Abstract
Yeast and human Tgs1 are orthologous RNA cap (guanine-N2) methyltransferases that convert m7G caps into the 2,2,7-trimethylguanosine (TMG) caps characteristic of spliceosomal snRNAs. TMG caps are dispensable for vegetative yeast growth, but are essential in the absence of Mud2, the putative yeast homolog of human splicing factor U2AF. Here we exploited the synthetic lethal interactions of tgs1Δ and mud2Δ mutations to identify essential structural features of the Tgs1 and Mud2 proteins. Thirty-two new mutations were introduced into human Tgs1 and surveyed for their effects on function in vivo in yeast and on the two sequential guanine-N2 methylation reactions in vitro. The structure-function data highlight a strictly essential π-cation interaction between Trp766 and the m7G base and a network of important enzymic contacts to the cap triphosphate via Lys646, Tyr771, Arg807, and Lys836. Mud2 is a 527-amino acid polypeptide composed of a hydrophilic N-terminal domain and a C-terminal RRM domain. We found that the RRM domain is necessary but not sufficient for Mud2 function in complementing growth of tgs1Δ mud2Δ and mud1Δ mud2Δ strains. Other changes in Mud2 elicited distinct phenotypes in tgs1Δ versus mud1Δ backgrounds. mud2Δ also caused a severe growth defect in cells lacking the Tgs1-binding protein encoded by the nonessential gene YNR004w (now renamed SWM2, synthetic with mud2Δ). Mud2 mutational effects in the swm2Δ background paralleled those for mud1Δ. The requirements for Mud2 function are apparently more stringent when yeast cells lack TMG caps than when they lack Mud1 or Swm2.
Keywords: RRM domain, TMG caps, methyltransferase, snRNPs
INTRODUCTION
A 2,2,7-trimethylguanosine (TMG) cap is a signature feature of the U1, U2, U4, and U5 small nuclear (sn) RNAs that direct pre-mRNA splicing in eukarya (Busch et al. 1982). The enzyme Tgs1 is responsible for TMG capping (Mouaikel et al. 2002; Komonyi et al. 2005; Lemm et al. 2006; Hausmann et al. 2007). Purified yeast and human Tgs1 catalyze two successive methyltransfer reactions from AdoMet to the N2 atom of guanosine nucleotides via a distributive mechanism (Hausmann and Shuman 2005a; Hausmann et al. 2008; Benarroch et al. 2010). Their activity in vitro is strictly dependent on guanine-N7 methylation, signifying that guanine-N2 methylation in vivo is likely confined to RNAs that already have an m7G cap. It is thought that the U snRNAs are targeted for TMG capping in vivo by interactions of Tgs1 with protein components of the U snRNPs (Mattaj 1986; Plessel et al. 1994; Mouaikel et al. 2002).
Mutational studies of several Tgs enzymes identified a suite of essential amino acid side chains that were predicted to form the binding pockets for the AdoMet donor and m7G acceptor (Mouaikel et al. 2003; Hausmann and Shuman 2005b; Hausmann et al. 2007, 2008; Simoes-Barbosa et al. 2008; Benarroch et al. 2009). Most of the substrate interactions inferred from mutational studies and modeling exercises were confirmed by the crystal structure of human Tgs1 (hTgs1) in a ternary complex with m7GTP and AdoHcy (Fig. 1; Monecke et al. 2009). hTgs1 has a classical AdoMet-binding site composed of 694VVDAFCGVGGN704 and 717IAIDI721 motifs, the underlined residues of which make main chain or side chain contacts to AdoHcy. The m7G binding site comprises a 763SPPWGG768 motif, the underlined residues of which make main chain contacts to the N2, N1, O6, and C7 atoms on the edge of the m7G base; the Trp766 side chain (which is conserved in all Tgs enzymes) stacks on the purine base. The hTgs1 crystal structure also revealed an extensive network of enzymic contacts to the triphosphate moiety of m7GTP (Fig. 1A) that had not been anticipated by prior experimental or in silico studies (Monecke et al. 2009). Two of the triphosphate-binding residues, Lys646 and Tyr647, are part of a unique α-helical module (amino acids 634–674) at the N terminus of the minimal hTgs1 catalytic domain (amino acids 631–846). Deletion of the segment from amino acids 631 to 661 that includes these residues abolishes hTgs1 methyltransferase activity (Hausmann et al. 2008). One presumes that the distinctive triphosphate-binding site of hTgs1 contributes to its activity and cap specificity. In the present study, we exploit the hTgs1 crystal structure to complete a definitive mutational analysis of the substrate binding sites, especially the triphosphate site, with in vivo and in vitro readouts of hTgs1 function.
FIGURE 1.
