Abstract
Influenza virus remains a significant health concern with current circulating strains that affect millions each year plus the threat of newly emerging strains, such as swine-origin H1N1 and avian H5N1. Our hypothesis is that influenza-derived HLA-Class I-restricted epitopes can be identified for use as a reagent to monitor and quantitate human CD8+ T cell responses and for vaccine development to induce protective cellular immunity. Protein sequences from influenza A virus strains currently in circulation, agents of past pandemics and zoonotic infections of man were evaluated for sequences predicted to bind to alleles representative of the most frequent HLA-A and -B (Class I) types worldwide. Peptides that bound several different HLA molecules and were conserved among diverse influenza subtypes were tested for their capacity to recall influenza-specific immune responses using human donor PBMC. Accordingly, 28 different epitopes antigenic for human donor PBMC were identified and 25 were 100% conserved in the newly emerged swine-origin H1N1 strain. The epitope set defined herein should provide a reagent applicable to quantitate CD8+ T cell human responses irrespective of influenza subtype and HLA composition of the responding population. Additionally, these epitopes may be suitable for vaccine applications directed at the induction of cellular immunity.
Keywords: Influenza virus, CTL, HLA class I, cellular immunity, vaccine
1. Introduction
Influenza viruses are single-stranded RNA viruses of the family Orthomyxoviridae, of which 3 types (A, B, C) are recognized; only influenza A and B viruses occur in highly pathogenic forms and only the A virus is known to cause pandemics. Seasonal epidemics affect up to 15% of the world’s population resulting in 3-5 million cases of severe illness and approximately 500,000 deaths per year [1]. The young, immunologically naïve, and elderly, undergoing immune senescence, are most susceptible to infections. During epidemics, approximately 90% of all influenza-related deaths occur among people 65 years of age or older [2]. Although vaccination in the elderly, in particular, is associated with both a reduction in hospitalizations and death, it is clear that development of more effective vaccines for the elderly is needed [3].
Pandemics of influenza A viruses continue to occur at sporadic intervals in human populations. Four have occurred in the last 100 years in 1918, 1957, 1968 and 2009 [4-6]. Some of these pandemics are noted for their high mortality. For example, it is estimated that approximately 50 million people died in the 1918 pandemic and at least 1.5 million people in the 1957 and 1968 outbreaks combined [7]. Since 1996, several novel avian subtypes, H7N7, H5N1, H9N2, H7N2, H7N1 and H7N3 have crossed the species barrier from domestic poultry to humans and have caused a spectrum of mild to severe and even fatal human disease [8-12]. The recent outbreak and subsequent pandemic caused by a novel swine-origin H1N1 influenza virus (S-OIV) has been at the forefront of news and healthcare agency agendas [13-18]. In contrast to epidemic influenza, a majority of deaths (April-October 17. 2009) attributed to the emerging HIN1 infections were in the 18-64 age group, i.e., approximately 2,920 of 3,900 total deaths [19].
While hemagglutinin (HA)-specific antibody responses are most commonly used to evaluate vaccine protection, additional surrogate markers currently under study include matrix2-(M2)-specific antibodies and CD8+ and CD4+ T cell responses [20, 21]. A majority of information describing the role of cellular immunity in influenza infection was obtained in the murine model [for a review see ref. 22]. In the mouse, recovery from infection correlated with virus-specific CD8+ CTL activity [23] and lack of CD8+ CTL activity was associated with delayed viral clearance and increased mortality [24]. A DNA vaccine encoding the viral nucleoprotein (NP) and matrix (M) gene proteins, respectively, [25, 26] induced CD8+ CTL that provided cross-strain protection.
Further studies suggest that cellular immune responses also play a role in controlling influenza infection in humans. McMichael and colleagues inoculated 63 volunteers intranasally with live unattenuated influenza A/Munich/1/79 virus and found that all subjects with demonstrable T-cell responses cleared virus effectively [27]. The protective effect of cellular immune responses may be particularly relevant in the elderly where studies suggest that cellular immune responses may be an important predictor of protection [28]. Recent studies have also supported a correlation of cellular immune responses with protection against culture-confirmed influenza virus in young children [29].
Our hypothesis is that influenza-derived HLA-Class I-restricted epitopes can be prospectively identified for use as a reagent universally applicable to monitor and quantitate human CD8+ CTL responses irrespective of strain and HLA composition of the responding population. Additionally, these epitopes may be applicable for vaccine development to induce protective cellular immunity.
2. Materials and methods
2.1. HLA binding predictions
Computer-based algorithm [30-33] scans of influenza proteins were performed to obtain sequences predicted to bind with high affinity to HLA-A1, -A2, -A3, -A24, -B7 and -B44 Class I supertypes, which are representative of the most frequent HLA-A and -B types worldwide [30]. A method frequently used to predict MHC binding affinity is the matrix or linear coefficients method. Average Relative Binding (ARB) matrix is based on the assumption that each residue along the peptide molecule independently contributes to binding affinity. The effect of each of the 20 possible amino acids at each position along the peptide sequence is estimated by a matrix of coefficients. The overall binding of each peptide is then estimated by a simple polynomial function and expressed as an algorithm score. The ARB score-based matrix method generates binding affinity as ranked predictions specific for a given peptide size and MHC allele. To predict binding affinity (IC50 values), transformation functions were derived and utilized to fit nondimensional matrix scores to measured IC50 values [33].
2.2. Peptide synthesis
Peptides were synthesized initially in small amounts as crude material by Mimotopes (Minneapolis, MN) or Pepscan Systems (Lelystad, Netherlands). Larger quantities of selected peptides were synthesized at Pharmexa-Epimmune using an Applied Biosystems (Foster City, CA) 430A peptide synthesizer and F-moc solid phase methods or purchased from A & A labs (San Diego, CA). The purity of the peptides was substantiated by mass spectrometry and generally found to be greater than 95%. Peptides were dissolved in dimethyl sulfoxide at a concentration of 20 mg/ml, stored frozen at -20 °C, and diluted with phosphate-buffered saline (PBS) before use.
