Summary
Background
Polyphosphate (a linear polymer of inorganic phosphate) is secreted from platelet dense granules and we recently showed that it accelerates factor V activation by thrombin.
Objective
To examine the interaction of polyphosphate with thrombin.
Methods and Results
Thrombin, but not prothrombin, altered the electrophoretic migration of polyphosphate in gel mobility assays. Thrombin binding to polyphosphate was influenced by ionic strength and was evident even in plasma. Two positively charged exosites on thrombin mediate its interactions with other proteins and accessory molecules: exosite I (mainly with thrombin substrates), and exosite II (mainly with certain anionic polymers). Free thrombin, thrombin in complex with hirudin’s C-terminal dodecapeptide, and gamma-thrombin all bound polyphosphate similarly, excluding exosite I involvement. Mutations within exosite II, but not exosite I, the Na+-binding site or hydrophobic pocket, weakened thrombin binding to polyphosphate as revealed by NaCl-dependence. Surface plasmon resonance demonstrated tight interaction of polyphosphate with thrombin (Kd ~5 nM) but reduced interaction with a thrombin exosite II mutant. Certain glycosaminoglycans, including heparin, only partially competed with polyphosphate for binding to thrombin, and polyphosphate did not reduce heparin-catalyzed inactivation of thrombin by antithrombin.
Conclusion
Polyphosphate interacts with thrombin’s exosite II at a site that partially overlaps, but is not identical to, the heparin binding site. Polyphosphate interactions with thrombin may be physiologically relevant, since polyphosphate concentrations achievable following platelet activation are far above the ~5 nM Kd for the polyphosphate-thrombin interaction.
Keywords: exosite II, prothrombin, polyphosphate, thrombin
Introduction
Thrombin interacts with many substrates, modulatory proteins and accessory molecules. As a procoagulant, thrombin converts fibrinogen into fibrin, activates platelets via PAR-1 and PAR-4, and proteolyzes clotting factors V, VII, VIII, XI and XIII. Thrombin activity is tightly regulated by a number of mechanisms, including binding to thrombomodulin (switching it to an anticoagulant protease), and direct inhibition by plasma protease inhibitors like antithrombin and heparin cofactor II. Thrombin’s exquisite specificity is largely determined by its anion-binding exosites I and II, both of which lie distal to the active site. Exosite I interacts with protein substrates and modulators such as fibrinogen [1], P A R-1 [2], thrombomodulin [3] and the C-terminal dodecapeptide tail of hirudin [4], while exosite II interacts with platelet GPIbα̣ [5], protease nexin I [6], prothrombin activation fragment 2 [7], heparin [8, 9] and the chondroitin sulfate moiety of thrombomodulin [10].
Inorganic polyphosphate (polyP), a linear polymer of orthophosphate residues linked via phosphoanhydride bonds, plays important and diverse roles in nature. Dense granules of human platelets contain polyP which is secreted upon platelet activation [11]. We recently showed that polyP is a potent modulator of blood clotting, and in particular that it enhances thrombin generation by accelerating conversion of factor V to Va [12]. These results led to the hypothesis that polyP interacts directly with thrombin. In this study we demonstrate that polyP associates with thrombin’s exosite II by way of tight electrostatic interactions.
Materials and Methods
Materials
Materials were from the following suppliers: polyP (sold under the name “sodium phosphate glass”) of mean polymer lengths 25 (polyP25) and 75+ (polyP75), toluidine blue O, unfractionated heparin (from porcine intestinal mucosa, 140 USP/mg; average chain length 17–19 kDa), heparan sulfate (from bovine kidney), chondroitin sulfate A (from bovine tracheae), B (from porcine intestine), and C (from shark cartilage), hyaluronic acid, and heparin-agarose beads, Sigma (St. Louis, MO, USA); dextran sulfate (average Mr 500,000), Fisher Scientific (Pittsburgh, PA, USA); human prothrombin and α-thrombin, Enzyme Research Laboratories (South Bend, IN, USA); biotin-d-Phe-Pro-Arg chloromethylketone, antithrombin and γ-thrombin, Haematologic Technologies (Essex Junction, VT, USA); polyclonal anti-prothrombin antibody, Affinity Biologicals (Ancaster, ON, Canada); pooled normal human plasma, George King Bio-Medical (Overland Park, KS, USA); Spectrozyme TH, American Diagnostica (Stamford, CT, USA); zirconia beads, ZirChrom Separations (Anoka, MN, USA); Microcon YM10 concentrators, Millipore (Billerica, MA, USA); polyacrylamide Ready Gels, Bio-Rad (Hercules, CA, USA); Handee mini-spin columns, polyvinylidene fluoride membranes, avidin-horseradish peroxidase and Gelcode Blue stain, Pierce (Rockford, IL, USA); ECL plus detection system, Amersham Biosciences (Pittsburgh, PA, USA); streptavidin sensor chips, BIAcore (Piscataway, NJ, USA); and phospholipids, Avanti Polar Lipids (Alabaster, AL, USA). Vesicles composed of 80% phosphatidylcholine/20% phosphatidylserine (PCPS) were prepared by sonication. Alanine-substituted mutants of thrombin, numbered from the amino terminus of the B-chain (with chymotrypsinogen numbering in curly braces in some cases) were expressed and purified as previously described [13, 14]. Unless otherwise indicated, polyP concentrations are given throughout this paper in terms of the concentration of phosphate monomer (monomer formula: NaPO3).
Gel mobility shift assays
PolyP interaction with thrombin was investigated using native gel electrophoresis. Thrombin (10 µg) was preincubated with polyP25 (100 µg) for 10 min at ambient temperature before addition of sample buffer (60 mM Tris-HCl pH 6.8, 10% glycerol, 0.01% bromophenol blue) and resolving on 10% polyacrylamide gels. Gels were stained for protein with Gelcode Blue, or for polyP with 0.25% toluidine blue O in 25% methanol, 5% glycerol for 10 min and destained in the same solution without dye.
Thrombin binding to immobilized polyP
PolyP was immobilized onto porous zirconia beads essentially as described [15]. Briefly, 250 mg zirconia beads were incubated with 10 mg/ml polyP75 in water for 20 h at 37°C, washed with distilled water, blocked with 10% BSA for 15 h at ambient temperature, then dried in vacuum at 80°C for 2 h. Control beads were treated with water and BSA only. To conduct binding assays, polyP-zirconia beads (10 mg dry weight) were washed twice with Binding Buffer (50 mM Tris-HCl pH 7.5, 50 mM NaCl, 0.1% BSA), resuspended in 200 µl Binding Buffer with 27 pmol thrombin (=135 nM thrombin) and incubated at ambient temperature for 30 min, after which the mixture was centrifuged in mini-spin columns at 1677 × g for 30 s to collect the flow-through. Beads were washed with 200 µl Binding Buffer followed by 200 µl Elution Buffer (50 mM Tris-HCl pH 7.5, 1 M NaCl, 0.1% BSA), and thrombin recovery was quantified by measuring rates of Spectrozyme TH hydrolysis compared to a standard curve. Salt sensitivities of thrombin-polyP interactions were investigated by modifying either the Binding or Elution Buffer to contain 50–1000 mM NaCl. For competition studies, thrombin was preincubated for 30 min in Binding Buffer containing heparin, heparan sulfate, dextran sulfate, hyaluronic acid or chondroitin sulfate A, B, or C (all 100 µg/ml) before adding the mixture to polyP-zirconia beads and analyzing thrombin binding as above. Binding of 143 pmol prothrombin to immobilized polyP was analyzed in a similar manner except that prothrombin was detected by western blots probed with peroxidase-conjugated anti-prothrombin antibody.