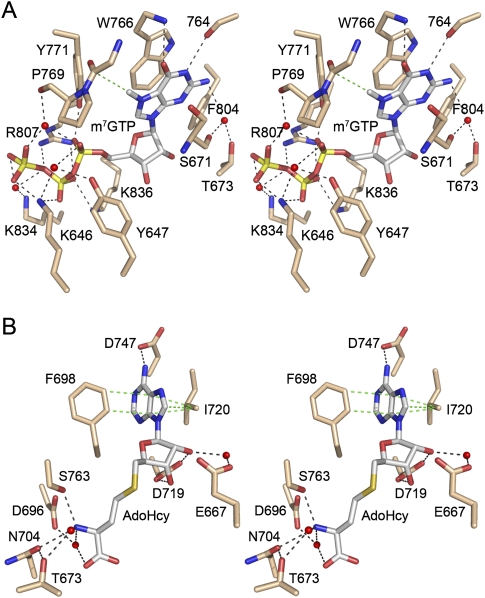
Methyl acceptor and donor sites of human Tgs1. The figure shows a stereo view of the active site of human Tgs1 in complex with m7GTP (A) and AdoHcy (B) (from PDB 3GDH). (A) Highlighted are the hTgs1 side chains that contact the cap methyl acceptor, either directly, or via waters (depicted as red spheres). Hydrogen bonds and electrostatic interactions are denoted by black dashed lines. A single van der Waals contact between a main chain Cα and the guanine-N7 methyl group is depicted as a dashed green line. The m7G base is sandwiched between Trp766 and Ser671. (B) Highlighted are the hTgs1 side chains that contact the methyl donor. The adenine base of AdoHcy is sandwiched between Phe698 and Ile720, which make van der Waals contacts denoted by the dashed green lines.
A genetic assessment of Tgs1 activity is complicated by the fact that Tgs1 and TMG caps are dispensable for vegetative growth of budding and fission yeast (Mouaikel et al. 2002; Hausmann et al. 2007). Moreover, silencing of hTgs1 by RNAi has little or no effect on the growth of HeLa cells in culture (Lemm et al. 2006). The initially surprising conclusion that fungi and human somatic cells grow in the absence of Tgs1 suggested there might be alternative means to ensure the function of the many essential TMG-capped RNAs when the TMG modification is missing. This was confirmed via synthetic enhancement genetics in budding yeast, which highlighted a role of the TMG cap in spliceosome assembly, insofar as the output of a genome-wide screen (of nonessential yeast genes) for mutational enhancement of tgs1Δ was highly enriched in proteins implicated in U1 and U2 snRNP function during pre-mRNA splicing (Hausmann et al. 2008).
The strongest genetic interactor with Tgs1 was Mud2. Whereas ablation of Mud2 per se has no apparent effect on yeast cell growth, the tgs1Δ mud2Δ combination resulted in unconditional synthetic lethality (Hausmann et al. 2008). Mud2 was initially identified by the Rosbash lab in a “MUD screen” (mutant U1 die) for yeast mutations that cause synthetic lethality with otherwise viable mutations in the U1 snRNA (Abovich et al. 1994). It is noteworthy that two other nonessential genes identified in the MUD screen, NAM8(MUD15) and MUD1 (Liao et al. 1993; Gottschalk et al. 1998), are also either synthetic lethal or synthetic sick with tgs1Δ (Hausmann et al. 2008). Nam8 and Mud1 are components of the U1 snRNP. Mud2 interacts with the pre-mRNA/U1snRNP commitment complex, in a manner that depends on the branchpoint sequence of the intron and the association of Mud2 with the yeast branchpoint binding protein (BBP), and then facilitates recruitment of the U2 snRNP (Abovich et al. 1994; Abovich and Rosbash 1997; Rain et al. 1998; Rutz and Seraphin 1999; Wang et al. 2008). Mud2 and Mud1 appear to be functionally overlapping, insofar as the mud1Δ mud2Δ double mutation is lethal (Abovich et al. 1994).
Mud2 figures prominently in the present study of Tgs1. We rely on the synthetic lethality of the tgs1Δ mud2Δ mutant to survey mutational effects on hTgs1 function in vivo. Prior studies showed that expression of the hTgs1(631–853) catalytic domain in budding yeast fully rescued the growth of tgs1Δ mud2Δ cells and that genetic complementation depended on methyltransferase activity (Hausmann et al. 2008). Thus, yeast tgs1Δ mud2Δ provides a convenient surrogate system for genetic analysis of Tgs orthologs from other taxa.
