Abstract
Background
Antiretroviral therapy (ART) has become more available throughout the developing world during the past five years. The World Health Organization recommends nonnucleoside reverse transcriptase inhibitor-based regimens as initial ART. However, their efficacy may be compromised by resistance mutations selected by single-dose nevirapine (sdNVP) used to prevent mother-to-child-transmission of HIV-1 (PMTCT). There is no simple and efficient method to detect such mutations at initiation of ART.
Methods
181 women participating in a PMTCT clinical trial who started NVP-ART after they had received sdNVP or placebo were tested for nevirapine-resistance point-mutations (K103N, Y181C, and G190A) using 100 copies of HIV-1 DNA with a sensitive oligonucleotide ligation assay (OLA) able to detect mutants at low concentrations (≥5% of the viral population). Virologic failure was defined as plasma HIV-1 RNA confirmed >50 copies/mL between 6–18 months of NVP-ART.
Results
At initiation of NVP-ART, resistance mutations were identified in 26% of 148 participants given sdNVP (K103N-13%, Y181C-5%, G190A-19%; ≥2 mutations-10%) at a median 9.3 months after sdNVP. The risk of virologic failure was .62 (95% confidence interval (CI), 0.46–0.77) in women with ≥1 resistance mutation, compared to 0.25 (95% CI, 0.17–0.35) in those without detectable resistance mutations (P<.0001). Failure was independently associated with resistance, an interval of <6 months between sdNVP and NVP-ART initiation, and a viral load above the median at NVP-ART initiation.
Conclusions
Access to simple and inexpensive assays to detect low-concentrations of NVP-resistant HIV-1 DNA prior to the initiation of ART could help improve the outcome of first-line antiretroviral therapy.
Keywords: HIV-1, resistance mutations, nevirapine, HAART, oligonucleotide ligation assay, developing countries
Introduction
HIV-1 drug resistance testing is currently recommended before starting antiretroviral therapy (ART) in developed countries. However, in low resource settings, although ART has become gradually more available during the past five years, resistance testing is not routinely available. This is concerning because the World Health Organization (WHO) recommends including nonnucleoside reverse transcriptase inhibitors (NNRTI) such as nevirapine or efavirenz in initial, or “first-line”, ART regimen, and yet single-dose nevirapine (sdNVP) is used extensively to prevent mother-to-child-transmission of HIV-1 (PMTCT) and has been associated with the selection of resistance mutations and a higher risk of NNRTI-based ART failure [1–2]. This creates a diagnostic challenge because NVP-resistant mutants decay to concentrations not detected by consensus sequencing over 6 to 18 months following sdNVP [3]. Studies have observed that longer the period between sdNVP and initiation of ART, the more effective NNRTI-based ART is at achieving suppression HIV-1 replication [2, 4], the selection and decay rate of NVP-resistant viruses appear to vary by subtype [5], codon [6], between individuals [3, 7–9], and by methods used to detect mutations [7, 10, 11]. Recent studies suggest that detection of persistent resistance mutations would help identify individuals with a higher risk of virologic failure [12–16]. Because NVP-resistance mutations may persist longer in cellular DNA compared to plasma RNA [17, 18], we hypothesized that, in women who previously received sdNVP or placebo, resistance detected using a sensitive assay, capable of detecting point-mutations at concentrations as low as ≥5% of the viral population, applied to HIV-1 DNA at the time ART is initiated, would be associated with virologic failure of NVP-ART.
Methods
Study population and design
Women participating in PHPT-2 [19] (ClinicalTrials.gov Identifier: NCT00398684), a randomized, placebo-controlled trial of adding sdNVP in labor to ongoing zidovudine, who subsequently started NVP + stavudine and lamivudine (NVP-ART) were studied. We evaluated whether detection of NVP-resistance in the pre-ART specimen was associated with the virologic response to NVP-ART.
Banked plasma-poor whole blood specimens collected from women immediately prior to initiating NVP-ART were retrospectively assessed for three HIV-1 pol mutations conferring high-level resistance to nevirapine (K103N, Y181C, and G190A). The personnel testing the specimens did not have access to the participants’ clinical data or study randomization.
DNA extraction and quantification
DNA was extracted from plasma-depleted whole blood using QiAmp DNA Midi kits (Qiagen Inc., Valencia, CA). DNA was quantified using a Hoefer DyNA Quant 200 Fluorometer (Amersham Pharmacia Biotech, UK).
HIV-1 quantification and amplification
The HIV-1 DNA concentration in each participant’s genomic DNA was determined using real-time PCR amplification of HIV-1 long-terminal-repeat (LTR) [20]. DNA containing a total of 100 copies of HIV-1 was split between two separate nested PCR, then tested by OLA [21].
Oligonucleotide Ligation Assay (OLA)
The oligonucleotide ligation assay we developed uses an enzyme-linked immunoassay to detect HIV-1 point-mutations associated with resistance to antiretroviral agents [21]. Two PCR-amplicons from each participant were evaluated in duplicate by OLA for mutations in three codons (K103N, Y181C and G190A) associated with resistance to NVP, using reagents adapted to HIV-1 CRF01_AE viruses. OLA results were interpreted by comparison to subtype- and codon-specific controls. The assay was repeated in case of discordance between duplicates, and resistance was determined by majority. Samples testing positive for a mutant codon in either or both of the two PCR performed on each woman were classified as NVP-resistant, on the assumption that rare drug resistance mutants would be distributed randomly across independent PCR. Samples in which neither the wild-type nor mutant oligonucleotides produced a positive signal were considered indeterminate.