2.3. Sequence conservancy
Amino acid sequence conservancy was evaluated using 69 diverse subtype strains; 1) currently in circulation (H1N1, H3N2), 2) potential components of a pandemic influenza virus including agents of past pandemics (H1N1, H2N2, H3N2) and zoonotic influenza infections of man (H5N1, H1N1, H7N2, H7N3, H7N7, H9N2), see Table 1. The conservancy of each epitope was defined as the number of the strains containing the epitope at a 100% identity level divided by the total number of strains, i.e., 69. Due to the recent pandemic outbreak of S-OIV H1N1, conservancy analysis was also completed using 10 S-OIV strains isolated from diverse geographic regions of the world (A/Canada-AB/RV1532/2009; A/California/04/2009; A/England/195/2009; A/Bayern/63/2009; A/Fuzhou/01/2009; A/GuangzhouSB/1/2009; A/Hyogo/1/2009; A/Indiana/09/2009; A/Italy/05/2009; A/Mexico/InDRE4114/2009). The conservancy tool used for this analysis may be found in the Immune Epitope Database Analysis Resource (http://tools.immuneepitope.org/main/).
Table 1.
Sixty-nine diverse influenza strains used for epitope conservancy analysis
| H1N1 | H2N2 | H3N2 |
|---|---|---|
| A/Brevig Mission/1/1918 | A/Leningrad/134/17/57 | A/Hong Kong/1/68 |
| A/PR/8/34 | A/Singapore/1/57 | A/Albany/1/76 |
| A/Fort Monmouth/1/47 | A/Ann Arbor/6/60 | A/Moscow/10/99 |
| A/USSR/90/1977 | A/Berlin/3/64 | A/Hong Kong/1774/99 |
| A/New Caledonia/20/1999 | A/Tokyo/3/67 | A/Wisconsin/67/2005 |
| A/California/UR06-0125/2007 | A/Brisbane/10/2007 | |
| A/District of Columbia/WRAMC-1154047/08 | ||
| H5N1 | H7N1 & H7N2 | H7N3 & H7N7 |
| A/goose/Guangdong/1/1996 | A/turkey/Italy/3675/99 (N1) | A/fowl/Dobson/1927 (N7) |
| A/Hong Kong/483/97 | A/chicken/Italy/2335/2000 (N1) | A/equine/San Paulo/4/76 (N7) |
| A/Hong Kong/156/97 | A/turkey/Virginia/55/02 (N2) | A/chicken/Victoria/1/1985 (N7) |
| A/Hong Kong/213/03 | A/New York/107/03 (N2) | A/Netherlands/219/2003 (N7) |
| A/Vietnam/1203/04 | A/chicken/New York/21211-2/05 (N2) | A/chicken/Brit. Col./GSC human B/04 (N3) |
| A/Indonesia/5/2005 | A/duck/Tasmania/277/07 (N2) | A/Canada/rv504/04 (N3) |
| A/turkey/Turkey/1/2005 | A/ruddy turnstone/Delaware Bay/108/07 (N3) | |
| A/Bar-headed Goose/Qinghai/12/05 | A/northern shoveler/California/HKWF2031/08 (N3) | |
| A/Egypt/902782/2006 | ||
| A/China/GD01/2006 | ||
| A/Laos/Nong Khai 1/2007 | ||
| A/Indonesia/CDC1031/2007 | ||
| A/chicken/Laos/17/2008 | ||
| H9N2 | H4N6 | H6N1 & H6N2 |
| A/chicken/Beijing/1/94 | A/swine/Ontario/01911-1/99 | A/teal/Hong Kong/W312/97 (N1) |
| A/chicken/Hong Kong/G9/1997 | A/environment/Maryland/1101/2006 | A/chicken/Taiwan/0204/05 (N1) |
| A/quail/Hong Kong/G1/1997 | A/mallard/Interior Alaska/3/2007 | A/American wigeon/California/HKWF1174/2007 (N1) |
| A/Hong Kong/1073/1999 | A/ring-necked duck/California/HKWF402/2007 (N1) | |
| A/Korea/KBNP-0028/00 | A/chicken/California/139/01 (N2) | |
| A/chicken/Jiangsu/L1/2004 | A/northern shoveler/California/HKWF268/2007 (N2) | |
| A/ferret/Maryland/P10-UMD/2008 | ||
| A/chicken/Israel/292/2008 | ||
| A/chicken/Pakistan/UDL-02/2008 | ||
| H8N4 & H10N3 & H10N7 | H11N1 | H12N5 & H16N3 |
| A/northern shoveler/California/HKWF1203/07 | A/mallard/Ohio/1851/2005 | A/northern shoveler/Interior Alaska/1/2007 |
| A/northern shoveler/California/HKWF1005/07 (N3) | A/shorebird/Delaware/168/06 | |
| A/greater white fronted goose/California/HKWF446/07 (N7) | ||
2.4. Peptide-binding assays
Previous studies indicated that most epitopes bind Class I molecules with a binding affinity of 500 nM or less [31]. Therefore, we measured the peptide binding affinity for purified HLA molecules representative of the allelic variants most frequently found in each supertype. Peptide-HLA binding measurements were completed using a standard binding competition format. Radiolabeled reference peptides with known binding affinity were used and the capacity of the uncharacterized test peptides to compete for binding to HLA molecules was measured [32]. The percentage of HLA molecule-bound radioactivity was determined by capturing HLA/peptide complexes on anti-HLA antibody-coated Optiplates (Packard Instrument, Meriden, CT) and measuring bound cpm using the TopCount (Packard Instrument) microscintillation counter. In appropriate stoichiometric conditions, the IC50 nM of an unlabeled test peptide to the purified HLA molecule is a reasonable approximation of the affinity of interaction (Kd).
2.5. Population coverage calculations
Predicted population coverage for the epitope package was calculated using peptide binding affinities and reported gene frequencies for HLA-A and -B, and molecules to indicate the fraction of individuals expected to respond to a given vaccine epitope set [34]. Correspondence between different alleles sharing the same serological antigens and a representative allele for which peptide binding assays have been developed was assumed in certain instances.
