Abstract
Babesia bovis, an intraerythrocytic parasite of cattle, establishes persistent infections of extreme duration. This is accomplished, at least in part, through rapid antigenic variation of a heterodimeric virulence factor, the variant erythrocyte surface antigen-1 (VESA1) protein. Previously, the VESA1a subunit was demonstrated to be encoded by a 1α member of the ves multigene family. Since its discovery the 1β branch of this multigene family has been hypothesized to encode the VESA1b polypeptide, but formal evidence for this connection has been lacking. Here, we provide evidence that products of ves1β genes are rapidly variant in antigenicity and size-polymorphic, matching known VESA1b polypeptides. Importantly, the ves1β-encoded antigens are co-precipitated with VESA1a during immunoprecipitation with anti-VESA1a monoclonal antibodies, and antisera to ves1β polypeptide co-precipitate VESA1a. Further, the ves1β-encoded antigens significantly co-localize with VESA1a on the infected-erythrocyte membrane surface of live cells. These characteristics all match known properties of VESA1b, allowing us to conclude that the ves1β gene divergently apposing the ves1α gene within the locus of active ves transcription (LAT) encodes the 1b subunit of the VESA1 cytoadhesion ligand. However, the extent and stoichiometry of VESA1a and 1b co-localization on the surface of individual cells is quite variable, implicating competing effects on transcription, translation, or trafficking of the two subunits. These results provide essential information facilitating further investigation into this parasite virulence factor.
Keywords: Babesia bovis, antigenic variation, VESA1b, ves multigene family, ves1β gene, cytoadhesion ligand
Introduction
Babesial parasites infect animals of many species, including humans. The bovine hemoparasites, Babesia bovis and Babesia bigemina, have a major impact internationally on livestock development in most tropical and subtropical regions of the world [1]. Animals infected with B. bovis suffer an extremely severe acute infection, hallmarks of which include high fevers, severe anemia and hemoglobinuria, an acute respiratory distress syndrome-like condition, and sequestration of mature parasites in the deep vasculature [2,3]. Occasionally, development of a cerebral form of babesiosis may occur which is almost uniformly lethal once initiated. B. bovis shares these traits with Plasmodium falciparum, the most virulent and frequently lethal of the human malarial parasites. As with P. falciparum, parasites which have initiated erythrocytic infection are not able to infect other tissues in the mammalian host yet are able to establish persistent infections of long duration [4–6], and those animals surviving the acute infection go on to develop asymptomatic but persistent B. bovis infections of extreme duration [7–10], perhaps even for the remaining lifespan of the animal.
At least two interrelated mechanisms are thought to play key roles in the persistence of these two parasites. The first mechanism involves sequestration [11–17] of infected erythrocytes (IE) that is mediated by cytoadhesion of IE [18–22] to the capillary and post-capillary venous endothelium. This phenomenon is thought to benefit the parasite by enabling it to avoid splenic passage, and likely facilitates parasite development by ensuring that most development occurs under microaerophilic conditions. The second mechanism, rapid antigenic variation of the parasite-derived components responsible for cytoadhesion [10,23–26], is thought to protect the adhesion function from the ongoing host adaptive immune response (reviewed elsewhere [27,28]).
Antigenic variation and cytoadhesion in B. bovis are mediated through the variant erythrocyte surface antigen-1 (VESA1) [10,19,23,29,30]. The recently described SmORF proteins and genes also appear to be candidates for rapid antigenic variation based upon key differences in genome and transcriptome sequence data [31], but this possibility currently remains without direct experimental support. VESA1 is a size-polymorphic, heterodimeric protein comprised of two subunits approximately 105–115 and 120–135 kDa in mass, depending upon the isolate and clonal line examined [10,32]. The larger subunit, VESA1a, has been shown previously to be encoded by the ves1α gene [29], but the gene encoding the smaller VESA1b subunit was unknown. It was discovered that the actively transcribed ves1α gene resides in a divergently organized “locus of active ves transcription” (LAT) opposite a member of a second, clearly related but structurally distinct branch of the ves gene family, ves1β [30]. The members of the 1β branch are quite variable in both size and exon/ intron structure [31]. Similarly to ves1α, the ves1β gene is modified in situ through a mechanism of segmental gene conversion, and this seems to be the primary mechanism by which B. bovis varies these genes [30]. Despite their functional and behavioral similarities, this contrasts sharply with variation of the var genes encoding the cytoadhesion ligand, PfEMP1, of P. falciparum, which appears to utilize epigenetically-regulated in situ transcriptional switching as the primary mechanism [33,34]. Rather, the situation in B. bovis resembles more the variation of VSG genes encoding the variant surface glycoprotein in African trypanosomes because of its heavy reliance upon gene conversion mechanisms [35–37]. Like the African trypanosomes, however, B. bovis is likely to take advantage of alternative mechanisms, such as in situ transcriptional switching [38], but with much lower frequency. Preliminary evidence regarding the functionality of transcriptionally silent ves loci is consistent with this possibility (unpublished data).
The organization of ves1β genes together with ves1α genes, their shared mechanism of variation, and the possibility of their convenient co-regulation as subunits of VESA1 through their coupled organization led us to hypothesize that the ves1β gene encodes the VESA1b subunit. Despite these conceptual links, direct evidence linking ves1β genes to VESA1b polypeptides has been lacking. To test for such a connection, antibodies generated to segments derived from ves1β gene-encoded polypeptides were used to characterize the encoded products and their interacting partners, and to determine their cellular location. The results of these studies, presented here, establish that the 1β branch of the ves multigene family encodes VESA1b polypeptides.
Materials and Methods
Parasites
B. bovis parasites derived from the Mexico isolate were used in this study. The origins of the antigenically variant, clonally-derived B. bovis lines MO7, C9.1, B9, C8, H10, CD7, and CE11 are described elsewhere [10,19,39]. The parasites were grown in vitro under microaerophilous stationary phase culture conditions, as described [32].
Generation of antibodies
Mouse anti-VESA1a monoclonal antibodies
The development of the B. bovis C9.1 line VESA1a-specific mouse monoclonal antibody (mAb), 4D9.1G1, has been described previously [23]. This mAb also reacts with the cytoadhesive CD7 line due to co-selection of this line for that trait [19].
Bovine infection serum
The derivation of bovine B2442 immune infection serum has been described previously [10].
Rabbit antisera to ves1β polypeptide, amino acids 1–765 (Ra-v1β765)
Sequences from nucleotide 70 to 2364 of ves1β-encoding phagemid2-2 cDNA (accession number DQ267447.1) [30] were amplified by PCR, using primers DA98-NdeI (GGTA[CATATG]TCTCAGAGTACCTGGCAAC) and DA99-XhoI (GTTC[CTCGAG]GTGAGCAGGGTACTTGTTATTG). The sequences in brackets represent NdeI and XhoI restriction endonuclease sites added for cloning purposes. The product was cut with NdeI and XhoI and inserted into pET41b vector (Novagen, Inc.), resulting in the expression of a truncated ves1β-encoded polypeptide (amino acids 1–765; Figure 1, red arrow) with a C-terminal 6x-His fusion. The construct was transfected into E. coli DH5α-T1R and sequenced to confirm construction. Recombinant protein was expressed in E. coli BL21(DE3), by 3 hours induction with 0.3 mM IPTG at 37°C. The recombinant protein was released by disruption of E. coli with BugBuster reagent (Novagen, Inc.), 1 mg ml−1 lysozyme and 25 U ml−1 benzonase (Sigma Chemical, St. Louis, MO), followed by centrifugation at 16,000 × g for 20 min for clarification. The recombinant protein was found to be heavily cross-linked and very difficult to solubilize. Therefore, insoluble materials were sedimented at 16,000 ×g for 20 minutes, and the pellet treated again with BugBuster and lysozyme for 20 minutes to disrupt remaining cellular material. Insoluble materials were sedimented at 5,000 × g for 15 min. at 4°C, then pellets were washed into PBS by centrifugation as above. The pellets were disrupted with 8 M urea, 10 mM sodium phosphate, 150 mM NaCl, pH 8.0 (8 M urea-PBS) in a Dounce homogenizer. Insoluble materials were collected by sedimentation at 5000 × g for 15 min. at 4°C, then dissolved in 20 mM dithiothreitol/ urea-PBS. Residual insoluble materials were sedimented at 2700 × g for 10 min. at 4°C, and the supernatant containing solubilized recombinant ves1β polypeptide collected. The protein was sequentially dialyzed into 0.9 mM DTT/ 0.8 M urea-PBS, 0.1 mM DTT/ 0.2 M urea/PBS, and finally PBS alone. Any precipitates were again sedimented by centrifugation at 2700 × g for 10 min., and the supernatant was equilibrated with physiological saline by ultrafiltration for immunization. Rabbits were immunized by injection with 100 µg of protein emulsified into Freunds complete adjuvant, followed by three booster immunizations, at 2 week intervals, with 50 µg of protein in incomplete adjuvant. Immunizations were conducted through Cocalico Biologicals, Inc. (Reamstown, PA) under U.S. Department of Agriculture license #23-R-0089, Animal Welfare Assurance #A-3669-01. The antisera were diluted 1/10 in PBS and pre-adsorbed four times against E. coli DH5α-T1R expressing the corresponding region of VESA1a polypeptide encoded by the p9.6.2 cDNA and expressed as described above for ves1β polypeptide. Pre-adsorption was performed as described [40].