Statistical Analysis
The primary analysis was performed using the cases previously reported [1] for whom there were OLA data from specimens collected at ART initiation. The primary endpoint was a plasma HIV-1 RNA above 50 copies/mL between 6 and 18 months following initiation of NVP-ART, confirmed as sustained viremia by a second viral load above 50 copies/mL in the next blood specimen drawn from the subject. Observations were censored at time of switch to a protease inhibitor-based regimen or the last visit in cases of discontinuous follow-up. A secondary analysis was performed using the plasma HIV-1 RNA threshold of 400 copies/mL, and sensitivity analyses evaluated the outcome when death was classified as failure, and when specimens yielding insufficient HIV-1 DNA to perform OLA were assumed to have no NVP-resistance.
To compare distributions of categorical data and continuous variables, we used Fisher’s exact test and the Wilcoxon rank-sum test, respectively. For multivariate analyses, variables were selected on the basis of the results of univariate analyses (P<.20). A multivariate logistic regression model was used to study the association of NNRTI resistance mutations by consensus sequencing at 10 days post partum, time interval between sdNVP and ART initiation, and HIV RNA load and CD4 cell count at time of ART initiation with the detection of resistance mutations by OLA at ART initiation. Kaplan-Meier survival estimates of 18-month virologic failure rate were calculated and Cox proportional hazards models were used to assess the respective role on failure of NVP resistance by OLA and resistance mutations detected by sequencing 10 days after sdNVP, time from sdNVP to initiation of therapy, and plasma HIV RNA, CD4 cell count, and resistance mutations detected by OLA at initiation of therapy. Statistical analyses were performed using Stata 10.1 (StataCorp LP, Texas, USA).
Ethics
The PHPT-2 and PHPT Cohort Study protocols and their amendments received ethical clearance from the Thai Ministry of Public Health and Chiang Mai University Faculty of Medical Associated Sciences Ethics Committees.
Results
Study population
OLA results were obtained from 181 of the 269 PHPT-2 trial women (148 of the 221 women exposed to sdNVP and 33 of the 48 unexposed women) who started NVP-ART (P=.87) at a median 9.3 months after sdNVP. Eighty-eight women were not analyzed by OLA, primarily because the available plasma-poor whole blood specimens yielded insufficient HIV-1 DNA (see Figure 1). The characteristics of the 181 women tested by OLA were compared to the 88 (73 exposed to sdNVP and 15 unexposed) who were not tested by OLA (Table 1). Median age, CD4 cell count during pregnancy and at ART initiation, interval between sdNVP and NVP-ART, proportion exposed to sdNVP and risk of virologic failure between 6 and 18 months of therapy were similar between the two groups. However, median plasma HIV-1 RNA during pregnancy and at the initiation of NVP-ART was ~0.5 log10 copies/mL greater in those assessed by OLA. The risk of virologic failure was 0.31 in the group of women with OLA results and 0.33 in those without (P=.72). CRF 01_AE HIV-1 subtype was identified in 96% of the 253 subjects with no differences in the two groups.
Figure 1.
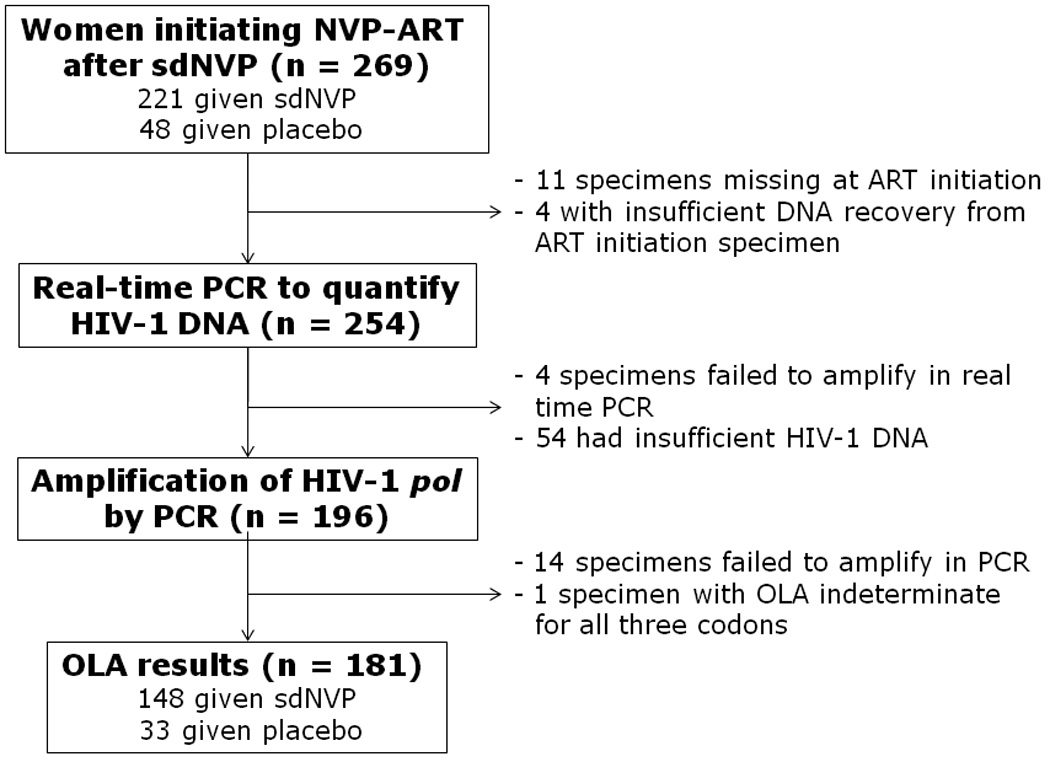
Women’s specimens analyzed by Oligonucleotide Ligation Assay (OLA) at initiation of nevirapine based antiretroviral treatment (NVP-ART).