2.6. Measurement of human memory Class I-restricted T cell responses
In the initial screens, Class I typed donor PBMCs from leukapheresis were obtained from HemaCare Corporation, Van Nuys, CA, purified from blood using gradient centrifugation with Histopaque-1077 (Sigma, St Louis, MO) and stored in liquid N2. Frozen donor PBMC were thawed and rested overnight in media containing RPMI 5% AB human serum/complete media followed by a seven day expansion of peptide-specific T cells using a pool of approximately 10 peptides, 1 μg/ml final concentration of each peptide. Both influenza-specific and negative control Plasmodium falciparum-, HBV- and HIV-specific peptides were used in this step. On days 1 and 3, T cells were fed with fresh media and IL2 (100U/ml). On day 7, CD8+ T cells were enriched by depleting CD4+ lymphocytes (anti-CD4+ coated magnetic beads, Miltenyi Biotec, Auburn, CA) prior to their use in an IFN-γ ELISPOT assay.
2.7. Human IFN-γ ELISPOT assay
PBMC responses to the panel of Class I binding peptides were evaluated using an IFN-γ ELISPOT assay. Briefly, membrane-based 96-well plates (Millipore, Bedford, MA) were coated overnight at 4 °C with the murine mAb specific for human IFN-γ (clone 1-D1k: Mabtech, Cincinnati, OH) at the concentration of 5 μg/ml. After washing with PBS, RPMI + 10% heat-inactivated human AB serum was added to each well and incubated at 37 °C for at least 1 h to block membranes. The Class I binding peptides were added to irradiated autologous PBMCs in triplicate wells (100,000 cells/well) in a volume of 50 μl at the final concentration of 10 μg/ml. Enriched CD8+ PBMC were resuspended in RPMI at a concentration of 1 × 106 PBMC/ml, and dispensed in 50 μl volume into test wells (50,000/well). The assay plates were incubated at 37 °C for 18 h, after which they were washed with PBS + 0.5% Tween 20. To each well, 100 μl of biotinylated mAb specific for human IFN-γ (clone 7-B6-1; Mabtech), at the concentration of 2 μg/ml, was added and plates were incubated at 37°C for 2 h. The plates were again washed, avidin-peroxidase complexes (Vectastain Elite kit) was added to each well, and the plates were incubated at room temperature for 1 h. The plates were developed using AEC (3-amino-9-ethylcarbazole: Sigma-Aldrich), washed, and dried. Data are shown as IFN-γ spot forming cells (SFC)/million CD8+ enriched PBMC. Spots were counted using an AID ELISPOT Reader System (Straβberg, Germany). HLA Class I-restricted peptide-specific responses from HIV, HCV and Plasmodium falciparum were used to determine background responses.
2.8. Statistical analysis
In IFN-γ ELISPOT assays using human PBMC we knew the donors were not previously exposed to HIV, HCV and assume lack of infection with P. falciparum. Therefore, responses to peptides derived from these pathogens were used to determine assay background. A mean influenza-specific response was considered significant if the net-SFC number was greater than the mean response of the background plus 2 standard deviations. In addition, a response greater than 100 IFN-γ SFC/106 CD8+ cells was considered the minimum for a positive response.
3. Results
3.1. Identification of influenza peptides predicted to bind with high affinity to Class I molecules
Influenza protein sequences were scanned to identify peptides predicted to bind with high affinity to alleles representative of the HLA-A1, -A2, -A3, -A24, -B7 and -B44 supertypes. A total of 427 peptide sequences predicted to bind to HLA alleles were subsequently synthesized for use in in vitro peptide binding assays. Specifically, the number of peptides identified for each protein is as follows: HA (21); neuraminidase (NA) (37); NP (56); M1 (19); M2 (11); nonstructural protein 1 (NS1) (20); nonstructural protein 2 (NS2) (20); polymerase acid (PA) (78); polymerase basic 1 (PB1) (85) and polymerase basic 2 (PB2) (80).
Amino acid sequence conservancy was also examined, using 69 different strains (Table 1). The conservancy of each peptide was defined as the number of the strains containing the peptide at a 100% identity level divided by the total number of strains, i.e., 69. The influenza strains used in conservancy analysis were derived from a diverse set of subtypes, selected using the following criteria; 1) currently in circulation (H1N1, H3N2), 2) agents of past pandemics (H1N1, H2N2, H3N2) and zoonotic influenza infections of man (H1N1, H5N1, H7N2, H7N3, H7N7, H9N2). The majority of peptides, 268 of 427 (63%), were conserved in 70% or higher of the 69 diverse influenza isolates. However, at this stage all 427 peptides were analyzed for peptide binding affinity.
3.2. In vitro peptide-HLA molecule binding and recall immunogenicity using human donor PBMC
We measured the peptide binding affinity for purified HLA molecules representative of the allelic variants most frequently found in each supertype. This analysis identified 291 (177 ≥ 70% conserved) of 427 peptides that bound multiple related HLA molecules within a supertype with high affinity. Specifically, we identify 21 (9 ≥ 70% conserved) A1; 53 (27 ≥ 70% conserved) A2; 78 (61 ≥ 70% conserved) A3; 33 (23 ≥ 70% conserved) A24; 17 (10 ≥ 70% conserved) B7; and 89 (47 ≥ 70% conserved) B44 supertype binders. The peptides that were highly conserved and bound multiple alleles within a HLA supertype were next evaluated for immunogenicity using human donor PBMC.
The 177 conserved peptides were next evaluated for their capacity to elicit influenza-specific recall immune responses. At least three to four different donors for each of the Class I supertype representatives were used in this analysis. Specifically, donors with A*0101, A*0201, A*0301 and A*1101, A*2402, B*0702 and B44 alleles were used. Peptide-specific responses obtained after one peptide in vitro restimulation for 3 different HLA-A1-positive donors, are given in Fig. 1. In the case of donor X757, 3 recall HLA-A1-restricted responses of approximately 800 to 1,500 SFC/106 Cells were observed, NP.44, PB1.590 and PB2.523. Significant peptide-specific responses are also depicted for the HLA-A1-positive donors X1257 and X716; NP.273, PB1.30 and NP.140. Peptide-specific recall responses identified for other representative HLA Class I supertype donors are depicted in Fig. 2. Specifically, responses induced for the HLA-A2-positive donor AC04 were M1.58, M1.59, NP.275, NP.458, NS1.9, NS2.173, PA.46 and PB1.413; for the HLA-A24-positive donor BB24 responses induced were NP.39, PA.130 and PB1.496; for the HLA-A3/A11-positive donor 3501 responses induced were M1.125, M2.45, NA.96, NP.407, NP.413, NS1.122, NS2.106, PA.104, PB2.323, PB1.471, PB1.689 and PB2.322; also for the HLA-B7-positive donor 3501 response induced was NP.172; for the HLA-B44-positive donor 114 response induced was M1.196.