Here we extended use of the tgs1Δ mud2Δ synthetic lethality to interrogate the important structural features of yeast Mud2, which connects multiple players involved in TMG cap function and spliceosome assembly. Mud2, a 527-amino acid polypeptide composed of a hydrophilic N-terminal domain and a single C-terminal RRM domain, is the putative yeast homolog of human splicing factor U2AF65 (Abovich et al. 1994). However, Mud2 differs substantially from U2AF65, which consists of three tandem RRM domains and an N-terminal arginine-serine-rich (RS) domain. The RRM fold comprises a four-stranded β-sheet flanked on one side by two α-helices. The RRM1 and RRM2 domains of U2AF65 bind pyrimidine-rich RNA along the exposed surface of the β-sheet opposite the α-helices (Sickmier et al. 2006). The central β1 and β3 strands of the sheet (referred to as the RNP2 and RNP1 motifs, respectively) make most of the atomic contacts with the bound RNA. The C-terminal RRM3 domains of human U2AF65 and yeast Mud2 are implicated in protein–protein interactions with BBP rather than, or in addition to, RNA binding (Berglund et al. 1998; Rain et al. 1998; Selenko et al. 2003). RRM3-like domains, renamed “U2AF homologs motifs” (UHMs), have now been identified in many other proteins, where they are proposed to comprise a protein interaction module (Kielkopf et al. 2004). We report that the yeast Mud2 RRM3/UHM domain and components of the hydrophilic N-terminal domain are both critical for complementation of the tgs1Δ mud2Δ mutant. Yet, the requirements for Mud2 function differ when assayed in a lethal mud1Δ background or in tandem with a mutation that ablates the Tgs1-binding protein YNR004w.
RESULTS AND DISCUSSION
New mutational analysis of human Tgs1
We used the crystal structure of the hTgs1–m7GTP–AdoHcy ternary complex (Monecke et al. 2009) to guide a new round of structure–function studies of the methyl donor and acceptor sites (Fig. 1). AdoHcy is an inhibitory product of the methylation reaction that binds to hTgs1 with similar affinity as the AdoMet substrate (Hausmann et al. 2008). AdoHcy/AdoMet-binding residues Asp696, Asn704, and Asp719 were identified previously as essential for hTgs1 activity in vitro and in vivo (Hausmann et al. 2008). Here we tested the effects of alanine mutations at Phe698 and Ile720 (which form a hydrophobic sandwich on the AdoHcy adenine), Asp747 (which coordinates the adenine N6 atom), and Glu667 (which makes a water-mediated contact to the ribose O2′) (Fig. 1B). The methyl acceptor pocket consists of distinct docking sites for the m7G nucleoside and the cap triphosphate, respectively. The m7G site is predominantly hydrophobic and includes an essential π-cation interaction with Trp766 (Fig. 1A; Hausmann et al. 2008; Benarroch et al. 2010). Presently, we targeted two other constituents of the m7G site for alanine scanning: Ser671 and Phe804. The cap triphosphate site of hTgs1 is predominantly hydrophilic and entails a network of electrostatic and hydrogen-bonding interactions with the phosphate oxygens involving Lys646, Tyr647, Tyr771, Arg807, Lys834, and Lys836 (Fig. 1A). All six of these residues were subjected to alanine scanning.
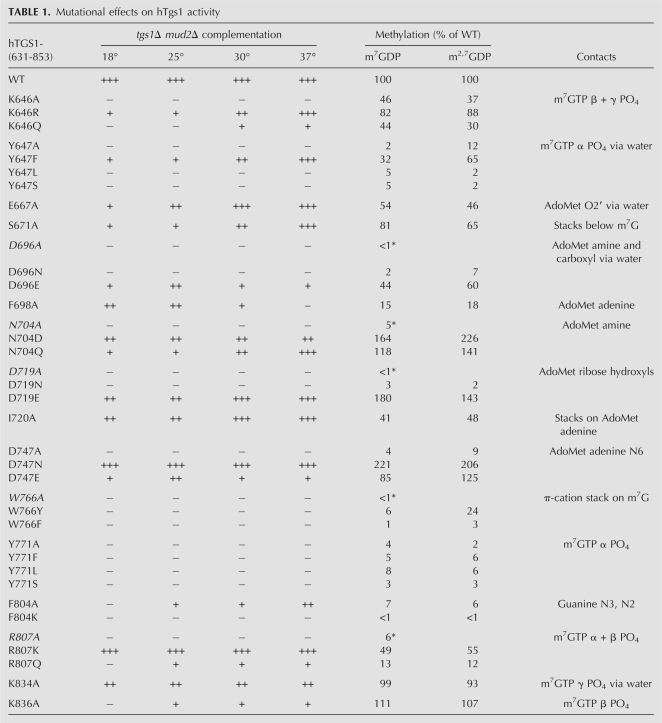
hTgs1 function in vivo was gauged by complementation of the Saccharomyces cerevisiae tgs1Δ mud2Δ double mutant. 2μ LEU2 plasmids bearing wild-type hTGS1-(631–853) or mutated hTGS1-(631–853)-Ala genes under the control of the constitutive yeast TPI1 promoter were transformed into tgs1Δ mud2Δ cells carrying a CEN URA3 yTGS1 plasmid to sustain viability. If the hTgs1 mutant is active in TMG cap formation in vivo, the yeast cells can lose the URA3 yTGS1 plasmid and grow on medium containing 5-fluoroorotic acid (FOA), a drug that selects against URA3. The results of the plasmid shuffle assay are compiled in Table 1. Lethal mutations were those that failed to support the appearance of FOA-resistant colonies at 18°C, 25°C, 30°C, or 37°C (scored as “−” at all temperatures). We found that alanine substitutions at four positions were lethal, thereby defining Lys646, Tyr647, Asp747, and Tyr771 as essential for Tgs1 activity in vivo (Table 1). Lys646, Tyr647, and Tyr771 are constituents of the cap triphosphate binding site.