Table 1.
Comparison of characteristics of women assessed by OLA versus those excluded from OLA testing due primarily to insufficient HIV-1 DNA in their specimen from the initiation of NVP-ART (all 269 women initiated nevirapine-based antiretroviral treatment after they had participated in a randomized double-blinded trial of adding single-dose nevirapine or placebo to zidovudine starting in the third trimester of pregnancy [1]).
| Characteristic | OLA results available (n =181) |
OLA results not available (n = 88) |
P* |
|---|---|---|---|
| Age, median years (interquartile range) | 28.8 (25.4 – 32.5) |
28.2 (24.9 – 33.1) |
.94 |
| CD4+ cell count during pregnancy, median cells/uL (interquartile range) |
186 (146 – 242) |
187 (125 – 236) |
.46 |
| Plasma HIV-1 RNA during pregnancy, median log10 copies/mL (interquartile range) |
4.63 (4.08 – 4.96) |
4.25 (3.64 – 4.88) |
.003 |
| Administered intrapartum single-dose NVP (percentage) |
148 (82%) |
73 (83%) |
.87 |
| HIV-1 subtype CRF01_AE (253 sequences available) | 169 (96%) |
74 (96%) |
1.0 |
| Interval between single-dose NVP and initiation of NVP-ART, median months (interquartile range) |
9.3 (3.8 – 15.9) |
6.2 (2.7 – 14.0) |
.10 |
| Started HAART less than 6 months after delivery (percentage) |
74 (41%) |
43 (49%) |
.24 |
| CD4+ cell count at initiation of NVP-ART, median cells/uL (interquartile range) |
166 (83 – 217) |
184 (99 – 231) |
.47 |
| Plasma HIV-1 RNA at initiation of NVP-ART – median log10 copies/mL (interquartile range) |
4.81 (4.27 – 5.17) |
4.27 (3.46 – 4.84) |
.0001 |
| Risk of virologic failure between 6 and 18 months of therapy (95% confidence interval) |
0.32 (0.26 – 0.40) |
0.33 (0.23 – 0.45) |
.90 |
P value for comparison of distributions using the Wilcoxon rank-sum test for continuous variables, Fisher’s exact test for categorical data, log rank for risk of failure
NVP resistance mutations detected by OLA
At least one of the three resistance mutations was detected at NVP-ART initiation in 26% of the 148 women exposed to sdNVP with OLA results: 13% with the K103N mutation, 5% with the Y181C, and 19% with the G190A. At least two mutations were detected in 10%. None of the 33 women unexposed to sdNVP had resistance mutations at NVP-ART initiation. The OLA was indeterminate for all three codons in one woman, excluded from analysis (Figure 1), and for a single codon in four other women (one at position 103, one 181, and two 190 including one with an identified K103N mutation). OLA of duplicate PCR reactions from the same DNA sample were discordant in 7% of samples, suggesting low concentrations of mutant. Replicate OLA results from individual PCR reactions were discordant in 6% of samples mostly with readings near the 5% limit of detection and across plates, suggesting inter-assay variability.
Factors associated with the detection of mutations by OLA
Of the 148 women with available OLA results, 145 had previously been tested for NNRTI resistance by consensus sequencing 10 days after sdNVP exposure. Women with NVP resistance mutations detected by consensus sequencing were more likely to have resistance mutations detected by OLA at the initiation of ART. Specifically, 50% (24 of 48) of the women with NNRTI resistance detected by consensus sequencing 10 days after sdNVP had at least one mutations by OLA, compared to 14% (14 of 97) of those who had no mutations by consensus sequencing (P<.001). More women initiating ART within 6 months of sdNVP had at least one NVP mutation detected by OLA compared to women initiating ART more than 6 months after sdNVP (37% (26 of 70) versus 15% (12 of 78), P=.004). Nevirapine mutations by OLA were not significantly associated with a higher viral load or a lower CD4 cell counts at time of ART start (P=.09 and P=.71, respectively).
Upon multivariate logistic regression, only NVP-mutations at 10 days postpartum and initiation of ART within 6 months of delivery remained independently associated with the detection of mutations by OLA (adjusted odds ratios 5.7; 95% CI, 2.5–13.0, P<.001, and 3.0; 95% CI, 1.3– 7.1, P=.01, respectively) and no significant interaction was detected between these two factors.
NVP-ART outcome according to resistance mutations detected by OLA
In the group of 148 women exposed to sdNVP and with available OLA results, three women (2%) died (7, 7, and 10 months after NVP-ART initiation), four (3%) withdrew from the study because they moved to another province, and 16 (11%) were lost to follow-up. In the group of 48 women unexposed to sdNVP, these figures were two (4%), one (2%) and one (2%), respectively. There were no cases of switch to protease inhibitor regimen before failure.
Among the 148 women administered sdNVP with available OLA results, the risk of virologic failure was 0.62 (95% CI, 0.46–0.77) in women with at least one resistance mutation detected by OLA at the time of NVP-ART initiation, compared to 0.25 (95% CI, 0.17–0.35) in those with no detectable resistance mutations (P<.0001) (Figure 2). The detection of two or more mutant codons was associated with a higher rate of virologic failure (HR 5.6; 95% CI, 2.9–11) (Table 2). Among the 33 women unexposed to sdNVP with available OLA result, the risk of failure was 0.13 (95% CI, 0.05–0.32) (Figure 2).