Fig. 1.
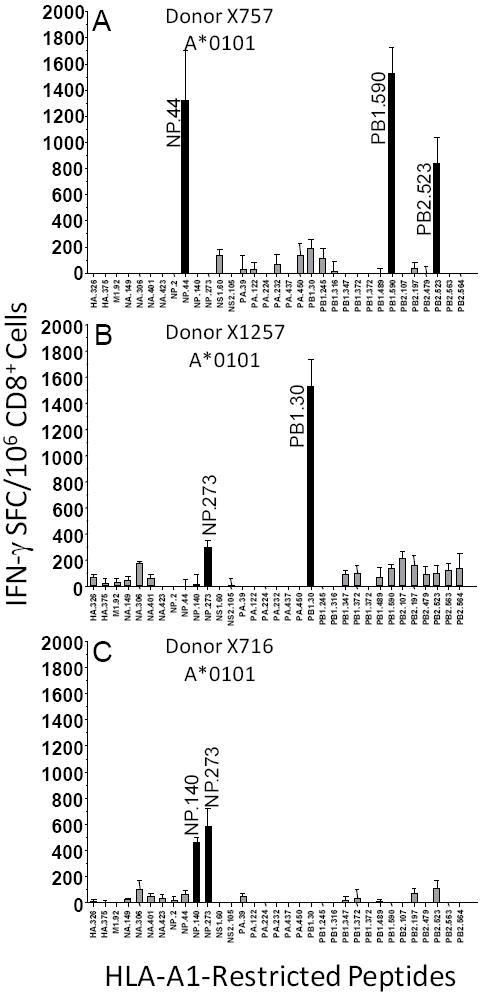
HLA-A*0101-restricted influenza recall responses from human donors. Human donor X757 (A), donor X1257 (B) and donor X716 (C). CD8+ T cells were purified for IFN-γ ELISPOT assays from normal human donors. The experimental values are expressed as the mean IFN-γ net SFC/106 CD8+ T cell lymphocytes ± SEM for each peptide; tests were completed using triplicate wells. Significant responses from the indicated epitopes were defined as responses > than mean of background responses + (2.0 × Std. Dev.). Background responses were determined using supertype binding peptides from pathogens to which donors were not exposed, i.e., HIV, HCV and Plasmodium falciparum.
Fig. 2.
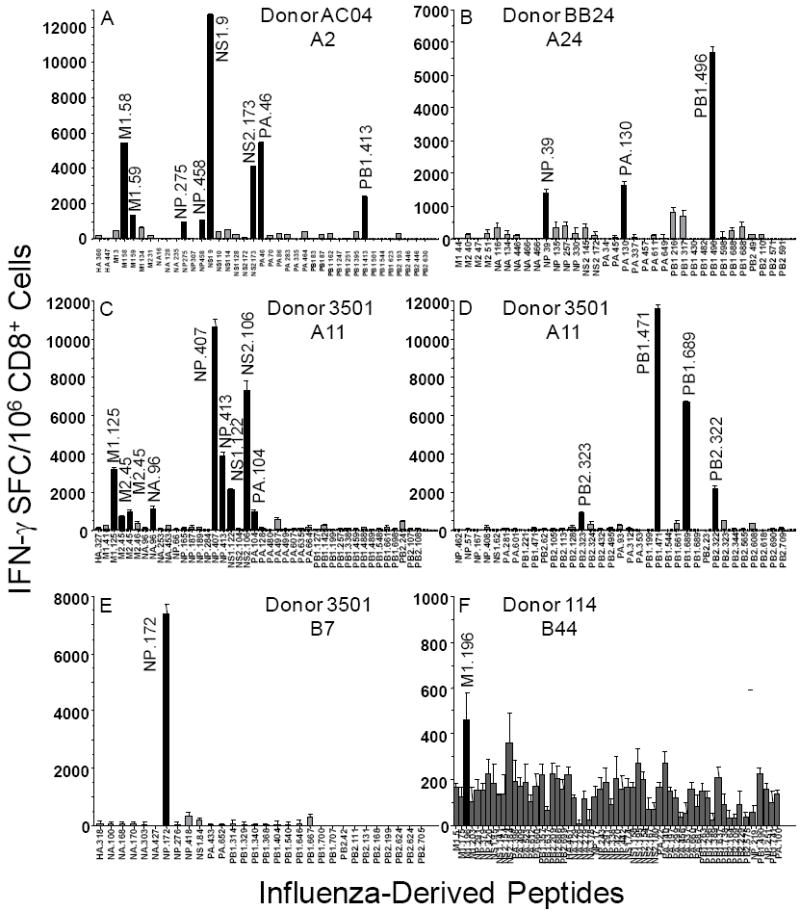
HLA-A*0201, -A*2402, -A*1101, -B*0702 and –B44-restricted influenza recall responses from representative human donors. Human HLA-A*0201-positive donor AC04 (A), HLA-A*2402-positive donor BB24 (B), HLA-A*1101-positive donor 3501 (C) and (D), HLA-B*0702-positive donor 3501 (E) and HLA-B44-positive donor 114 (F). Experimental design as described in Materials and Methods and also Fig. 1.
In total, positive peptide-specific responses were detected for 28 different Class I binding peptides. A summary is provided in Table 2, including frequency and magnitude of IFN-γ responses, epitope conservancy, and peptide binding affinity to the various HLA supertype molecules. A majority of epitopes, 18 of 28 were active in at least two individual donors.
Table 2.