Binding of thrombin to heparin-agarose beads
Thrombin (27 pmol) was incubated for 30 min at ambient temperature in Binding Buffer containing 55 µg/ml heparin (3 µM, assuming an average chain length of 18 kDa) or 133 µM polyP75 (equivalent to ~1.8 µM polymer), then incubated with 215 µg heparin-agarose beads for 30 min, after which thrombin was eluted and detected as described for polyP-zirconia beads.
Binding of thrombin to polyP-zirconia in plasma
Thrombin (50 µM) was biotinylated at its active site by reacting at ambient temperature for 30 min with 500 µM biotin-d-Phe-Pro-Arg chloromethylketone, after which it had undetectable enzymatic activity by Spectrozyme TH hydrolysis. Unreacted inhibitor was removed by repeated centrifugation using Microcon YM10 filters. PolyP-zirconia beads were preincubated with plasma or 20 mM HEPES-NaOH pH 7.4, 150 mM NaCl, 50 mg/ml BSA, after which 27 pmol biotin-thrombin was added to the plasma or BSA-containing buffer. Thrombin binding assays were performed as described above, with biotin-thrombin being detected by probing western blots with avidin-peroxidase.
Thrombin inhibition by antithrombin
Heparin-catalyzed thrombin inhibition by antithrombin was carried out at the following reactant conditions: 25 nM thrombin, 50 nM antithrombin, 16 mM HEPES-NaOH pH 7.4, 120 mM NaCl, 0.08% BSA, 0–1000 U/ml (0–400 µM) heparin, with or without 37.5 µM polyP75. Alternatively, reactions contained 0 to 375 µM polyP75 at a constant heparin concentration (0.01 U/ml, or 4 nM). In some experiments the heparin and polyP75 concentrations were constant (0.01 U/ml or 4 nM and 37.5 µM respectively) and the salt concentration of the buffer was varied from 50 mM – 500 mM. Reactions proceeded for 10 min at 37°C before the remaining thrombin activity was quantified (using Spectrozyme TH hydrolysis).
Surface plasmon resonance
Biotin-thrombin, prepared as described above using active-site-specific labeling, was bound to streptavidin sensor chips (in a BIAcore 3000 instrument) until a response of 2000 resonance units (RU) was achieved. Various concentrations of polyP25 or polyP75 in 20 mM HEPES-NaOH pH 7.4, 0.005% P-20, at either 50 mM or 150 mM NaCl, were flowed over the chip at 10 µl/min until a steady-state was reached. Maximal (steady-state) RU values were plotted versus polyP concentration, to which the single-site ligand binding equation was fitted. Regeneration of the thrombin-chip surface was achieved by flowing 50 mM HEPES-NaOH pH 7.4, 1 M NaCl, 0.005% P-20 until the response returned to baseline.
Results
We previously showed that polyP accelerates thrombin-catalyzed conversion of factor V to Va [12]. This exhibited a bell-shaped concentration dependence, with an optimal polyP75 concentration of 75 µM (expressed in terms of phosphate monomer; data not shown), consistent with a template mechanism in which both factor V and thrombin likely bind to polyP. In this study, we therefore examined how polyP interacts with thrombin.
PolyP binds to thrombin but not prothrombin
We used gel mobility-shift assays in native gels to investigate binding of polyP to prothrombin or thrombin. Toluidine blue staining revealed a pronounced retardation of polyP25 (Fig. 1A) and polyP75 (not shown) migration in the presence of thrombin but not prothrombin. We further examined thrombin-polyP interactions using polyP75 immobilized on zirconia beads. When incubated with polyP-zirconia in 50 mM NaCl, thrombin was undetectable in the flow-though (by western blots; Fig. 1B, lane 2) but appeared in the 1M NaCl eluate (Fig. 1B, lane 3). PolyP therefore binds to thrombin with an ionic component to the interaction. In contrast, prothrombin appeared in the flow-through but not the 1M NaCl eluate, indicating a lack of polyP75 binding (Fig. 1B). In quantitative experiments (n=6), thrombin failed to bind to BSA-zirconia beads (25.84 ± 1.3 pmol thrombin in the flow-through, compared to 27 pmol thrombin initially added to the beads) but bound well to polyP-zirconia beads (0.02 ± 0.01 pmol thrombin in the flow-through). High salt washes eluted negligible thrombin from BSA-zirconia beads (0.06 ± 0.01 pmol thrombin), but eluted thrombin essentially quantitatively from the polyP-zirconia beads (23.08 ± 1.14 pmol thrombin).
Fig. 1.
PolyP binds to thrombin but not prothrombin. (A) Gel-shift mobility assay in which 1 µg thrombin (T) or prothrombin (PT) were incubated ± 100 µg polyP25, resolved on native PAGE and stained with toluidine blue to detect polyP. Free polyP25 migrated at the dye front. (B) Thrombin (27 pmol) or prothrombin (143 pmol) were incubated with polyP-zirconia after which fractions were resolved on SDS-PAGE and probed with anti-prothrombin antibody. Lanes: 1, starting material; 2, flow-through; and 3, high salt (1 M NaCl) eluate. (C) Zirconia beads with (+) or without (−) attached polyP75 were incubated with biotin-thrombin in plasma or buffer. Bound thrombin was eluted with high salt buffer, resolved on SDS-PAGE and probed with avidin-peroxidase (representative of four blots).
Plasma has a very high protein concentration, including proteins that might compete with thrombin for binding to polyP. We therefore evaluated thrombin binding to polyP in plasma. Biotin-thrombin mixed with plasma or a buffer containing 50 mg/ml BSA was incubated with polyP-zirconia or BSA-zirconia beads. After washing and elution with high salt buffer, biotin-thrombin was recovered equally well from plasma or buffer (Fig. 1C), demonstrating that the thrombin-polyP interaction occurs efficiently in a plasma milieu.
