Abstract
We studied differential splicing of nebulin, a giant filamentous F-actin binding protein (Mr ~700–800 kDa) that is found in skeletal muscle. Nebulin spans the thin filament length, its C-terminus is anchored in the Z-disc, and its N-terminal region is located toward the thin filament pointed end. Various lines of evidence indicate that nebulin plays important roles in thin filament and Z-disc structure in skeletal muscle. In the present work we studied nebulin in a range of muscle types during postnatal development and performed transcript studies with a mouse nebulin exon microarray, developed by us, whose results were confirmed by RT-PCR. We also performed protein studies with high-resolution SDS-agarose gels and Western blots, and structural studies with electron microscopy. We found during postnatal development of the soleus muscle major changes in splicing in both the super-repeat region and the Z-disc region of nebulin; interestingly, these changes were absent in other muscle types. Three novel Z-disc exons, previously described in the mouse gene, were upregulated during postnatal development of soleus muscle and this was correlated with a significant increase in Z-disc width. These findings support the view that nebulin plays an important role in Z-disc width regulation. In summary, we discovered changes in both the super-repeat region and the Z-disc region of nebulin, that these changes are muscle-type specific, and that they correlate with differences in sarcomere structure.
Introduction
Major changes in the structural and functional characteristics of skeletal muscle take place during postnatal development, as reflected by the altered expression of isoforms of many of the sarcomeric proteins (1). Recently it was shown that during postnatal development of skeletal muscle, passive stiffness increases through a decrease in titin isoform size, due mainly to a marked restructuring of its spring region (2). Because it is likely that the function of titin is coordinated with that of nebulin, we focused here on nebulin expression during postnatal development of various skeletal muscles of the mouse.
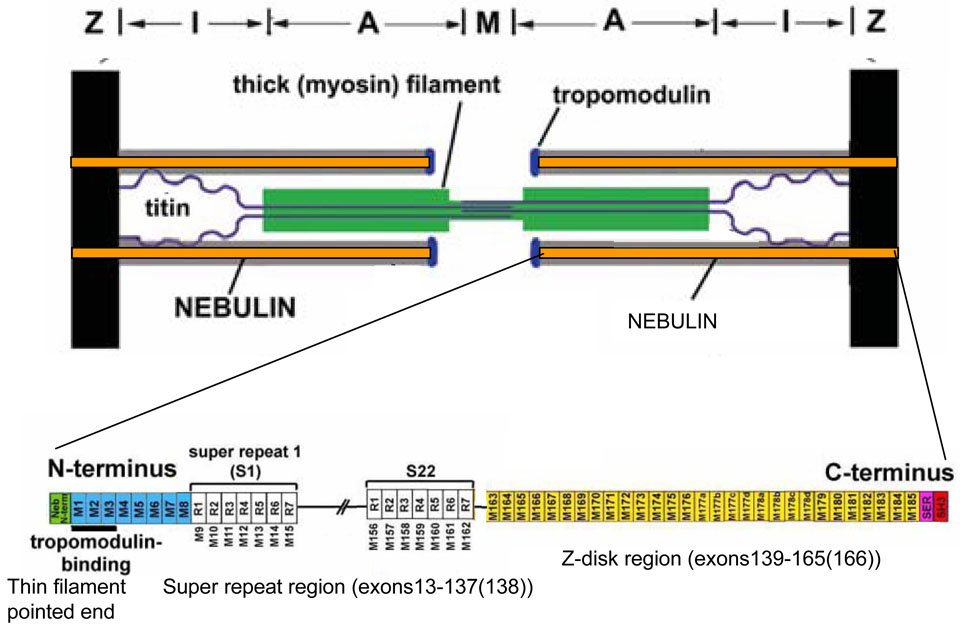
Nebulin is a giant filamentous F-actin binding protein (Mr ~700–800 kDa) that is one of the least well understood major muscle proteins (3,4). Nebulin spans the thin filament length, with its C-terminus anchored in the Z-disc, and its N-terminal region located toward the thin filament pointed end (5,6). The majority of nebulin’s sequence is comprised of ~35 aa modules, with the central M9 through M162 modules arranged into seven-module super-repeats (figure 1) (7–10). This arrangement enables a single nebulin module to interact with a single actin monomer, and each nebulin super-repeat to associate with a single Tropomyosin(Tm)/Troponin(Tn) complex (4,11–13). Evidence for a role for nebulin in establishing thin filament length was obtained in earlier studies that revealed that the electrophoretic mobility of nebulin from different muscle types correlates with thin filament length (5,6). The subsequent analysis of nebulin’s cDNA sequence corroborated this notion because it revealed that the bulk of the molecule is comprised of modules that are organized into super-repeats that match the repeat of the actin filament (8). Further support was obtained in studies on nebulin knockout (KO) mice that had thin filament lengths that were shorter than normal (14,15).
Figure 1.
Top: Layout of nebulin in the sarcomere. Bottom: domain composition of nebulin.
Immunoelectron-microscopy has shown that the C-terminal ~30 kDa of nebulin integrates into the Z-disc lattice and c-DNA sequencing of rabbit nebulin has demonstrated that the Z-disc region of nebulin is differentially expressed (16). Several different isoforms were identified that result from the skipping of various combinations of the modules M176 to M182 (16). It has therefore been suggested that nebulin is one of several proteins that are important for Z-disc width regulation (16).
Recent work revealed that mutations in the nebulin gene are a common cause of nemaline myopathy (NM), accounting for ~50% of all NM cases (17), further supporting the importance of nebulin in muscle function. We have shown that similar to the nebulin KO mouse model, human NM patients with nebulin-deficiency also have shorter and non-uniform thin filament lengths, and that this can partly account for the observed muscle weakness in nebulin-based NM (18–21). Considering the evidence of nebulin's importance in thin filament and Z-disc structure in skeletal muscle, and the possibility for extensive differential splicing, we studied in the present work nebulin during postnatal development, using a range of muscle types. We performed transcript studies with a novel mouse nebulin exon microarray, validated results with quantitative real time PCR (qPCR), carried out protein studies with high-resolution SDS-agarose gels and Western blots, and performed structural studies with electron microscopy.
Methods
Tissue harvesting
BL6 mice of various ages were anesthetized using isoflurane and the m. soleus (SOL), tibialis cranialis (TC), extensor digitorum longus (EDL), gastrocnemius (GAST), quadriceps (QUAD), and diaphragm (DIAPH) were rapidly dissected. For microarray and qPCR studies, muscles were dissected and stored in RNAlater (Ambion Inc., Austin, TX, USA); for gel-electrophoresis studies, muscles were quick-frozen in liquid nitrogen and stored at −80°C. For electron microscopy studies the soleus and TC were rapidly dissected in oxygenated HEPES (NaCl, 133.5 mM; KCl, 5mM; NaH2PO4, 1.2mM; MgSO4, 1.2mM; HEPES, 10mM). The tissue was skinned in relaxing solution (BES 40 mM, EGTA 10 mM, MgCl2 6.56 mM, ATP 5.88 mM, DTT 1 mM, K-propionate 46.35 mM, creatine phosphate 15 mM, pH 7.0) (chemicals from Sigma-Aldrich, MO, USA) with 1% Triton-X-100 (Pierce, IL, USA) overnight at 4°C. Muscles were then washed thoroughly with relaxing solution and stored at −20°C in relaxing solution containing 50% (v/v) glycerol. All solutions contained protease inhibitors (phenylmethylsulfonyl fluoride (PMSF), 0.5 mM; Leupeptin, 0.04 mM; E64, 0.01 mM). All experiments were approved by IACUC and followed the NIH Guidelines “Using Animals in Intramural Research” for animal use.