TABLE 1.
Mutational effects on hTgs1 activity
The seven viable Ala mutants that gave rise to FOA-resistant colonies at one or more temperatures were tested for growth on rich medium (YPD agar) at 18°C, 25°C, 30°C, and 37°C. Growth was scored as follows: “+++” indicates colony size indistinguishable from strains bearing wild-type hTGS1-(631–853); “++” denotes reduced colony size; “+” indicates that only pinpoint colonies were formed; and “−” indicates no growth. A hierarchy of conditional growth defects was observed, predominantly entailing cold sensitivity (Table 1). F804A and K836A cells had the severest phenotypes, with + or ++ growth at 37°C, + growth at 30°C and 25°C, and no growth at 18°C. Other mutants had milder cs defects: E667A cells were +++ at 37°C and 30°C, ++ at 25°C, and + at 18°C. S671A cells were +++ at 37°C, ++ at 30°C, and + at 25°C and 18°C. I720A cells were +++ at 37°C and 30°C, but ++ at 25°C and 18°C. In contrast, F698A cells were sensitive to elevated temperatures, showing ++ growth at 18°C and 25°C, + growth at 30°C, and no growth at 37°C. K834A cells grew ++ at all temperatures tested (Table 1).
Effects of alanine mutations on methyltransferase activity in vitro
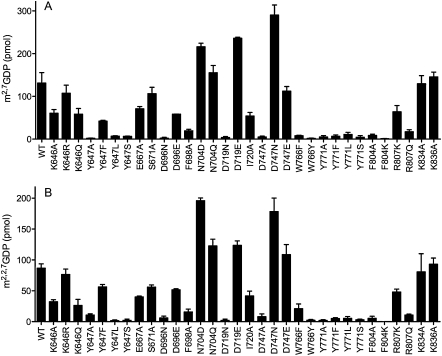
Wild-type hTgs1-(631–853) and the 11 Ala mutants were produced in Escherichia coli as His10-Smt3-tagged fusions, purified from soluble bacterial extracts by adsorption to nickel-agarose and elution with imidazole (Supplemental Fig. S1; Hausmann et al. 2008), and then tested in parallel for their ability to perform the first and second methylation steps of TMG synthesis in vitro (Benarroch et al. 2010). To assay the first methylation step (DMG synthesis), the proteins were incubated with 100 μM [3H-CH3]-AdoMet and 1 mM m7GDP for 30 min at 37°C. To assay the second methylation step (TMG synthesis), the proteins were reacted with 100 μM [3H-CH3]-AdoMet and 1 mM m2,7GDP for 30 min at 37°C. The reaction products were then analyzed by PEI-cellulose TLC to separate the anionic 3H-labeled m2,7GDP (step 1) or m2,2,7GDP (step 2) product from the cationic 3H-AdoMet substrate. The radioactivity in each species was quantified by scintillation counting of the excised segments of the TLC plates containing substrate and product (localized by UV illumination), and the extents of product formation were plotted in Figure 2A (DMG synthesis) and Figure 2B (TMG synthesis). Each datum is the average of three separate experiments ±SEM. The methyltransferase activities of the Ala mutants were normalized to that of wild-type hTgs1-(631–853) and compiled as such in Table 1.
FIGURE 2.
Methyltransferase activities of hTgs1 mutants. The extents of 3H-methyl transfer from 100 μM [3H-CH3]AdoMet to 1 mM m7GDP (A) or m2,7GDP (B) by 0.1 μg of wild-type (WT) or mutant hTgs1-(631–853) are plotted. Each datum is the average of three independent assays ± SEM.
The hTgs1 mutants that were unconditionally lethal in vivo had the most severely reduced methyltransferase activities in vitro: Y647A (2% and 12% of wild type in steps 1 and 2, respectively), D747A (4% and 9%), and Y771A (4% and 2%). hTgs1 mutants that displayed slow growth and conditional lethality in vivo also had significantly reduced methyltransferase activities in vivo, specifically F804A (7% and 6% of wild type in steps 1 and 2, respectively) and F698A (15% and 18%). (We regard a fivefold decrement in methyltransferase activity as the criterion for a significant mutational effect.) In contrast, four of the Ala mutants that supported +++ and/or ++ yeast growth at 30°C and 37°C—E667A, S671A, I720A, and K834A—displayed wild-type or near wild-type methyltransferase activities in vitro (Table 1). Thus, for the alanine mutants cited above, there was a reasonably good correlation between the in vivo growth phenotype and catalytic function in vitro.