Figure 2.
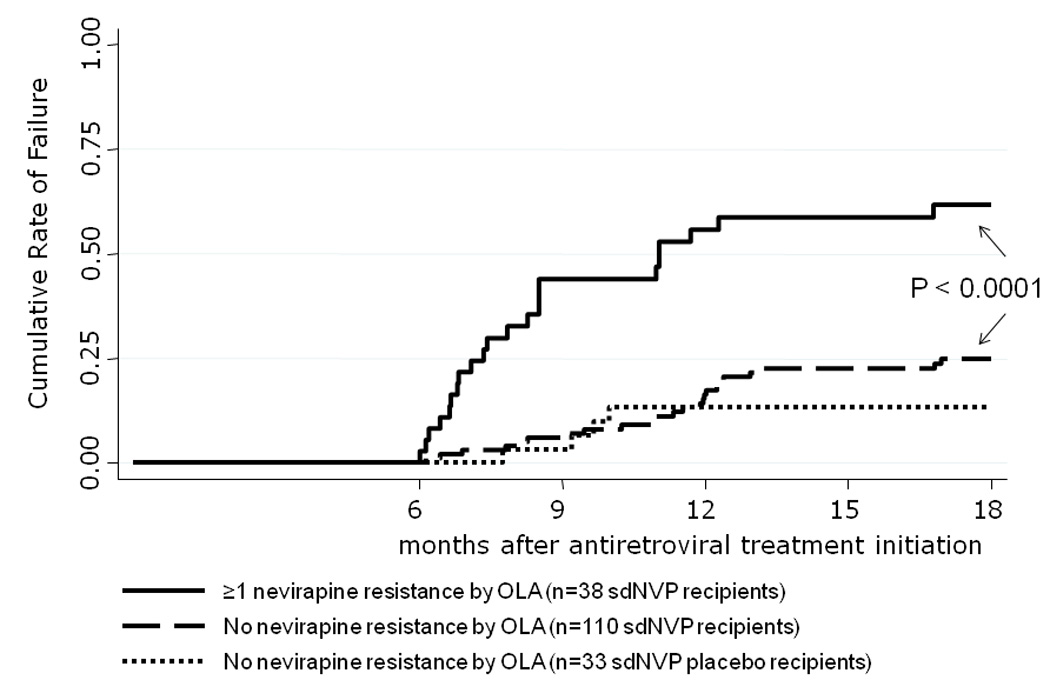
Kaplan-Meier estimates of virologic failure (confirmed viral load above 50 copies/mL) between 6 and 18 months after initiating a nevirapine based antiretroviral treatment in women who were or were not given single-dose nevirapine for prevention of mother-to-child-transmission of HIV-1,[19] and with or without resistance mutations detected by Oligonucleotide Ligation Assay (OLA).
Table 2.
Risk factors for virologic failure† in the 148 women given single dose nevirapine and tested for nevirapine resistant HIV-1 by OLA prior to initiation of nevirapine-based antiretroviral therapy.
| Univariate Analysis | Multivariate Analysis | |||
|---|---|---|---|---|
| Factor | Hazard ratio for failure (95% CI) |
P value | Adjusted Hazard Ratio (95% CI) |
P value |
| K103N detected by OLA* | 4.9 (2.6–9.4) | < .001 | - | |
| Y181C detected by OLA* | 6.1 (2.6–14) | < .001 | - | |
| G190A detected by OLA* | 2.8 (1.5–5.3) | .001 | - | |
| ≥ 1 NVP-resistance mutations (K103N, Y181C and/or G190A) by OLA |
3.8 (2.1–6.8) | < .001 | 2.5 (1.3–4.5) | .004 |
| ≥ 2 NVP-resistance mutations (K103N, Y181C and/or G190A) by OLA * |
5.6 (2.9–11) | < .001 | - | |
| Plasma HIV-1 RNA | - | |||
| - above median (4.58 log10 copies/ml) during pregnancy * |
2.4 (1.3–4.4) | .004 | ||
| - above median (4.77 log10 copies/ml) at initiation of NVP-ART |
2.2 (1.2–4.1) | .01 | 2.4 (1.3–4.5) | .007 |
| CD4 cell count |
- | |||
| - below median (183 cells/uL) during pregnancy* |
2.0 (1.1–3.7) | .02 | ||
| - below median (153 cells/uL) at initiation of NVP-ART |
1.2 (0.65–2.1) | .61 | - | |
| Time from intrapartum single dose nevirapine to antiretroviral therapy initiation ≤ 6 months |
3.4 (1.8–6.3) | < .001 | 3.2 (1.7–6.2) | .001 |
| NNRTI resistance mutations detected 10 days post partum |
1.4 (0.8–2.5) | .28 | ||
defined as confirmed plasma HIV-1 RNA >50 copies/mL between 6 and 18 months of ART.
Variables not included in the multivariate analysis because of co-linearity with other variables selected in the multivariate analysis
Other risk factors associated with virologic failure
Virologic failure was also increased (Table 2) with the detection by OLA of each of the three mutations (K103N and Y181C: P<.0001; G190A: P=.001); a viral load above the median at enrollment during pregnancy (4.58 log10 copies/mL) or at initiation of NVP-ART (4.77 log10 copies/mL) (P=.004 and .01, respectively); a CD4 cell count below the median (183 cells/uL) during pregnancy (P=.02) but not at initiation of NVP-ART (median 153 cells/uL, P=.61); and with an interval of less than 6 months between delivery and NVP-ART initiation (P<.0001). Notably, women with one or more resistance mutations detected by consensus sequencing at 10 days postpartum did not have a higher risk of virologic failure, compared to women with no mutations (P=.28).