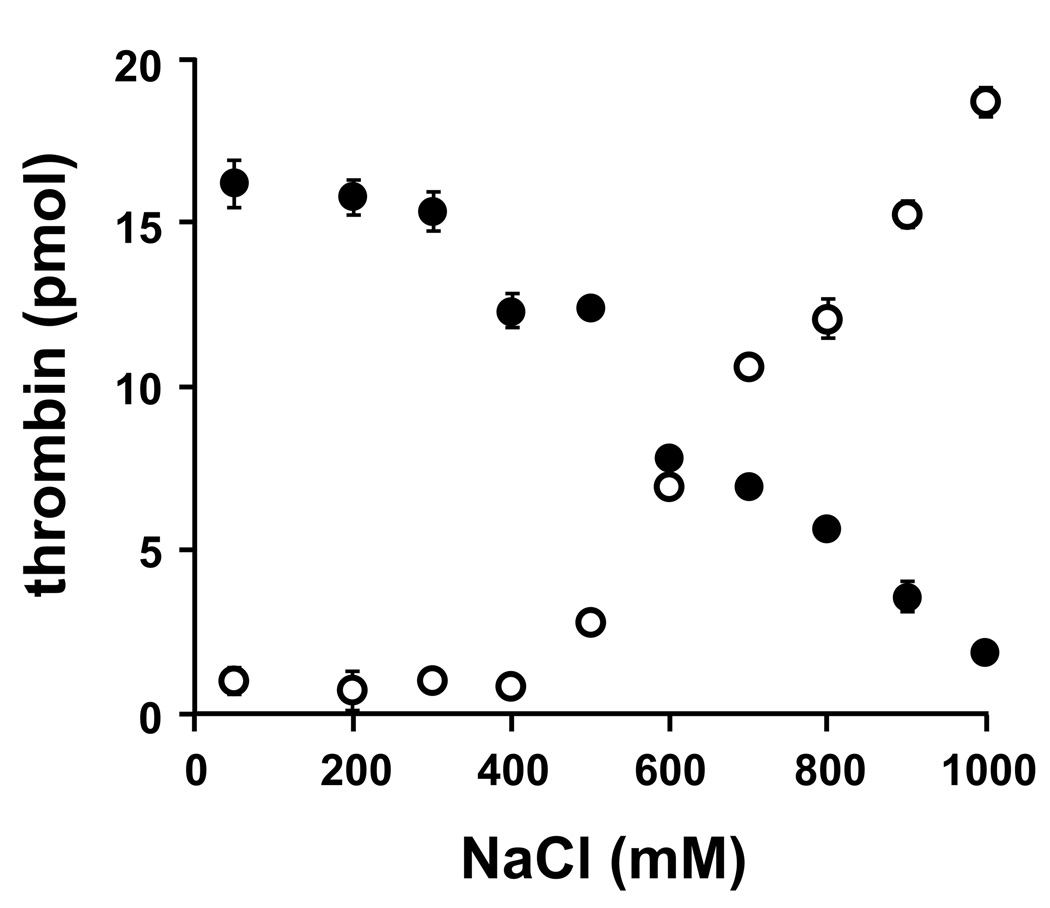
We investigated the salt dependence of polyP-thrombin interactions by incubating thrombin with immobilized polyP at increasing NaCl concentrations, yielding half-maximal thrombin-polyP binding at ~700 mM NaCl (Fig. 2, open symbols). Similarly, thrombin bound to polyP75 at 50 mM NaCl showed half-maximal elution at ~700 mM NaCl (Fig. 2, closed symbols). PolyP interacts with divalent metal ions, but we found that including 2.5 mM CalCl2 had no discernable effect on binding of thrombin to, or elution from, immobilized polyP75 (not shown). Plasma Ca2+ concentrations therefore do not interfere with thrombin-polyP interactions.
Fig. 2.
PolyP interaction with thrombin is influenced by salt concentration. Thrombin was incubated with polyP-zirconia beads at varying NaCl concentrations, after which the activity of unbound thrombin was quantified in the flow-through (○). Alternatively, thrombin was incubated with polyP-zirconia beads in binding buffer containing 50 mM NaCl, and the activity of bound thrombin was quantified after elution with increasing NaCl concentrations (●). Data represent mean ± S.D. (n = 3).
Location of the polyP binding site on thrombin
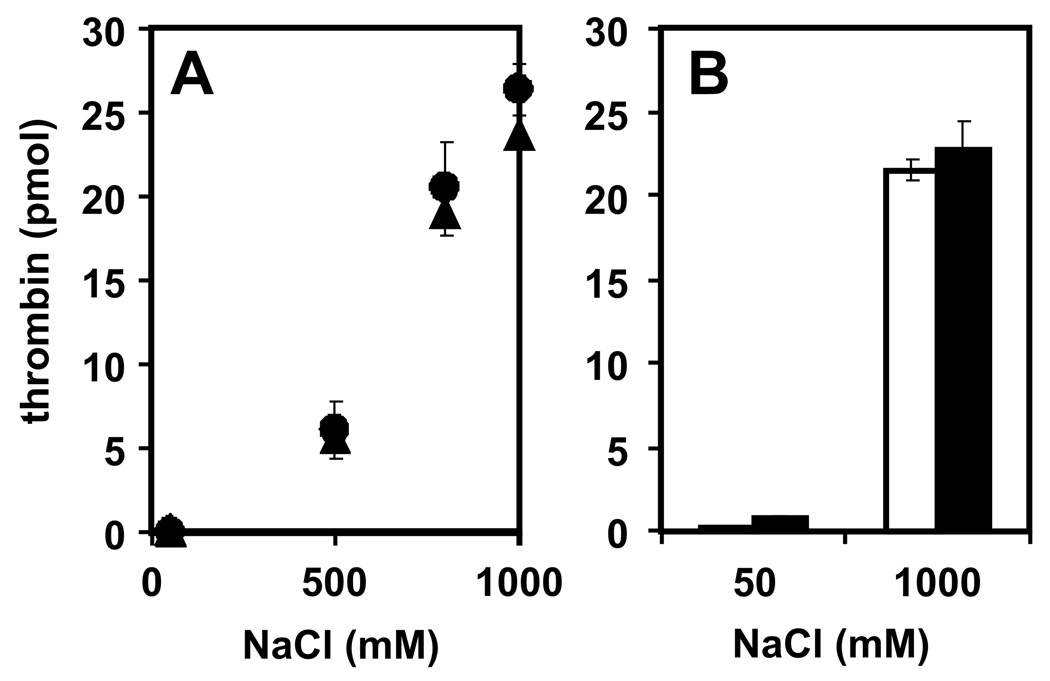
PolyP is highly anionic, so we investigated the possibility that it interacts with one or both of the anion-binding exosites on thrombin. Binding of thrombin but not prothrombin to polyP indicates the binding site is masked in the zymogen. Proexosite I in prothrombin is only partially exposed, exhibiting ~100-fold decreased affinity ligands compared to α-thrombin [16, 17]. Exosite II is completely buried within prothrombin, being fully exposed only upon activation to α-thrombin [10, 17, 18]. We observed comparable binding of both α-thrombin and γ-thrombin to immobilized polyP75, including nearly identical salt sensitivities (Fig. 3A). Similarly, preincubating thrombin with the C-terminal dodecapeptide of hirudin (a high-affinity exosite I ligand) had no discernable effect on thrombin binding to immobilized polyP75 (Fig. 3B). These data exclude a role for exosite I in polyP binding.
Fig. 3.
Exosite I is not involved in polyP binding to thrombin. (A) α-thrombin (●) or γ-thrombin (▲), both at 27 pmol were incubated with polyP-zirconia beads in binding buffer with varying NaCl concentrations, after which the flow-through was analyzed for thrombin activity. (B) Thrombin (27 pmol) was preincubated for 30 min with vehicle alone (open bars) or with the C-terminal dodecapeptide of hirudin (solid bars; 5 µg/ml), after which the mixtures were incubated with polyP-zirconia beads in binding buffer containing 50 mM or 1 M NaCl. Thrombin activity was quantified in the flow-through. Data represent mean ± S.D. (n = 3).