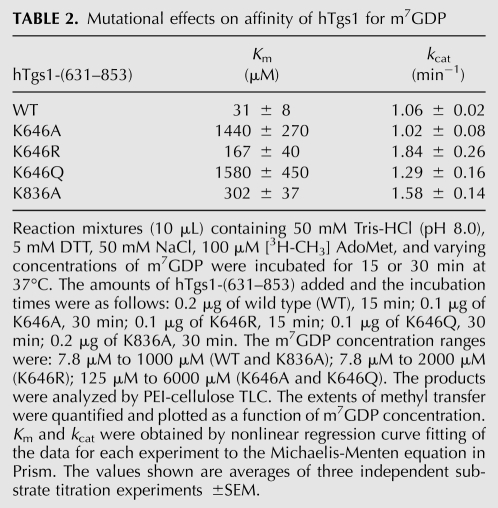
We noted two exceptions whereby Ala mutants that were either conditionally lethal (K836A) or unconditionally lethal (K646A) in vivo retained substantial methyltransferase activity in vitro. K836A displayed 111% and 107% of wild-type activity in steps 1 and 2, respectively, whereas K646A had 46% and 37% activity (Table 1). Lys836 and Lys646 are both components of the cap triphosphate binding site (Fig. 1A). We considered the possibility that K836A and K646A might have defects in substrate binding that were not revealed by the screening assays in which the concentration of m7GDP (1 mM) was in vast excess of the Km value for hTgs1 for the step 1 methyl acceptor (Hausmann et al. 2008). To address this point, we determined kinetic parameters for wild-type hTgs1-(631–853) and the K646A and K836A mutants by titrating m7GDP at a fixed concentration of AdoMet (100 μM). The results are summarized in Table 2. The wild-type enzyme had a Km of 31 μM m7GDP and a kcat of 1.06 min−1. The instructive finding was that the K646A mutation increased the Km value to 1440 μM m7GDP (46-fold higher than wild type), with no impact on kcat (1.02 min−1). This severe decrement in affinity for m7GDP is consistent with the atomic contacts made by Lys646 to the m7GTP β-phosphate (direct) and α-phosphate (water-mediated) in the hTgs1 crystal structure (Fig. 1A). Considering that Lys646 also directly contacts the γ-phosphate of m7GTP (Fig. 1A), it is likely that the loss of these several contacts to the cap accounts for the lethality of K646A in yeast. The K836A mutation increased the Km for m7GDP to 302 μM (10-fold higher than wild type), while eliciting a slight increase in kcat to 1.58 min−1 (Table 2). Lys836 makes a single contact to the m7GTP β-phosphate (Fig. 1A).
TABLE 2.
Mutational effects on affinity of hTgs1 for m7GDP
Structure–function relationships gleaned from effects of conservative mutations
Conservative amino acid substitutions were introduced in lieu of the four residues defined as unconditionally essential in vivo in the present alanine scan (Lys646, Tyr647, Asp747, Tyr771) and the five essential residues identified previously (Asp696, Asn704, Asp719, Trp766, Arg807) (Hausmann et al. 2008). The conservative mutants were tested for activity in vivo in the plasmid shuffle assay of yeast tgs1Δ mud2Δ complementation (Table 1). The mutant proteins were also produced in E. coli as His10Smt3 fusions, purified from soluble extracts (Supplemental Fig. S1), and assayed for DMG and TMG synthesis in vitro as described above for the Ala mutants (Fig. 2; Table 1). The findings for the cap triphosphate, m7G, and AdoMet/AdoHcy-binding residues are considered separately below.
Triphosphate binding site
The lethal effects of the K646A mutation were reversed in vivo by arginine, which conferred +++ growth at 37°C, ++ at 30°C, and + at 25°C and 18°C. The K646Q allele supported only + growth at 37°C and 30°C and was lethal at 25°C and 18°C (Table 1). We conclude that positive charge at position 646 is important for hTgs1 activity in vivo. The K646R change increased the in vitro methyltransferase activities (to 82% and 88% of wild-type in step 1 and step 2, respectively) compared to K646A, whereas the K646Q mutant (44% and 30%) was no better than K646A (Table 1). The kinetic parameters for the K646R and K646Q mutants were determined by m7GDP titration (Table 2). K646R had a Km of 167 μM m7GDP, representing a ninefold gain in affinity compared to K646A. In contrast, the Km of K646Q (1580 μM m7GDP) was just as unfavorable as that of K646A. These findings emphasize the crucial role of the electrostatic contacts of Lys646 with the cap phosphates, which can be sustained by arginine, but evidently not by glutamine.
Arg807 makes bidentate contacts from its terminal guanidinium nitrogens to two β-phosphate oxygens of m7GTP (Fig. 1A). The R807A mutation was lethal in vivo in yeast, and the R807A protein had only 6% of wild-type activity in converting m7GDP to m2,7GDP (Hausmann et al. 2008). Here we found that replacing Arg807 with lysine restored +++ yeast growth at all temperatures tested and increased methylation activity in vitro to 49% and 55% of wild-type in steps 1 and 2, respectively (Table 1). The R807Q allele sustained + yeast growth at 25°C–37°C and no growth at 18°C; the R807Q protein was marginally more active than R807A in vitro (13% and 12% of wild type in steps 1 and 2, respectively) (Table 1). Here too, the mutational effects underscore the essentiality of the ionic interactions of Arg807 with the β-phosphate seen in the crystal structure, which can be fulfilled by lysine but not glutamine.
