Abstract
Phosphorylation of the cytochrome P450 aromatase has been proposed as a switch to rapidly modulate enzymatic activity and estrogen biosynthesis. Herein, we demonstrate that aromatase serine-118 is a potential phosphorylation site in mammalian cells. The amino acid context surrounding S118 is highly conserved among diverse animal species and suggests that an AGC-like kinase may phosphorylate aromatase. Mutation of S118 to Ala blocked phosphorylation. Mutation of S118 to either Ala or Asp destabilized aromatase, indicating an important structural role for S118. The phosphomimetic S118D mutant showed decreased specific enzymatic activity, decreased Vmax, and increased Km, while the S118A phospho-inhibiting mutant showed opposite effects. Our findings suggest that phosphorylation of S118 may decrease aromatase activity, presenting a mechanism whereby kinase signaling may modulate estrogen production and hormone balance.
Keywords: aromatase, phosphorylation, estrogen synthase, post-translational modification
1. Introduction
Aromatase is the rate-limiting cytochrome P450 enzyme in estrogen biosynthesis, converting C19 androgens to C18 estrogens. Expression of the CYP19 gene encoding aromatase is controlled by tissue-specific promoters, which modulate expression in diverse tissues including brain, testes, breast, ovary, placenta, adipose tissue, skin, and bone [1]. Aberrant promoter activation has been implicated in disease, including breast, prostate, and endometrial cancers (reviewed in [2]). For example, levels of cyclooxygenase-2 (COX-2) and aromatase are directly correlated in human breast cancers [3]. COX-2 produces prostaglandin E2 (PGE2), and COX-2 expression is induced by estrogens [4]. PGE2 stimulates aromatase expression via the cAMP-inducible promoter II [5]. The enhanced expression of aromatase by PGE2 increases estrogen production, thus resulting in a positive feedback loop to drive hormone-responsive breast cancer cell growth.
In contrast, the non-genomic mechanisms regulating aromatase enzymatic activity remain poorly understood. Studies have described conflicting changes in aromatase activity in different cell types upon the addition of growth factors [6-8] and kinase inhibitors [9-12], suggesting that aromatase is modulated by post-translational modification. Reports have also shown that aromatase can be glycosylated at the N-terminal membrane anchor domain, but there is arguably no effect on enzymatic activity [13-15]. Findings from studies in quail brain homogenates suggest that aromatase is phosphoregulated [16]. However, the site(s) of phosphorylation and its effects on aromatase are unclear.
Phosphoregulation of aromatase has been suggested to occur through a Ca2+-dependent mechanism involving PKA, PKC, or CAMK [17]. Additionally, inhibition of Map/Erk kinase-1 and -2 (MEK1/2) or phosphoinositide 3-kinase (PI3K) in aromatase-over-expressing breast cancer cells decreased aromatase activity [11]. The potential for kinase-mediated control of hormone balance and estrogen synthesis, both in normal and diseased settings, prompted us to further investigate aromatase phosphoregulation in situ.
2. Materials and Methods
2.1 Plasmids and cell lines
The murine aromatase cDNA was cut from pTG6 (a gift of R. Tekmal, Univ. of Texas San Antonio) [18] and subcloned into pcDNA3.1/myc-His-B (Invitrogen) to generate mArom-myc-his/pcDNA3.1. Single amino acid aromatase mutants (S118A, S118D, S238A, S247A) were generated using the QuickChange site-directed mutagenesis kit (Stratagene) and confirmed by DNA sequencing. The human aromatase cDNA vector was a gift of E. Simpson (Prince Henry's Inst., Australia).
HEK-293T, COS-7, and JEG-3 human choriocarcinoma cells were obtained from ATCC. MCF-7 human breast adenocarcinoma cells stably transfected with aromatase cDNA (MCF-7/CA) were a gift from R. Santen (Univ. of Virginia).
2.2 Immunoprecipitation & Western blotting
HEK-293T cells were transfected with mArom-myc-his/pcDNA3.1 (or pcDNA3.1 control) using Fugene6 (Roche). Cells were lysed at 2 days post-transfection using NP-40 buffer (0.5% NP-40, 50 mM Tris, pH 7.4, 120 mM NaCl, protease inhibitor cocktail [PIC, Sigma]). Fifty μg of protein was used for Western blotting. Alternatively, cells were lysed in 1% Triton buffer for immunoprecipitation (1% Triton X-100, 150 mM NaCl, 10 mM Na phosphate, pH 7.2, PIC, 20 mM NaF, 2 mM Na3VO4). Cleared lysates were incubated with protein G Dynal beads (Invitrogen) prebound with antibodies against myc (9E10, Upstate) or aromatase (677 mAb) [19]. Beads were washed, immunoprecipitated protein was eluted using reducing sample buffer, and protein was used for Western blotting. Blots were probed with Abs against myc (9E10), actin (Sigma), and aromatase (a gift of N. Harada, Shiga Univ., Japan).
2.3 32P labeling studies
HEK-293T cells were transfected with mArom-myc-his/pcDNA3.1 using Fugene6 (Roche). Phosphorylation studies are based on the methods described in [20]. The day following transfection, medium was changed to PO4-free DMEM + 10% dialyzed FBS (Gibco). The next morning, cells were metabolically labeled with 32P-orthophosphate (1-2.5 mCi/dish) in fresh PO4-free medium × 3-4 hrs. Cells were lysed in 1% Triton buffer and used for immunoprecipitation as above, followed by SDS-PAGE.
For phospho-amino acid analysis, protein was transferred to PVDF membrane. After autoradiography, bands corresponding to 32P-labeled aromatase were cut from the membrane. Hydrolyzed amino acids were mixed with non-radioactive phospho-amino acid controls and separated by electrophoresis on TLC plates (EMD chemicals). 32P-phospho-amino acid content of aromatase was determined by autoradiography and comparison to phospho-amino acid controls.