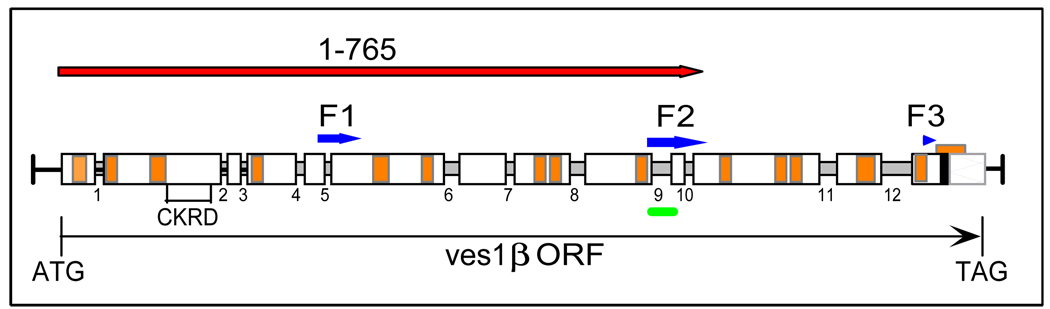
Figure 1. Structure of the LAT ves1β gene and recombinant immunogens.
The structure of the LAT-associated ves1β gene is shown in schematic. Exons which are a part of the translated open reading frame are shown as thick white bars and introns as numbered, narrow gray bars. The narrow black line represents 5'- and 3'-untranslated sequences. The orange segments within the exons are patches of sequence conserved among ves genes (adapted from [30]). The segments incorporated into recombinant immunogens are indicated as follows: fragments included in the r-v1β(f1-3) immunogen are indicated by blue arrows labeled “F1”, “F2”, and “F3”, whereas sequences contained in the r-v1β(765) immunogen are indicated by the red arrow labeled “1–765”. The position of the peptide incorporated in the v1β(peptide) immunogen is indicated by a green bar. Note that only exon-encoded sequences taken from a mature mRNA-derived cDNA were included in each immunogen. “CKRD” indicates the position of the cysteine-lysine-rich domain. The black bar near the 3' end indicates the predicted transmembrane domain, and “ATG” and “TAG” are the translational start and stop codons, respectively.
Chicken anti-ves1β recombinant fragments 1–3 (Ca-v1β(f1-3)) antibodies
To generate potentially pan-reactive anti-ves1β antibodies the mosaic recombinant polypeptide, r-v1βf1-3, was made by a crossover polymerase chain reaction (PCR) strategy [41], employing PCR primers Ves1b-F1 (GTGGCAG[GAA TTC]AAAAATGG), Ves1b-R1(GAT[CCCGGG]GTAGTCCTTACAGAG), Ves1b-F2 (TCAG[CCCGGG]AAAGACAATCACTTCAAAGATTTG), Ves1b-R2proper (GACTCCCTTCAAGGTCTCGCAGTGAGCAGGGTACTTG), Ves1b-F3 (CAGG[CTGCAG]GGAGACCTTGAAGGGAGTCC), and Ves1b-R3 (GATC[CTCGAG]CTGGTTTATTTCGTGGTGGAG) to recover sequences from the previously described cDNA, phagemid 2-2 [30]. The bracketed sequences are EcoRI, XmaI, PstI, and Xho I restriction sites added to the sequences to facilitate construction and insertion at vector junctions. Ves1b-F3 primer was originally designed for a different purpose but allowed crossover PCR to occur accurately despite the unnecessary 5' sequences. This construct encodes amino acid residues 325–362, 735–766, and 1021–1046 of the B. bovis C9.1 line ves1β-encoded polypeptide (Figure 1, blue arrows), representing three segments with sequences that are highly conserved among ves1β-encoded polypeptides [29,30]. The chimeric sequence was inserted into the pET41a expression vector (Novagen, Inc.; Darmstadt, Germany) for inducible expression in Escherichia coli BL21(DE3) as a C-terminal fusion with glutathione-S-transferase. Protein expression was induced with 1.0 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) at 37°C for two hours. Soluble protein was collected after disruption of recombinant E. coli as described earlier. Recombinant v1βf1-3 polypeptide was affinity-purified from the supernatant by chromatography on a GST-Bind Resin (Novagen, Inc.), following the manufacturer’s recommendations. The purified protein was washed into physiologic saline by ultrafiltration and adjusted to 1 mg ml−1. Immunization of chickens was performed by Aves Labs, Inc. (Tigard, OR). Chickens were given 200 µg antigen emulsified in Complete Freunds adjuvant for the primary immunization, whereas three booster immunizations, given at two-week intervals, included incomplete Freunds adjuvant. To reduce background reactivities, crude IgY was adsorbed with fixed E. coli BL21(DE3) expressing GST, as described [40]. This work was covered by Animal Protocol #3, approved by the US Department of Health and Human Services, Office for Laboratory Animal Welfare (accreditation #A-4232).
Rabbit anti-ves1β synthetic peptide antibodies (Ra-v1β(pep))
A synthetic peptide, CKDNHFKDLTTGSHK, was synthesized by Sigma Genosys (The Woodlands, Texas), using Fmoc chemistry. The peptide was purified by reverse-phase high pressure liquid chromatography and determined to be 86% pure by mass spectrometric analysis. This peptide corresponds to a consensus form of the sequence represented by amino acids 740–753 of the LAT ves1β gene-encoded polypeptide [30], positioned immediately C-terminal to ves-common sequence block IX [30] (Figure 1, green bar). Only sequences encoded by the spliced, mature mRNA were included. The N-terminal cysteine residue was added to the peptide to facilitate conjugation to keyhole limpet hemocyanin (KLH). Rabbits were immunized with 200 µg of conjugated KLH-peptide emulsified in complete Freund’s adjuvant, followed by five booster immunizations of 100 µg each, given in incomplete Freund’s adjuvant at two-week intervals, to yield Ra-v1β(pep) antiserum. All procedures followed standard operating protocol 1, approved by Sigma-Genosys IACUC review (Animal Welfare Act registration #93-R-0283).
Immunochemical assays
Western blots
Western blots were performed essentially as described previously [29]. When chicken IgY and rabbit antisera were used as the primary antibody, horseradish peroxidase-conjugated rabbit anti-chicken IgY antibody (Thermo-Pierce Inc.; Rockford, IL) or goat anti-rabbit IgG (H+L chains; Zymed, Inc.; San Diego, CA), respectively, was used for detection. Bound antibody was detected with SuperSignal West Pico luminescent substrate (Thermo-Pierce). For some experiments, blots were stripped by incubation with 0.2 N NaOH for 5 minutes at room temperature, followed by a water wash, blocking, and re-probing as above.
