]
- 18.Patwardhan AJ, Strittmatter EF, Camp DG, II, Smith RD, Pallavicini MG. Proteomics. 2006;6:2903. doi: 10.1002/pmic.200500582. [DOI] [PubMed] [Google Scholar]
- 19.Schneider LV, Hall MP. Drug Discov Today. 2005;10:353. doi: 10.1016/S1359-6446(05)03381-7. [DOI] [PubMed] [Google Scholar]
- 20.Fenselau C. J Chromatogr B. 2007;855:14. doi: 10.1016/j.jchromb.2006.10.071. [DOI] [PubMed] [Google Scholar]
- 21.Lill J. Mass Spectrom Rev. 2003;22:182. doi: 10.1002/mas.10048. [DOI] [PubMed] [Google Scholar]
- 22.Thiede B, Kretschmer A, Rudel T. Proteomics. 2006;6:614. doi: 10.1002/pmic.200500120. [DOI] [PubMed] [Google Scholar]
- 23.Everley PA, Krijgsveld J, Zetter BR, Gygi SP. Mol Cell Proteomics. 2004;3:729. doi: 10.1074/mcp.M400021-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Nat Biotech. 1999;17:994. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 25.Ross PL, Huang YLN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Mol Cell Proteomics. 2004;3:1154. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.DeSouza L, Diehl G, Rodrigues MJ, Guo JZ, Romaschin AD, Colgan TJ, Siu KWM. J Proteome Res. 2005;4:377. doi: 10.1021/pr049821j. [DOI] [PubMed] [Google Scholar]
- 27.Zieske LR. J Exp Bot. 2006;57:1501. doi: 10.1093/jxb/erj168. [DOI] [PubMed] [Google Scholar]
- 28.Hanash SM, Pitteri SJ, Faca VM. Nature. 2008;452:571. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 29.Horvatovich P, Govorukhina N, Bischoff R. The Analyst. 2006;131:1193. doi: 10.1039/b607833h. [DOI] [PubMed] [Google Scholar]
- 30.Veenstra TD. J Chromatogr B. 2007;847:3. doi: 10.1016/j.jchromb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 31.Listgarten J, Emili A. Drug Discov Today. 2005;10:1697. doi: 10.1016/S1359-6446(05)03645-7. [DOI] [PubMed] [Google Scholar]
- 32.Zolg W. Mol Cell Proteomics. 2006;5:1720. doi: 10.1074/mcp.R600001-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, Chan DW. Cancer Epidemiol Biomarkers Prev. 2005;14:2283. doi: 10.1158/1055-9965.EPI-05-0774. [DOI] [PubMed] [Google Scholar]
- 34.Qian W-J, Jacobs JM, Liu T, Camp DG, II, Smith RD. Mol Cell Proteomics. 2006;5:1727. doi: 10.1074/mcp.M600162-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lescuyer P, Hochstrasser D, Rabilloud T. J Proteome Res. 2007;6:3371. doi: 10.1021/pr0702060. [DOI] [PubMed] [Google Scholar]
- 36.Marko-Varga GA, Nilsson J, Laurell T. Electrophoresis. 2004;25:3479. doi: 10.1002/elps.200406109. [DOI] [PubMed] [Google Scholar]
- 37.Nedelkov D, Kiernan UA, Niederkofler EE, Tubbs KA, Nelson RW. Mol Cell Proteomics. 2006;5:1811. doi: 10.1074/mcp.R600006-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Azad NS, Rasool N, Annunziata CM, Minasian L, Whiteley G, Kohn EC. Mol Cell Proteomics. 2006;5:1819. doi: 10.1074/mcp.R600008-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Ahmed FE. Curr Genomics. 2006;7:399. [Google Scholar]
- 40.Hu S, Loo JA, Wong DT. Proteomics. 2006;6:6326. doi: 10.1002/pmic.200600284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polanski M, Anderson NL. Biomark Insights. 2006;1:1. [PMC free article] [PubMed] [Google Scholar]
- 42.Sarvaiya HA, Yoon JH, Lazar IM. Rapid Commun Mass Spectrom. 2006;20:3039. doi: 10.1002/rcm.2677. [DOI] [PubMed] [Google Scholar]
- 43.Griffin TJ, Xie HW, Bandhakavi S, Popko J, Mohan A, Carlis JV, Higgins L. J Proteome Res. 2007;6:4200. doi: 10.1021/pr070291b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garcia M, Platet N, Liaudet E, Laurent V, Derocq D, Brouillet J-P, Rochefort H. Stem Cells. 1996;14:642. doi: 10.1002/stem.140642. [DOI] [PubMed] [Google Scholar]
- 45.Liu YF, Xiao ZQ, Li MX, Li MY, Zhang PF, Li C, Li F, Chen YH, Yi H, Yao HX, Chen ZC. J Pathol. 2008;9999 doi: 10.1002/path.2429. n/a. [DOI] [PubMed] [Google Scholar]
- 46.Armenta JM, Hoeschele I, Lazar IM. J Am Soc Mass Spectrom. 2009;20:1287. doi: 10.1016/j.jasms.2009.02.029. [DOI] [PubMed] [Google Scholar]
- 47.Chang X-Z, Li D-Q, Hou Y-F, Wu J, Lu J-S, Di G-H, Jin W, Ou Z-L, Shen Z-Z, Shao Z-M. Breast Cancer Res. 2007;9:R76. doi: 10.1186/bcr1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morino M, Tsuzuki T, Ishikawa Y, Shirakami T, Yoshimura M, Kiyosuke Y, Matsunaga K, Yoshikumi C, Saijo N. In Vivo. 1997;11:17. [PubMed] [Google Scholar]
- 49.Bröer S, Bröer A, Hamprecht B. Biochem J. 1995;312:863. doi: 10.1042/bj3120863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alldridge L, Metodieva G, Greenwood C, Al-Janabi K, Thwaites L, Sauven P, Metodiev M. J Proteome Res. 2008;7:1458. doi: 10.1021/pr7007829. [DOI] [PubMed] [Google Scholar]
- 51.Sandhu C, Hewel JA, Badis G, Talukder S, Liu J, Hughes TR, Emili A. J Proteome Res. 2008;7:1529. doi: 10.1021/pr700836q. [DOI] [PubMed] [Google Scholar]
- 52.Anderson L, Hunter CL. Mol Cell Proteomics. 2006;5:573. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 53.Lin S, Shaler TA, Becker CH. Anal Chem. 2006;78:5762. doi: 10.1021/ac060613f. [DOI] [PubMed] [Google Scholar]
- 54.Unwin RD, Griffiths JR, Leverentz MK, Grallert A, Hagan IM, Whetton AD. Mol Cell Proteomics. 2005;4:1134. doi: 10.1074/mcp.M500113-MCP200. [DOI] [PubMed] [Google Scholar]
- 55.Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. PNAS. 2007;104:5860. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.