Gel-electrophoresis
SDS-agarose electrophoresis was performed as previously described (22,23). Briefly, muscle samples were solubilized in a urea and glycerol buffer and analyzed by vertical SDS-agarose electrophoresis. The 1% agarose gels were run at 15mA per gel for 3 hours and 20 minutes. The gels were stained with coomassie brilliant blue (CBB), and subsequently scanned and analyzed using One-D scan EX (Scanalytics Inc., Rockville, MD, USA) software. To determine the molecular weights of nebulin isoform and stoichiometry, samples of interest were co-electrophoresed together with titin, nebulin and myosin heavy chain (MHC) of known sizes from human soleus (titin 3.82 MDa; nebulin: 773kDa, MHC 223kDa (top MHC seen on the agarose gels) and human left ventricle (N2B titin 2.97 MDa) samples. Plotting the logarithm of the molecular weight vs. migration distance on the gel yields a linear relationship, as shown previously(24,25), and allowed us to estimate the Mw of the proteins of interest from their migration distance (see Figure 2 for further explanation). The integrated optical density of nebulin and MHC was determined as a function of the volume of solubilized protein sample that was loaded (a range of volumes was loaded on each gel). The slope of the linear range of the relationship between integrated optical density and loaded volume was obtained for each protein. To verify the nebulin mobility differences that were found, samples were fluorescently labeled using Amersham CyDye DIGE Fluors (GE healthcare). Two samples of interest, with different fluorescent labels, were run side-by-side as well as mixed on a 1% agarose gel; the gel was scanned using a Typhoon 9400 Scanner (GE Healthcare).
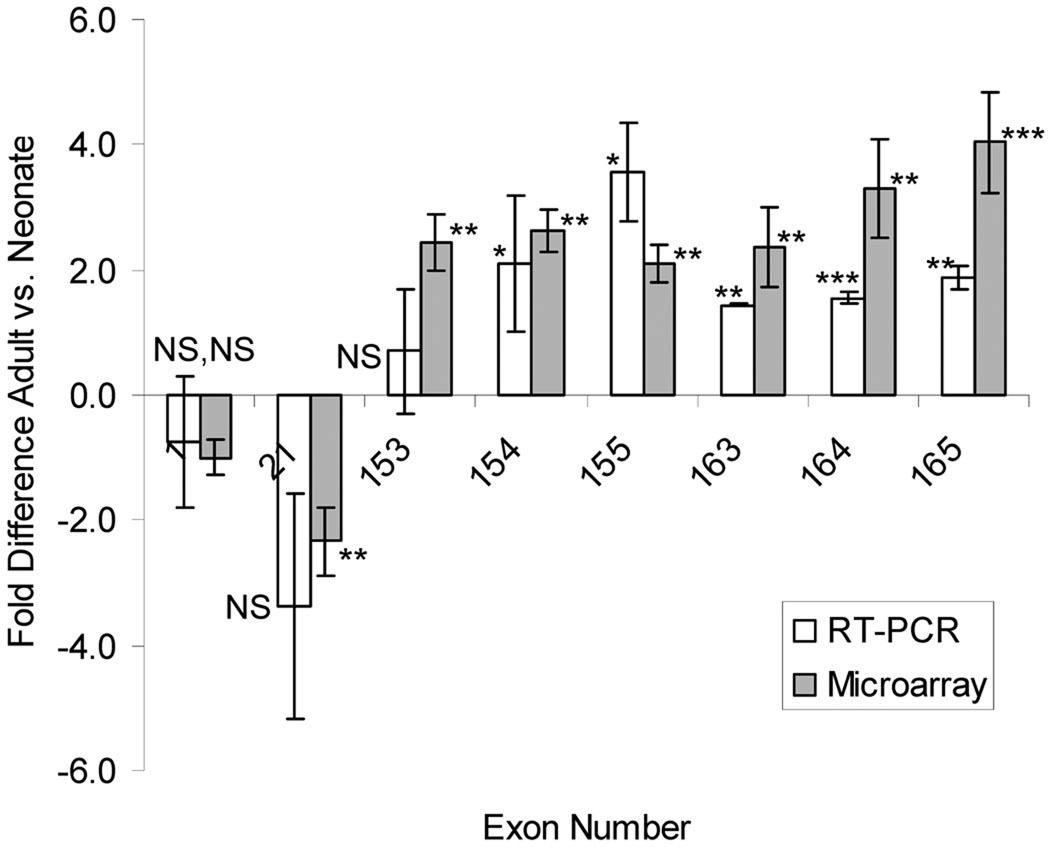
Figure 2.
qPCR experiments to validate microarray results. Exon 1 and 21 were selected for qPCR analysis because microarray analysis revealed no difference (exon 1) or down regulation in the adult vs. neonate (exon 21). Exons encoding the z-disc repeat (153–155), serine rich (exon 163 and 164), and SH3 (exon 165) domains were chosen because the microarray had revealed upregulation in the adult. Results with the two techniques are similar. (n=3 for neonate and adult).
Western Blot
For Western blotting of nebulin’s M176-M181 (exons 151–158), soleus samples were run on 1% agarose gels, and transferred to PVDF membrane using a semi-dry transfer unit (Bio-Rad, Hercules, CA). The blots were stained with PonceauS to visualize total transferred protein. The blots were then probed with rabbit polyclonal antibodies against M176-M181 which corresponds to exons 151–158 and antibodies against the serine rich domain exon 164 (9,16) To normalize for loading differences, M176-M181 labeling was normalized to total nebulin protein, determined from the PonceauS-stained membrane. Secondary antibodies conjugated with fluorescent dyes with infrared excitation spectra were used for detection. One-color IR western blots were scanned (Odyssey Infrared Imaging System, Li-Cor Biosciences, NE. USA) and the images analyzed with One-D scan EX.
Microarray studies
We dissected mouse soleus and TC muscles from neonatal (day 5) and adult (typically day 100). Due to the small size of the soleus muscle it was not practical to use mice younger than 5 day. We pooled muscles from 6 mice and did this 4 times (total number of mice 24) to provide us 4 independent pools per age group for soleus and 4 pools for TC. The microarray experiments were performed as described previously (2). Briefly, RNA was isolated using the Qiagen RNeasy Fibrous Tissue Mini Kit. RNA was amplified using the SenseAmp kit from Genisphere and Superscript III reverse transcriptase enzyme from Invitrogen. Reverse transcription and dye coupling (Alexa Fluor 555 and Alexa Fluor 647 were used) was done using Invitrogen’s superscript plus indirect cDNA labeling module. Half of each sample was incorporated with Alexa Fluor 555 and the other with Alexa Fluor 647. All the mouse nebulin (50mer oligonucleotides) were spotted in triplicate on Corning Ultra GAPS glass slides. Note that the arrays were made prior to the discovery by Donner et al(26) of a new exon, and that this exon is therefore not represented on the microarray. Donner et al. (26) numbered this exon 127 and incremented all upstream exon numbers by one. In this work we first show the old exon numbering based on Kazmierski et al (9)and in parenthesis the new numbers based on Donner et al. Slides were then baked at 90°C for 90 minutes and stored in a nonelectric desiccator. For each sample 750ng of cDNA (Nanodrop, Thermo scientific) from each sample was hybridized (Ambion: Slide-Hyb buffer #1) for 16 hours at 42°C after which slides were scanned at 595 nm and 685 nm with an Array WoRx scanner. Spot finding was done with SoftWoRx Tracker and spot analysis with CARMA (27). The analysis detects relative changes in the fluorescence of a probe (adult as compared to neonate). These changes are represented as a fold-difference and reflect the ratio of adult exon expression to neonate exon expression. A positive fold change (shown in the Results in green) indicates an increase in expression in adult relative to the neonate. A negative fold change (shown in the Results in red) indicates an increase in expression in neonate relative to the adult sample. To calculate the predicted molecular weight difference caused by the up/down regulated exons we assumed that the exon is expressed in both neonate and adult but at different levels, i.e., some of the nebulin molecules in a particular sample have the exon expressed and others do not (Western blots studies confirm this assumption, see Results). We set the expression level at 100% for the sample with the highest expression and calculated the expected molecular weight difference accordingly. For example, an exon that is two-fold upregulated in the adult and that encodes a 4 kDa protein results in a predicted molecular weight increase in the adult of 2 kDa, (i.e., 1.0 × 4 kDa – 0.5 × 4 kDa). (In contrast, if the exon were to be ‘on-off’ it would cause a molecular weight difference of 4 kDa.) Table 1 shows both the ‘on-off change in molecular weight and the predicted change in molecular weight following the calculation explained above.
Table 1.