Immunoprecipitations
Parasites were metabolically-labeled with L-[35S]-methionine at 100 µCi ml−1, and conventional and surface-specific immunoprecipitations (IPs) were performed essentially as described [32]. Recapture IPs were performed by initial capture of NP40-solubilized antigen with mAb 4D9.1G1 bound to rabbit anti-mouse IgG-coated Protein G-Sepharose. After washes to remove background, captured antigens were released with 8M urea in NP40 lysis buffer by incubation at 23°C for 5 minutes. The beads were removed and the antigen diluted 1/20 in NP40 lysis buffer, then re-incubated with Protein G-Sepharose beads to remove residual antibody. The cleared antigen was then re-incubated with Protein G-Sepharose beads pre-coated with either R6a-v1β765 or its preimmune serum (P6). After antigen capture and further washes to remove unbound materials, samples were heated in 5x SDS-PAGE sample buffer. Captured antigens were analyzed by sodium dodecylsulfate-polyacrylamide gel electrophoresis and fluorographic enhancement, as described previously [32,42]. Control samples included initial capture with mAb 4D9.1G1, mAb MBOC79B1 (anti-RAP1; a kind gift of G.H. Palmer), P6, or R6a-v1β765 only prior to analysis, and recapture with P6, as well as analysis of residual uncaptured antigens. Quantification of antigen capture was performed with the “Analyze/ Gels” function of ImageJ software (http://rsb.info.nih.gov/ij/).
Immunofluorescence
Dual-label live-cell immunofluorescence assays (live-IFA) were performed on IE [43], and the proportions of IE reactive with each antibody quantified, as described previously [10]. For quantification, a two-step amplification was employed to maximize sensitivity. When rabbit sera were used as primary antibody the signals were amplified by incubation with goat anti-rabbit IgG-biotin (Molecular Probes/ Invitrogen, La Jolla, CA), followed by visualization with streptavidin-Alexa 488 (Molecular Probes). When using mAb 4D9.1G1 as the primary antibody, signals were amplified with chicken anti-mouse IgG (H+L chains; Aves Labs, Inc.; Tigard, OR), and visualized with goat anti-chicken IgY-Alexa 594 (Molecular Probes). For colocalization studes, primary antibodies were directly detected with goat anti-mouse IgG (H & L chains)- Alexa 568 and goat anti-rabbit IgG- Alexa 488 (both from Molecular Probes) to minimize immune complex size and therefore localization uncertainty. Images were captured with a Retiga 1300B cooled monochromatic CCD camera (QImaging; Surrey, BC) on an Olympus BX50 microscope fitted with a 100× oil-immersion (NA 1.3) phase contrast objective. Images were processed using IP Lab (Scanalytics, Inc.; Rockville, MD) and ImageJ software. Black values were adjusted, based upon histogram analysis of the images, prior to merging images in order to reduce the non-specific background signal from autofluorescence. Co-localization was performed on RGB merged images using the ImageJ “RG2B Colocalization” plug-in.
Results and Discussion
To make a connection between the ves1β genes and their encoded products antisera were generated to predicted ves1β polypeptide sequences, either as synthetic peptide or as recombinantly expressed polypeptides. These were then used to characterize the parasite products with which they react and their locations.
Western blot analysis
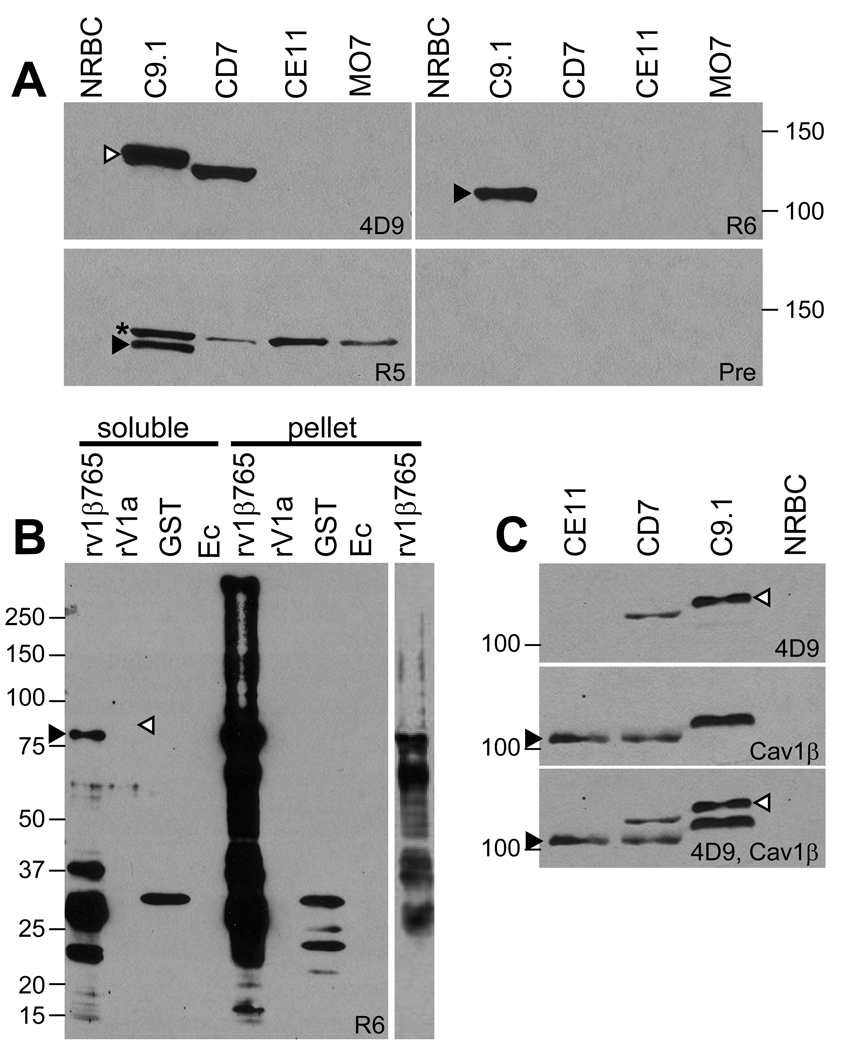
Rabbit antibodies raised to a recombinant polypeptide representing amino acids 1– 765 of the polypeptide encoded by the transcriptionally active C9.1 line LAT-associated ves1β gene (R5a-v1β765 and R6a-v1β765) and the anti-VESA1a monoclonal antibody, 4D9.1G1, were used to probe western blots of antigens derived from B. bovis C9.1, CD7, CE11, and MO7 line IE. As expected, mAb 4D9.1G1 reacted with bands of 128 and 123 kDa, respectively, on antigens from the C9.1 and CD7 lines, consistent with its known reactivity pattern with VESA1a in these parasite lines (Figure 2A, top left panel, white arrowhead) [19]. It was anticipated that cross-reactivity would occur between the ves1β-encoded polypeptide and VESA1a due to the inclusion of nine sequence segments shared between ves1α and ves1β genes [30]; therefore, the antiserum was pre-adsorbed on E. coli expressing recombinant VESA1a. Accordingly, R6a-v1β765 antiserum reacted with a single band of 113 kDa in the C9.1 line antigen only (Figure 2A, top right panel, filled arrowhead), consistent with the known size of the VESA1b polypeptide expressed by this clonal line. In addition, a weak reaction occurred with an erythrocyte antigen of about 40 kDa (not shown). The anti-erythrocyte reactivity was present in rabbit 6 preimmune serum, P6, as well, but no reaction with the ves1β-encoded polypeptide was evident. R5a-v1β765 similarly reacted with the 113 kDa band in C9.1 line antigen only, but also reacted with a size-invariant band of approximately 120 kDa in all four B. bovis lines (Figure 2A, bottom left panel, filled arrowhead and asterisk, respectively). The nature and significance of this antigen is not clear, as R5 failed to react with this antigen in non-denatured form during immunoprecipitations (Figure 3A), leaving the host/ parasite origin of this polypeptide in question. While the possibility that this band represents an additional VESA1 polypeptide expressed from another ves locus cannot be definitively ruled out, this possibility is unsupported by available evidence, and unlike VESA1a or 1b it is invariant in size and reactivity. No reaction was observed of pooled preimmune serum from these rabbits with any parasite antigens (Figure 2A, bottom right panel). It is clear that rabbits immunized with the N-terminal 765 amino acids of the ves1β-encoded polypeptide react, in a variant-specific manner, with a polypeptide consistent in size with the VESA1b polypeptide. The non-reaction of these pre-adsorbed anti-ves1β polypeptide antisera with VESA1a is further confirmed in Figure 2B, wherein reactivity is evident with the ves1β-encoded recombinant polypeptide but not with the corresponding segment of recombinant VESA1a polypeptide (Figure 2B, black and white arrowheads, respectively). In contrast, antibodies to the 6x-his tag demonstrated comparable amounts of each polypeptide (not shown).
Figure 2. Western blot reactivity of anti-ves1β polypeptide antibodies.