For 2-D phosphopeptide mapping studies, protein was transferred to nitrocellulose membrane. After autoradiography, 32P-labeled aromatase bands were cut from the membrane and incubated with 2 μg trypsin (TPCK, Promega) in 50 mM ammonium bicarbonate overnight. Peptides were oxidized with performic acid × 1 hour on ice, lyophilized, and separated on TLC plates in pH 1.9, 4.72, 6.5, and 8.9 electrophoresis buffers, followed by chromatography in phospho-chromatography buffer [20]. ε-Dinitrophenyllysine (dnpLys) and xylene cyanol FF were used as control dyes. Phosphopeptides were located by autoradiography. Relative electrophoretic (Emobil) and chromatographic (Cmobil) mobility values were calculated as described [21]. Briefly, Emobil of the aromatase phosphopeptide was calculated relative to Emobil of dnpLys (charge × mass-2/3 of dnpLys = 0.0217). Cmobil was calculated as the ratio of migration distance of the aromatase phosphopeptide relative to dnpLys.
A theoretical 2-D phosphopeptide map of an aromatase tryptic digest was generated using the Mobility 2006 program (www.genestream.org). Phosphorylation sites were also predicted using NetPhosK1.0 [22] and NetPhos2.0 [23] programs.
2.4 Aromatase activity assay
COS-7 cells were transfected in triplicate with mArom-myc-his/pcDNA3.1 using Fugene6. Aromatase activity was assayed using the 3H-water release assay [11, 24]. Cells were labeled with [1β-3H]-androst-4-ene-3,17-dione (AD, Perkin-Elmer, 0 to 100 nM AD in 1.5 mL medium × 3 hrs) in IMEM + 10% dextran-coated charcoal-treated (DCC)-FBS (Hyclone). Five-hundred μL of medium was harvested from each well, and the reaction was terminated by the addition of 150 μL 50% TCA. Samples were centrifuged × 1 min., and 500 μL of supernatant was added to 1 mL chloroform. Samples were mixed, centrifuged × 1 min., and 350 μL of supernatant was added to 350 μL of 2.5% activated charcoal suspension. Samples were incubated with agitation × 1 hour, centrifuged, 500 μL of supernatant was added to 5 mL of scintillation cocktail, and 3H-water content was measured with a scintillation counter. Cell monolayers were washed with PBS, lysed in NP-40 lysis buffer, centrifuged, and protein content was determined using BCA assay (Pierce). Aromatase activity was calculated as pmol of AD metabolized per mg of protein per hour. Whole cell lysates were used for Western blotting as above, and enzymatic activity was normalized to aromatase protein levels as determined by densitometry of Western blots. Normalized aromatase activities are plotted as mean of triplicates +/- SD (Fig. 6C). Km and Vmax values were calculated by extrapolation of Lineweaver-Burk plots (x-axis = 1/substrate concentration; y-axis = 1/velocity), where x-intercept = -1/Km, and y-intercept = 1/Vmax (Fig. S3).
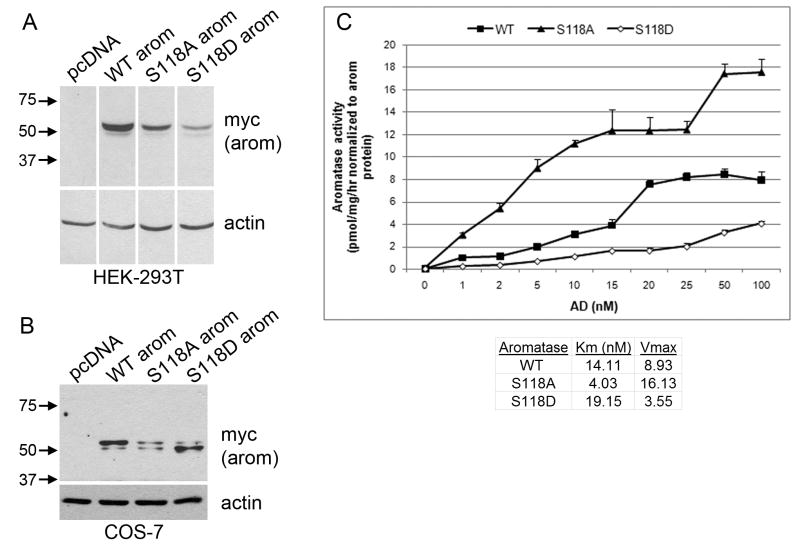
Fig. 6.
S118 mutants exhibit decreased stability and altered enzymatic activity. (A) HEK-293T cells were transfected with WT or mutant (S118A, S118D) aromatase. Cell lysates were used for Western blotting. Equal amounts of protein were loaded in each lane. Blots were probed for myc-tagged aromatase and actin. Images were taken from the same film and are exposure-matched. MW markers are shown at left. (B) COS-7 cells were transfected and analyzed by Western blotting as in (A). (C) COS-7 cells transfected at the same time as in (B) were used to measure aromatase enzymatic activity by the 3H-water release assay. Data are presented as pmol/mg/hr following normalization to aromatase protein levels as determined by densitometry of Western blots in (B), mean of triplicates +/- SD. Km and Vmax were calculated by extrapolation of Lineweaver-Burk plots as described in Methods.