In the multivariate analysis, the risk factors that remained independently associated with an increased rate of failure were an interval of less than 6 months between delivery and NVP-ART initiation (adjusted hazard ratio 3.2, CI 1.7 to 6.2, P=.001), the detection of any NVP resistance by OLA at the time of NVP-ART initiation (adjusted HR 2.5, 95% CI, 1.3–4.5, P=.004), and a viral load above median at initiation of NVP-ART (adjusted HR 2.4, 95% CI, 1.3–4.5, P=.007).
Secondary and sensitivity analyses
A secondary analysis used a confirmed viral load above 400 copies/mL as the threshold for virologic failure. In this model the hazard ratios for failure and the levels of statistical significance were very similar to those reported in the primary analysis, except that a CD4 cell count above median (183 cells/uL) during pregnancy was not significantly associated with virologic failure (P=.11). A sensitivity analysis that classified deaths with virologic failures also gave very similar results.
Discussion
In this study, genotypic NVP resistance in proviral HIV-1 DNA collected at the initiation of NVP-ART was strongly associated with virologic outcome. Specifically, women with a proviral population containing greater than 5% resistant mutants (K103N, Y181C or G190A) were significantly more likely to experience virologic failure. In contrast, the risk of virologic failure was not significantly associated with resistance detected by consensus sequencing in the period following exposure to sdNVP. The marked association between NVP-resistance detected by OLA and virologic failure provides a rationale for testing patients with previous exposure to NNRTI at the initiation of ART to guide selection of antiretrovirals for treatment. From a public health perspective, the selective use of alternative regimen, such as a protease inhibitor-based regimen, would improve the chance of successful suppression of HIV-1 replication in women with NVP-resistance. Importantly, women without mutations had a rate of virologic failure similar to those who had not been exposed to sdNVP, suggesting that it may be reasonable for these women to start NVP-based ART, a simpler, better-tolerated, and less expensive regimen. While there are many communities where testing for HIV-1 drug resistance is not feasible today, others may realize a cost benefit with resistance testing by OLA. OLA costs less than <$US20–40 (depending on whether multiplexed) for the three codons corresponding to the most prevalent NNRTI resistance mutations, while use of a protease inhibitor-based ART cost US$300 to US$400 more per year than an NNRTI based-ART in June 2009 [24].
Multiple studies [2, 4], including this study, suggest that sdNVP selects NVP-resistant viruses that then decay over the subsequent months, declining to clinically insignificant levels in many women. The detection of HIV-1 drug resistance mutations identified by more-sensitive assays are associated with virologic failure of ART in case reports and in a few larger studies [12–16], although one study found no association between low-frequency mutations and ART outcome [22]. In our study, women with ≥5% NVP-resistant mutants in cellular DNA had a high risk (0.62) of virologic failure. Yet, some subjects with detected resistance experienced virologic success. This may be because resistance mutations can be detected as a part of an integrated, yet non-functional proviral genome [23], falsely indicating a risk of virologic failure.
The persistence of NVP-resistance mutants for more than six months following sdNVP in association with virologic failure, suggest that the effects of sdNVP can linger for more than six months. The women with NVP-resistance persisting until the initiation of NVP-ART did not have lower levels of CD4 lymphocytes or higher concentrations of plasma HIV-1 RNA at the initiation of NVP-ART; however, these women had a shorter interval between sdNVP and treatment. A threshold for the effect of time between sdNVP and initiation of NVP-ART could not be identified, similar to the analysis combining cohorts from Zambia, Kenya and Thailand (not inclusive of our subjects)[4]. Both our and the combined cohort [4] detect an effect of sdNVP beyond the 6-month interval, previously suggested as the outer limit for the effect of sdNVP on the virologic outcome of NVP-ART [2].
Several novel and important observations come from our study. First, drug-resistant HIV-1 variants appear to decay at different rates across individuals, therefore mutations present immediately prior to ART appear more relevant to treatment outcome than those detected postpartum. Second, we assayed PBMC-derived HIV-1 DNA rather than plasma-derived HIV-1 RNA. Comparative studies have shown that HIV-1 resistance mutations are detectable for longer in peripheral blood cellular DNA compared to plasma RNA, and using the OLA compared to consensus sequencing, both in women following sdNVP [18] and individuals failing ART [17]. Although others observed resistance to persist for longer in HIV-1 RNA compared to DNA [9, 14], these findings may have been affected by sampling too few copies of HIV-1 DNA. Third, OLA that detected mutants present at ≥5% of the total viral population, which is considerably more sensitive than consensus sequencing, capturing a large part of the risk for virologic failure. More sensitive methods, such as allele-specific PCR, may be able to detect more resistance, but the concentration of clinically relevant mutant is uncertain and, in the case of our study, it is not likely that a more sensitive assay would have improved the predictive power.
A limitation of our study is that a significant number of samples could not be analyzed by OLA, mostly because of insufficient available DNA. While the exclusion of these subjects could have the potential to bias our findings, the only difference between these and the group analyzed by OLA was slightly higher plasma HIV-1 RNA among the latter. Nevertheless, the rate of virologic failure among the 88 excluded subjects was similar to those included in the study, suggesting that the study was not biased towards women more likely to fail therapy.