Transcript analysis in soleus and TC muscle. Adult expression is shown relative to that of the neonate with exons that are up-regulated shown in green (and with positive values) and exons that are down-regulated in red (and with negative values). The protein modules encoded by the exons colored in correspondence to the regions of nebulin in Figure 1 bottom; their molecular weights are shown in kDa. Nomenclature adapted from (37) and(9). (Exon numbers in parenthesis are according to (26), see Methods for details.) The expected differences in the average molecular weight of adult nebulins is calculated based on the fold difference (See Methods for details). According to these results, the nebulin transcript is larger in the adult than in the neonate and is predicted to result in a nebulin protein that is 13.1 kDa larger in the soleus and 3.6 kDa in the TC.
| SOLEUS | ||||
|---|---|---|---|---|
| Exon # | Encodes | Mw (kDa) | Fold | Expected Contrbution to Mw (kDa) Ad vs neonate |
| 8 | M3/M4 | 4.08 | 2.199 | 2.23 |
| 11 | M6/M7 | 4.11 | 2.063 | 2.12 |
| 13 | M8/S1R1 | 4.41 | −2.199 | −2.40 |
| 14 | S1 R1/R2 | 4.20 | −2.301 | −2.38 |
| 18 | S1 R5/R6 | 4.08 | −2.180 | −2.21 |
| 19 | S1 R6/R7 | 4.04 | −2.324 | −2.30 |
| 21 | S2 R1/R2 | 4.86 | −2.336 | −2.78 |
| 25 | S2 R5/R6 | 4.02 | −2.166 | −2.16 |
| 26 | S2 R6/R7 | 4.04 | −2.053 | −2.07 |
| 33 | S3 R7/ S4 R1/R2/R3 | 12.33 | −2.136 | −6.56 |
| 45 | S6 R7/ S7/R1/R2/R3 | 12.12 | 2.191 | 6.59 |
| 133 (134) | S22 R3/R4 | 4.09 | 2.031 | 2.08 |
| 134 (135) | S22 R4/R5 | 3.92 | 2.098 | 2.05 |
| 135 (136) | S22 R5/R6 | 4.32 | 2.527 | 2.61 |
| 136 (137) | S22 R6/R7 | 4.33 | 2.101 | 2.27 |
| 137 (138) | S22 R7/M163 | 3.79 | 2.000 | 1.90 |
| 138 (139) | M163/M164 | 3.91 | 2.307 | 2.22 |
| 145 (146) | M170/M171 | 4.34 | −2.336 | −2.48 |
| 146 (147) | M171/M172 | 3.96 | −2.213 | −2.17 |
| 153 (154) | M177/M178b | 3.58 | 2.428 | 2.10 |
| 154 (155) | M177/M178c | 3.55 | 2.617 | 2.19 |
| 155 (156) | M177/M178d | 3.48 | 2.084 | 1.81 |
| 163 (164) | M185/Ser | 5.31 | 2.373 | 3.07 |
| 164 (165) | Ser | 3.82 | 3.309 | 2.67 |
| 165 (166) | SH3 | 6.32 | 4.034 | 4.75 |
|
| ||||
| Cummulative Difference: | 13.1 kDa | |||
| TC | ||||
| Exon # | Encodes | Mw (kDa) | Fold | Expected Contrbution to Mw (kDa) 5d/Ad |
| 127 (128) | S21 R5/R6 | 4.16 | 7.465 | 4.72 |
| Cummulative Difference: | 3.6 kDa | |||
RT-PCR
Primers were designed to single exons of nebulin. Other than a smaller product size range of 60bp to 100bp, the standard settings were used with Primer3 software (Code available at http://www-genome.wi.mit.edu/genome_software/other/primer3.html.). Amplified RNA (prepared in the same way as microarray) was converted to cDNA using SuperScript™ III First-Strand Synthesis System (Invitrogen). The cDNA was used in the qPCR reaction kit, Maxima SYBR Green qPCR (Fermentas), and placed in the Roto Gene Q machine (Qiagen). Roto Gene 6000 software (Qiagen) was used to process the qPCR reactions and results. The standard software settings were used for all reactions and GAPDH was used as a reference to normalize each reaction set.
Electron Microscopy
Skinned skeletal muscle bundles were stretched (at ~0.25 µm/sarcomere) in relaxing solution to 120% of their original length, held in that state for 5 minutes and then fixed, embedded, and processed for electron microscopy as explained earlier (28,29). Briefly, the samples were fixed in 3% paraformaldehyde/PBS solution for 30 minutes at 4°C, then washed in PBS plus inhibitors for 3 × 15 minutes at 4°C. The samples were then fixed in 3% glutaraldehyde + 2% tannic acid in PBS for 1 hour at 4°C, rinsed in PBS 3 times for 5 minutes each, then washed in PBS + inhibitors overnight again at 4°C. The next morning, samples were postfixed in 1% OsO4 in PBS for 3 minutes at 4°C and then washed and embedded in araldite. Sections were cut using a Leica microtome, stained with 2% potassium permanganate and lead citrate, and imaged at 88,000–110,000X using a Phillips CM-12 electron microscope. Z-disc width measurements were made by importing the Z-disc image into ImageJ and generating a histogram plot of the grey value versus the distance in pixels. The histogram was then fit by a Gaussian curve in a program created in LabView. The full width at the baseline of the Gaussian curve was determined to be the Z-disc width.
Statistics
Data are presented as mean ± SEM. Significance was defined using the Student’s t-test and probability values <0.05 were taken as significant and are indicated on Figures as: * p<0.05; ** p<0.01; *** p<0.001. ANOVA with a linear model that incorporates terms to account for experimental variability was used in analysis of microarray data (for details see(27)).
Results
Recently it was discovered that large changes take place in transcript splicing of the sarcomeric protein titin (2). Like titin, nebulin is a candidate for functional modification through differential splicing (the nebulin gene contains 166 exons in mouse (9) and 183 in human (10)); here we address whether changes in nebulin due to differential splicing take place during neonatal development (this is when extensive changes in splicing takes place in titin). For this work we developed a microarray which represents all of the murine nebulin exons. The layout of nebulin in the sarcomere is shown in figure 1. (The color coding of the various regions in the schematic are carried over to colored areas of Table 1, for easier identification.) We first studied the soleus muscle and compared 5 day old neonatal with adult mice. The array included probes that represent the nebulin binding proteins, CapZ, desmin, myopalladin and tropomodulin (both tropomodulin 1 and tropomodulin 4). None of these were significantly up/down regulated. Of the nebulin exons, we observed 25 exons with significantly different expression levels expression levels all of which were >2 fold different between neonatal and adults (table 1). This indicates that the adult expression relative to the neonate expression is 2 or more times greater (indicated by positive values, exons are highlighted in green) or 2 or more times less (exons indicated in red). The differential exons appear to be grouped in clusters with a cluster of exons down regulated in the adult, demarcated by exons 13 and 26, encoding exons for super repeat 1 and 2, and several clusters up regulated in the adult, exons 133 – 138(139), exons 153(154)-155(156), and 163(164)-165(166), encoding super repeat 22, Z-disc repeats, and serine rich/SH3 domains respectively (Table 1). (The first shown exon number is based on Kazmierski et al (9) and the number in parenthesis is based on Donner et al. (26), see Methods for details).
qPCR was used to validate representative results obtained in the microarray studies. (To keep the experiments manageable we studied 8 exons, representing exons that were unchanged, reduced, or increased on the microarray). Obtained results were overall similar to those obtained with the microarray (Figure 2). Of the tested exons, upregulated in the adult were exons 154(155), 155(156), 163(164), 164(165), and 165(166). We further verified these findings at the protein levels by performing Western Blot studies with an antibody raised against nebulin repeats M176–181 (encoded by Z-disc repeat exons) and the serine-rich domain. Results reveal significant upregulation in the adult (Fig. 3), consistent with the transcript findings.