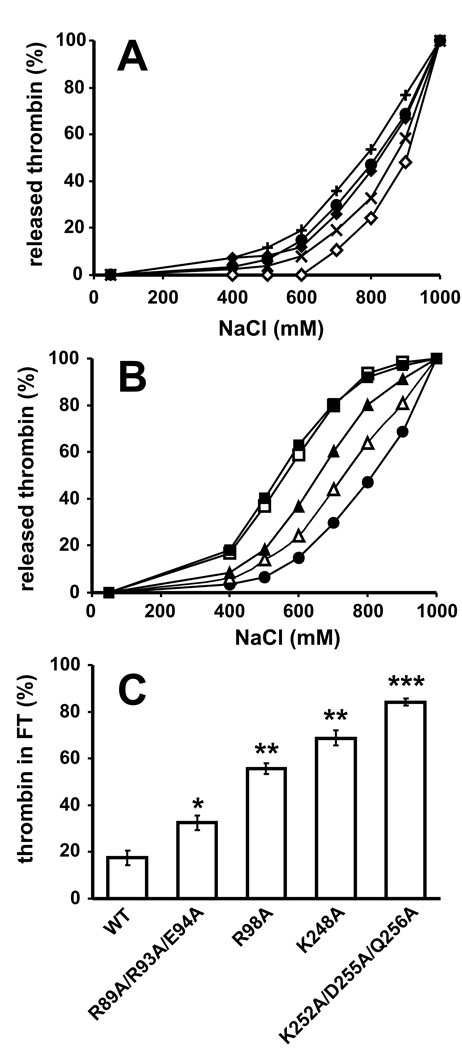
Thrombin mutants with alanine substitutions in key regions were employed to investigate the polyP binding site on thrombin. Wild-type (WT) and mutant thrombins were incubated with immobilized polyP75 and eluted with increasing NaCl concentrations. Thrombins with point mutations in the Na+ binding site (E229A), the 50-insertion loop that defines the active site hydrophobic pocket (W50A), or exosite I (Y71A; H66A) eluted at essentially the same (W50A; H66A) or slightly higher (E229A; Y71A) NaCl concentration than WT thrombin (Fig. 4A). In contrast, four mutants with substitutions in exosite II (R89A/R93A/E94A; R98A; K248A; K252A/D255A/Q256A) eluted from immobilized polyP75 at lower NaCl concentrations than WT thrombin, with K248A and K252A/D255A/Q256A showing the greatest shift in salt sensitivity (Fig. 4B). At 500 mM NaCl, a significant reduction in binding was observed with all four mutants (P<0.05 in all cases) when compared to WT (Fig. 4C), with the weakest binding observed with K248A (69% ± 3 unbound; P< 0.005) and the exosite II triple mutant K252A/D255A/Q256A (84% ± 2 unbound; P< 0.0005), compared to WT (18 ± 3% unbound).
Fig. 4.
Exosite II mutants of thrombin show reduced binding to immobilized polyP. (A) 27 pmol WT (●), H66A (◆), Y71A (◇), E229A (×), or W50A (+) thrombins were incubated with polyP-zirconia beads, and bound enzyme was sequentially eluted with increasing NaCl concentrations (plotted as cumulative thrombin recovery). (B) 27 pmol WT (●), R89A/R93A/E94A (△), R98A (▲), K248A (□), or K252A/D255A/Q256A (■) thrombins were incubated and eluted as in panel A. (C) 27 pmol nM WT, R89A/R93A/E94A, R98A, K248A, or K252A/D255A/Q256A thrombins were incubated with polyP-zirconia beads in binding buffer containing 500 mM NaCl, with unbound thrombin in the flow-through expressed as percent of the starting thrombin concentration. The data shown in (A) and (B) are representative graphs of four separate experiments performed in duplicate and the values shown in (C) represent mean ± S.D. (n = 3). *P< 0.05, **P< 0.005, ***P< 0.0005.
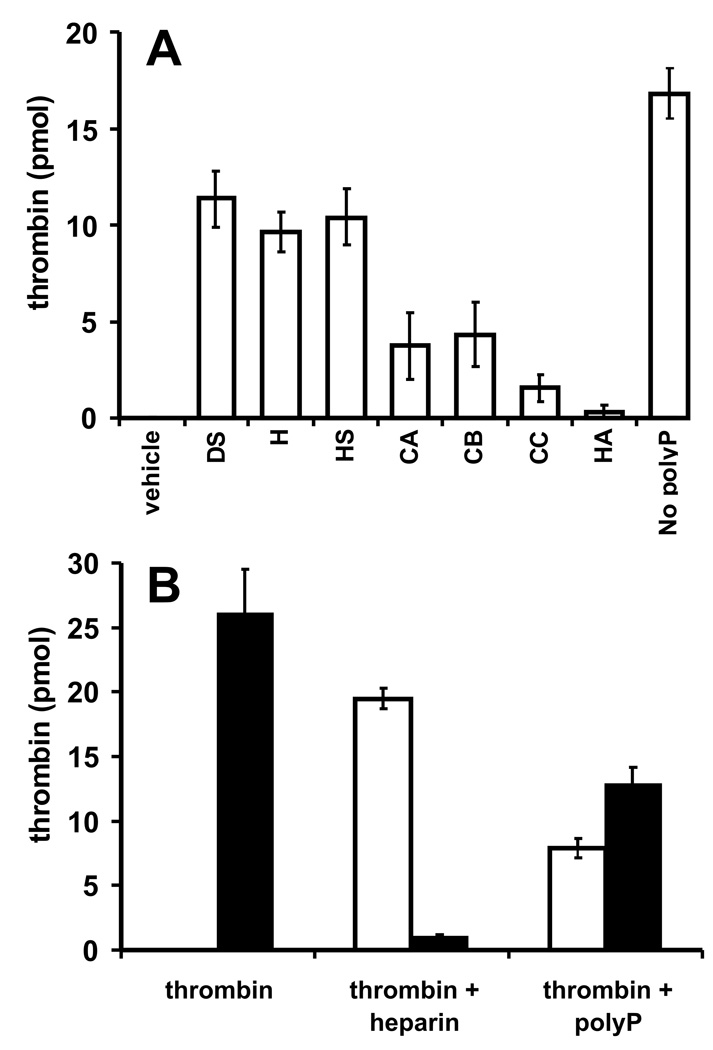
Thrombin’s exosite II binds certain glycosaminoglycans and was originally known as the heparin binding site [19]. We therefore investigated whether glycosaminoglycans competed with polyP for binding to thrombin. We preincubated thrombin with glycosaminoglycans before adding immobilized polyP75, after which the flow-through was analyzed for thrombin activity (Fig. 5A). Dextran sulfate, heparin and heparan sulfate partially competed with polyP75 for binding to thrombin, with approximately half of the thrombin remaining unbound. On the other hand, chondroitin sulfate A, B and C were relatively weak competitors, while hyaluronic acid showed negligible competition. In another experiment, thrombin was preincubated with heparin or polyP75, then added to heparin-agarose beads (Fig. 5B). In the absence of competitor, all the thrombin bound to heparin-agarose and was recovered by elution with 1 M NaCl. As expected, heparin abrogated thrombin binding to heparin-agarose. In contrast, polyP75 only partially diminished thrombin binding to heparin-agarose, with 29% of the thrombin remaining in the flow-through and 48% recovered in the high salt eluate.