3. Results
3.1 Aromatase is phosphorylated at a single serine residue in situ
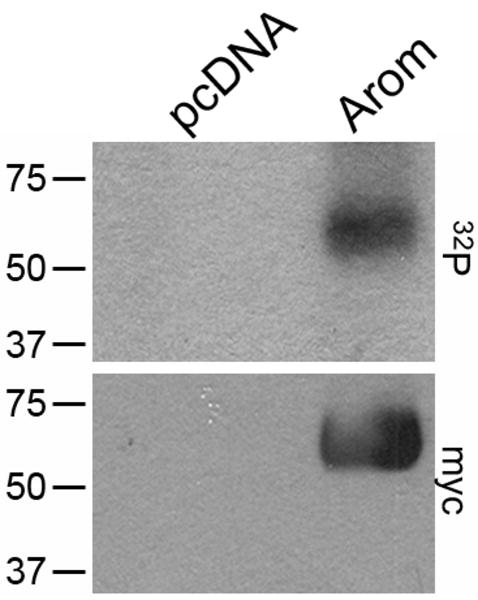
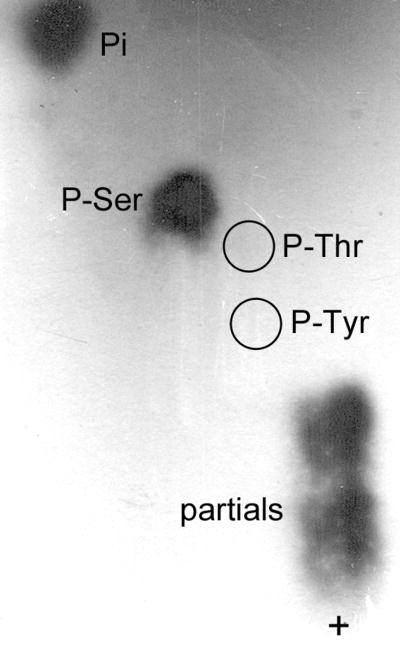
We analyzed myc-tagged murine aromatase immunoprecipitated from transfected HEK-293T cells metabolically labeled with 32P-orthophosphate. As depicted in Fig. 1A, a 32P-labeled protein was detected at the correct MW for aromatase-myc (54 kD by Western blotting, Fig. 1B) in cells transfected with a vector encoding aromatase-myc (lane 2). Cells transfected with an empty vector control (lane 1) showed no 32P-labeled, immunoprecipitated protein in the region of 50 kD. We further explored this 32P-labeled aromatase protein by phospho-amino acid analysis to determine which amino acid is phosphorylated. Comparison with non-radioactive controls indicated that aromatase contains phosphoserine (Fig. 2).
Fig. 1.
Aromatase is phosphorylated in intact mammalian cells. HEK-293T cells transiently transfected with a pcDNA control vector (lane 1) or a vector encoding myc-tagged murine aromatase (lane 2) were metabolically labeled with 32P-orthophosphate. Myc-aromatase was immunoprecipitated and subjected to SDS-PAGE. Autoradiogram is shown on top. Membrane was then used for Western blotting and probed with myc Ab (bottom). MW markers are noted at left.
Fig. 2.
Aromatase contains phosphoserine. HEK-293T cells expressing aromatase were labeled with 32P as in Fig. 1. 32P-labeled aromatase was used for phospho-amino acid analysis. Autoradiogram of 32P-labeled amino acids is shown. Non-radioactive phospho-amino acids were used as controls, and their locations are noted (P-Tyr, P-Thr, P-Ser). ‘Pi’ represents free phosphate, and ‘partials’ denotes partially hydrolyzed phosphopeptides. + denotes site of sample application.
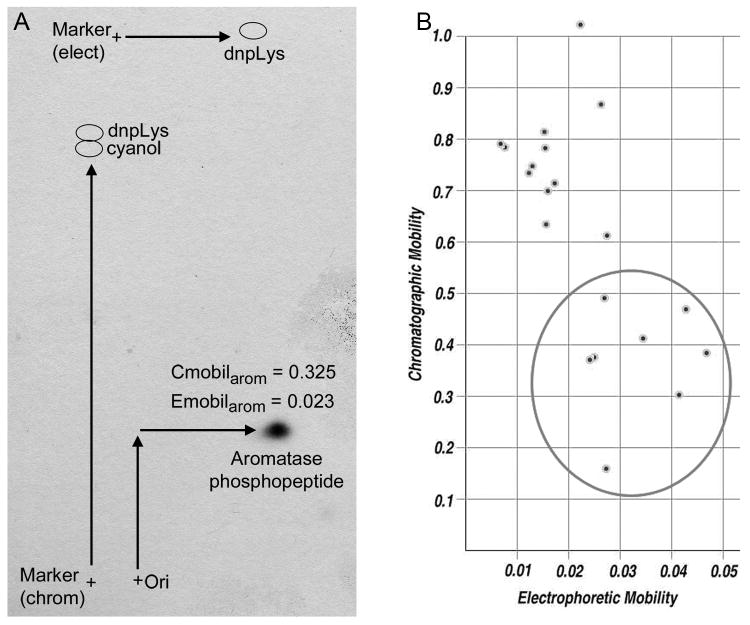
To determine the number of phosphorylation sites on aromatase, 2-D phosphopeptide mapping of trypsin-digested, 32P-labeled aromatase was used. The presence of a single 32P-labeled spot (Fig. 3A) suggests that aromatase is phosphorylated at a single site; however, this approach does not rule out the possibility of multiple proximal phosphoserine sites within the same phosphopeptide.
Fig. 3.
Aromatase is phosphorylated at a single site. (A) HEK-293T cells expressing aromatase were labeled with 32P as in Fig. 1. 32P-labeled aromatase was digested with trypsin and used for 2-D phosphopeptide mapping. Autoradiogram of 32P-labeled peptide is shown. Dnp-Lys and xylene cyanol dyes were used as controls for electrophoresis and chromatography. Locations of spotting of tryptic digest (‘ori’ +) and marker dyes (+) are noted. Relative electrophoretic and chromatographic mobility values (Emobil, Cmobil) were calculated as in Materials & Methods. (B) Theoretical Emobil and Cmobil of potential phospho-serine-containing peptides were calculated using Mobility 2006 program. Candidate phospho-peptides with predicted mobilities similar to those found in (A) are encircled (include serines: S110, S114, S118, S247, S267, S478, S493, S497).
3.2 Serine-118 is a candidate aromatase phosphorylation site
Predictive and experimental approaches were used to determine the site of aromatase phosphorylation. A theoretical 2-D phosphopeptide map was generated from a tryptic digest of aromatase using the Mobility 2006 program (Fig. 3B). The serine-containing peptides with predicted electrophoretic (Emobil) and chromatographic (Cmobil) mobility values similar to those of the aromatase phosphopeptide (Fig. 3A) are encircled and include residues S110, S114, S118, S247, S267, S478, S493, S497.