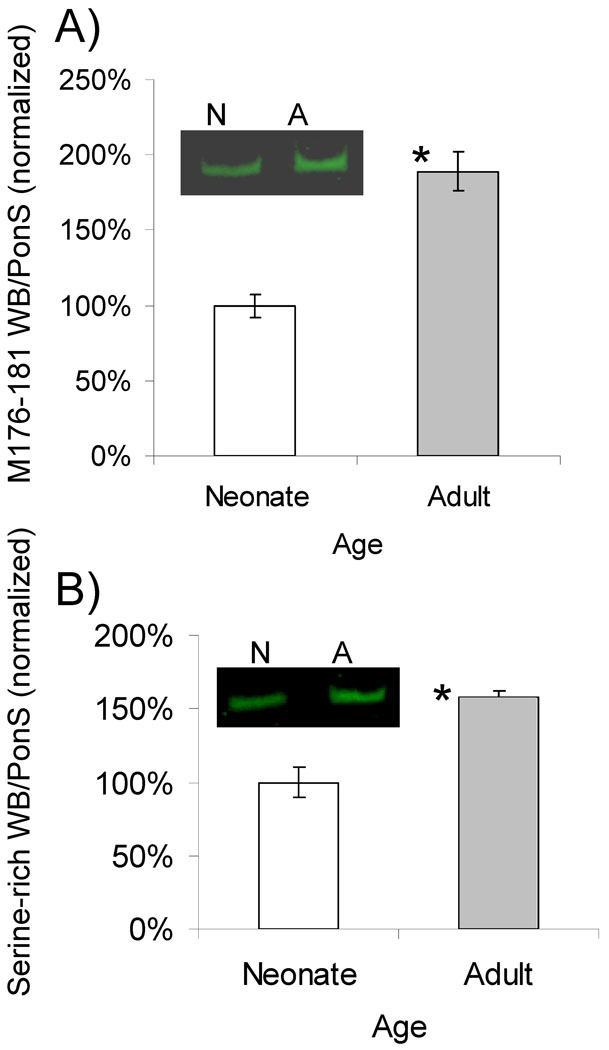
Figure 3.
Western blot against A) nebulin Z-repeats M176–M181 and B) serine rich domain, performed on neonatal day 5 samples (n=4) versus adult (4 mo. n=6). Both M176–181 and the serine rich domain are significantly upregulated in the adult. Shown are results normalized to the nebulin protein level, as obtained by the PonceauS-stained membrane (see Methods), with the average value for the neonates set to 100%. Example results are shown in insets. N = neonate, A = adult.
Although it is difficult to correlate transcript changes with changes in protein, if we assume that the differential exons are ‘on-off’ in the protein, the combined predicted effect of the identified differentially expressed exons is 20.4 kDa. However the measured expression ratios are only 2–4 fold (‘on-off’ would result in much larger ratios) and therefore we calculated the change in molecular weight taking the measured expression ratio into account. We obtained a 13.1 kDa larger protein in the adult (Table 1). This value was calculated assuming that exons are expressed in both neonate and adult but at different levels (Western blots studies confirm thus assumption, see below), with the calculation carried out as explained in the Methods section (under heading Microarray studies). Thus the effect of the identified exons with different expression levels in neonate and adult is 20.4 kDa when assuming that in the protein they are ‘on-off’ and 13.1 kDa assuming that in the protein they have an expression ratio as identified for the transcript.
Next, we investigated whether the changes in transcript found in the soleus also occur in different muscle types, and studied the tibialis cranialis (TC) muscle, a fast twitch muscle. Surprisingly, only exon 127 (128) was upregulated (~7 fold) in the adult vs 5 day old TC mice (figure 1C, bottom).
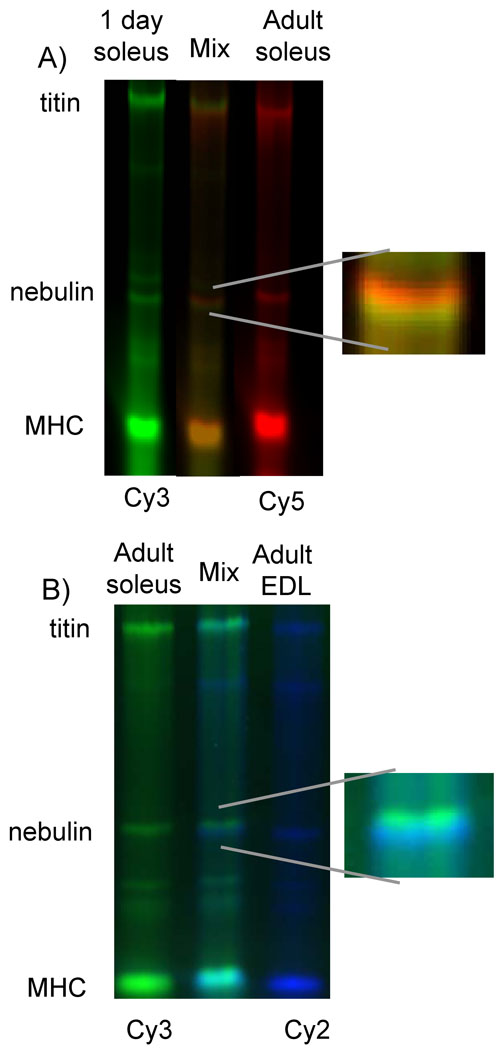
The transcript results suggest that differential splicing of nebulin has a limited effect on the molecular mass of nebulin. Because this contrasts with previous protein work where large differences in the size of nebulin were detected (5), we also performed a protein analysis of nebulin expression, using 1% agarose gels. We co-electrophoresed human soleus (HS) and human heart (HH) standards on the same gel because they express titin and nebulin of known molecular weights, providing molecular weight markers. Figure 4 shows an example of a gel used for analysis of nebulin expression in mouse soleus (MS) muscle (additional examples are shown in the supplemental figure S1 and figure S2). Note that at day 1, MS titin has a mobility that is indistinguishable from adult HS titin and that MS titin has a much faster mobility than HS in adults (reflecting massive differential splicing in titin’s spring region(2)). Nebulin of both neonatal and adult mice was much smaller in size than HS nebulin, with little difference visually noticeable between day 1 and adult MS muscles. Densitometry, however, revealed that the adult soleus nebulin had a slightly lower mobility than neonatal samples. Additionally, neonatal day 1 and adult (4 mo.) were fluorescently labeled with Cy 3 and Cy 5, respectively, and co-electrophoresed on a 1% agarose gel. Two distinct nebulin bands were observed (Fig. 5A). Analysis of neonatal and adult samples revealed that this slight mobility difference corresponds to a molecular mass difference between the neonatal and adult muscles of 16 kDa (Table 2) which is slightly larger than that predicted from the transcript data (see Discussion for details). We also studied several other skeletal muscle types and included the TC (studied by microarray), as well as the extensor digitorum longus (EDL), gastrocnemius (GAST), quadriceps (QUAD), and diaphragm (DIAPH). We found that all of the muscle types had only small differences is molecular weight when comparing adult vs. neonatal samples and that they were typically 10–20 kDa smaller in the adult (Table 2; supplementary figure 3 ??). It is also of interest to note that in adult mice, TC, EDL, GAST, QUAD, muscles all express nebulin that is similar in size (~700–710 kDa) but that nebulin size in the slow twitch soleus muscle and the DIAPH is somewhat larger (740–750 kDa). This can be visualized in figure 5B which shows a co-electrophoresis of Cy3 labeled adult soleus and Cy2 labeled adult EDL.