Tyr771 donates a hydrogen bond from the phenolic hydroxyl to a nonbridging α-phosphate oxygen of m7GTP (Fig. 1A). Replacing Tyr771 with phenylalanine, leucine, or serine was unconditionally lethal in yeast and uniformly deleterious to methyltransferase activity in vitro (Table 1). These results highlight the essentiality of the polar contact to the α-phosphate and the requirement for proper spacing of the hydroxyl from the protein main chain (i.e., serine could not function in lieu of tyrosine).
Different structure–activity relations were seen at Tyr647, an essential residue that makes a water-bridged hydrogen bond to the α-phosphate of m7GTP (Fig. 1A), to wit: activity in vivo and in vitro was restored when Tyr647 was replaced by phenylalanine (+++ growth at 37°C), whereas leucine and serine had no salutary effect relative to the alanine mutant (Table 1). We surmise that the tyrosine hydroxyl is not strictly essential, although its subtraction in the Y647F mutant elicited a cold-sensitive phenotype in yeast (with + growth at 18°C–25°C) and a modest decrement in methyltransferase activity (32% and 65% of wild-type in steps 1 and 2, respectively) that did not meet our fivefold criterion of significance. These findings, whereby Phe suffices but Leu does not, implicate either the aromatic quality of Tyr647 or the Cɛ and Cζ atoms of the phenyl ring in promoting Tgs1 function. The crystal structure shows that Tyr647 is sandwiched between Phe670 and Lys646 and that the Tyr647 Cɛ2 and Cζ atoms make multiple van der Waals contacts. Specifically, Tyr647 packs closely against Phe670 so that Ty647 Cɛ2 contacts Phe670 Cɛ1 (3.53 Å) and Ty647 Cζ contacts Phe670 Cδ1 (3.42 Å). The Tyr647 Cɛ2 and Cζ atoms also make van der Waals contacts (3.8 Å) to the Cγ and Cɛ atoms of Lys646, respectively. These interactions likely stabilize the hydrophobic core of the distinctive hTgs1 N-terminal helical module that binds the cap triphosphate.
m7G binding site and the critical role of the Trp766-m7G π-cation interaction
The m7G binding site includes two aromatic residues important for methyltransferase activity, Trp766 and Phe804. Previous studies had shown that the W766A mutation was lethal in yeast and the W766A protein had <1% of wild-type methyltransferase activity in vitro (Hausmann et al. 2008). Here we found that Trp766 is strictly essential for hTgs1 activity in vivo, insofar as substitutions with phenylalanine or tyrosine were unconditionally lethal (Table 1). The W776F protein was severely defective in vitro (1% and 3% of wild-type methyltransferase activity in steps 1 and 2, respectively), and the W776Y mutant was defective in step 1 methylation (6% of wild type), although its activity in converting DMG to TMG (24% of wild type) exceeded our cut-off for a significant mutational effect. This was the only instance in this study in which a disparity between mutational effects (significant vs. not) on the two methylation steps was observed (Table 1). Note that the conversion of m7G to m2,7G is certain to be a limiting step in sustaining the viability of tgs1Δ mud2Δ cells, insofar as the lethality of the double mutation can be rescued by expressing mimivirus Tgs, a guanine-N2 methyltransferase that is exclusively capable of DMG synthesis in vitro (Benarroch et al. 2009). Thus, we surmise that the lethality of W766Y reflects its feeble activity in forming m2,7G.
The present mutational data fortify conclusions from studies using cap analogs (Benarroch et al. 2010) that the π-cation stacking interactions of Trp766 and m7G are crucial for hTgs1 activity. The guanine-N7 methyl makes only a single van der Waals contact to protein in the hTgs1 structure (to the Gly770 Cα atom) (Fig. 1A), thus its contribution to the cap specificity of the guanine-N2 methyltransferase reaction likely derives from the positive charge it imparts to the purine ring, resulting in a π-cation stack in addition to a π–π stack. Because hTgs1 efficiently methylates benzyl7GTP and benzyl7GpppG but is inert with GTP or GpppG (Benarroch et al. 2010), it is safely concluded that the cationic N7-alkylated guanine, not the methyl group per se, is the key property of the methyl acceptor. From the present mutational data, we see that only tryptophan can productively sustain the π-cation contacts for DMG synthesis. Trp has a larger planar surface than either Tyr or Phe, and, in the active site of hTgs1, the Trp766 indole makes van der Waals contacts to all nine purine-ring atoms of m7G plus the O6 atom at distances of 3.21 to 3.64 Å. Yet, it is likely that the strength of the π-cation interaction of tryptophan is decisive. Ab initio calculations of electrostatic potential surfaces highlight indole(Trp) as superior to either benzene(Phe) or phenol(Tyr) in forming model π-cation interactions (Mecozzi et al. 1996). The primacy of tryptophan in π-cation interactions for tight binding to m7G has been noted previously for vaccinia virus cap-specific nucleoside-2′-O-methyltransferase VP39 and the cap-binding translation initiation factor eIF4E. Whereas native VP39 sandwiches the m7G cap between a tyrosine and a phenylalanine, changing both residues to tryptophan elicited a 40-fold increase in affinity for m7G (Hu et al. 2002). Conversely, native eIF4E binds m7G between two tryptophans (Trp56 and Trp102 in mammalian eIF4E) and changing Trp102 to tyrosine or phenylalanine virtually abolished m7GDP binding (Hsu et al. 2000).