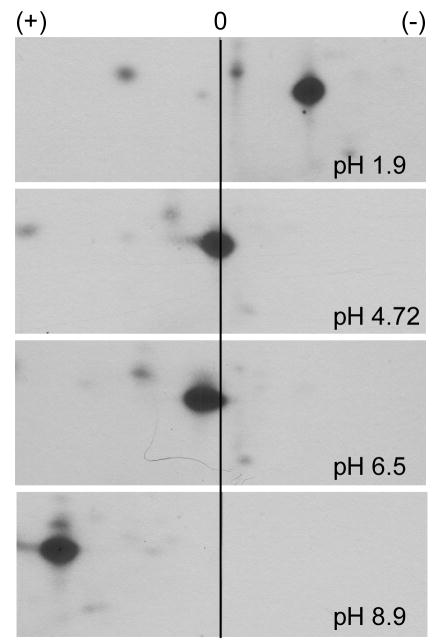
To further define the aromatase phosphopeptide, we performed 2-D phosphopeptide mapping using electrophoresis buffers of different pH. The charge of a peptide may change and can be predicted at a given pH. At pH 1.9, 4.72, 6.5, and 8.9, the aromatase phosphopeptide was positively charged, neutral, slightly negatively charged, and negatively charged, respectively (Fig. 4). This pattern of charge behavior was compared to the charges of peptides predicted from trypsin digestion (Mobility 2006 program) (Fig. S1). Candidate phosphopeptides include the serinyl residues S72, S118, S238, and S247. We also used the NetPhos2.0 program [23] to predict phosphorylation sites on aromatase (Fig. S2), where residues S90, S101, S118, S167, S182, S247, S267, S478, S493, and S497 scored highly.
Fig. 4.
Charge behavior of aromatase phosphopeptide changes with pH. HEK-293T cells expressing aromatase were labeled with 32P as in Fig. 1. 32P-labeled aromatase was digested with trypsin and used for 2-D phosphopeptide mapping, using buffers of different pH (1.9, 4.72, 6.5, 8.9) for the electrophoretic dimension. Autoradiograms of 32P-labeled peptide are shown. Cathode (+) and anode (-) are shown at top, neutral (0) is marked with center line.
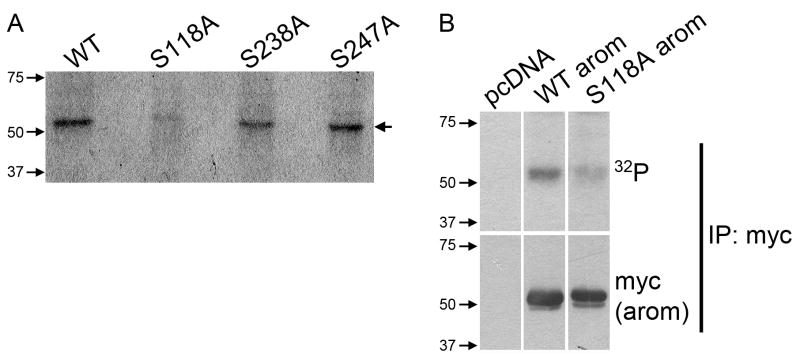
We selected the most likely serine phosphorylation sites based on the above findings and made phospho-inhibiting mutants by changing Ser→Ala at S118, S238, and S247. HEK-293T cells transiently expressing wild-type (WT) or mutant aromatase were metabolically labeled with 32P as in Fig. 1. The S118A aromatase mutant showed a drastic decrease in 32P incorporation, suggesting that S118 is critical for aromatase phosphorylation (Fig. 5A). The S238A and S247A mutations had no effect on 32P incorporation. We verified that the S118A mutation decreased aromatase phosphorylation but still permitted protein expression by Western blotting prior to autoradiography (Fig. 5B).
Fig. 5.
Serine-118 is a candidate aromatase phosphorylation site. (A) HEK-293T cells expressing wild-type (WT) or mutant (S118A, S238A, S247A) aromatase were labeled with 32P as in Fig. 1. Immunoprecipitated aromatase was subjected to SDS-PAGE. Autoradiogram is shown. MW markers are noted at left. Arrow denotes myc-aromatase. (B) Same as in (A), except Western blotting for myc-tagged aromatase was performed prior to exposure for autoradiogram. Images were taken from the same film and are exposure-matched.
3.3 Phosphomimetic mutation of Serine118 decreases aromatase enzymatic activity
Mutating phosphorylation sites can be used to mimic or inhibit the effects of phosphorylation on protein function. The phospho-inhibiting Ser→Ala mutation blocks the effects of phosphorylation, while the phosphomimetic Ser→Asp/Glu mutation has a more limited usefulness and only sometimes mimics phosphorylation effects [25]. We therefore analyzed the effects of Ser→Ala and Ser→Asp mutations at S118 on aromatase. Western blotting of lysates from HEK-293T cells transiently expressing WT or mutant aromatase showed that both S118A and S118D mutations conferred decreased protein stability (Fig. 6A). This suggests that the S118 residue may play a more general, structural role.
We then performed kinetic analyses of the effects of S118A and S118D mutations on aromatase enzymatic activity. We expressed WT, S118A, and S118D aromatase cDNA in E. coli [26-28], but only the WT aromatase preparation yielded protein, indicating that the S118 mutants were also unstable in bacteria. Instead, we evaluated enzymatic activity in transfected COS-7 cells, and normalized enzymatic activity to total amounts of aromatase protein as determined by densitometry of Western blots (Fig. 6B-C). We found that the S118A mutant showed increased specific enzymatic activity relative to WT aromatase control, while the S118D mutant showed decreased activity vs. WT control. Km and Vmax were calculated by extrapolation of Lineweaver-Burk plots (Fig. S3). The S118A mutant had lower Km (4.03 nM) and increased Vmax (16.13 pmol/mg/hr) compared to the WT enzyme (Km = 14.11 nM; Vmax = 8.93 pmol/mg/hr), while the S118D mutant showed a higher Km (19.15 nM) and decreased Vmax (3.55 pmol/mg/hr). Therefore, phosphomimetic mutation of S118 decreases aromatase activity and may decrease androstenedione substrate affinity. Alternatively, the S118 mutant proteins may fold differently than WT aromatase, which could also affect function.