Influenza CD8+ epitope summary
| Sequence | Source | Cons. %a) | IFN-γ Respsonsesb) | Supertype | A*0101 | A*2601 | A*2902 | A*3002 | Conserved % S-OIVc) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HSNLNDATY | NP.140 | 80 | 450; 1000 | A1 | 177d) | 791 | 5896 | 24 | 100 | ||
| KSCLPACVY | NP.273 | 99 | 590; 1200; 300 | A1 | 359 | --e) | -- | 43 | 100 | ||
| YSHGTGTGY | PB1.30 | 99 | 12500; 180; 1500 | A1 | 199 | 5 | 486 | 131 | 100 | ||
| LVSDGGPNLYf) | PB1.590 | 91 | 1100; 3800; 1500; 1000 | A1 | 399 | 156 | 479 | 57 | 100 | ||
| Sequence | Source | Cons. (%) | Supertype | A*0201 | A*0202 | A*0203 | A*0206 | A*6802 | |||
| GLFGAIAGFI | HA.345 | 89 | 1800 | A2 | 41 | 0.4 | 1 | 2539 | 1531 | 100 | |
| GILGFVFTLg) | M1.58 | 93 | 8090; 5400; 760; 6700 | A2 | 1 | 2 | 21 | 1 | 522 | 100 | |
| FQVDCFLWHV | NS1.9 | 79 | 1460; 3200; 12700; 3900 | A2 | 4 | 4 | 3 | 1 | 219 | 0 | |
| CLPACVYGL | NP.275 | 73 | 350; 986; 2200 | A2 | 79 | 4 | 143 | 95 | 3771 | 100 | |
| FQGRGVFEL | NP.458 | 91 | 800; 1000; 1200 | A2 | 143 | 7 | 307 | 7 | -- | 100 | |
| FMYSDFHFIh) | PA.46 | 99 | 690; 320; 5400; 1300; 260 | A2 | 5 | 1 | 2 | 6 | 14 | 100 | |
| NMLSTVLGVi) | PB1.413 | 99 | 1100; 400; 2400 | A2 | 4 | 5 | 11 | 3 | 1061 | 100 | |
| Sequence | Source | Cons. (%) | Supertype | A*0301 | A*1101 | A*3101 | A*3301 | A*6801 | |||
| ASCMGLIYNR | M1.125 | 99 | 560; 3170 | A3 | 816 | 32 | 7 | 445 | 391 | 100 | |
| RLFFKCIYRR | M2.45 | 71 | 240; 970 | A3 | 135 | 294 | 5 | 238 | 88 | 100 | |
| SVQPTFSVQR | NP.407 | 79 | 3670; 10600 | A3 | -- | 37 | 21 | 181 | 84 | 100 | |
| SVQRNLPFER | NP.413 | 70 | 350;3870 | A3 | -- | 411 | 104 | 321 | 19 | 100 | |
| KFLPDLYDYK | PA.104 | 97 | 970 | A3 | 255 | 22 | 2406 | -- | 507 | 100 | |
| KLVGINMSKK | PB1.471 | 89 | 11500 | A3 | 14 | 315 | 518 | -- | 1614 | 100 | |
| GTFEFTSFFY | PB1.488 | 99 | 7900 | A3 | 16 | 1 | 242 | 5411 | 100 | 100 | |
| SFSFGGFTFKj) | PB2.322 | 97 | 13200; 2150 | A3 | 74 | 7 | 19 | 85 | 60 | 100 | |
| VLRGFLILGK | PB2.690 | 93 | 6150 | A3 | 10 | 460 | 85 | -- | 1161 | 100 | |
| Sequence | Source | Cons. (%) | Supertype | A*2301 | A*2402 | A*2902 | A*3002 | ||||
| FYIQMCTELk) | NP.39 | 93 | 1200; 3600 | A24 | 8 | 24 | 1561 | -- | 100 | ||
| YYLEKANKI | PA.130 | 99 | 100; 250; 1600 | A24 | 33 | 32 | -- | -- | 100 | ||
| FYRYGFVANF | PB1.496 | 97 | 550; 475; 1100; 1020; 5800 | A24 | 13 | 17 | 812 | -- | 100 | ||
| Sequence | Source | Cons. (%) | Supertype | B*0702 | B*3501 | B*5101 | B*5301 | B*5401 | |||
| LPRRSGAAGAl) | NP.172 | 99 | 7390 | B7 | 2 | -- | -- | -- | 47 | 100 | |
| Sequence | Source | Cons. (%) | Supertype | B*1801 | B*4001 | B*4002 | B*4402 | B*4403 | B*4501 | ||
| TEVETYVLSI | M1.5 | 97 | 260 | B44 | 136 | 133 | 195 | 6880 | 247 | 504 | 100 |
| SEQAAEAMEV | M1.196 | 86 | 460 | B44 | 1900 | 2670 | 85 | 2637 | 7667 | 35 | 100 |
| GERQNATEI | NP.17 | 89 | 130 | B44 | -- | 199 | 53 | -- | -- | 304 | 0 |
| QEIRTFSFQL | NS2.184 | 70 | 360 | B44 | 515 | 135 | 68 | 102 | 431 | 380 | 0 |
amino acid conservancy among H1N1, H2N2, H3N2, H5N1, H7N7, H9N2, etc. (69 strains, Table 1)
IFN-γ responses: IFN-γ SFC/106 CD8+ cells; each number indicates a different donor
amino acid conservancy among 10 swine origin H1N1 strains (A/Canada-AB/RV1532/2009; A/California/04/2009; A/England/195/2009; A/Bayern/63/2009; A/Fuzhou/01/2009; A/GuangzhouSB/1/2009; A/Hyogo/1/2009; A/Indiana/09/2009; A/Italy/05/2009; A/Mexico/InDRE4114/2009)
IC50 nM to purified HLA (In general, IC50 nM < 500 is immunogenic)
--indicates IC50 nM > 10,000
Refs (f, Dibrino 1993; g, Gotch 1987 and Lalvani 1997; h, i, l Giafrani 2000; j, Assarsson 2008; k, Deng 1997)
3.3. Epitope characterization in terms of protein source and predicted population coverage
Next, the protein source and population coverage were evaluated. In regard to protein representation, a large percentage (15 of 28) of the Class I binding epitopes was identified from the NP and PB1 proteins (Table 2). Between 1 and 4 epitopes were selected from the M1, PA, PB2, NS1, NS2 and M2 proteins. Only one conserved epitope was identified from the HA protein, probably reflecting the lack of conservation of the surface proteins across influenza subtypes.