In summary, testing women previously exposed to sdNVP just prior to the initiation of NVP-ART identified NVP-resistant HIV-1 and the women with detected resistance were at increased risk of virologic failure. Identification of two or more NVP-resistance mutations conferred an extremely high likelihood of virologic failure. These results may also be of interest for patients who previously discontinued a NVP based regimen for toxicity and are later candidates for an efavirenz-based ART, or even for antiretroviral naïve patients starting a NNRTI-based regimen as NVP-resistant viruses have also been detected in patients not treated with sdNVP [14, 16], presumably due to the transmission of drug-resistant viruses. These observations call for further investigations to determine the thresholds above which drug-resistant viruses are clinically relevant, and warrant access to simple and inexpensive assays that evaluate low-concentrations of drug-resistant HIV-1.
Summary
In women with a history of single dose nevirapine administration to prevent perinatal transmission of HIV, the detection of low concentration mutations at the time of antiretroviral therapy initiation is strongly associated with virologic failure within the following 18 months.
Acknowledgments
Program for HIV Prevention and Treatment (PHPT) study group, Thailand
Rayong: Si. Ariyadej (Int.), Su. Ariyadej (Obst.), C. Pinyowittayakool (Dir.), C. Tantiyawarong (Dir.); Nakornping: P. Leenasirimakul (Int.), V. Gomuthbutra (Obst.), S. Kahintapongs (Dir.), C. Sirinirundr (Dir.); Health Promotion Region 10, Chiang Mai: A. Limtrakul (Obst.); Prapokklao: N. Chuachamsai (Int.), M. Techapornroong (Int.), P. Yuthavisuthi (Obst.), D. Sinthuvanich (Dir.); Chonburi: C. Bowonwatanuwong (Int.), N. Chotivanich (Obst.), P. Kittikoon (Dir.), C. Tantiyawarong (Dir.); Phayao Provincial: G. Halue (Int.), W. Leongjiranothai (Int.), J. Hemvuttiphan (Obst.), S. Sangsawang (Obst.), S. Attawibool (Dir.); Nakhonpathom: O. Kamsao (Int.), R. Pittayanon (Int.), V. Chalermpolprapa (Obst.), P. Hirunchote (Dir.); Hat Yai: A. Nilmanat (Int.), S. Lamlertkittikul (Obst.), T. Jarupanich (Obst.), K. Veerapradist(Dir.); Chacheongsao: P. Wittayapraparat(Int.), A. Kanjanasing(Obst.), C. Jirawison (Dir.), V. Latdhivongsakorn (Dir.); Bhumibol Adulyadej: S. Prommas (Obst.), K. Kengsakul (Obst.), P. Prateeprat (Dir.), Y. Vonglertvit (Dir.); Mae Sai: C. Jongpipan (Int.), K. Kongsing (Int.), S. Kunkongkapan (Dir.); Samutsakorn: A. Chutanunta (Int.), T. Sukhumanant (Obst.), C. Pinsuwan (Dir.); Chiangrai Prachanukroh: P. Kantipong (Int.), J. Achalapong (Obst.), R. Srismith (Dir.), S. Yanpaisan (Dir.); Chiang Kham: Y. Buranawanitchakorn (Int.), C. Putiyanun (Obst.), C. Kulkolakan (Dir.); Phan: S. Suwan (Int.), S. Jungpichanvanich (Obst.), T. Changchit (Dir.); Ratchaburi: W. Panitsuk (Int.), P. Sang-a-gad (Int.), T. Chonladarat (Obst.), N. Pinyotrakool (Obst.), M. Jittwatanakorn (Dir.), P. Bunjongjit (Dir.); Pranangklao: S. Pipatnakulchai (Obst.), S. Hongyok (Dir.); Lamphun: N. Luekamlung (Wirayutwatthana)(Int.), W. Matanasaravoot (Obst.), C. Wannalit (Dep. Dir.), S. Yanpaisan (Dir.); Somdej Prapinklao: P.Wongsarot (Int.), S. Suphanich (Obst.), P. Kanchanakitsakul (Obst.), P. Sunalai (Dir.); Samutprakarn: N. Eiamsirikit (Int.), P. Sabsanong (Obst.), C. M. Hongsawinitkul (Dir.); Mae Chan: S. Buranabanjasatean(Int.), S. Piyaworawong(Dir.); Banglamung: K. Boonrod (Int.), J. Ithisuknanth (Obst.), P. Jittiwattanapongs (Dir.); Kalasin: P. Thaingamsilp (Int.), B. Suwannachat (Obst.), S. Nitpanich (Dir.); Buddhachinaraj: S. Tunsupasawasdikul (Int.), W. Wannapira (Obst.), P. Thanomrat (Obst.), W. Boonyawatana (Dir.); Mahasarakam: C. Thundee (Int.), C. Churaree (Int.), S. Nakhapongse (Obst.), S. Tonmat (Obst.), W. Worngsatthanaphong (Dir.); Somdej Pranangchao Sirikit: V. Attakornwatana (Int.), B. Aumpaporn (Int.), W. Pornkitprasarn (Obst.), W. Rutirawat (Dir.); Khon Kaen: W. Susaengrat (Int.), J. Ratanakosol (Obst.), V. Jarupoonphol (Dir.); Nong Khai: N. Yuthakasaemsan (Int.), N. P. ruttana-Aroongorn (Obst.), T. Wichatrong (Dir.); Health Promotion Region 6 Khon Kaen: N. Winiyakul (Obst.), W. Sinchai(Dir.); Chiang Khong Royal Crown Prince: S. Monchit (Dir.); Klaeng: B. Chetanachan (Int.), S. Sungpapan (Int.), S. Techapalokul (Obst.); Phaholpolphayuhasena: P. Chirawatthanaphan (Int.), Y. Srivarasat (Obst.), T. Buddhaboriwan (Dir.); Prajaksilapakom Army: P. Nakchun (Int.), D. Langkafa (Dir.); Bamrasnaradura: S. Tunsupasawaskul (Int.); Nopparat Rajathanee: J. Wongchinsri (Int.), S. Surawongsin (Obst.), T. Chanpoo (Obst.), N. Thamanavat (Obst.), P. Hotrarapavanond (Dir.); Phayamengrai: S. Kamsrisuk (Int.); Srinagarind: P. Chetchotisakd (Int.), C. Sakondhavat (Obst.), W. Laupattarakasem (Dir.), S. Kraitrakul (Dir.); Kranuan Crown Prince: S. Benchakhanta (Int.), R. Thongdej (Obst.), T. Chaiyabut, P. Kovit(Dir.), A. Rattanaparinya(Dir.); Roi-et: B. Jeerasuwannakul (Int.), W. Atthakorn (Obst.), W. Supanchaimat (Dir.).