3.4 Phosphorylation of endogenous aromatase in JEG-3 cells
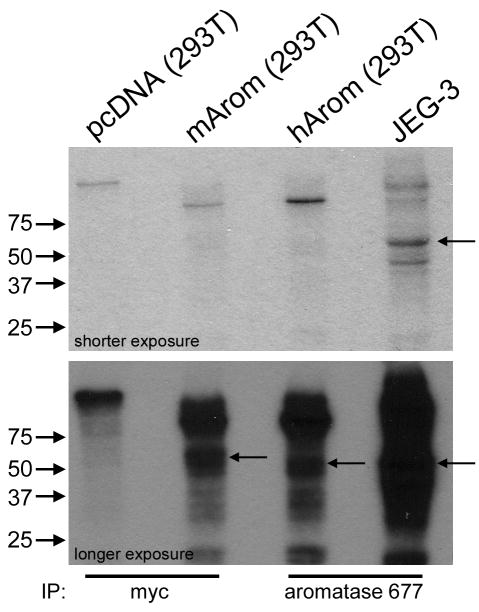
We sought to determine whether endogenous aromatase is also phosphorylated in mammalian cells. We immunoprecipitated myc-tagged murine aromatase or untagged human aromatase from 32P-labeled, transfected HEK-293T cells, or from JEG-3 human choriocarcinoma cells which endogenously express aromatase. Untagged aromatase is expected to run at a slightly lower MW (50 kD, Fig. S4) than myc-tagged aromatase (54 kD). We detected 32P-labeled proteins in lanes 2, 3, and 4 which are at the correct MW of respective aromatases (Fig. 7), while lane 1 (IP from vector control-transfected cells) showed no 32P-labeled protein. Although we demonstrated that the aromatase Ab selectively immunoprecipitates aromatase (Fig. S4), we cannot exclude the possibility that this Ab non-specifically immunoprecipitated another phosphorylated, 50 kD protein from JEG-3 cells. These data indicate that untagged human aromatase is phosphorylated in transfected cells, and endogenous aromatase may also be phosphorylated in JEG-3 cells.
Fig. 7.
Phosphorylation of endogenous aromatase. HEK-293T cells transiently transfected with vectors encoding WT murine (myc-tagged), untagged human aromatase, or vector control (pcDNA), and JEG-3 choriocarcinoma cells were metabolically labeled with 32P as in Fig. 1. Myc-tagged murine aromatase (myc Ab) and untagged human aromatase (aromatase 677 Ab) were immunoprecipitated and subjected to SDS-PAGE. Autoradiogram is shown. MW markers are noted at left. Arrows denote aromatase.
4. Discussion
Post-translational modifications, including phosphorylation, glycosylation, ubiquitination, and nitration, have been described for cytochromes P450 [29]. Phosphorylation of several P450 enzymes (CYP2B1, CYP2B2, CYP2E1, CYP2B4, and CYP reductase) has been proposed as a means to promptly modulate enzymatic activity or target them for degradation (reviewed in [30]). A role for rapid modulation of aromatase activity is suggested by its localization to neuronal presynaptic terminals [16], where estrogens have a putative role as neurotransmitters [31]. Rapid changes in local estrogen levels in mouse brain have been linked to aromatase activity, where injection of aromatase inhibitors quickly altered male sexual behavior [32]. Estrogens have also been shown to modulate non-genomic intracellular signaling pathways [33, 34], which may require strict control of local estrogen levels. In contrast, a role for aromatase phosphorylation in targeting protein for degradation seems unlikely. Mutation of S118 to either Ala or Asp destabilized aromatase. Given that the half-life of aromatase in cells is rather long (28 hours) [35], it is improbable that phosphorylation is a major mechanism in aromatase turnover.
Predictive modeling suggests that S118 lies within the β1-5 β-strand of aromatase [36], adjacent to the putative substrate recognition site-1. Cytochromes P450 have six common substrate recognition sites which comprise the active site of each enzyme [37]. Analysis of the substrate-bound aromatase-androstenedione complex showed that K119 is part of the active site [36]. It is therefore conceivable that phosphorylation at S118 may affect the ability of aromatase to bind substrate and/or catalyze estrogen synthesis. Indeed, the phosphomimetic S118D aromatase mutant showed decreased specific enzymatic activity, with reduced Vmax and increased Km relative to WT aromatase, while the S118A phospho-inhibiting mutant showed the opposite properties (Fig. 6C). Wild-type aromatase showed Vmax and Km values in between those of the S118A and S118D mutants, suggesting that phosphorylated and non-phosphorylated WT protein species may exist simultaneously. However, we cannot exclude the possibilities that the S118 mutations altered enzymatic activity through effects on protein folding, or that the S118D mutation is not an accurate mimic of S118 phosphorylation [25]. Alignment of the aromatase sequence with that of bacterial cytochromes P450BM3 and P450cam, having known 3-D structures, suggests that F116 lies in the active site. Site-directed mutagenesis of F116 rendered aromatase inactive, suggesting that mutation of this region may cause improper protein folding or substrate binding. Alternatively, mutation of F116 may inhibit aromatase binding to reductase, since the putative reductase binding site is on this side of aromatase [38]. These observations support our findings that S118 lies within a critical region of aromatase, and mutation of S118 alters enzyme stability and activity.
Here we provide the first demonstration that aromatase is phosphorylated in cells at S118. Balthazart et al. detected quail aromatase phosphorylation in vitro in brain homogenates at Ser, Thr, and Tyr residues by Western blotting using phospho-amino acid-specific antibodies [17]. Our use of metabolic 32P-labeling of aromatase may have generated a different pattern of phosphorylation, or there may be different kinases involved in our intracellular system. Also, the interspecies amino acid sequence differences may explain in part the observed differences in phosphorylated amino acids. Although the aromatase amino acid sequence is fairly conserved [39], interspecies differences affect glycosylation, substrate affinity, and substrate turnover [13].