Fig. 5.
Some glycosaminoglycans partially compete with polyP for binding to thrombin. (A) 27 pmol thrombin was preincubated with vehicle (V) or glycosaminoglycans (all at 100 µg/ml): DS, dextran sulfate; H, heparin; HS, heparan sulfate; CA, chondroitin sulfate A; CB, chondroitin sulfate B; CC, chondroitin sulfate C; or HA, hyaluronic acid. Mixtures were incubated with polyP-zirconia or zirconia beads only (labeled “no polyP”), after which unbound thrombin in the flow-through was quantified. (B) 27 pmol thrombin was preincubated with buffer alone, or with 55 µg/ml heparin (equivalent to 3 µM) or 130 µM polyP75 (equivalent to 1.8 µM polymer). The mixtures were then incubated with heparin-agarose beads (50 µl) and thrombin activity in the flow-through (open bars) and high salt eluate (closed bars) was quantified.
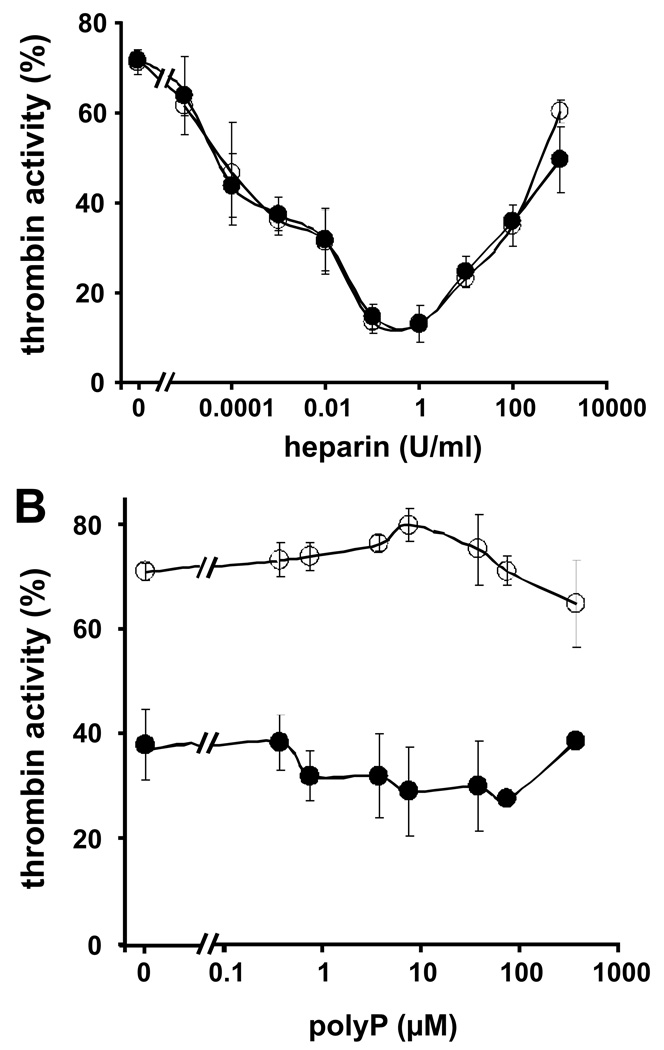
Functional analyses of heparin-catalyzed antithrombin inhibition of thrombin revealed little or no interference by polyP75 over a range of heparin concentrations (Fig. 6A) indicating that polyP does not affect either aspect of the template mechanism. Similarly, a range of polyP75 concentrations failed to interfere with heparin-catalyzed antithrombin inhibition of thrombin (Fig 6B). Experiments performed over a range of salt concentrations (50–500 mM NaCl) did not reveal any additional interference of polyP75 in the heparin-catalyzed antithrombin inhibition of thrombin (data not shown). These findings are consistent with the experiments above showing that polyP does not block heparin binding to thrombin.
Fig. 6.
PolyP does not interfere with heparin-catalyzed inhibition of thrombin by antithrombin. (A) Residual thrombin activity (A405/min) was measured after incubating 25 nM thrombin with 50 nM antithrombin and 0–1000 U/ml heparin in the absence (○) or presence (●) of 37.5 µM polyP75. The zero point refers to antithrombin inhibition of thrombin without heparin. (B) Residual thrombin activity (A405/min) was measured after incubating 25 nM thrombin with 50 nM antithrombin and 0–375 µM PolyP75 in the absence (○) or presence (●) of 0.01 U/ml heparin. The zero points refer to antithrombin inhibition of thrombin (± heparin) in the absence of polyP75. Data represent mean ± S.D. (n = 3).
Kd for polyP binding to thrombin
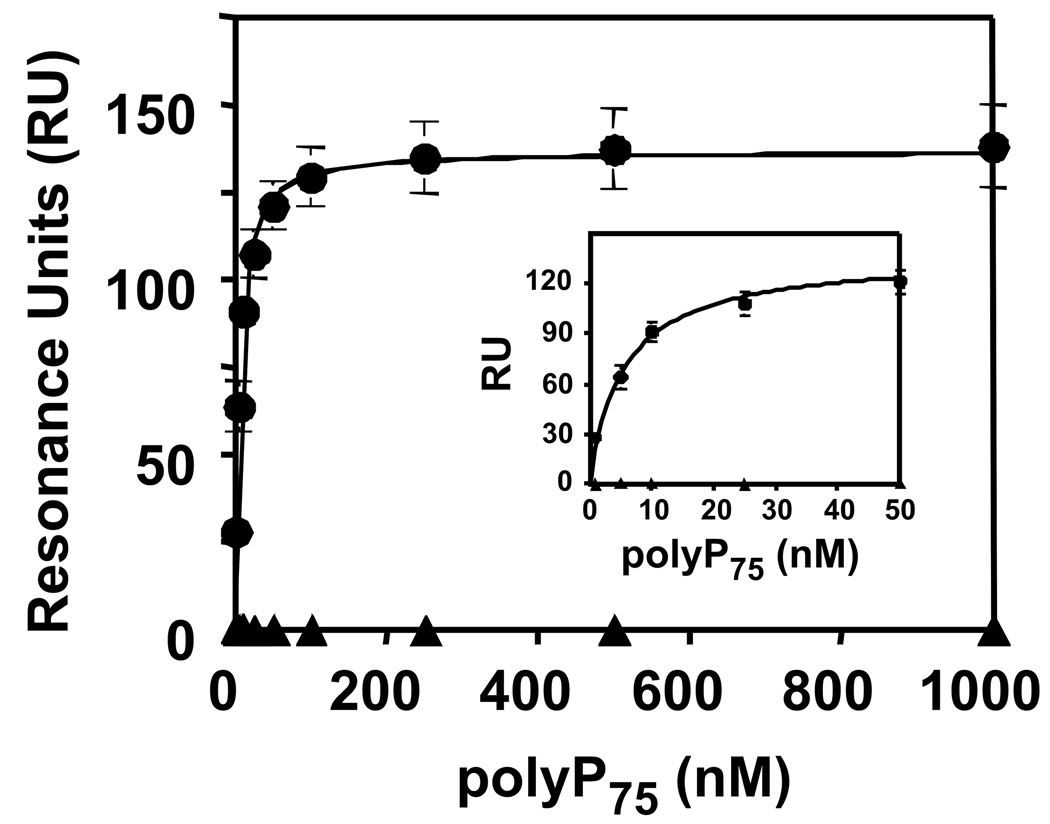
WT and the K248A {236} exosite II mutant of thrombin were biotinylated at their active sites and immobilized onto streptavidin sensor chips for surface plasmon resonance studies. Increasing polyP concentrations were flowed over the surface of the chip until steady-state binding was achieved (Fig. 7). From these data, binding constants for polyP association with thrombin were derived. In the presence of 50 or 150 mM NaCl, polyP75 bound with very high affinity to WT thrombin (Table 1), and similar binding isotherms were obtained with polyP25 (not shown). Binding of polyP75 to the K248A {236} exosite II mutant of thrombin was reduced at 50 mM NaCl compared to WT, resulting in an approximately 2.5-fold higher Kd value (P<0.005). Interestingly, binding of polyP75 to the K248A mutant was undetectable at 150 mM NaCl, supporting the observations that polyP interactions with thrombin are highly dependent on the NaCl concentration.