Antigen prepared from non-infected erythrocytes, IE materials from B. bovis clonal lines C9.1, CD7, CE11, and MO7, or recombinant E. coli were reacted with the antisera to identify the sizes and variant-specificity of the recognized antigens. (A) Top panel contains antigens reacted with mAb 4D9.1G1 (left half; “4D9”) and R6a-v1β765 (right half; “R6”); bottom panel contains antigens reacted with R5a-v1β765 (left half; “R5”) and pooled preimmune from rabbits R5 and R6 (right half; “Pre”). Lanes are labeled based upon the B. bovis line antigen contained, or “NRBC” for normal, uninfected red blood cell antigens. (B) R6a-v1β765 antiserum was reacted with E. coli expressing various polypeptides, either as soluble antigens or from the BugBuster-insoluble pellet. Samples contained lysate from E. coli expressing recombinant ves1β(1–765) polypeptide (rv1β765); recombinant VESA1a polypeptide (rV1a); glutathione-S-transferase (GST); or non-recombinant E. coli (Ec). The right-most lane shows a lighter exposure of the internal pellet “rv1β765” sample to facilitate visualization of major bands contained within this sample. (C) The top panel shows recognition of VESA1a in various antigenically variant B. bovis clonal lines by mAb 4D9.1G1 (4D9), whereas reactivity of the Chicken a-v1β(f1-3) with the same blot following stripping is shown in the middle panel (Cav1β). The bottom panel is a superimposition of exposures from the two probings. Gel lanes contain antigen from B. bovis IE of the CE11, CD7, and C9.1 lines, or NRBC. Note the size-polymorphism of both VESA1a and the ves1β-encoded proteins. In all panels, black arrowheads indicate VESA1b-size polypeptides, white arrowheads indicate VESA1a polypeptides, and the asterisk indicates an unexpected, size-invariant, IE-associated antigen recognized only by R5a-v1β765. Numbers to the left or right of the figures refer to molecular mass standards (in kDa).
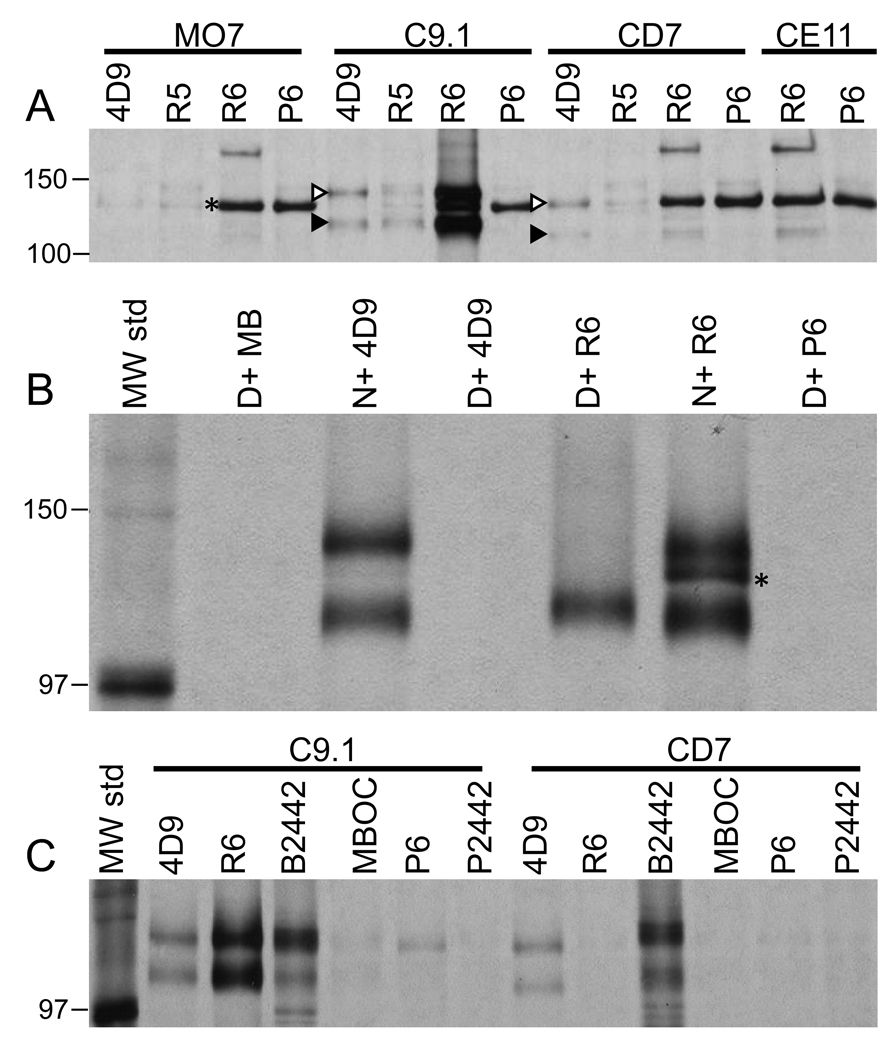
Figure 3. Immunoprecipitation (IP) analysis of metabolically labeled B. bovis IE antigens.
(A) Conventional IP analysis of variant B. bovis IE antigens solubilized in NP40 lysis buffer prior to addition of antibody. Above each set of samples is labeled the variant B. bovis line involved (MO7, C9.1, CD7, and CE11), whereas gel lanes are labeled according to the antibody used in the immunoprecipitation. (B) Conventional IP analysis of non-denatured and 8M urea-denatured B. bovis C9.1 line IE antigens. Lanes are labeled to indicate reaction between non-denatured (N) or denatured (D) antigen, and the antibody source. (C) Surface-specific IP analysis of C9.1 and CD7 variant B. bovis clonal line IE. Parasite line is indicated above each sample set. Lanes are labeled according to the antibody used during immunoprecipitation. In all three panels only the region of the gel around the VESA1 bands is shown. The asterisk indicates a presumably unrelated parasite antigen captured from all lines by antibodies present in R6 serum prior to immunization. The white and black arrowheads indicate the VESA1a and 1b subunits, respectively, which were co-precipitated by mAb 4D9.1G1. The antibodies used in this figure include: 4D9, mAb 4D9.1G1; R5, R5a-v1β765, R6, R6a-v1β765; P6, rabbit 6 preimmune serum; MB, anti-Rap1 mAb MBOC79B1; B2442, bovine #2442 infection serum to B. bovis MO7; and P2442, bovine #2442 preimmune serum. “MW std” indicates molecular mass standards. Numbers on the left indicate positions of molecular mass standards (in kDa).
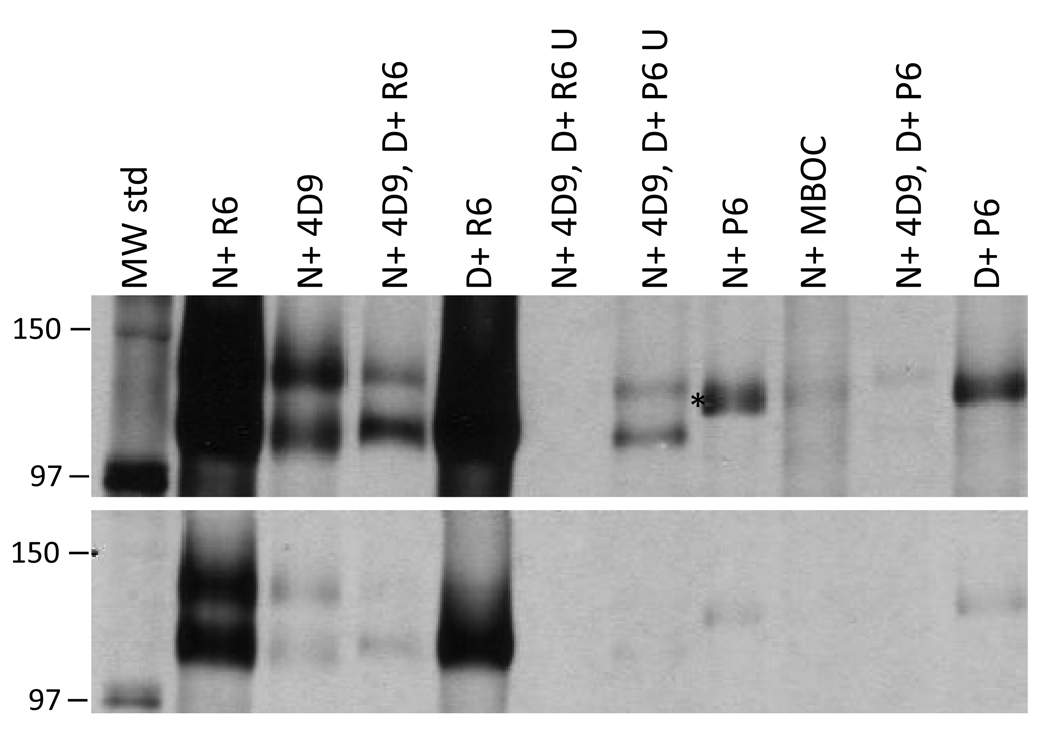
Figure 4. Confirmation of VESA1b identity through re-capture IP of VESA1 subunits.