The use of supertype-restricted epitopes, those capable of binding with substantial affinity to multiple related HLA alleles, provides a means to address HLA polymorphism and population coverage. Predicted population coverage for the HLA-A1, -A2, -A3/A11, -A24, -B7 and –B44 supertypes epitope package was calculated using peptide binding affinities and reported gene frequencies for HLA-A, -B, molecules to indicate the fraction of individuals expected to respond to the vaccine epitope set. The theoretical average population coverage for all ethnic populations studied, Australia, Europe, North Africa, North America, North-East Asia, Oceania, South America South-East Asia, South-West Asia and Sub-Saharan Africa was approximately 96.2% with the average minimum number of epitope hits/HLA combinations recognized by 90% of the population of greater than 3, Fig.3.
Fig. 3.
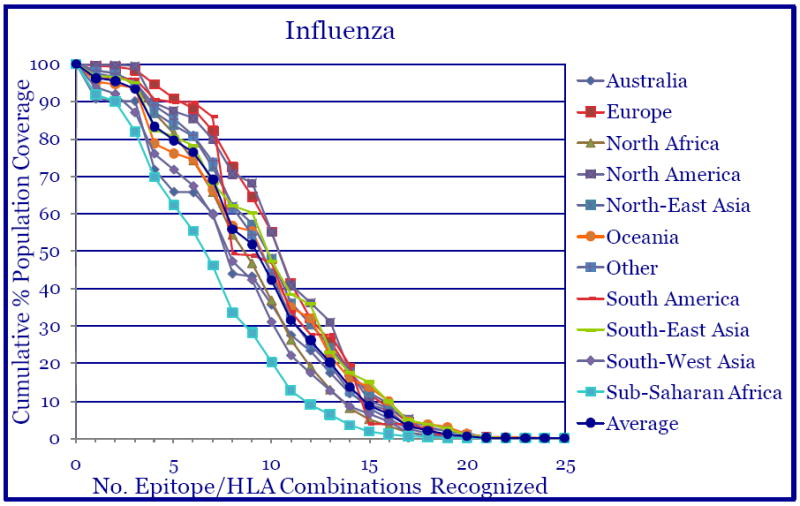
The identified 28 Class I-restricted epitopes provide broad population coverage. Based on the binding data for each Class I-restricted epitope, theoretical population coverage was calculated for each ethnic population, Australia, Europe, etc. The average population coverage of all areas/populations was approximately 96% and the average minimum number of epitope hits/HLA combinations recognized by 90% of the populations was greater than 3.
3.4. Epitope conservation in new and emerging strains
The studies described herein were initiated in year 2007, and as such no sequence from 2009 was included in our conservancy analysis. Our hypothesis is that selection of highly conserved epitopes should provide prospective coverage of new and emerging strains, thus alleviating or eliminating the need to select new epitopes from emerging strains. To address this point we examined the conservation of our epitope set in 10 additional sequences, encompassing S-OIV H1N1 isolates from the 2009 period, Table 2. Of 28 CD8+ epitopes, 25 are totally conserved in 100% of the S-OIV H1N1 isolates considered. These results demonstrate that the conserved epitopes should provide coverage of new and emerging strains, and in particular S-OIV H1N1.
4. Discussion
Our goal is to develop a “universal” set of CD8+ T cell influenza epitopes derived from portions of influenza viral proteins highly conserved in past, current and potentially emerging viral strains. The epitope set defined herein should provide a reagent universally applicable to monitor and quantitate human CD8+ T cell responses irrespective of strain and HLA composition of the responding population. In addition, these epitopes may be suitable for vaccine applications directed at the induction of cellular immunity. It is anticipated that cellular immunity will contribute to the control and clearance of the influenza infection during the course of the disease. However, it is not known how many epitope-specific responses would be required to perform this function. In our study, the range of epitope-specific responses detected per allele ranged from one (HLA-B7) to 13 (HLA-A11). It can be speculated that one dominant epitope-specific response may be sufficient for protection, as in the case of the matrix 58 epitope (GILGFVFTL) in a mouse study [35]. However, multiple responses, potentially from multiple proteins may be more likely to be efficient at control and clearance of influenza infection.
The CD8+ T cell epitopes described in this study were identified based on several criteria. Epitopes predicted to bind with high affinity to HLA supertype molecules and highly conserved among diverse influenza strains were tested in recall assays using human PBMC, thus identifying epitopes associated with responses following natural influenza infection. However, it is also possible that in part, recall responses were due to seasonal influenza vaccination inducing responses specific for contaminating internal influenza virus proteins.
Because degenerate binding and the sequence conservancy were used as preselection criteria, we expected only a partial overlap with epitopes previously identified by other studies that utilized different strategies for epitope identification [36-42]. Of note is the study of Assarsson et al., which identified HLA Class I-restricted T cell epitopes also using the methods of peptide/HLA binding assays, conservancy analysis among various different subtype strains and recall responses with human donor PBMC [36]. We suspect that differences in influenza-derived epitope identification may be in part due to (1) different set of influenza strains used to identify conserved epitopes and (2) a 7-day peptide-stimulated expansion step in the PBMC culture by us. The expansion culture step may have augmented T cell populations that were minor in vivo but could be useful as vaccine candidates. Indeed, 7 of the 28 epitopes were previously described and thus so indicated by reference in Table 2. To the best of our knowledge, 21 of 28 CD8+ T cell epitopes are newly described herein. The identification of such a large number of new influenza epitopes points to the extreme breadth of human responses to this pathogen. As noted in Fig. 1, three different HLA-A1 donors respond to different HLA-A1-restricted epitopes again supporting the breadth of T cell response to this pathogen.
Considering the hundreds of HLA alleles present in the human population, it might seem difficult to identify sufficient numbers of epitopes allowing coverage, irrespective of the HLA composition of the responding population. We have exploited the observation that a large fraction of HLA Class I molecules have overlapping repertoires of binding specificity that can be grouped into broad supertype families [30]. In spite of the fact that various HLA molecules occur at significantly different frequencies in different ethnicities, the overall frequency of each supertype is relatively consistent among the various ethnicities. Accordingly, we have identified 28 different epitopes from 6 different supertypes, and we show that the predicted population coverage is, as expected, fairly balanced amongst several different common ethnicities.