Fig. 7.
PolyP binds to WT thrombin with high affinity, as measured by surface plasmon resonance. Maximal steady-state binding of polyP75 to biotinylated WT thrombin (●) or K248A mutant thrombin (▲) in 150 mM NaCl was plotted versus polyP75 polymer concentration, to which the single-site ligand binding equation was fitted. Inset shows the same data plotted from 0–50 nM polyP. Data represent mean ± S.D. (n = 3).
Table 1.
Thrombin binds polyP with high affinity
| Kd (nM) at 50 mM NaCl* | Kd (nM) at 150 mM NaCl* | |
|---|---|---|
| WT thrombin | 4.9 ± 0.5 | 5.4 ± 0.4 |
| K248A {236} mutant | 12.2 ± 1.6 | NB† |
Binding of polyP75 to immobilized biotin-thrombin (WT and mutant) was quantified in surface plasmon resonance studies at 50 mM and 150 mM NaCl. Kd values in this table are given in terms of polyP polymer concentration (mean ± S.D.; n = 3).
NB, negligible binding.
Discussion
We previously showed that polyP accelerates factor V activation by thrombin [12]. PolyP is highly anionic, leading us to hypothesize that it binds to one of thrombin’s anion-binding exosites. We found clear evidence that polyP interacts with high affinity with exosite II, the more basic of thrombin’s two exosites and which is known to bind a number of anionic glycosaminoglycans [19]. Interaction of polyP with exosite I was ruled out, as polyP bound equally well to thrombin in complex with the C-terminus of hirudin [4] and to γ-thrombin, a proteolytic derivative in which exosite I is impaired [20]. Experiments using thrombins mutated in exosites I and II, and also using competitors that bind to these regions, showed that polyP binding to thrombin requires exosite II. The interaction clearly has an ionic component, as high NaCl concentrations weakened thrombin binding to polyP.
Another similarity between polyP and known exosite II ligands lies in the cofactor role polyP plays in the positive feedback of thrombin-catalyzed factor V activation. Both exosites on thrombin have been implicated in factor V activation [21, 22], and we propose that polyP binding to exosite II promotes bridging of thrombin to factor V, thereby facilitating cleavage and factor Va formation. This “approximation” phenomenon is well documented in terms of thrombin’s activation of factor XI via linkage with GPIb [23], factor XIII via fibrin [24] and both protein C [25, 26] and thrombin activatable fibrinolysis inhibitor [14] through interaction with thrombomodulin. This is in addition to the clearly characterized roles of glycosaminoglycan cofactors during thrombin inhibition by antithrombin and heparin cofactor II. The role of cofactors in the regulation of thrombin activity extends the enzymes substrate specificity and appears crucial to its lifespan in the body [27]. To date, no cofactor has been reported to be involved in the early reactions elicited by thrombin, i.e. cleavage of fibrinogen to fibrin, or activation of factor V or factor VIII [27]. Our data suggest that polyP may function in regulating thrombin activity at this time. During primary haemostasis circulating platelets will contact the damaged endothelium becoming activated; this will result in release of polyP and exposure of negatively charged phospholipids, required for assembly of the prothrombinase complex. During these initial stages of coagulation polyP is in prime position to bind newly generated thrombin and enhance back-activation of factor V to stimulate further thrombin formation. Further studies will be necessary to define how thrombin transitions through its different cofactors during its lifespan, which is most probably influenced by availability, location and binding affinities of the reactants.