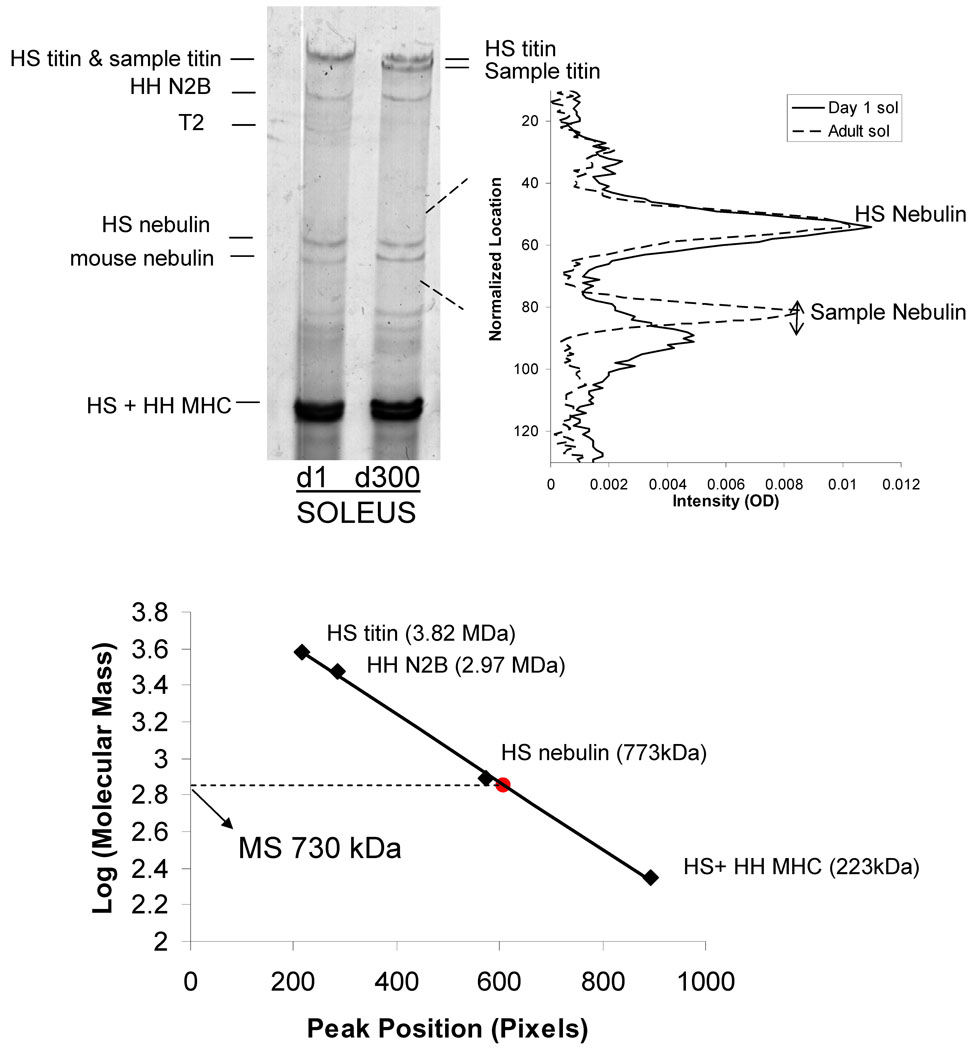
Figure 4.
Nebulin size analysis. Left Panel) SDS-agarose gel electrophoresis from day 1 (d1) and adult (d300) mouse soleus. The sample was co-electrophoresed with human soleus titin and human heart to compare nebulin isoform size. Right panel) Densitometry scan of two lanes (shown at the left) superimposed. Adult soleus nebulin (broken line) is larger in size than the neonate soleus (solid line) as observed by the reduced distance to human soleus nebulin. Bottom: distance of protein bands from top of gel vs log Mw. The known co-electrophoresed standards were used to determine the shown calibration line and this line was then used to estimate the Mw of the mouse soleus (MS) nebulins.
Figure 5.
Co-electrophoresis of two samples with different fluorescent labeling. A) Day 1 neonatal soleus (green) and adult soleus (red) were co-electrophoresed, and two distinct nebulin isoforms were observed with adult nebulin having a lower mobility than neonatal nebulin (see enlarged inset). B) Adult (4 month) soleus and EDL which have the highest and lowest nebulin molecular weight of the studied muscle types, respectively, were co-electrophoresed and show distinct differences in nebulin mobility.
Table 2.
Molecular weight estimates based on gel electrophoresis.
| Muscle Type | Neonate (d1) | Adult (d300) | Δ Mw |
|---|---|---|---|
| SOLEUS | 732 ± 2.4 | 748 ± 1.3 * | 16 |
| TC | 723 ± 1.2 | 715 ± 1.0 * | −8 |
| GAST | 719 ± 3.5 | 700 ± 1.8 * | −19 |
| EDL | 726 ± 4.2 | 701 ± 3.0 * | −25 |
| QUAD | 718 ± 0.4 | 710 ± 1.3 * | −10 |
| DIAPH | 731 ± 4.3 | 740 ± 1.1 | 9 |
P<0.05 neonates versus adults. (n=3–5 for neonatal samples; n=5–6 for adult samples).
We also analyzed the width of the nebulin bands to infer changes in isoform composition in the neonatal and adult muscle (with the idea that changes in band width reflect changes in isoform composition). We fitted the bands in soleus and TC muscle (n=4 each) with a Gaussian and determined the width at half height and at the base. No significant differences in the width were found between neonatal and adult muscle samples (results not shown). These results do not rule out that the band contains different isoforms and that during development the composition changes but the width stays the same.
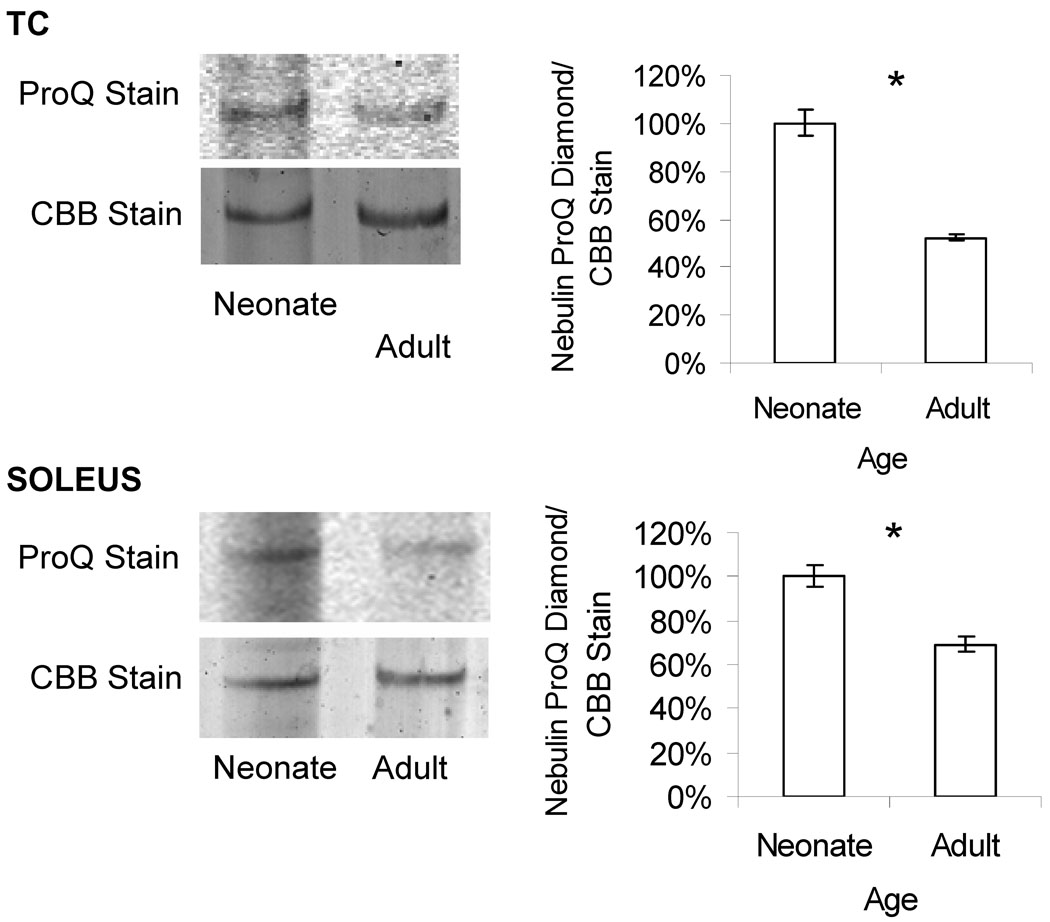
The agarose gel based data indicate that at the protein level soleus and TC are 16 kDa larger and 8 kDa smaller in the adult than the neonate, respectively. The transcript data predict 13 kDa larger and 3.6 kDa larger, respectively. One hypothesis for the difference in the observed molecular weight on protein gels versus the expected molecular weight observed from our microarray is that the difference in size of the protein is due to post-translational modifications. Phosphorylation is a good starting point for this hypothesis as the nebulin sequence is predicted to have ~200 phosphorylation sites (Buck et al. unpublished observations). To determine if some of the discrepancies in the molecular weight might be due to differences in phosphorylation we evaluated the phosphorylation status of nebulin using the ProQ Diamond stain. In both soleus and TC muscle there was a significant decrease in the relative level of phosphorylation in the adult versus the neonate (Figure 6). This decrease can partially explain why in the TC muscle the estimated molecular weight of nebulin is slightly less in the adult than in the neonate (Table 2), whereas transcript data predict a slight increase (Table 1, bottom). However, for the soleus muscle the reduced phosphorylation of the adult is expected to decrease the molecular weight of the adult protein and, thus, phosphorylation can not explain why the protein-based increase (16 kDa) is larger than that based on the microarray (13 kDa). Thus, changes in phosphorylation status do not provide a unifying explanation for the protein and transcript differences.