We would like to thank the patients who participated in the PHPT-2 study, and the following colleagues who contributed to this project in many critical ways: K. McIntosh, S. Kanchanavanit, P. Matangkhasombut, H. D’Oriano, R. Lefait-Robin, N. Mann, and all members of the hospital teams.
We are grateful for the advice and assistance from the Thai Ministry of Public Health: Office of the Permanent Secretary, Departments of Health, Communicable Diseases Control, Food and Drug Administration, Health Sciences, and Provincial Hospitals Division and especially, V. Thaineua, S. Kanshana, M. Teeratantikanont, A. Chitwarakorn, S. Thanprasertsuk, V. Chokevivat, T. Siraprapasiri, T. Suntrajarn, P. Sirinirund, P. Sirivongrungson, S. Bhakeecheep, P. Satasit, T Liewsaree, W. Liewsaree; from Chiang Mai University: U. Haesungcharern, D. Romcai.
Footnotes
Financial support
National Institute of Allergy and Infectious Disease (NIAID), National Institutes of Health, 5 R01 AI058723 (LMF, Reservoirs of Resistance) and 1 U01 AI068632 (LMF, Developmental Virology subcontract)
Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, 5 R01 HD 39615 (ML, PHPT-2 study)
French National Agency for Research on AIDS and Viral Hepatitis (ANRS), 12–08 (ML, PHPT-2 study)
Global Fund to fight AIDS, Tuberculosis and Malaria, Thailand, PR-A-N-008
Institut de Recherche pour le Développement (IRD), Marseilles, France
The sponsors had no role in the design and conduct of the study, collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
The authors have no other potential conflicts of interest to disclose.
References
- 1.Jourdain G, Ngo-Giang-Huong N, Le Coeur S, et al. Intrapartum exposure to nevirapine and subsequent maternal responses to nevirapine-based antiretroviral therapy. N Engl J Med. 2004 Jul 15;351(3):229–240. doi: 10.1056/NEJMoa041305. [DOI] [PubMed] [Google Scholar]
- 2.Lockman S, Shapiro RL, Smeaton LM, et al. Response to antiretroviral therapy after a single, peripartum dose of nevirapine. N Engl J Med. 2007 Jan 11;356(2):135–147. doi: 10.1056/NEJMoa062876. [DOI] [PubMed] [Google Scholar]
- 3.Eshleman SH, Mracna M, Guay LA, et al. Selection and fading of resistance mutations in women and infants receiving nevirapine to prevent HIV-1 vertical transmission (HIVNET 012) Aids. 2001 Oct 19;15(15):1951–1957. doi: 10.1097/00002030-200110190-00006. [DOI] [PubMed] [Google Scholar]
- 4.Weidle P, Stringer J, McConnell M, et al. Effectiveness of NNRTI-containing ART in Women Previously Exposed to a Single Dose of Nevirapine: A Multi-country Cohort Study; 15th Conference on Retroviruses and Opportunistic Infections.2008. [Google Scholar]
- 5.Eshleman SH, Hoover DR, Chen S, et al. Nevirapine (NVP) resistance in women with HIV-1 subtype C, compared with subtypes A and D, after the administration of single-dose NVP. J Infect Dis. 2005 Jul 1;192(1):30–36. doi: 10.1086/430764. [DOI] [PubMed] [Google Scholar]
- 6.Eshleman SH, Guay LA, Mwatha A, et al. Comparison of nevirapine (NVP) resistance in Ugandan women 7 days vs. 6–8 weeks after single-dose nvp prophylaxis: HIVNET 012. AIDS ResHumRetroviruses. 2004;20(6):595–599. doi: 10.1089/0889222041217518. [DOI] [PubMed] [Google Scholar]
- 7.Flys T, Nissley DV, Claasen CW, et al. Sensitive drug-resistance assays reveal long-term persistence of HIV-1 variants with the K103N nevirapine (NVP) resistance mutation in some women and infants after the administration of single-dose NVP: HIVNET 012. J Infect Dis. 2005 Jul 1;192(1):24–29. doi: 10.1086/430742. [DOI] [PubMed] [Google Scholar]
- 8.Palmer S, Boltz V, Martinson N, et al. Persistence of nevirapine-resistant HIV-1 in women after single-dose nevirapine therapy for prevention of maternal-to-fetal HIV-1 transmission. Proc Natl Acad Sci U S A. 2006 May 2;103(18):7094–7099. doi: 10.1073/pnas.0602033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loubser S, Balfe P, Sherman G, Hammer S, Kuhn L, Morris L. Decay of K103N mutants in cellular DNA and plasma RNA after single-dose nevirapine to reduce mother-to-child HIV transmission. AIDS. 2006 Apr 24;20(7):995–1002. doi: 10.1097/01.aids.0000222071.60620.1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becker-Pergola G, Mellquist JL, Eshleman JR, Jackson JB, Eshleman SH. Improved detection of human immunodeficiency virus type 1 variants by analysis of replicate amplification reactions: relevance to studies of human immunodeficiency virus type 1 vertical transmission. Mol Diagn. 1999 Dec;4(4):261–268. doi: 10.1016/s1084-8592(99)80001-0. [DOI] [PubMed] [Google Scholar]