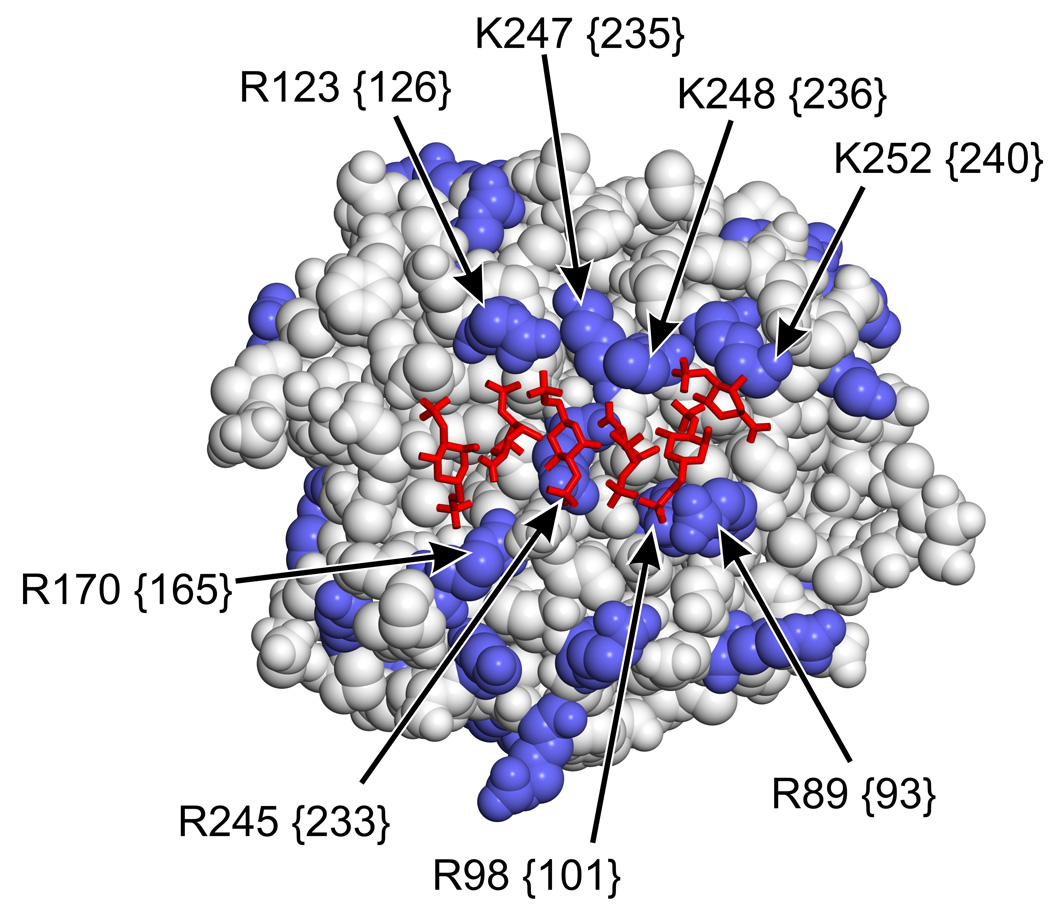
Binding of heparin to thrombin has been localized through mutagenesis studies [19, 28–30] and confirmed by high-resolution crystallography [31]. The interaction is ionic in nature and involves residues R89 {93}, K248 {236}, K252 {240}, R98 {101} and R245 {233} (in decreasing order of contribution to heparin binding) [31]. The data presented here indicate that, while some overlap exists in the binding sites for heparin and polyP within exosite II, they are not identical. Interaction of both heparin and polyP with thrombin depends on some common residues like K248 {236} and K252 {240}, but importantly, mutating R89 {93} -- which is most crucial for heparin’s interaction with thrombin -- had the least impact on polyP interaction with thrombin. A similar situation is observed with heparin and prothrombin fragment 2, with a degree of overlap in the binding sites of these molecules on thrombin but with only one common residue in exosite II involved [32]. He et al. [33] illustrated that interactions occurring at exosite II in thrombin are mediated by different arginine residues. The unique specificity of residues in this region and the weak influence of R89 {93} and R98 {101} on polyP binding to thrombin help explain the lack of competition with heparin. We observed only partial competition between heparin and polyP for binding to exosite II, and furthermore polyP did not appreciably interfere with heparin catalyzed thrombin inhibition by antithrombin. This suggests that both ligands can bind to the same thrombin molecule. In the crystal structure of heparin bound to exosite II of thrombin (Fig. 8), residues R89 {93} and R98 {101} (which were the least important in polyP binding but the most important in heparin binding) lie on one side of the bound heparin molecule, while residues K248 {236} and K252 {240} (which are very important for polyP binding) lie on the opposite side of the bound heparin. Furthermore, these latter residues appear to be more solvent-accessible even in the presence of bound heparin. Further mutagenesis and crystallographic studies are required to resolve the extent of the overlap in polyP and heparin binding. Competition between polyP and chrondroitin sulfate was even less apparent than heparin, despite its known binding to exosite II [10]. The residues important in this interaction are assumed to be similar to heparin, but have not been extensively defined. It is possible that the chondrotin sulfate-thrombin interactin has a lower affinity than the heparin-thrombin interaction, thereby explaining the reduced competition with polyP.
Fig. 8.
Thrombin-heparin co-crystal structure. Thrombin is rendered as space-filling with Lys and Arg side chains in blue, while heparin is rendered as red wires. Key Lys and Arg residues implicated in heparin and/or polyP binding are indicated by arrows using thrombin numbering, with chymotrypsinogen equivalents in curly braces. Structure is from PDB file 1XMN [31].
The concentration of polyP75 in whole blood may reach 3 µM following platelet activation and much higher levels are expected in platelet-rich thrombi [11, 12]. This polyP concentration is far above the 5 nM Kd of the polyP-thrombin interaction, suggesting that complex formation is favored in vivo. PolyP may therefore act as a physiologic modulator of thrombin function.
Acknowledgments
We thank Collin Waters for excellent technical assistance. This work was supported in part by NIH/NHLBI grants HL47014 (to J.H. Morrissey) and HL057530 (to L. Leung), and by Roy J. Carver Charitable Trust grant 06-2328 (to J.H. Morrissey).
Footnotes
Disclosure of Conflicts of Interest
N.J. Mutch and J.H. Morrissey are coinventors on pending patent applications covering some of the technologies discussed in this article.
REFERENCES
- 1.Naski MC, Shafer JA. Alpha-thrombin-catalyzed hydrolysis of fibrin I. Alternative binding modes and the accessibility of the active site in fibrin I-bound alpha-thrombin. J Biol Chem. 1990;265:1401–1407. [PubMed] [Google Scholar]
- 2.Bouton MC, Jandrot-Perrus M, Moog S, Cazenave JP, Guillin MC, Lanza F. Thrombin interaction with a recombinant N-terminal extracellular domain of the thrombin receptor in an acellular system. Biochem J. 1995;305((Pt 2)):635–641. doi: 10.1042/bj3050635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsiang M, Lentz SR, Dittman WA, Wen D, Scarpati EM, Sadler JE. Equilibrium binding of thrombin to recombinant human thrombomodulin: effect of hirudin, fibrinogen, factor Va, and peptide analogues. Biochemistry. 1990;29:10602–10612. doi: 10.1021/bi00499a005. [DOI] [PubMed] [Google Scholar]
- 4.Rydel TJ, Tulinsky A, Bode W, Huber R. Refined structure of the hirudin-thrombin complex. J Mol Biol. 1991;221:583–601. doi: 10.1016/0022-2836(91)80074-5. [DOI] [PubMed] [Google Scholar]
- 5.De Cristofaro R, De Candia E, Landolfi R, Rutella S, Hall SW. Structural and functional mapping of the thrombin domain involved in the binding to the platelet glycoprotein Ib. Biochemistry. 2001;40:13268–13273. doi: 10.1021/bi010491f. [DOI] [PubMed] [Google Scholar]
- 6.Leung LL, Hall SW. Dissociation of thrombin's substrate interactions using site-directed mutagenesis. Trends Cardiovasc Med. 2000;10:89–92. doi: 10.1016/s1050-1738(00)00047-5. [DOI] [PubMed] [Google Scholar]
- 7.Arni RK, Padmanabhan K, Padmanabhan KP, Wu TP, Tulinsky A. Structures of the noncovalent complexes of human and bovine prothrombin fragment 2 with human PPACK-thrombin. Biochemistry. 1993;32:4727–4737. doi: 10.1021/bi00069a006. [DOI] [PubMed] [Google Scholar]
- 8.Church FC, Pratt CW, Noyes CM, Kalayanamit T, Sherrill GB, Tobin RB, Meade JB. Structural and functional properties of human alpha-thrombin, phosphopyridoxylated alpha-thrombin, and gamma T-thrombin. Identification of lysyl residues in alpha-thrombin that are critical for heparin and fibrin(ogen) interactions. J Biol Chem. 1989;264:18419–18425. [PubMed] [Google Scholar]