Figure 6.
Phosphorylation analysis using ProQ Diamond stain. The ratio of ProQ Diamond florescence staining over total nebulin protein staining (with coomassie blue) in the soleus and TC muscle indicate that there is a significantly higher level of phosphorylation in the neonates as compared to the adults. (n=4 neonates, n=5 adults.)
Finally, we also used gel electrophoresis to determine the expression ratio of nebulin relative to MHC (the main protein visible on the agarose gels; actin was run off). This was explored in the muscle types of soleus, TC, EDL, GAST, QUAD, and DIAPH and in various ages day 1 to adult (~10 ages). We found that the nebulin:MHC ratio was not dependent on age or on muscle type (Supplemental figure 3) and that the ratio was on average 0.06. Thus our protein analysis revealed only minor differences in nebulin size during neonatal development, that in the adult animal nebulin size is similar in fast twitch (EDL), slower twitch (DIAPH), and mixed muscles (QUAD and GAST), but slightly larger in slow twitch muscle (soleus) and that the stoichiometry of nebulin does not vary with development stage nor with muscle type.
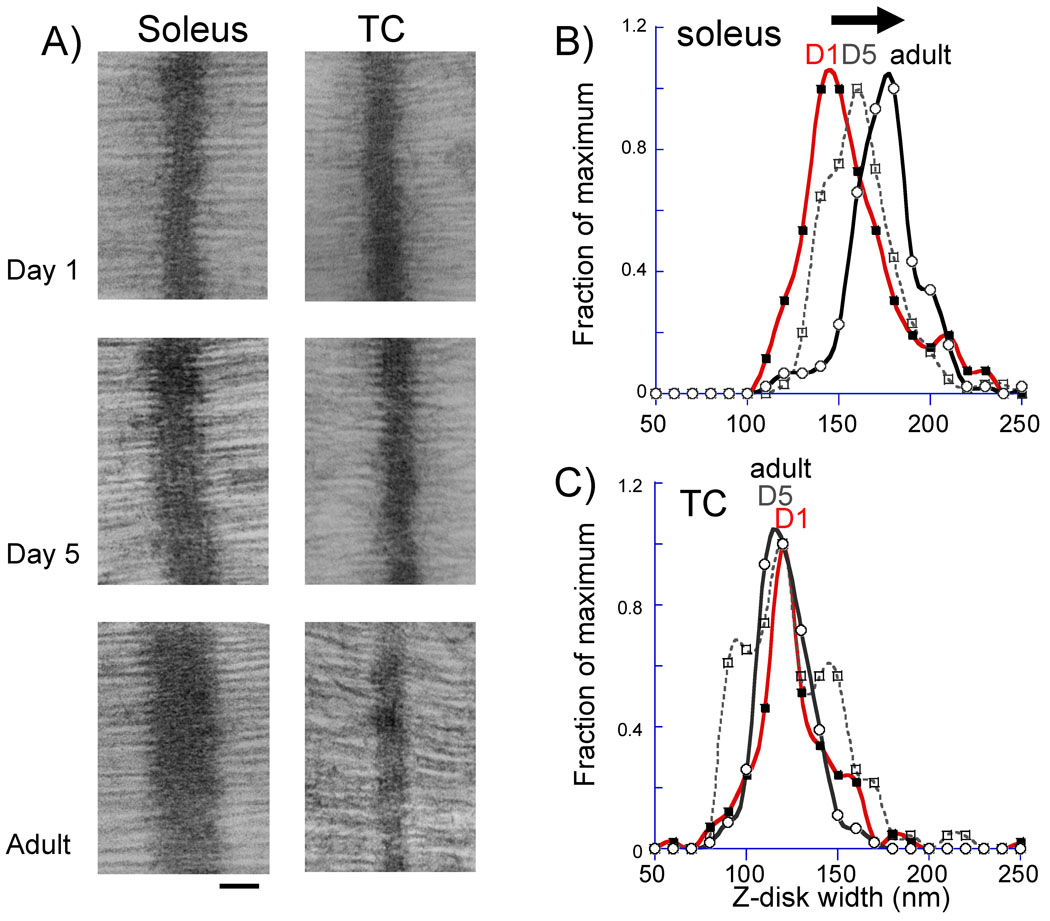
Since we had found differential splicing in the Z-disc region of nebulin in soleus muscle, and because it had been previously proposed that nebulin plays a role in setting the width of the Z-disc (16), we performed electron microscopy on soleus muscle from neonatal day 1, 5, and adult mice and measured Z-disc widths. Representative results are shown in figure 7A, left and analyzed results in 7B. Interestingly, the soleus Z-disc is wider in the adult than in the neonates, see example micrographs of figure 7A bottom and the histograms of measurements in figure 7B. The mean width of the Z-disc increased from 151 nm in day 1 soleus to 156 nm in day 5 soleus and 170 nm in the adult (p<0.001) (Table 3). To further test the correlation between differential Z-disc exon splicing and Z-disc width we also studied the TC muscle, where few differences exist in Z-disc exon composition when comparing neonates with adult muscle (see above). Data are shown in figure 7A right, figure 7C and Table 2. The TC muscle had a mean Z-disc width of 120 nm in the day 1 neonate, 120 nm in the day 5 neonate and 117 nm in the adult (differences are not significant). This data therefore confers a correlation between Z-disc width and the isoform shift in nebulin consistent with previous work.
Figure 7.
Z-disc width measurement in neonatal (day 1 and day 5) and adult mice. A) Examples of micrographs for soleus (left) and TC (right) muscle. Bar is 100 nm. B and C) histograms of Z-disc width measurements. Data were binned in 10 nm wide Z-disc bins and each value represents the number of measurements that falls within the bin. (The values are expressed as a fraction of the value of the bin with the most measurements).
Table 3.
Z-disc width measurements in soleus and TC muscles.
| Sample | Z-disc Width | N Size | |
|---|---|---|---|
| SOLEUS | Day 1 | 151 ± 28 nm *** | 139 |
| Day 5 | 156 ± 20 nm *** | 280 | |
| Adult | 170 ± 20 nm | 179 | |
| TC | Day 1 | 120 ± 24 nm | 138 |
| Day 5 | 120 ± 26 nm | 124 | |
| Adult | 117 ± 14 nm | 164 |
P<0.001 neonates (day 1 or 5) versus adults.
Discussion
We recently reported that due to extensive differential splicing during postnatal development of skeletal muscle, titin is ~50 exons smaller (representing ~200 kDa in protein) in adult mice than in neonates (2). Similar to titin, nebulin also contains a large number of exons (166 in mice) and here we studied differential splicing of nebulin during skeletal muscle development, using a novel home-made nebulin exon microarray. We focused on two different muscle types, the soleus and tibialis cranialis (TC) muscles, representatives of slow and fast twitch muscle types, respectively, and studied muscles from neonatal and adult mice. The soleus muscle had 25 exons differentially spliced between 5 day old neonates and adults, with differences in the super-repeat region and the Z-disc region of nebulin. Far fewer changes were found in the TC muscle where only one exon was differentially spliced (comparing 5 day old and adult mice). These differences in splicing between soleus and TC muscle during development correlate with changes in Z-disc width, as shown by electron microscopy. Below we discuss these findings in detail.
Nebulin’s N-terminal modules (M1-M8) contain binding sites for the thin filament pointed-end capping protein tropomodulin, and the C-terminal modules M163 through M185 are located in and near the Z-disc (4). The large central region of the nebulin gene contains exons that code for a highly modular structure, the so-called SDXXYK-repeat modulus that are each thought to represent individual actin-binding motifs. These motifs are organized into seven-module super-repeats (figure 1A), characterized by the Tm/Tn binding motif, WLKGIGW, that match the 38.5 nm repeat of the actin filament (4,8–10).