hTgs1 residue Phe804 lines the hydrophobic methyl acceptor pocket and makes two van der Waals contacts to the m7G base, from Cɛ2 to guanine N3 (3.93 Å) and from Cδ2 to guanine N2 (3.88 Å). Our results imply that these contacts are important for hTgs1 activity, insofar as the F804A mutant was only 7% as active as wild type in m7GDP methylation in vitro and displayed cold-sensitive lethality in yeast (Table 1). It had been speculated that Phe804 might play a special role in the second guanine-N2 methylation step of TMG synthesis, by accommodating the first N2 methyl within a hydrophobic pocket lined by Ser763, Pro764, Trp766, and Phe804 (Monecke et al. 2009; Benarroch et al. 2010). Indeed, we find here that the F804A mutant is defective in converting DMG to TMG (6% of wild-type activity). Phe804 is conserved in Schizosaccharomyces, Entamoeba, and Trypanosoma Tgs1 proteins, each of which has biochemically documented TMG synthase activity (Hausmann and Shuman 2005a; Simoes-Barbosa et al. 2008), but is substituted by a lysine in Giardia Tgs2 and MimiTgs, which are DMG-only synthases (Hausmann and Shuman 2005b; Benarroch et al. 2010). A simple speculation is that the hydrophobic-to-charged substitution might disrupt the putative m2,7G substrate docking site for the second transmethylation reaction. We attempted to test this idea by replacing hTgs1 Phe804 with lysine, in the hope that it might convert hTgs1 into a DMG-only synthase. However, we found that the F804K was unconditionally lethal in yeast and abolished both steps of the guanine-N2 methyltransferase reaction in vitro (Table 1). We surmise that the imposed positive charge is deleterious per se (clearly more so than the alanine change) in the context of the hTgs1 active site.
Methyl donor site
Four hTgs1 residues that bind AdoHcy in the crystal structure are essential for hTgs1 function in vivo: Asp696, Asp719, Asp747, and Asn704 (Fig. 1B). Asp719 receives bidentate hydrogen bonds to its carboxylate oxygens from the AdoHcy ribose 2′ and 3′ hydroxyls. Both contacts are apparently critical, insofar as the asparagine change was lethal in yeast and had no salutary effect on methyltransferase activity in vitro. However, introducing glutamate restored yeast growth (+++ at 37°C and 30°C; ++ at 25°C and 18°C) and full methyltransferase activity in vitro (Table 1), signifying that a carboxylate is crucial and the hTgs1 active site can accommodate the longer linker from the main chain in Glu versus Asp. It is noteworthy that the ribose-binding residue corresponding to hTgs1 Asp719 is naturally a glutamate in the Tgs homologs of fission yeast and Giardia. In Giardia Tgs, replacing this glutamate by alanine or glutamine reduced m7GDP methylation in vitro to <1% of the wild-type activity, whereas aspartate partially revived activity to 8% of wild type (Hausmann et al. 2007).
Asp696 participates in a hydrogen-bonding network via two waters, each coordinated by one of the carboxylate oxygens. Asp696 Oδ2 interacts via water with the AdoHcy amine and carboxylate (Fig. 1B), the Cys699 main chain carbonyl, and the Thr705 main chain amide. Asp Oδ2 also makes direct hydrogen bonds to the Thr705 Oγ and main chain amide. Asp696 Oδ1 interacts via another water with the Ser763 Oγ, the Ser763 and Phe698 main chain amides, and the Phe761 main chain carbonyl. Thus, Asp696 likely plays a role in ensuring the proper conformation of the AdoMet-binding loop/helix segment 694VVDAFCGVGGNT705 and the loop's proximity to Ser763 of the m7G-binding motif, in addition to binding the methionine component of AdoMet. Here we found that whereas asparagine was inactive in lieu of Asp696, glutamate substitution revived activity in vivo (++ growth at 25°C) and in vitro (44% and 60% of wild type in steps 1 and 2, respectively) (Table 1). Different structure–activity relationships were seen at Asp747, which coordinates the AdoHcy adenine N6 (Fig. 1B). The introduction of asparagine restored full hTgs1 function in vivo and in vitro, whereas glutamate restored function partially (Table 1). We surmise that hydrogen bond acceptance from adenine N6 is the key function of Asp747, which can be performed as well by the isosteric amide of asparagine, but is suboptimal when the spacing of the carboxylate from the main chain is increased in the case of glutamate.