The aromatase amino acid context surrounding S118 is highly conserved across diverse animal species (Fig. S5, and [39]). The conserved Arg at the -3 position relative to S118 suggests that an AGC-like kinase (cAMP-dependent protein kinase/ protein kinase G/ protein kinase C extended family) may phosphorylate aromatase (e.g. PKA, PKB/AKT, PKC). The NetPhosK1.0 program [22] predicted S118 to be a PKC site. We treated cells with chelerythrine Cl or phorbol 12-myristate 13-acetate (PMA) to respectively inhibit or activate PKC, but mild effects on aromatase activity varied between cell types, and PMA did not induce phosphorylation (not shown). Balthazart et al. found that Ca2+-dependent PKC (and possibly PKA and CAMK) activity can downregulate aromatase activity in brain homogenates [17]. Their results agree with our finding that the S118D aromatase phosphomimetic mutant showed decreased specific enzymatic activity (Fig. 6C).
We have shown for the first time that aromatase is post-translationally modified by phosphorylation in intact cells. The roles of estrogens in diverse non-genomic, membrane-initiated, and neurotransmitter signaling mechanisms [40, 41] present a case for rapid modulation of estrogen levels via phosphorylation-dependent regulation of intracellular aromatase function and/or protein levels. Our findings bring to light two important questions: which kinase(s) is implicated in aromatase phosphorylation, and what role(s) does phosphorylation play in aromatase regulation? Addressing these issues will further our understanding of how cells control estrogen production, and provide insight into disease mechanisms whereby kinase signaling pathways may alter aromatase phosphoregulation and subsequent hormone levels.
Supplementary Material
Fig. S1. Charges of potential aromatase phosphopeptides. Charges of serine-containing phosphopeptides following a theoretical tryptic digest of aromatase at pH 1.9, 4.72, 6.5, and 8.9 were predicted using Mobility 2006 program (Genestream). A single phosphorylation site was assumed. All electrophoretic separations in Fig. 4 were performed with the same voltage, time, and distance between electrodes so that relative migratory distances could be directly compared to relative pH-induced changes in charge. Boxes indicate candidate peptides whose pattern of charge behavior mimic that of the aromatase phosphopeptide observed by 2-D mapping.
Fig. S2. Aromatase phosphorylation sites predicted by NetPhos2.0 program. The murine aromatase sequence was used to query NetPhos2.0. High-scoring potential phosphoserine sites include S90, S101, S118, S167, S182, S247, S267, S478, S493, S497.
Fig. S3. Lineweaver-Burk plots of enzymatic activities of WT and S118 mutant aromatases. Plots were generated from data in fig. 6C. Km and Vmax values were calculated by extrapolation (x-axis = 1/AD substrate concentration (nM); y-axis = 1/velocity (pmol/mg/hr normalized to aromatase protein levels). x-intercept = -1/Km, y-intercept = 1/Vmax.
Fig. S4. Aromatase antibody immunopreciptates aromatase. The 677 monoclonal Ab was used to immunoprecipitate untagged human aromatase from lysate of stably transfected MCF-7/CA cells. Immunoprecipitate was used for Western blotting (IP), control lane contained 677 Ab alone. Whole cell lysates (WCL) of HEK-293T cells transiently transfected with pcDNA control or untagged human aromatase cDNA vector were used as controls. Blot was probed using rabbit-anti-aromatase. Images were taken from the same film and exposure-matched. MW markers are shown at left.
Fig. S5. Aromatase amino acid sequence near S118 phosphorylation site is conserved across species. Amino acid sequences of diverse animal species were compared. S118 is boxed. The presence of Arg at -3 position is suggestive of an AGC-like kinase motif. Accession ID numbers from Pubmed are listed.
Acknowledgments
We thank Genbin Shi and Mark de Caestecker for assistance with 32P studies. This work was supported in part by NIH R01 grants CA80195 (CLA) and GM37942 (MRW, NK), NIH F32 grant CA121900 (TWM), Breast Cancer Specialized Program of Research Excellence (SPORE) grant P50 CA98131, and Vanderbilt-Ingram Comprehensive Cancer Center Support grant P30 CA68485.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bulun SE, Sebastian S, Takayama K, Suzuki T, Sasano H, Shozu M. The human CYP19 (aromatase P450) gene: update on physiologic roles and genomic organization of promoters. J Steroid Biochem Mol Biol. 2003;86:219–24. doi: 10.1016/s0960-0760(03)00359-5. [DOI] [PubMed] [Google Scholar]
- 2.Bulun SE, Noble LS, Takayama K, Michael MD, Agarwal V, Fisher C, Zhao Y, Hinshelwood MM, Ito Y, Simpson ER. Endocrine disorders associated with inappropriately high aromatase expression. J Steroid Biochem Mol Biol. 1997;61:133–9. [PubMed] [Google Scholar]
- 3.Brueggemeier RW, Quinn AL, Parrett ML, Joarder FS, Harris RE, Robertson FM. Correlation of aromatase and cyclooxygenase gene expression in human breast cancer specimens. Cancer Lett. 1999;140:27–35. doi: 10.1016/s0304-3835(99)00050-6. [DOI] [PubMed] [Google Scholar]
- 4.Egan KM, Lawson JA, Fries S, Koller B, Rader DJ, Smyth EM, Fitzgerald GA. COX-2-derived prostacyclin confers atheroprotection on female mice. Science. 2004;306:1954–7. doi: 10.1126/science.1103333. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Y, Agarwal VR, Mendelson CR, Simpson ER. Estrogen biosynthesis proximal to a breast tumor is stimulated by PGE2 via cyclic AMP, leading to activation of promoter II of the CYP19 (aromatase) gene. Endocrinology. 1996;137:5739–42. doi: 10.1210/endo.137.12.8940410. [DOI] [PubMed] [Google Scholar]