- 9.Bode W, Turk D, Karshikov A. The refined 1.9-A X-ray crystal structure of D-Phe-Pro- Arg chloromethylketone-inhibited human alpha-thrombin: structure analysis, overall structure, electrostatic properties, detailed active-site geometry, and structure-function relationships. Protein Sci. 1992;1:426–471. doi: 10.1002/pro.5560010402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu LW, Rezaie AR, Carson CW, Esmon NL, Esmon CT. Occupancy of anion binding exosite 2 on thrombin determines Ca2+ dependence of protein C activation. J Biol Chem. 1994;269:11807–11812. [PubMed] [Google Scholar]
- 11.Ruiz FA, Lea CR, Oldfield E, Docampo R. Human platelet dense granules contain polyphosphate and are similar to acidocalcisomes of bacteria and unicellular eukaryotes. J Biol Chem. 2004;279:44250–44257. doi: 10.1074/jbc.M406261200. [DOI] [PubMed] [Google Scholar]
- 12.Smith SA, Mutch NJ, Baskar D, Rohloff P, Docampo R, Morrissey JH. Polyphosphate modulates blood coagulation and fibrinolysis. Proc Natl Acad Sci U S A. 2006;103:903–908. doi: 10.1073/pnas.0507195103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Myles T, Church FC, Whinna HC, Monard D, Stone SR. Role of thrombin anion-binding exosite-I in the formation of thrombin-serpin complexes. J Biol Chem. 1998;273:31203–31208. doi: 10.1074/jbc.273.47.31203. [DOI] [PubMed] [Google Scholar]
- 14.Hall SW, Nagashima M, Zhao L, Morser J, Leung LL. Thrombin interacts with thrombomodulin, protein C, and thrombin-activatable fibrinolysis inhibitor via specific and distinct domains. J Biol Chem. 1999;274:25510–25516. doi: 10.1074/jbc.274.36.25510. [DOI] [PubMed] [Google Scholar]
- 15.Lorenz B, Marme S, Muller WE, Unger K, Schroder HC. Preparation and use of polyphosphate-modified zirconia for purification of nucleic acids and proteins. Anal Biochem. 1994;216:118–126. doi: 10.1006/abio.1994.1015. [DOI] [PubMed] [Google Scholar]
- 16.Anderson PJ, Nesset A, Dharmawardana KR, Bock PE. Characterization of proexosite I on prothrombin. J Biol Chem. 2000;275:16428–16434. doi: 10.1074/jbc.M001254200. [DOI] [PubMed] [Google Scholar]
- 17.Wu Q, Picard V, Aiach M, Sadler JE. Activation-induced exposure of the thrombin anion-binding exosite. Interactions of recombinant mutant prothrombins with thrombomodulin and a thrombin exosite-specific antibody. J Biol Chem. 1994;269:3725–3730. [PubMed] [Google Scholar]
- 18.Stubbs MT, Bode W. A player of many parts: the spotlight falls on thrombin's structure. Thromb Res. 1993;69:1–58. doi: 10.1016/0049-3848(93)90002-6. [DOI] [PubMed] [Google Scholar]
- 19.Sheehan JP, Sadler JE. Molecular mapping of the heparin-binding exosite of thrombin. Proc Natl Acad Sci U S A. 1994;91:5518–5522. doi: 10.1073/pnas.91.12.5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rydel TJ, Yin M, Padmanabhan KP, Blankenship DT, Cardin AD, Correa PE, Fenton JW, 2nd, Tulinsky A. Crystallographic structure of human gamma-thrombin. J Biol Chem. 1994;269:22000–22006. [PubMed] [Google Scholar]
- 21.Esmon CT, Lollar P. Involvement of thrombin anion-binding exosites 1 and 2 in the activation of factor V and factor VIII. J Biol Chem. 1996;271:13882–13887. doi: 10.1074/jbc.271.23.13882. [DOI] [PubMed] [Google Scholar]
- 22.Myles T, Yun TH, Hall SW, Leung LL. An extensive interaction interface between thrombin and factor V is required for factor V activation. J Biol Chem. 2001;276:25143–25149. doi: 10.1074/jbc.M011324200. [DOI] [PubMed] [Google Scholar]
- 23.Yun TH, Baglia FA, Myles T, Navaneetham D, Lopez JA, Walsh PN, Leung LL. Thrombin activation of factor XI on activated platelets requires the interaction of factor XI and platelet glycoprotein Ib alpha with thrombin anion-binding exosites I and II, respectively. J Biol Chem. 2003;278:48112–48119. doi: 10.1074/jbc.M306925200. [DOI] [PubMed] [Google Scholar]
- 24.Philippou H, Rance J, Myles T, Hall SW, Ariens RA, Grant PJ, Leung L, Lane DA. Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition for cofactor sites on thrombin determines its fate. J Biol Chem. 2003;278:32020–32026. doi: 10.1074/jbc.M305364200. [DOI] [PubMed] [Google Scholar]
- 25.Rezaie AR, Yang L. Thrombomodulin allosterically modulates the activity of the anticoagulant thrombin. Proc Natl Acad Sci U S A. 2003;100:12051–12056. doi: 10.1073/pnas.2135346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu G, Chhum S, Krishnaswamy S. The affinity of protein C for the thrombin.thrombomodulin complex is determined in a primary way by active site-dependent interactions. J Biol Chem. 2005;280:15471–15478. doi: 10.1074/jbc.M500881200. [DOI] [PubMed] [Google Scholar]
- 27.Lane DA, Philippou H, Huntington JA. Directing thrombin. Blood. 2005;106:2605–2612. doi: 10.1182/blood-2005-04-1710. [DOI] [PubMed] [Google Scholar]
- 28.Ye J, Rezaie AR, Esmon CT. Glycosaminoglycan contributions to both protein C activation and thrombin inhibition involve a common arginine-rich site in thrombin that includes residues arginine 93, 97, and 101. J Biol Chem. 1994;269:17965–17970. [PubMed] [Google Scholar]
- 29.Gan ZR, Li Y, Chen Z, Lewis SD, Shafer JA. Identification of basic amino acid residues in thrombin essential for heparin-catalyzed inactivation by antithrombin III. J Biol Chem. 1994;269:1301–1305. [PubMed] [Google Scholar]
- 30.Tsiang M, Jain AK, Gibbs CS. Functional requirements for inhibition of thrombin by antithrombin III in the presence and absence of heparin. J Biol Chem. 1997;272:12024–12029. doi: 10.1074/jbc.272.18.12024. [DOI] [PubMed] [Google Scholar]
- 31.Carter WJ, Cama E, Huntington JA. Crystal structure of thrombin bound to heparin. J Biol Chem. 2005;280:2745–2749. doi: 10.1074/jbc.M411606200. [DOI] [PubMed] [Google Scholar]
- 32.Bock P, Panizzi P, Verhamme I. Exosites in the substrate specificity of blood coagulation reactions. J Thromb Haemost. 2007;5:81–94. doi: 10.1111/j.1538-7836.2007.02496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He X, Ye J, Esmon CT, Rezaie AR. Influence of Arginines 93, 97, and 101 of thrombin to its functional specificity. Biochemistry. 1997;36:8969–8976. doi: 10.1021/bi9704717. [DOI] [PubMed] [Google Scholar]