B. bovis C9.1 line VESA1 was immunoprecipitated directly from non-denatured (N) or 8M urea-denatured (D) antigen. Alternatively, antigen was captured from non-denatured antigen with mAb 4D9.1G1, then was released by denaturation with 8M urea, diluted to allow antibody binding, and re-captured with a second antibody of different specificity. The order of reagents used in this experiment is indicated, as well as the nature of the antigens. The designation “U” refers to uncaptured antigens left over after recapture with the indicated antibodies. Only the region of the gel around the VESA1 complex is shown. The lower panel is a lighter exposure of the same region to facilitate visualization of bands in more heavily exposed samples. Antibodies used include: 4D9, mAb 4D9.1G1; R6, R6a-v1β765; P6, rabbit 6 preimmune serum; MBOC, mAb MBOC79B1. Asterisk and numbering on the left have the same meanings as in Figure 3
Because we were interested in obtaining antibodies which were pan-reactive with ves1β-encoded polypeptides in multiple assays and the rabbits had reacted primarily to immunodominant variant-specific epitopes, we attempted to focus the immune response on less immunogenic common epitopes by immunizing with smaller segments of the polypeptide. Chickens were immunized with a recombinant polypeptide containing three short conserved segments from the predicted polypeptide, generating Ca-v1β(f1-3). When Ca-v1β(f1-3) IgY was used in western blot assays, size-polymorphic antigens of 113 kDa in the C9.1 line and 107–109 kDa in the CD7 and CE11 lines were detected, consistent with the known sizes and polymorphism of the VESA1b polypeptide (Figure 2C, middle and bottom panels; black arrowheads). These bands were not detected by preimmune IgY from the same chicken (not shown). The chicken IgY also reacted lightly with a doublet of approximately 70 kDa of erythrocyte origin (not shown). This result confirmed our initial observation made with Ra-v1β765 regarding the size of the IE-related bands and extended it to include other clonal lines. Unfortunately, the chicken IgY failed to react usefully in other immunoassays. Another attempt was therefore made to focus the response on a ves1β-common sequence, without competition from variant epitopes. This was done by immunizing rabbits with a synthetic peptide representing amino acids 740–763 of the phagemid2-2 form of the LAT-associated C9.1 line ves1β gene (Figure 1, green bar) [30]. This sequence is well but imperfectly conserved among different predicted ves1β gene products, and is predicted to be immunogenic based upon its composition and characteristics [44]. The peptide was conjugated to keyhole limpet hemocyanin and used to immunize rabbits, yielding the rabbit anti-ves1β(peptide) antisera, Ra-v1β(pep). When Ra-v1β(pep) was employed in western blots it, too, gave a variant-specific reaction with a polypeptide the size of VESA1b (not shown). Unexpectedly, the reaction occurred strongly with CE11 line antigen and weakly with CD7, but not with C9.1 or MO7. Again, preimmune serum failed to react specifically with any parasite antigens (not shown). Although this result was not expected it is also not unreasonable: although well-conserved, the segment of protein represented by this peptide is not perfectly conserved. Phagemid2-2, whose sequence was used, carries one of several slightly different forms of the ves1β gene observed to be present in the LAT among members of the B. bovis C9.1 line population after having been in culture for an extended period after in vitro cloning. Thus, this peptide likely more closely matched the major ves1β sequence expressed by the CE11 and CD7 line populations than it did that of the C9.1 or other clonal line populations. Regardless, antibodies generated to a ves1β-encoded polypeptide again reacted only with a polypeptide band the same apparent size as VESA1b. Thus, whether immunizing with a short polypeptide only, a chimera of three longer peptide segments, or a truncated version of the protein containing nearly 70% of the predicted sequence, all antibodies reacted specifically with bands consistent with the known sizes of VESA1b subunits, and these reactions were variant-specific among the clonal B. bovis lines. Regrettably, both the Ca-v1β(f1-3) and Ra-v1β(pep) antibodies either failed to react or gave uninterpretable reactions in any assays besides western blotting, and so could not be used in other immunoassays.
Immunoprecipitation
The host-parasite origin of the antigens recognized by rabbit a-v1β765 sera was determined by IP of antigens from L-[35S]-methionine metabolically-labeled B. bovis IE. In conventional IP assays, R6a-v1β765 antiserum captured an antigen doublet from B. bovis C9.1 line IE antigen that exactly co-migrated with that captured by the VESA1a-specific mAb 4D9.1G1 (Figure 3A, white and black arrowheads). Previously, this doublet was demonstrated to contain the VESA1a and 1b subunits [23]. This result confirmed the parasite-derived nature of the precipitated antigens and suggested that both subunits of the VESA1 complex were being captured. Importantly, the antigen doublet was captured by R6a-v1β765 only from C9.1 line antigen, again indicating its variant-specific nature. This is in contrast to the pattern for mAb 4D9.1G1, which recognizes CD7 line VESA1a and captured the VESA1 doublet from that line. Neither antibody captured VESA1 antigen from MO7 or CE11 line IE. R6a-v1β765 and its preimmune (P6) both precipitated a presumably unrelated parasite antigen of about 120 kDa. In addition, R6a-v1β765 appears to specifically capture a size-constant antigen of about 180 kDa after immunization. However, this reaction was observed only when it failed to capture the VESA1-sized doublet, suggesting the induction of a weakly cross-reactive specificity that is overwhelmed in the presence of the specific heavy reaction with the VESA1-sized doublet (e.g., C9.1 antigen, R6 sample). The identity of this antigen is not clear, but it is not recognized on western blots (not shown) and variant-common reactions on live-cell IFAs were not observed, suggesting that its presence in this experiment does not conflict with our identification of the ves1β-encoded polypeptides.
To establish that capture of the VESA1-sized doublet with R6a-v1β765 occurred through reactivity with the lower band specifically, samples were solubilized in ordinary lysis buffer, or in lysis buffer containing 8M urea. The denatured antigen was then diluted 1/20 in the same lysis buffer without urea, and used for IP analysis. The results demonstrate that R6a-v1β765 captures only the lower of the two bands when the antigen is first denatured to dissociate the subunits (Figure 3B and Figure 4, samples “D+ R6”). Consistent with prior unpublished observations, mAb 4D9.1G1 recognized VESA1a only very poorly following full denaturation of this antigen.
The VESA1 protein is known to be expressed on the IE surface [10,32]. Therefore, a surface-specific IP assay was performed on L-[35S]methionine-labeled parasites to ask whether the antigen could be found there. A protein doublet of 128 and 113 kDa was captured from B. bovis C9.1 line IE with R6a-v1β765, co-migrating with that captured by mAb 4D9.1G1 or bovine immune serum (Figure 3C). In contrast, whereas 4D9.1G1 and bovine immune serum captured a protein doublet from the surface of B. bovis CD7 line IE that was similar in size but slightly smaller than that of C9.1, R6a-v1β765 failed to capture these antigens (Figure 3C). This result confirms the apparent specificity of antigen recognition observed by western blotting. As R6a-v1β765 specifically captures the smaller subunit, this provides the first direct evidence for the surface disposition of the ves1β-encoded antigen, which we propose is the VESA1b subunit.