- 6.Ryde CM, Nicholls JE, Dowsett M. Steroid and growth factor modulation of aromatase activity in MCF7 and T47D breast carcinoma cell lines. Cancer Res. 1992;52:1411–5. [PubMed] [Google Scholar]
- 7.Richards JA, Petrel TA, Brueggemeier RW. Signaling pathways regulating aromatase and cyclooxygenases in normal and malignant breast cells. J Steroid Biochem Mol Biol. 2002;80:203–12. doi: 10.1016/s0960-0760(01)00187-x. [DOI] [PubMed] [Google Scholar]
- 8.Mendelson CR, Corbin CJ, Smith ME, Smith J, Simpson ER. Growth factors suppress and phorbol esters potentiate the action of dibutyryl adenosine 3′,5′-monophosphate to stimulate aromatase activity of human adipose stromal cells. Endocrinology. 1986;118:968–73. doi: 10.1210/endo-118-3-968. [DOI] [PubMed] [Google Scholar]
- 9.Kinoshita Y, Chen S. Induction of aromatase (CYP19) expression in breast cancer cells through a nongenomic action of estrogen receptor alpha. Cancer Res. 2003;63:3546–55. [PubMed] [Google Scholar]
- 10.Shozu M, Sumitani H, Murakami K, Segawa T, Yang HJ, Inoue M. Regulation of aromatase activity in bone-derived cells: possible role of mitogen-activated protein kinase. J Steroid Biochem Mol Biol. 2001;79:61–5. doi: 10.1016/s0960-0760(01)00129-7. [DOI] [PubMed] [Google Scholar]
- 11.Yue W, Wang JP, Conaway MR, Li Y, Santen RJ. Adaptive hypersensitivity following long-term estrogen deprivation: involvement of multiple signal pathways. J Steroid Biochem Mol Biol. 2003;86:265–74. doi: 10.1016/s0960-0760(03)00366-2. [DOI] [PubMed] [Google Scholar]
- 12.Balthazart J, Baillien M, Charlier TD, Cornil CA, Ball GF. Multiple mechanisms control brain aromatase activity at the genomic and non-genomic level. J Steroid Biochem Mol Biol. 2003;86:367–79. doi: 10.1016/s0960-0760(03)00346-7. [DOI] [PubMed] [Google Scholar]
- 13.Jo Corbin C, Mapes SM, Lee YM, Conley AJ. Structural and functional differences among purified recombinant mammalian aromatases: glycosylation, N-terminal sequence and kinetic analysis of human, bovine and the porcine placental and gonadal isozymes. Mol Cell Endocrinol. 2003;206:147–57. doi: 10.1016/s0303-7207(02)00422-7. [DOI] [PubMed] [Google Scholar]
- 14.Moslemi S, Vibet A, Papadopoulos V, Camoin L, Silberzahn P, Gaillard JL. Purification and characterization of equine testicular cytochrome P-450 aromatase: comparison with the human enzyme. Comp Biochem Physiol B Biochem Mol Biol. 1997;118:217–27. doi: 10.1016/s0305-0491(97)00033-3. [DOI] [PubMed] [Google Scholar]
- 15.Sethumadhavan K, Bellino FL, Thotakura NR. Estrogen synthetase (aromatase). The cytochrome P-450 component of the human placental enzyme is a glycoprotein. Mol Cell Endocrinol. 1991;78:25–32. doi: 10.1016/0303-7207(91)90182-r. [DOI] [PubMed] [Google Scholar]
- 16.Balthazart J, Ball GF. Is brain estradiol a hormone or a neurotransmitter? Trends Neurosci. 2006;29:241–9. doi: 10.1016/j.tins.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 17.Balthazart J, Baillien M, Charlier TD, Ball GF. Calcium-dependent phosphorylation processes control brain aromatase in quail. Eur J Neurosci. 2003;17:1591–606. doi: 10.1046/j.1460-9568.2003.02598.x. [DOI] [PubMed] [Google Scholar]
- 18.Tekmal RR, Ramachandra N, Gubba S, Durgam VR, Mantione J, Toda K, Shizuta Y, Dillehay DL. Overexpression of int-5/aromatase in mammary glands of transgenic mice results in the induction of hyperplasia and nuclear abnormalities. Cancer Res. 1996;56:3180–5. [PubMed] [Google Scholar]
- 19.Sasano H, Anderson TJ, Silverberg SG, Santen RJ, Conway M, Edwards DP, Krause A, Bhatnagar AS, Evans DB, Miller WR. The validation of new aromatase monoclonal antibodies for immunohistochemistry--a correlation with biochemical activities in 46 cases of breast cancer. J Steroid Biochem Mol Biol. 2005;95:35–9. doi: 10.1016/j.jsbmb.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 20.van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by electrophoresis and chromatography on thin-layer cellulose plates. Electrophoresis. 1994;15:544–54. doi: 10.1002/elps.1150150173. [DOI] [PubMed] [Google Scholar]
- 21.Melsenhelder J, Hunter T, van der Geer P. Phosphopeptide mapping and identification of phosphorylation sites. In: Ausubel FM, editor. Current Protocols in Molecular Biology. John Wiley & Sons; Hoboken, N.J.: 1999. pp. 18.9.1–18.9.28. [DOI] [PubMed] [Google Scholar]
- 22.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–49. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 23.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–62. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 24.Thompson EA, Jr, Siiteri PK. Utilization of oxygen and reduced nicotinamide adenine dinucleotide phosphate by human placental microsomes during aromatization of androstenedione. J Biol Chem. 1974;249:5364–72. [PubMed] [Google Scholar]
- 25.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–74. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kagawa N, Hori H, Waterman MR, Yoshioka S. Characterization of stable human aromatase expressed in E. coli. Steroids. 2004;69:235–43. doi: 10.1016/j.steroids.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Kagawa N, Cao Q, Kusano K. Expression of human aromatase (CYP19) in Escherichia coli by N-terminal replacement and induction of cold stress response. Steroids. 2003;68:205–9. doi: 10.1016/s0039-128x(02)00168-x. [DOI] [PubMed] [Google Scholar]