Furthermore, we have utilized sequence conservancy in different influenza subtypes as a preselection criteria for our epitope identification studies. Our basic hypothesis was that targeting conserved epitopes should generate an epitope set applicable to newly emerging influenza virus strains. The new emergence of S-OIV H1N1 strains provided an opportunity to test this hypothesis. The result of the analysis of the epitope conservancy in S-OIV H1N1 sequences demonstrated that 25 of 28 epitopes were highly conserved in these strains and thus provided a conceptual validation of our approach.
Acknowledgments
We thank Barbara Stewart, Kerin Arora, Charlie Huang, David Seiber for technical assistance and data analysis. This work was partially supported by NIH-NIAID contract number N01 AI-30039.
Abbreviations
- PBMC
peripheral blood mononuclear cells
- HBV
Hepatitis B virus
- HCV
Hepatitis C virus
- SFC
Spot Forming Cells
- IFN-γ
interferon-gamma
- HA
hemagglutinin
- NA
neuraminidase
- NP
nucleoprotein
- M1
matrix 1
- M2
matrix 2
- NS1
nonstructural protein 1
- NS2
nonstructural protein 2
- PA
polymerase acid
- PB1
polymerase basic 1
- PB2
polymerase basic 2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.WHO. Influenza Fact Sheet No. 211. [November, 2009]; http://www.who.int/mediacentre/factsheets/fs211/en/
- 2.Simonsen L, Reichert TA, Viboud C, Blackwelder WC, Taylor RJ, Miller MA. Impact of influenza vaccination on seasonal mortality in the US elderly population. Arch Intern Med. 2005;165:265–72. doi: 10.1001/archinte.165.3.265. [DOI] [PubMed] [Google Scholar]
- 3.Treanor JD. Influenza-the goal of control. N Engl J Med. 2007;357:1439–41. doi: 10.1056/NEJMe078140. [DOI] [PubMed] [Google Scholar]
- 4.Beveridge WI. The chronicle of influenza epidemics. Hist Philos Life Sci. 1991;13:223–34. [PubMed] [Google Scholar]
- 5.Kilbourne ED. Influenza pandemics of the 20th century. Emerg Infect Dis. 2006;12:9–14. doi: 10.3201/eid1201.051254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potter CW. A history of influenza. J Appl Microbiol. 2001;91:572–579. doi: 10.1046/j.1365-2672.2001.01492.x. [DOI] [PubMed] [Google Scholar]
- 7.Oxford JS. Influenza A pandemics of the 20th century. Rev Med Virol. 2000;10:119–33. doi: 10.1002/(sici)1099-1654(200003/04)10:2<119::aid-rmv272>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 8.Alexander DJ. An overview of the epidemiology of avian influenza. Vaccine. 2007;25:5637–44. doi: 10.1016/j.vaccine.2006.10.051. [DOI] [PubMed] [Google Scholar]
- 9.Belser JA, Blixt O, Chen LM, Pappas C, Maines TR, Van Hoeven N, Donis R, Busch J, McBride R, Paulson JC, Katz JM, Tumpey TM. Contemporary North American influenza H7 viruses possess human receptor specificity: Implications for virus transmissibility. Proc Natl Acad Sci USA. 2008;105:7558–63. doi: 10.1073/pnas.0801259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butt KM, Smith GJ, Chen H L, Zhang LJ, Leung YH, Xu KM, Lim W, Webster RG, Yuen KY, Peiris JS, Guan Y. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J Clin Microbiol. 2005;43:5760–7. doi: 10.1128/JCM.43.11.5760-5767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perdue ML, Swayne DE. Public health risk from avian influenza viruses. Avian Dis. 2005;49:317–27. doi: 10.1637/7390-060305R.1. [DOI] [PubMed] [Google Scholar]
- 12.Webster RG, Peiris M, Chen H, Guan Y. H5N1 outbreaks and enzootic influenza. Emerg Infect Dis. 2006;12:3–8. doi: 10.3201/eid1201.051024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belshe RB. Implications of the emergence of a novel H1 influenza virus. N Engl J Med. 2009;360:2667–8. doi: 10.1056/NEJMe0903995. [DOI] [PubMed] [Google Scholar]
- 14.Gatherer D. The 2009 H1N1 influenza outbreak in its historical context. J Clin Virol. 2009;45:174–8. doi: 10.1016/j.jcv.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–15. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 16.Shinde V, Bridges CB, Uyeki TM, Shu B, Balish A, Xu X, Lindstrom S, Gubareva LV, Deyde V, Garten RJ, Harris M, Gerber S, Vagasky S, Smith F, Pascoe N, Martin K, Dufficy D, Ritger K, Conover C, Quinlisk P, Klimov A, Bresee JS, Finelli L. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005-2009. N Engl J Med. 2009;360:2616–25. doi: 10.1056/NEJMoa0903812. [DOI] [PubMed] [Google Scholar]
- 17.Smith GJD, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JSM, Guan Y, Rambaut A. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nat. 2009;459:1122–6. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 18.Wang TT, Palese P. Unraveling the mystery of swine influenza virus. Cell. 2009;137:983–5. doi: 10.1016/j.cell.2009.05.032. [DOI] [PubMed] [Google Scholar]
- 19. [Dec, 2009]; http://www.cdc.gov/h1n1flu/estimates_2009_h1n1.htm.