To confirm the presumptive VESA1b identity of the antigen recognized by western blotting and conventional IP, a re-precipitation assay was performed. Parasite antigens were first immunoprecipitated with mAb 4D9.1G1, released by incubation with 8M urea, then diluted and re-precipitated with R6a-v1β765. When this assay was performed, R6a-v1β765 re-captured the lower band of the denatured complex very efficiently, whereas only about 40% of the VESA1a was re-captured (Figure 4). Thus, the ratio of the higher (VESA1a) to lower (ves1β-encoded) mass bands was reduced from 1.38 after mAb 4D9.1G1 capture to 0.56 following R6a-v1β765 re-capture. Although R6a-v1β765 specifically captures VESA1b if antigen is first denatured, VESA1a is also re-captured following dilution and, presumably, partial renaturation and reassociation of VESA1a and 1b. The P6 preimmune serum failed to capture either band under any circumstance (Figure 4). Conversely, neither VESA1 subunit remained in the supernatant in detectable quantities following re-capture by R6a-v1β765, whereas both subunits remained following re-capture by preimmune (P6) serum. These results clearly indicate that the anti-ves1β polypeptide antiserum, R6a-v1β765, captures the lower mass subunit of the VESA1 protein, VESA1b, as previously defined [23], bringing down VESA1a with it.
Live-cell immunofluorescence
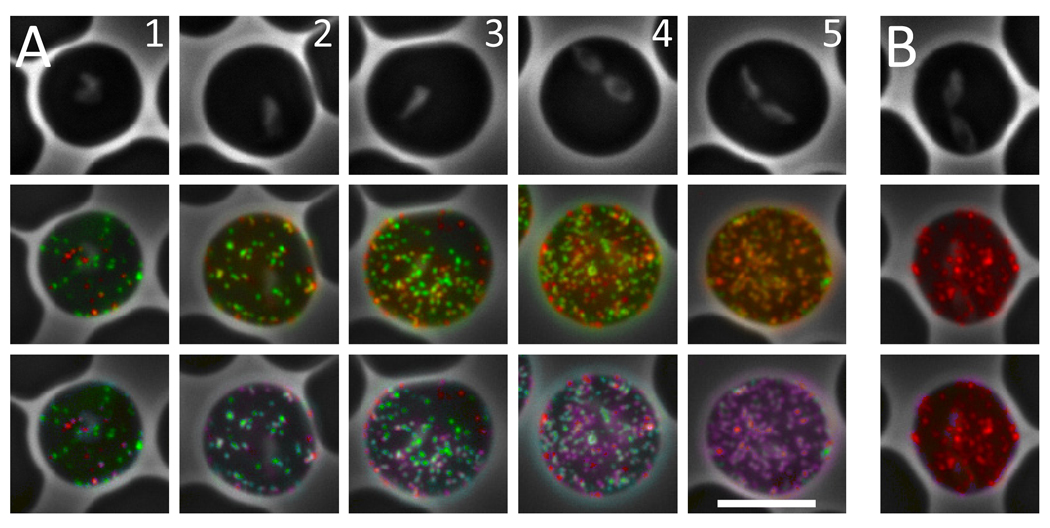
VESA1a was previously demonstrated by immunoelectron microscopy to localize at the tips of small, ridge-like deformations of the IE membrane [19]. It was presumed that the intact VESA1 complex was similarly situated, but there was no direct evidence for VESA1b location. When observed by live-cell IFA with mAb 4D9.1G1, B. bovis IE are immunoreactive primarily in a finely punctate pattern consistent with localization of most antigen in such ridge-like structures (Figure 5B). A live-cell IFA [43] was therefore performed to ask whether the polypeptides recognized by the anti-ves1β antisera were similarly distributed on the IE surface. The rabbit anti-peptide antisera, chicken anti-ves1βf1-3 antisera, and R5a-v1β765 antiserum failed to react significantly with native antigen on the surface of live cells (not shown). This may be due to a failure to recognize epitopes in their proper 3-dimensional conformation, or to a lack of surface exposure on the native antigen of linear epitopes recognized by these sera. In contrast, R6a-v1β765 antiserum reacted very strongly with its target antigen on the surface of live B. bovis C9.1 line cells, in a distinct, finely punctate pattern indistinguishable from that of VESA1a (Figure 5A). Preimmune serum failed to react with IE. Significantly, extensive co-localization of VESA1a and VESA1b occurred in reactions with mature C9.1 line-parasitized erythrocytes when using mAb 4D9.1G1 and R6a-v1β765, respectively, as primary antibodies. However, co-localization is not absolute and the ratio of the two antibodies bound, and presumably of the two target antigens, is not consistent over individual foci. This effect is particularly obvious on IE with low overall antigen content, for example Figure 5A, column 1. With parasite maturation and increasing antigen density there is a tendency for progressively greater signal colocalization (Figure 5A, columns 1–5). The varying ratios are evident in the range of pseudocolor applied by the co-localization software (ImageJ, RG2B Colocalization plug-in), ranging from red or green (one signal only) to purple, lavender, or cyan, depending upon signal ratios (Figure 5, lower panels). A calculated signal ratio cutoff of 0.10 was used, below which only the major signal (i.e., red or green) is displayed. These results both confirm the surface exposure of VESA1b on the IE surface, and demonstrate its extensive co-localization with VESA1a [10,32]. Previously, it was demonstrated that cytoadhesion can be both inhibited and reversed by the mAbs 4D9.1G1 (used here) and 3F7.1H11, each of which recognizes the VESA1a subunit specifically [19]. The failure to quantitatively co-localize the antigens, and the variability in their recognition over individual foci, raises several intriguing possibilities. One possibility is that the bidirectional promoter driving expression of both subunits may not be capable of transcribing in both directions simultaneously, resulting in temporal segregation of subunit expression. Alternatively, formation of VESA1 heterodimeric complex may occur within the IE membrane rather than earlier in the trafficking pathway due to a lack of co-accessibility prior to that point. The inconsistent subunit stoichiometry has potential ramifications for formation of functional VESA1 and thus for its adhesive function. For example, are both subunits necessary in complexed form-or at all-to achieve adhesion? If individual subunits can mediate cytaodhesion, do VESA1a and 1b bind to different endothelial receptors? These questions deserve further investigation to clarify the developmental timing of expression and transport of each subunit to the IE surface.
Figure 5. Live-cell immunofluorescence of B. bovis C9.1 line IE.
Mature stage B. bovis C9.1 line IE were reacted with mAb 4D9.1G1 and R6α-v1β765 (panel A; five individual cells of increasing parasite maturity and antigen density are shown) or with mAb 4D9.1G1 and P6 (panel B). Localization of VESA1a by mAb 4D9.1G1 is shown in red and reactivity of the rabbit antisera with VESA1b is shown in green. The top row shows phase contrast images of the individual IE, whereas the second row shows patterns of immunoreactivity superimposed upon the phase contrast images. Note the finely punctate pattern of labeling by rabbit antisera, which is indistinguishable from that of the monoclonal antibody. Co-localization of the two antigens is shown in the bottom row by the superimposition of blue color where red and green signals significantly coincide (ratio ≥0.10 of other signal). Note also the clearly variable ratios of the two antigens, and the occasional signals apparently derived from one antigen only. Single signal foci show up as red or green, whereas co-localized signals appear in varying shades of purple, lavender, and cyan, depending upon the calculated signal ratios. The white bar represents 5 µm; magnification is identical in each image.
Despite co-localization of VESA1a and 1b on the IE surface, the mechanistic involvement of VESA1b in adhesion remains to be established. Unfortunately, the available monospecific antiserum, R6a-v1β765 cannot be used to determine this directly, as it reacts variant-specifically with the C9.1 and B9 lines (Supplementary Figure 1), neither of which is cytoadhesive. The lack of R6a-v1β765 reactivity with lines CD7, CE11, C8, and H10 is consistent with the identification of its target as VESA1b. Similarly, the presence of reactivity with the B9 line is not inconsistent with this interpretation. B9 is an immediate descendant of the C9.1 line, having been cloned from cultures of parasites taken from a calf on day 41 post-infection with the C9.1 line [10]. Further, at the genetic level B9 is very similar to C9.1, hybridizing on Southern blots with oligonucleotide probes specific for the C9.1 LAT, whereas other variants did not [29,30]. In light of the segmented gene conversion mechanism of variation, we consider the most parsimonious interpretation of these results to be that sequences encoding one or more epitopes found in the C9.1 VESA1b polypeptide were retained during events leading to the variant B9 genotype. This does not diminish our interpretation of VESA1b being encoded by ves1β.