This arrangement enables a single nebulin module to interact with a single actin monomer, and each nebulin super-repeat to associate with a single Tm/Tn complex (4,11,13). We found that most of the exons that code for super-repeat 1 and 2 are down regulated in adult soleus muscle (figure 1C), suggesting that the N-terminal end of nebulin’s super-repeat region might be 2 × 38.5 nm shorter in the adult soleus muscle. On the other hand, most exons that represent super-repeat 22 are upregulated in adult soleus muscle (figure 1C), indicating that the near Z-disc region of nebulin’s super-repeat region is 38.5 nm longer. The combined effect of these two differentially spliced regions is a super-repeat region that is expected to be 38.5 nm shorter in adult soleus muscle.
The differences in the super-repeat region of soleus nebulin suggest that the thin filaments are slightly shorter in adult soleus muscle. Thin filament length is not an intrinsic property of actin filaments (actin monomers assemble in vitro to highly variable polymer lengths), and nebulin has been long considered a likely candidate that is involved in thin filament length control (6,7). Evidence for a role for nebulin in establishing thin filament length was obtained in earlier studies that revealed that the electrophoretic mobility of nebulin from different muscle types correlates with thin filament length (5). The recent work on nebulin KO models revealed that in nebulin-deficient muscle the thin filaments are on average shorter, thus further supporting a role for nebulin in the in vivo regulation of thin filament length (14,15). Thin filament length is a key aspect of muscle function and the extent of overlap between thick and thin filaments determines the sarcomere’s force generating capacity (30,31). For muscles that work on the descending limb of the force-sarcomere length relationship short thin-filaments reduce overlap and this lowers force generation, which is disadvantageous(31). However for muscles that work on the ascending limb, shorter thin filaments can increase force production, because the overlap zone of thin filaments in the center of the sarcomere will be reduced (force production is greatly reduced in the thin filament overlap region, see(31)). Thus, the reduction in thin filament length during postnatal development of soleus muscle, that is implied by our microarray data, might reflect a reduction in the working range of the sarcomere (note that a reduction in working range is consistent with the reduction in the length of the titin’s spring region that occurs during postnatal development (2)). Whether these changes in nebulin also relate to the changes in myosin isoform expression which take place during postnatal development (32) remains to be established.
Because no major changes were found in the super-repeat region of the nebulin gene in the TC muscle changes in thin filament length during postnatal development are unlikely to take place in this muscle type. It is noteworthy that exon 127(128) was greatly upregulated (>7-fold) in adult TC when compared to neonatal TC, and that this was absent in the soleus. This exon was also identified by Donner et al (26) as differentially spliced during murine muscle development. Donner et al (26) refer to this exon as exon 128 (their numbering is shown in our manuscript in parenthesis), because of a new exon that they identified and numbered exon 127. (Note that their exon 127 is not represented on our array because it was discovered after the array was designed.) Donner et al (26) showed that exon 127(128) in TC muscle is >10-fold increased during the first 6 weeks after birth, whereas in the soleus it is less than 2-fold increased. Our microarray findings are consistent with the proposal of Donner et al. that exon 127(128) has a muscle-specific regulatory function that is utilized during muscle development.
Additionally, we found that the stoichiometry of nebulin was independent of muscle type and developmental stage, and that the nebulin:MHC ratio was on average 0.06 (supplemental Fig. 3). Considering that there are 300 MHC molecules per half sarcomere (33), the measured nebulin:MHC ratio of 0.06, the 223 kDa molecular mass for MHC and ~710 kDa for nebulin (Table 1), and that there are twice as many thin than thick filament(33), makes it possible to calculate that there are ~2.8 nebulin molecules per thin filament. Because of the two-fold symmetry of F-actin (33) it seems likely that the number of molecules of nebulin bound to F-actin is an even number and we assume therefore that the real number is 2 (the somewhat higher value that we found could be explained by, for example, a binding affinity for Coommassie Blue that is higher for nebulin than MHC, or by a thin:thick filament ratio that is slightly higher than 2). Our finding of a nebulin stoichiometry that is independent of muscle type and developmental stage is consistent with a structural function for nebulin that needs to be constant during muscle development.
We found a ~20 nm increase in the Z-disc width during postnatal development of soleus muscle, but no changes in the Z-disc of the TC muscle (Table 2). Nebulin’s Z-disc modules M176 to M181 are known to be differentially spliced and different isoforms have previously been identified that result from the skipping of various combinations of the M176–181 modules(16). Importantly, the number of modules has been shown to be positively correlated with the Z-disc width of different rabbit and human muscle types (16). We earlier proposed that these nebulin modules play a role in terminating the Z-disc structure and establishing the transition to the I-band region of the sarcomere (16). In this differentially spliced region, the mouse nebulin gene has 3 additional exons inserted that have been named M177/M178b, M177/M178c, and M177/M178d(9). Interestingly all three novel exons are upregulated in the adult soleus relative to 5 day old neonates (table 1), whereas they are not differential in the TC. These findings are consistent with our earlier reported positive correlation between the number of M176-M82 modules and the Z-disc width. Thus our present findings support the view that differentially expressed Z-disc modules of nebulin play a critical role in determining the Z-disc width of the sarcomere. Future work is needed to directly test this by gene targeting experiments that delete nebulin Z-disc modules.
Both microarray analysis and qPCR show an increase in expression of the SH3 domain in the adult soleus muscle. Interestingly, a mouse model in which the SH3 domain is deleted shows that the SH3 domain is dispensable for muscle development and isometric stress production (34). The functional significance for the altered expression during development remains to be established.
The Z-disc plays a fundamental role in transmission of active and passive forces that are generated within the sarcomere; differences in Z-disc width in different muscle types have been proposed to reflect differences in mechanical stress levels experienced by their Z-discs (35). It seems likely therefore that the increase in the Z-disc width of the soleus muscle (Fig. 7) reflects the weight bearing function of this muscle and the increasing stress exerted on this muscle due to the rapid increase in body weight during postnatal development. Furthermore, these stresses have to be withstood for long periods of time and, thus, the ‘stress-time integral’ is high for the soleus muscle, in contrast to fast twitch muscles such as the TC which are typically active for short periods only. Thus we propose that the increased Z-disc width of the soleus muscle that we found reflects the increasing mechanical demands on solues muscle during postnatal development.
In summary, transcript studies with a newly developed mouse nebulin exon microarray and the supportive qPCR, revealed during postnatal development muscle-type specific changes in splicing in both the super-repeat region and the Z-disc region of nebulin. Three novel Z-disc exons were upregulated during postnatal development of soleus muscle and this was correlated with a significant increase in Z-disc width. These findings suggest that nebulin plays an important role in Z-disc width regulation by terminating the Z-disc and establishing the transition to the I-band region of the sarcomere. Consistent with this view are the pathologically wide Z-discs found in muscle from nebulin deficient mice(14) and nebulin deficient nemaline myopathy patients (36). Thus, we discovered during postnatal development changes in both the super-repeat region and the Z-disc region of nebulin, that these changes are muscle-type specific, and that they correlate with differences in sarcomere structure.
Supplementary Material
S1. Left Panel) SDS-agarose gel electrophoresis from day 1 (d1) and adult (d300) mouse soleus (top left) TC (top right), EDL (bottom left) and GAST (bottom right). The samples were co-electrophoresed with human soleus titin and human heart to compare nebulin isoform size. Right panels: Densitometry scan of the nebulin region of the two lanes (shown at the left) superimposed (as done in fig 4)
S2. Neonatal and adults samples from a range of muscles electrophoresed on the same gel . Samples were mixed with human soleus (HS) and human heart (HH) to have size markers.
S3. Nebulin expression relative to myosin heavy chain (MHC) of soleus, TC, EDL , GAST, and QUAD. Neonates (light grey, 1-5 days old), juvenile (white, 8-50 days old), and adult (dark grey, 100-300 days old). The expression ratios did not vary significantly with age or muscle type.