- 28.Tosha T, Kagawa N, Ohta T, Yoshioka S, Waterman MR, Kitagawa T. Raman evidence for specific substrate-induced structural changes in the heme pocket of human cytochrome P450 aromatase during the three consecutive oxygen activation steps. Biochemistry. 2006;45:5631–40. doi: 10.1021/bi060094a. [DOI] [PubMed] [Google Scholar]
- 29.Aguiar M, Masse R, Gibbs BF. Regulation of cytochrome P450 by posttranslational modification. Drug Metab Rev. 2005;37:379–404. doi: 10.1081/dmr-46136. [DOI] [PubMed] [Google Scholar]
- 30.Oesch-Bartlomowicz B, Oesch F. Cytochrome-P450 phosphorylation as a functional switch. Arch Biochem Biophys. 2003;409:228–34. doi: 10.1016/s0003-9861(02)00558-1. [DOI] [PubMed] [Google Scholar]
- 31.McEwen BS. Invited review: Estrogens effects on the brain: multiple sites and molecular mechanisms. J Appl Physiol. 2001;91:2785–801. doi: 10.1152/jappl.2001.91.6.2785. [DOI] [PubMed] [Google Scholar]
- 32.Taziaux M, Keller M, Bakker J, Balthazart J. Sexual behavior activity tracks rapid changes in brain estrogen concentrations. J Neurosci. 2007;27:6563–72. doi: 10.1523/JNEUROSCI.1797-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hisamoto K, Ohmichi M, Kurachi H, Hayakawa J, Kanda Y, Nishio Y, Adachi K, Tasaka K, Miyoshi E, Fujiwara N, Taniguchi N, Murata Y. Estrogen induces the Akt-dependent activation of endothelial nitric-oxide synthase in vascular endothelial cells. J Biol Chem. 2001;276:3459–67. doi: 10.1074/jbc.M005036200. [DOI] [PubMed] [Google Scholar]
- 34.Simoncini T, Mannella P, Fornari L, Caruso A, Varone G, Genazzani AR. Genomic and non-genomic effects of estrogens on endothelial cells. Steroids. 2004;69:537–42. doi: 10.1016/j.steroids.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Chen S. Aromatase destabilizer: novel action of exemestane, a food and drug administration-approved aromatase inhibitor. Cancer Res. 2006;66:10281–6. doi: 10.1158/0008-5472.CAN-06-2134. [DOI] [PubMed] [Google Scholar]
- 36.Karkola S, Holtje HD, Wahala K. A three-dimensional model of CYP19 aromatase for structure-based drug design. J Steroid Biochem Mol Biol. 2007;105:63–70. doi: 10.1016/j.jsbmb.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 37.Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- 38.Amarneh B, Corbin CJ, Peterson JA, Simpson ER, Graham-Lorence S. Functional domains of human aromatase cytochrome P450 characterized by linear alignment and site-directed mutagenesis. Mol Endocrinol. 1993;7:1617–24. doi: 10.1210/mend.7.12.8145767. [DOI] [PubMed] [Google Scholar]
- 39.Conley A, Hinshelwood M. Mammalian aromatases. Reproduction. 2001;121:685–95. doi: 10.1530/rep.0.1210685. [DOI] [PubMed] [Google Scholar]
- 40.Moriarty K, Kim KH, Bender JR. Minireview: estrogen receptor-mediated rapid signaling. Endocrinology. 2006;147:5557–63. doi: 10.1210/en.2006-0729. [DOI] [PubMed] [Google Scholar]
- 41.Ronnekleiv OK, Malyala A, Kelly MJ. Membrane-initiated signaling of estrogen in the brain. Semin Reprod Med. 2007;25:165–77. doi: 10.1055/s-2007-973429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Charges of potential aromatase phosphopeptides. Charges of serine-containing phosphopeptides following a theoretical tryptic digest of aromatase at pH 1.9, 4.72, 6.5, and 8.9 were predicted using Mobility 2006 program (Genestream). A single phosphorylation site was assumed. All electrophoretic separations in Fig. 4 were performed with the same voltage, time, and distance between electrodes so that relative migratory distances could be directly compared to relative pH-induced changes in charge. Boxes indicate candidate peptides whose pattern of charge behavior mimic that of the aromatase phosphopeptide observed by 2-D mapping.
Fig. S2. Aromatase phosphorylation sites predicted by NetPhos2.0 program. The murine aromatase sequence was used to query NetPhos2.0. High-scoring potential phosphoserine sites include S90, S101, S118, S167, S182, S247, S267, S478, S493, S497.
Fig. S3. Lineweaver-Burk plots of enzymatic activities of WT and S118 mutant aromatases. Plots were generated from data in fig. 6C. Km and Vmax values were calculated by extrapolation (x-axis = 1/AD substrate concentration (nM); y-axis = 1/velocity (pmol/mg/hr normalized to aromatase protein levels). x-intercept = -1/Km, y-intercept = 1/Vmax.
Fig. S4. Aromatase antibody immunopreciptates aromatase. The 677 monoclonal Ab was used to immunoprecipitate untagged human aromatase from lysate of stably transfected MCF-7/CA cells. Immunoprecipitate was used for Western blotting (IP), control lane contained 677 Ab alone. Whole cell lysates (WCL) of HEK-293T cells transiently transfected with pcDNA control or untagged human aromatase cDNA vector were used as controls. Blot was probed using rabbit-anti-aromatase. Images were taken from the same film and exposure-matched. MW markers are shown at left.
Fig. S5. Aromatase amino acid sequence near S118 phosphorylation site is conserved across species. Amino acid sequences of diverse animal species were compared. S118 is boxed. The presence of Arg at -3 position is suggestive of an AGC-like kinase motif. Accession ID numbers from Pubmed are listed.