The data provided here present a direct connection and provide a clear argument for the designation of polypeptides encoded by ves1β genes as the 1b subunit of the VESA1 holoprotein. These connections include correct apparent molecular masses and appropriate size polymorphisms of parasite-encoded polypeptides, incorporation into the same heterodimeric complex as VESA1a, and variant-specificity of the epitopes recognized by both the anti-peptide and anti-polypeptide antisera. Further, the ves1β-encoded polypeptide co-localized extensively with VESA1a on the IE membrane surface where VESA1 is involved in cytoadhesion [18,19]. The polypeptides recognized by ves1β polypeptide-specific antibodies are parasite-synthesized and, importantly, co-precipitate the VESA1a polypeptide. Based on this body of evidence, we conclude that ves1β genes encode the 1b polypeptide subunit of the VESA1 cytoadhesion ligand.
Prior work has demonstrated that a ves1α gene encoding the expressed VESA1a polypeptide [29] and a ves1β gene, here shown to encode the expressed VESA1b polypeptide, are co-localized within the locus of active ves transcription (LAT) [30]. These genes are divergently-oriented, with a shared promoter region and overlapping 5'-UTR sequences, a basic organization that is frequently repeated elsewhere in the genome [31]. Interestingly, preliminary data from transient transfection experiments indicates the functional equivalence of 5' regulatory sequences from many such loci (Wang, X. and Allred, D.R., unpublished data), suggesting the possibility of epigenetic control over the activity of individual loci. This possibility is supported by observations of steady state RNA by RT-PCR employing “universal primers” to the ves1α gene which are highly consistent with transcriptional activity of one ves locus at a time [45]. The divergent organization of the ves1α and ves1β genes of the LAT conceptually would be beneficial for regulating the coordinated expression of both subunits of VESA1. This possibility was difficult to pursue previously because the identity of the polypeptides encoded by the 1β branch of the ves multigene family was not known. With the establishment of the identity of ves1β gene-encoded products as VESA1b polypeptides, this gap in our knowledge may finally be closed, and it is now possible to consider modeling the regulation of expression of both subunits of the VESA1 virulence factor. However, immunolocalization data provided herein intriguingly bring the concept of closely coordinated expression and trafficking into question, despite the nature of the gene organization. This information will be important in devising strategies to reduce or prevent cytoadhesion through control of parasite transcriptional or protein trafficking activities, perhaps providing novel avenues for the alleviation of cerebral babesiosis and similar maladies induced by other parasites.
Supplementary Material
Supplementary Figure 1 Variant-specificity of rabbit anti-v1β765 polypeptide antisera with B. bovis clonal variants Rabbit antisera were reacted with IE carrying the genetically-related but antigenically variant B. bovis clonal lines C9.1, B9, C8, H10, CD7, and CE11. The results are plotted as the proportion of IE reactive with each primary antibody (error bars indicate ranges from repeated experiments). R5, R5a-v1β765 antiserum; R6, R6a-v1β765 antiserum; P5 and P6, rabbit 5 and rabbit 6 preimmune sera.
Acknowledgments
The authors thank Alexia Berg, Allison Vansickle, Jimmy Kidwell, and Brady Pratt for animal handling, and the anonymous reviewers for their fair and conscientious efforts to improve this manuscript. Supported by grant #R01 AI055864 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kuttler KL. World-wide impact of babesiosis. In: Ristic M, editor. Babesiosis of Domestic Animals and Man. Boca Raton: CRC Press, Inc.; 1988. pp. 1–22. [Google Scholar]
- 2.Wright IG, Goodger BV, Clark IA. Immunopathophysiology of Babesia bovis and Plasmodium falciparum infections. Parasitol Today. 1988;4:214–218. doi: 10.1016/0169-4758(88)90161-5. [DOI] [PubMed] [Google Scholar]
- 3.Wright IG, Goodger BV, Buffington GD, Clark IA, Parrodi F, Waltisbuhl DJ. Immunopathophysiology of babesial infections. Trans R Soc Trop Med Hyg. 1989;83 Supplement:11–13. doi: 10.1016/0035-9203(89)90596-8. [DOI] [PubMed] [Google Scholar]
- 4.Jeffery GM, Eyles DE. The duration in the human host of infections with a Panama strain of Plasmodium falciparum. Am J Trop Med Hyg. 1954;2:219–224. doi: 10.4269/ajtmh.1954.3.219. [DOI] [PubMed] [Google Scholar]
- 5.Bottius E, Guanzirolli A, Trape JF, Rogier C, Konate L, Druilhe P. Malaria: even more chronic in nature than previously thought; evidence for subpatent parasitemia detectable by the polymerase chain reaction. Trans R Soc Trop Med Hyg. 1996;90:15–19. doi: 10.1016/s0035-9203(96)90463-0. [DOI] [PubMed] [Google Scholar]
- 6.Schneider P, Bousema JT, Gouagna LC, Otieno S, Van de Vegte-Bolmer M, Omar SA, Sauerwein RW. Submicroscopic Plasmodium falciparum gametocyte densities frequently result in mosquito infection. Am J Trop Med Hyg. 2007;76:470–474. [PubMed] [Google Scholar]
- 7.Mahoney DF, Wright IG, Mirre GB. Bovine babesiasis: the persistence of immunity to Babesia argentina and B. bigemina in calves (Bos taurus) after naturally acquired infection. Ann Trop Med Parasitol. 1973;67:197–203. doi: 10.1080/00034983.1973.11686877. [DOI] [PubMed] [Google Scholar]
- 8.Mahoney DF, Wright IG, Goodger BV. Immunity in cattle to Babesia bovis after single infections with parasites of various origin. Aust Vet J. 1979;55:10–12. doi: 10.1111/j.1751-0813.1979.tb09535.x. [DOI] [PubMed] [Google Scholar]
- 9.Calder JAM, Reddy GR, Chieves L, Courtney CH, Littell R, Livengood JR, Norval RAI, Smith C, Dame JB. Monitoring Babesia bovis infections in cattle by using PCR-based tests. J Clin Microbiol. 1996;34:2748–2755. doi: 10.1128/jcm.34.11.2748-2755.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allred DR, Cinque RM, Lane TJ, Ahrens KP. Antigenic variation of parasite-derived antigens on the surface of Babesia bovis-infected erythrocytes. Infect Immun. 1994;62:91–98. doi: 10.1128/iai.62.1.91-98.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright IG. An electron microscopic study of intravascular agglutination in the cerebral cortex due to Babesia argentina infection. Intl J Parasitol. 1972;2:209–215. doi: 10.1016/0020-7519(72)90008-2. [DOI] [PubMed] [Google Scholar]
- 12.Wright IG. Ultrastructural changes in Babesia argentina-infected erythrocytes in kidney capillaries. J Parasitol. 1973;59:735–736. [PubMed] [Google Scholar]
- 13.Wright IG, Goodger BV, McKenna RV, Mahoney DF. Acute Babesia bovis infection: A study of the vascular lesions in kidney and lung. Z Parasitenk. 1979;60:19–27. [Google Scholar]
- 14.Nevils MA, Figueroa JV, Turk JR, Canto GJ, Le V, Ellersieck MR, Carson CA. Cloned lines of Babesia bovis differ in their ability to induce cerebral babesiosis in cattle. Parasitol Res. 2000;86:437–443. doi: 10.1007/s004360050691. [DOI] [PubMed] [Google Scholar]
- 15.Miller L. Distribution of mature trophozoites and schizonts of Plasmodium falciparum in the organs of Aotus trivirgatus, the night monkey. Am J Trop Med Hyg. 1969;18:860–865. doi: 10.4269/ajtmh.1969.18.860. [DOI] [PubMed] [Google Scholar]
- 16.Luse SA, Miller LH. Plasmodium falciparum malaria: ultrastructure of parasitized erythrocytes in cardiac vessels. Am J Trop Med Hyg. 1971;20:655–660. [PubMed] [Google Scholar]