- 11.Shi C, Eshleman SH, Jones D, et al. LigAmp for sensitive detection of single-nucleotide differences. Nat Methods. 2004 Oct 21;1(2):141–147. doi: 10.1038/nmeth713. [DOI] [PubMed] [Google Scholar]
- 12.Kuritzkes DR, Lalama CM, Ribaudo HJ, et al. Preexisting resistance to nonnucleoside reverse-transcriptase inhibitors predicts virologic failure of an efavirenz-based regimen in treatment-naive HIV-1-infected subjects. J Infect Dis. 2008 Mar 15;197(6):867–870. doi: 10.1086/528802. [DOI] [PubMed] [Google Scholar]
- 13.Johnson JA, Li JF, Wei X, et al. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med. 2008 Jul 29;5(7):e158. doi: 10.1371/journal.pmed.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coovadia A, Hunt G, Abrams EJ, et al. Persistent Minority K103N Mutations among Women Exposed to Single-Dose Nevirapine and Virologic Response to Nonnucleoside Reverse-Transcriptase Inhibitor-Based Therapy. Clin Infect Dis. 2009 Jan 9; doi: 10.1086/596486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Metzner KJ, Giulieri SG, Knoepfel SA, et al. Minority quasispecies of drug-resistant HIV-1 that lead to early therapy failure in treatment-naive and -adherent patients. Clin Infect Dis. 2009 Jan 15;48(2):239–247. doi: 10.1086/595703. [DOI] [PubMed] [Google Scholar]
- 16.Simen BB, Simons JF, Hullsiek KH, et al. Low-Abundance Drug-Resistant Viral Variants in Chronically HIV-Infected, Antiretroviral Treatment-Naive Patients Significantly Impact Treatment Outcomes. J Infect Dis. 2009 Mar 1;199(5):693–701. doi: 10.1086/596736. [DOI] [PubMed] [Google Scholar]
- 17.Ellis GM, Mahalanabis M, Beck IA, et al. Comparison of oligonucleotide ligation assay and consensus sequencing for detection of drug-resistant mutants of human immunodeficiency virus type 1 in peripheral blood mononuclear cells and plasma. J Clin Microbiol. 2004 Aug 42;(8):3670–3674. doi: 10.1128/JCM.42.8.3670-3674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kress C, Wagner T, Ngo-Giang-Huong N, et al. Dynamics of Nevirapine-resistant HIV-1 following Single-dose NVP Assessed in Plasma and Cells by Consensus Sequencing and Oligonucleotide Ligation Assay; 15th Conference on Retroviruses and Opportunistic Infections.2008. [Google Scholar]
- 19.Lallemant M, Jourdain G, Le Coeur S, et al. Single-dose perinatal nevirapine plus standard zidovudine to prevent mother-to-child transmission of HIV-1 in Thailand. N Engl J Med. 2004 Jul 15;351(3):217–228. doi: 10.1056/NEJMoa033500. [DOI] [PubMed] [Google Scholar]
- 20.Arvold ND, Ngo-Giang-Huong N, McIntosh K, et al. Maternal HIV-1 DNA load and mother-to-child transmission. AIDS Patient Care STDS. 2007 Sep 21;(9):638–643. doi: 10.1089/apc.2006.0169. [DOI] [PubMed] [Google Scholar]
- 21.Beck IA, Crowell C, Kittoe R, et al. Optimization of the oligonucleotide ligation assay, a rapid and inexpensive test for detection of HIV-1 drug resistance mutations, for non-North American variants. J Acquir Immune Defic Syndr. 2008 Aug 1;48(4):418–427. doi: 10.1097/QAI.0b013e31817ed7d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peuchant O, Thiebaut R, Capdepont S, et al. Transmission of HIV-1 minority-resistant variants and response to first-line antiretroviral therapy. AIDS. 2008 Jul 31;22(12):1417–1423. doi: 10.1097/QAD.0b013e3283034953. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Kappes JC, Conway JA, Price RW, Shaw GM, Hahn BH. Molecular Characterization of Human Immunodeficiency Virus Type-1 Cloned Directly From Uncultured Human Brain: Identification of Replication Competent and Defective Viral Genomes. JVirol. 1991;65:3973–3985. doi: 10.1128/jvi.65.8.3973-3985.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Médecins sans Frontières. [accessed on January 21, 2010];Untangling the Web of Antiretroviral Price Reductions: A pricing guide for the purchase of ARVs for developing countries. Upcoming 12th Edition - Pre-publication price analysis - July 2009. http://www.msfaccess.org/fileadmin/user_upload/diseases/hiv-aids/09_28_UTWPricingGuide_IAS_LowRes_FINAL.pdf.