Acknowledgements
We are grateful to Luann Wyly, Arturo Garcia-Montano, Adam Hoying, Kevin Greer, and Gina Zhang for technical assistance. We thank Dr. Carol Gregorio for stimulating discussions and help with Western Blot experiments. Danielle Buck received support from the University of Arizona UBRP undergraduate program (HHMI 52003749). Supported by National Institutes of Health, AR-053897 (HG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schiaffino S, Reggiani C. Physiol Rev. 1996;76:371–423. doi: 10.1152/physrev.1996.76.2.371. [DOI] [PubMed] [Google Scholar]
- 2.Ottenheijm CA, Knottnerus AM, Buck D, Luo X, Greer K, Hoying A, Labeit S, Granzier H. Biophys J. 2009;97:2277–2286. doi: 10.1016/j.bpj.2009.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang K. Adv Exp Med Biol. 1984;170:285–305. doi: 10.1007/978-1-4684-4703-3_25. [DOI] [PubMed] [Google Scholar]
- 4.McElhinny AS, Kazmierski ST, Labeit S, Gregorio CC. Trends Cardiovasc Med. 2003;13:195–201. doi: 10.1016/s1050-1738(03)00076-8. [DOI] [PubMed] [Google Scholar]
- 5.Kruger M, Wright J, Wang K. J Cell Biol. 1991;115:97–107. doi: 10.1083/jcb.115.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Labeit S, Gibson T, Lakey A, Leonard K, Zeviani M, Knight P, Wardale J, Trinick J. FEBS Lett. 1991;282:313–316. doi: 10.1016/0014-5793(91)80503-u. [DOI] [PubMed] [Google Scholar]
- 7.Wang K, Knipfer M, Huang QQ, van Heerden A, Hsu LC, Gutierrez G, Quian XL, Stedman H. J Biol Chem. 1996;271:4304–4314. doi: 10.1074/jbc.271.8.4304. [DOI] [PubMed] [Google Scholar]
- 8.Labeit S, Kolmerer B. J Mol Biol. 1995;248:308–315. doi: 10.1016/s0022-2836(95)80052-2. [DOI] [PubMed] [Google Scholar]
- 9.Kazmierski ST, Antin PB, Witt CC, Huebner N, McElhinny AS, Labeit S, Gregorio CC. J Mol Biol. 2003;328:835–846. doi: 10.1016/s0022-2836(03)00348-6. [DOI] [PubMed] [Google Scholar]
- 10.Donner K, Sandbacka M, Lehtokari VL, Wallgren-Pettersson C, Pelin K. Eur J Hum Genet. 2004;12:744–751. doi: 10.1038/sj.ejhg.5201242. [DOI] [PubMed] [Google Scholar]
- 11.Ogut O, Hossain MM, Jin JP. J Biol Chem. 2003;278:3089–3097. doi: 10.1074/jbc.M205853200. [DOI] [PubMed] [Google Scholar]
- 12.Jin JP, Wang K. J Biol Chem. 1991;266:21215–21223. [PubMed] [Google Scholar]
- 13.Jin JP, Wang K. FEBS Lett. 1991;281:93–96. doi: 10.1016/0014-5793(91)80366-b. [DOI] [PubMed] [Google Scholar]
- 14.Witt CC, Burkart C, Labeit D, McNabb M, Wu Y, Granzier H, Labeit S. Embo J. 2006;25:3843–3855. doi: 10.1038/sj.emboj.7601242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bang ML, Li X, Littlefield R, Bremner S, Thor A, Knowlton KU, Lieber RL, Chen J. J Cell Biol. 2006;173:905–916. doi: 10.1083/jcb.200603119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Millevoi S, Trombitas K, Kolmerer B, Kostin S, Schaper J, Pelin K, Granzier H, Labeit S. J Mol Biol. 1998;282:111–123. doi: 10.1006/jmbi.1998.1999. [DOI] [PubMed] [Google Scholar]
- 17.Pelin K, Hilpela P, Donner K, Sewry C, Akkari PA, Wilton SD, Wattanasirichaigoon D, Bang ML, Centner T, Hanefeld F, Odent S, Fardeau M, Urtizberea JA, Muntoni F, Dubowitz V, Beggs AH, Laing NG, Labeit S, de la Chapelle A, Wallgren-Pettersson C. Proc Natl Acad Sci U S A. 1999;96:2305–2310. doi: 10.1073/pnas.96.5.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ottenheijm CA, Hidalgo C, Rost K, Gotthardt M, Granzier H. J Mol Biol. 2009;393:10–26. doi: 10.1016/j.jmb.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ottenheijm CA, Hooijman P, Dechene ET, Stienen GJ, Beggs AH, Granzier H. J Struct Biol. 2009 doi: 10.1016/j.jsb.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandra M, Mamidi R, Ford S, Hidalgo C, Witt C, Ottenheijm C, Labeit S, Granzier H. J Biol Chem. 2009;284:30889–30896. doi: 10.1074/jbc.M109.049718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ottenheijm CA, Witt CC, Stienen GJ, Labeit S, Beggs AH, Granzier H. Hum Mol Genet. 2009;18:2359–2369. doi: 10.1093/hmg/ddp168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warren CM, Krzesinski PR, Greaser ML. Electrophoresis. 2003;24:1695–1702. doi: 10.1002/elps.200305392. [DOI] [PubMed] [Google Scholar]
- 23.Lahmers S, Wu Y, Call DR, Labeit S, Granzier H. Circ Res. 2004;94:505–513. doi: 10.1161/01.RES.0000115522.52554.86. [DOI] [PubMed] [Google Scholar]
- 24.Granzier HL, Irving TC. Biophys J. 1995;68:1027–1044. doi: 10.1016/S0006-3495(95)80278-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Granzier HL, Wang K. Electrophoresis. 1993;14:56–64. doi: 10.1002/elps.1150140110. [DOI] [PubMed] [Google Scholar]
- 26.Donner K, Nowak KJ, Aro M, Pelin K, Wallgren-Pettersson C. Genomics. 2006;88:489–495. doi: 10.1016/j.ygeno.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 27.Greer KA, McReynolds MR, Brooks HL, Hoying JB. BMC Bioinformatics. 2006;7:149. doi: 10.1186/1471-2105-7-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Granzier H, Kellermayer M, Helmes M, Trombitas K. Biophys J. 1997;73:2043–2053. doi: 10.1016/S0006-3495(97)78234-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trombitas K, Granzier H. Am J Physiol. 1997;273:C662–C670. doi: 10.1152/ajpcell.1997.273.2.C662. [DOI] [PubMed] [Google Scholar]
- 30.Granzier HL, Pollack GH. J Physiol. 1990;421:595–615. doi: 10.1113/jphysiol.1990.sp017964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Granzier HL, Akster HA, Ter Keurs HE. Am J Physiol. 1991;260:C1060–C1070. doi: 10.1152/ajpcell.1991.260.5.C1060. [DOI] [PubMed] [Google Scholar]
- 32.Agbulut O, Noirez P, Beaumont F, Butler-Browne G. Biol Cell. 2003;95:399–406. doi: 10.1016/s0248-4900(03)00087-x. [DOI] [PubMed] [Google Scholar]
- 33.Squire JM. Curr Opin Struct Biol. 1997;7:247–257. doi: 10.1016/s0959-440x(97)80033-4. [DOI] [PubMed] [Google Scholar]
- 34.Gokhin DS, Zhang J, Bang ML, Chen J, Lieber RL. Biophyscial Journal. 1999;96:213a. [Google Scholar]
- 35.Vigoreaux JO. J Muscle Res Cell Motil. 1994;15:237–255. doi: 10.1007/BF00123477. [DOI] [PubMed] [Google Scholar]
- 36.Pelin K, Donner K, Holmberg M, Jungbluth H, Muntoni F, Wallgren-Pettersson C. Neuromuscul Disord. 2002;12:680–686. doi: 10.1016/s0960-8966(02)00066-4. [DOI] [PubMed] [Google Scholar]
- 37.Pfuhl M, Winder SJ, Castiglione Morelli MA, Labeit S, Pastore A. J Mol Biol. 1996;257:367–384. doi: 10.1006/jmbi.1996.0169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1. Left Panel) SDS-agarose gel electrophoresis from day 1 (d1) and adult (d300) mouse soleus (top left) TC (top right), EDL (bottom left) and GAST (bottom right). The samples were co-electrophoresed with human soleus titin and human heart to compare nebulin isoform size. Right panels: Densitometry scan of the nebulin region of the two lanes (shown at the left) superimposed (as done in fig 4)
S2. Neonatal and adults samples from a range of muscles electrophoresed on the same gel . Samples were mixed with human soleus (HS) and human heart (HH) to have size markers.
S3. Nebulin expression relative to myosin heavy chain (MHC) of soleus, TC, EDL , GAST, and QUAD. Neonates (light grey, 1-5 days old), juvenile (white, 8-50 days old), and adult (dark grey, 100-300 days old). The expression ratios did not vary significantly with age or muscle type.