- 17.David PH, Hommel M, Miller LH, Udeinya IJ, Oligino LD. Parasite sequestration in Plasmodium falciparum malaria: Spleen and antibody modulation of cytoadherence of infected erythrocytes. Proc Natl Acad Sci (USA) 1983;80:5075–5079. doi: 10.1073/pnas.80.16.5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Connor RM, Long JA, Allred DR. Cytoadherence of Babesia bovis-infected erythrocytes to bovine brain capillary endothelial cells provides an in vitro model for sequestration. Infect Immun. 1999;67:3921–3928. doi: 10.1128/iai.67.8.3921-3928.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Connor RM, Allred DR. Selection of Babesia bovis-infected erythrocytes for adhesion to endothelial cells co-selects for altered variant erythrocyte surface antigen isoforms. J Immunol. 2000;164:2037–2045. doi: 10.4049/jimmunol.164.4.2037. [DOI] [PubMed] [Google Scholar]
- 20.Hutchings CL, Li An, Fernandez KM, Fletcher T, Jackson LA, Molloy JB, Jorgensen WK, Lim CT, Cooke BM. New insights into the altered adhesive and mechanical properties of red blood cells parasitized by Babesia bovis. Mol Microbiol. 2007;65:1092–1105. doi: 10.1111/j.1365-2958.2007.05850.x. [DOI] [PubMed] [Google Scholar]
- 21.Udeinya IJ, Schmidt JA, Aikawa M, Miller LH, Green I. Falciparum malaria-infected erythrocytes specifically bind to cultured human endothelial cells. Science. 1981;213:555–557. doi: 10.1126/science.7017935. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt JA, Udeinya IJ, Leech JH, Hay RJ, Aikawa M, Barnwell J, Green I, Miller LH. Plasmodium falciparum malaria: An amelanotic melanoma cell line bears receptors for the knob ligand on infected erythrocytes. J Clin Invest. 1982;70:379–386. doi: 10.1172/JCI110627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Connor RM, Lane TJ, Stroup SE, Allred DR. Characterization of a variant erythrocyte surface antigen (VESA1) expressed by Babesia bovis during antigenic variation. Mol Biochem Parasitol. 1997;89:259–270. doi: 10.1016/s0166-6851(97)00125-4. [DOI] [PubMed] [Google Scholar]
- 24.Leech JH, Barnwell JW, Miller LH, Howard RJ. Identification of a strain-specific malarial antigen exposed on the surface of Plasmodium falciparum-infected erythrocytes. J Exp Med. 1984;159:1567–1575. doi: 10.1084/jem.159.6.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magowan C, Wollish W, Anderson L, Leech J. Cytoadherence by Plasmodium falciparum-infected erythrocytes is correlated with the expression of a family of variable proteins on infected erythrocytes. J Exp Med. 1988;168:1307–1320. doi: 10.1084/jem.168.4.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biggs BA, Anders RF, Dillon HE, Davern KM, Martin M, Petersen C, Brown GV. Adherence of infected erythrocytes to venular endothelium selects for antigenic variants of Plasmodium falciparum. J Immunol. 1992;149:2047–2054. [PubMed] [Google Scholar]
- 27.Allred DR, Al-Khedery B. Antigenic variation and cytoadhesion in Babesia bovis and Plasmodium falciparum: different logics achieve the same goal. Mol Biochem Parasitol. 2004;134:27–35. doi: 10.1016/j.molbiopara.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 28.Allred DR. Babesiosis: persistence in the face of adversity. Trends Parasitol. 2003;19:51–55. doi: 10.1016/s1471-4922(02)00065-x. [DOI] [PubMed] [Google Scholar]
- 29.Allred DR, Carlton JMR, Satcher RL, Long JA, Brown WC, Patterson PE, O'Connor RM, Stroup SE. The ves multigene family of B. bovis encodes components of rapid antigenic variation at the infected erythrocyte surface. Mol Cell. 2000;5:153–162. doi: 10.1016/s1097-2765(00)80411-6. [DOI] [PubMed] [Google Scholar]
- 30.Al-Khedery B, Allred DR. Antigenic variation in Babesia bovis occurs through segmental gene conversion of the ves multigene family, within a bidirectional site of transcription. Mol Microbiol. 2006;59:402–414. doi: 10.1111/j.1365-2958.2005.04993.x. [DOI] [PubMed] [Google Scholar]
- 31.Brayton KA, Lau AOT, Herndon DR, Hannick L, Kappmeyer LS, Berens SJ, Bidwell SL, Brown WC, Crabtree J, Fadrosh D, Feldblum T, Forberger HA, Haas BJ, Howell JM, Khouri H, Koo H, Mann DJ, Norimine J, Paulsen IT, Radune D, Ren Q, Smith RK, Jr, Suarez CE, White O, Wortman JR, Knowles DP, Jr, McElwain TF, Nene VM. Genome sequence of Babesia bovis and comparative analysis of Apicomplexan hemoprotozoa. PLOS Pathogens. 2007;3:e148.1–e148.13. doi: 10.1371/journal.ppat.0030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allred DR, Hines SA, Ahrens KP. Isolate-specific parasite antigens of the Babesia bovis-infected erythrocyte surface. Mol Biochem Parasitol. 1993;60:121–132. doi: 10.1016/0166-6851(93)90035-v. [DOI] [PubMed] [Google Scholar]
- 33.Dzikowski R, Li F, Amulic B, Eisberg A, Frank M, Patel S, Wellems TE, Deitsch KW. Mechanisms underlying mutually exclusive expression of virulence genes by malaria parasites. EMBO Reports. 2007;8:959–965. doi: 10.1038/sj.embor.7401063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voss TS, Tonkin CJ, Marty AJ, Thompson JK, Healer J, Crabb BS, Cowman AF. Alterations in local chromain environment are involved in silencing and activation of subtelomeric var genes in Plasmodium falciparum. Mol Microbiol. 2007;66:139–150. doi: 10.1111/j.1365-2958.2007.05899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth C, Bringaud F, Layden RE, Baltz T, Eisen H. Active late-appearing variable surface antigen genes in Trypanosoma equiperdum are constructed entirely from pseudogenes. Proc Natl Acad Sci (USA) 1989;86:9375–9379. doi: 10.1073/pnas.86.23.9375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamper SM, Barbet AF. Surface epitope variation via mosaic gene formation is potential key to long-term survival of Trypanosoma brucei. Mol Biochem Parasitol. 1992;53:33–44. doi: 10.1016/0166-6851(92)90004-4. [DOI] [PubMed] [Google Scholar]
- 37.Marcello L, Barry JD. Analysis of the VSG gene silent archive in Trypanosoma brucei reveals that mosaic gene expression is prominent in antigenic variation and is favored by archive substructure. Genome Res. 2007;17:1344–1352. doi: 10.1101/gr.6421207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borst P. Antigenic variation and allelic exclusion. Cell. 2002;109:5–8. doi: 10.1016/s0092-8674(02)00711-0. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez SD, Buening GM, Green TJ, Carson CA. Cloning of Babesia bovis by in vitro cultivation. Infect Immun. 1983;42:15–18. doi: 10.1128/iai.42.1.15-18.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gruber A, Zingales B. Alternative method to remove antibacterial antibodies from antisera used for screening of expression libraries. BioTechniques. 1995;19:28–29. [PubMed] [Google Scholar]
- 41.Pont-Kindon G. Construction of chimeric molecules by a two-step recombinant PCR method. BioTechniques. 1994;16:1010–1011. [PubMed] [Google Scholar]
- 42.Allred DR, Sherman IW. Developmental modulation of protein synthetic patterns by the human malarial parasite Plasmodium falciparum. Can J Biochem Cell Biol. 1983;61:1304–1314. doi: 10.1139/o83-167. [DOI] [PubMed] [Google Scholar]
- 43.Allred DR. Immunochemical methods for identification of Babesia bovis antigens expressed on the erythrocyte surface. Methods. 1997;13:177–189. doi: 10.1006/meth.1997.0510. [DOI] [PubMed] [Google Scholar]
- 44.Hopp TP, Woods KR. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci (USA) 1981;78:3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zupanska AK, Drummond PB, Swetnam DM, Al-Khedery B, Allred DR. Universal primers suitable to assess population dynamics reveal apparent mutually exclusive transcription of the Babesia bovisves1α gene. Mol Biochem Parasitol. 2009;166:47–53. doi: 10.1016/j.molbiopara.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Variant-specificity of rabbit anti-v1β765 polypeptide antisera with B. bovis clonal variants Rabbit antisera were reacted with IE carrying the genetically-related but antigenically variant B. bovis clonal lines C9.1, B9, C8, H10, CD7, and CE11. The results are plotted as the proportion of IE reactive with each primary antibody (error bars indicate ranges from repeated experiments). R5, R5a-v1β765 antiserum; R6, R6a-v1β765 antiserum; P5 and P6, rabbit 5 and rabbit 6 preimmune sera.