- 20.Eichelberger M, Golding H, Hess M, Weir J, Subbarao K, Luke CJ, Friede M, Wood D. FDA/NIH/WHO public workshop on immune correlates of protection against influenza A viruses in support of pandemic vaccine development. Vaccine. 2008;26:4299–303. doi: 10.1016/j.vaccine.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 21.Rimmelzwaan GF, McElhaney JE. Correlates of protection: Novel generations of influenza vaccines. Vaccine. 2008;26S:D41–4. doi: 10.1016/j.vaccine.2008.07.043. [DOI] [PubMed] [Google Scholar]
- 22.Grebe KM, Yewdell JW, Bennink JR. Heterosubtypic immunity to influenza A virus: where do we stand? Microbes Infect. 2008;10:1024–9. doi: 10.1016/j.micinf.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackenzie CD, Taylor PM, Askonas BA. Rapid recovery of lung histology correlates with clearance of influenza virus by specific CD8+ cytotoxic T cells. Immunol. 1989;67:375–81. [PMC free article] [PubMed] [Google Scholar]
- 24.Bender BS, Croghan T, Zhang L, Small PA., Jr Transgenic mice lacking Class I MHC-restricted T cells have delayed viral clearance and increased mortality after influenza virus challenge. J Exp Med. 1992;175:1143–45. doi: 10.1084/jem.175.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okuda K, Ihata A, Watabe S. Protective immunity against influenza A virus induced by immunization with DNA plasmid containing influenza M gene. Vaccine. 2001;19:3681–91. doi: 10.1016/s0264-410x(01)00078-0. [DOI] [PubMed] [Google Scholar]
- 26.Ulmer JB, Fu TM, Deck RR, Friedman A, Guan L, DeWitt C, Liu X, Wang S, Liu MA, Donnelly JJ, Caulfield MJ. Protective CD4+ and CD8+ T cells against influenza virus induced by vaccination with nucleoprotein DNA. J Virol. 1998;72:5648–53. doi: 10.1128/jvi.72.7.5648-5653.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMichael AJ, Gotch FM, Noble GR, Beare PAS. CTL immunity to influenza. N Engl J Med. 1983;309:13–7. doi: 10.1056/NEJM198307073090103. [DOI] [PubMed] [Google Scholar]
- 28.McElhaney JE, Xie D, Hager WD, Barry MB, Wang Y, Kleppinger A, Ewen C, Kane KP, Bleackley RC. T cell responses are better correlates of vaccine protection in the elderly. J Immunol. 2006;176:6333–9. doi: 10.4049/jimmunol.176.10.6333. [DOI] [PubMed] [Google Scholar]
- 29.Forrest BD, Pride MW, Dunning AJ, Capeding MRZ, Chotpitayasunondh T, Tam JS, Rappaport R, Eldridge JH, Gruber WC. Correlation of cellular immune responses with protection against culture-confirmed influenza virus in young children. Clin Vaccine Immunol. 2008;15:1042–53. doi: 10.1128/CVI.00397-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA Class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sette A, Vitiello A, Reherman B, Fowler P, Nayersina R, Oseroff C, Yuan L, Ruppert J, Sidney J, del Guercio MF, Southwood S, Kubo RT, Chesnut RW, Grey HM, Chisari FV. The relationship between Class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J Immunol. 1994;153:5586–92. [PubMed] [Google Scholar]
- 32.Sette A, Sidney J, del Guercio MF, Southwood S, Ruppert J, Dahlberg C, Grey HM, Kubo RT. Peptide binding to the most frequent HLA-A Class I alleles measured by quantitative molecular binding assays. Mol Immunol. 1994;31:813–22. doi: 10.1016/0161-5890(94)90019-1. [DOI] [PubMed] [Google Scholar]
- 33.Bui HH, Sidney J, Peters B, Sathiamurthy M, Sinichi A, Purton K-A, Mothé BR, Chisari FV, Watkins DI, Sette A. Automated generation and evaluation of specific MHC binding predictive tools: ARB matrix applications. Immunogenetics. 2005;57:304–14. doi: 10.1007/s00251-005-0798-y. [DOI] [PubMed] [Google Scholar]
- 34.Bui HH, Sidney J, Ding K, Southwood S, Newman MJ, Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics. 2006;7:153. doi: 10.1186/1471-2105-7-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plotnicky H, Cyblat-Chanal D, Aubry J-P, Derouet F, Klinguer-Hamour C, Beck A, Bonnefory J-Y, Corvaia N. The immunodominant influenza matrix T cell epitope recognized in human induces influenza protection in HLA-A2/Kb transgenic mice. Virology. 2003;309:320–9. doi: 10.1016/s0042-6822(03)00072-2. [DOI] [PubMed] [Google Scholar]
- 36.Assarsson E, Bui HH, Sidney J, Zhang Q, Glenn J, Oseroff C I, Mbawuike IN, Alexander J, Newman MJ, Grey H, Sette A. Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans. J Virol. 2008;82:12241–51. doi: 10.1128/JVI.01563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng Y, Yewdell JW, Eisenlohr LC, Bennink JR. MHC affinity, peptide liberation, T cell repertoire, and immunodominance all contribute to the paucity of MHC Class I-restricted peptides recognized by antiviral CTL. J Immunol. 1997;158:1507–15. [PubMed] [Google Scholar]
- 38.DiBrino M, Tsuchida T, Turner RV, Parker KC, Coligan JE, Biddison WE. HLA-A1 and HLA-A3 T cell epitopes derived from influenza virus proteins predicted from peptide binding motifs. J Immunol. 1993;151:5930–5. [PubMed] [Google Scholar]
- 39.Gianfrani C, Oseroff C, Sidney J, Chesnut RW, Sette A. Human memory CTL response specific for influenza A virus is broad and multispecific. Hum Immunol. 2000;61:438–52. doi: 10.1016/s0198-8859(00)00105-1. [DOI] [PubMed] [Google Scholar]
- 40.Gotch F, Rothbard J, Howland K, Townsend A, McMichael A. Cytotoxic T lymphocytes recognize a fragment of influenza virus matrix protein in association with HLA-A2. Nature. 1987;326:881–2. doi: 10.1038/326881a0. [DOI] [PubMed] [Google Scholar]
- 41.Lalvani A, Brookes R, Hambleton S, Britton WJ, Hill AV, McMichael AJ. Rapid effector function in CD8+ memory T cells. J Exp Med. 1997;186:859–65. doi: 10.1084/jem.186.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang M, Lamberth K, Harndahl M, Roder G, Stryhn A, Larsen MV, Nielsen M, Lundegaard C, Tang ST, Dziegiel MH, Rosenkvist J, Pedersen AE, Buus S, Claesson MH, Lund O. CTL epitopes for influenza A including the H5N1 bird flu; genome-, pathogen-, and HLA-wide screening. Vaccine. 2007;25:2823–31. doi: 10.1016/j.vaccine.2006.12.038. [DOI] [PubMed] [Google Scholar]