Asn704 makes a bifurcated hydrogen bond from Oδ to the methionine amine of the methyl donor and to Thr673 Oγ (Fig. 1B). Whereas the N704A mutant was lethal in yeast and defective in m7GDP methylation in vitro (Hausmann et al. 2008), we found that activity in vivo was revived by introduction of either aspartate (++ growth at all temperatures) or glutamine (+++ at 37°C, with a + cs growth defect) and that both conservative changes supported full methyltransferase activities in vitro (Table 1). These results highlight hydrogen binding as the relevant property of the Asn704 side chain.
Mapping structural features required for Mud2 function in vivo
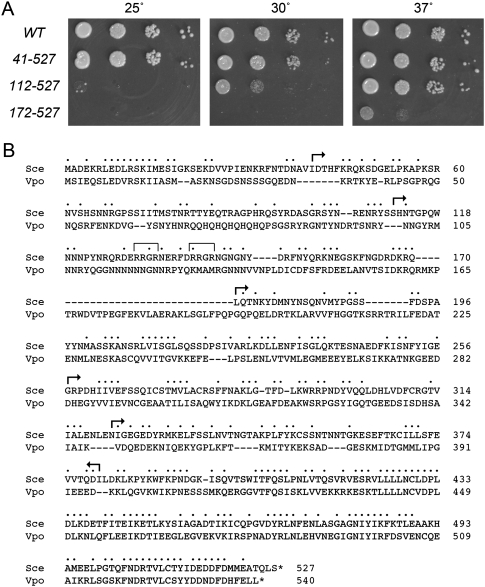
We exploited the lethality of the tgs1Δ mud2Δ strain to conduct a mutational analysis of the otherwise inessential yeast Mud2 protein. There are few close homologs of Mud2 that display primary structure similarity throughout most of the polypeptide; these are confined to a narrow range of fungi, including Vanderwaltozyma polyspora, Zygosaccharomyces rouxii, Candida glabrata, Candida albicans, Kluyveromyces lactis, Verticillium albo-atrum, Lachancea thermotolerans, and Ashbya gossypii. Alignment of the S. cerevisiae and V. polyspora Mud2 proteins (Fig. 3B) shows that the structural similarity is greatest in the C-terminal segment that includes the RRM3 domain. To delineate the proximal margins of functional Mud2, we constructed a series of incremental N-terminal deletions, Mud2-(41–527), Mud2-(112–527), Mud2-(172–597), Mud2-(258–527), and Mud2-(323–527), guided (in part) by the positions of gaps in the SceMud2 versus VpoMud2 alignment (Fig. 3B). CEN LEU2 plasmids bearing wild-type or truncated MUD2 alleles under the control of the native MUD2 promoter were tested by plasmid shuffle for complementation of tgs1Δ mud2Δ yeast. The MUD2-(258–527) and MUD2-(323–527) alleles failed to support growth on FOA agar at 25°C, 30°C, and 37°C and were therefore deemed lethal. The mutants that gave rise to FOA-resistant colonies at one or more temperatures were tested for growth on YPD agar (Fig. 3A). MUD2-(41–527) cells grew as well as “wild-type” MUD2 cells at 25°C, 30°C, and 37°C (scored as +++ growth in Table 3). The next truncation mutant, MUD2-(112–527), displayed a cold-sensitive phenotype, with +++ growth at 37°C, + growth at 30°C, and no growth 25°C (Fig. 3A). Finally, the MUD2-(172–527) strain formed only pinpoint colonies at 37°C and failed to grow at 30°C or 25°C (Fig. 3A). We conclude that, in the tgs1Δ background, the segment from amino acids 112 to 171 is critical for Mud2 function, while the N-terminal 40 amino acids are dispensable, and the segment from amino acids 41 to 111 is important for Mud2 function at ≤30°C.
FIGURE 3.
N- and C-terminal deletions of yeast Mud2. (A) The growth phenotypes of tgs1Δ mud2Δ cells harboring LEU2 plasmids with wild-type (WT) MUD2 or N-terminal truncation mutants MUD2-(41–527), MUD2-(112–527), or MUD2-(172–527) were compared by spotting 3-μL aliquots of serial 10-fold dilutions of cells (from liquid cultures grown to mid-log phase at 30°C and adjusted to A600 of 0.1) to YPD agar and incubating the plates at the indicated temperatures. (B) Alignment of the primary structures of S. cerevisiae and V. polyspora Mud2. (•) Positions of side chain identity/similarity; (—) gaps in the alignment. (Arrows) The margins of the N-terminal and C-terminal truncation mutants of S. cerevisiae Mud2. For the N-terminal deletions, the arrows specify the residues that were mutated to methionine to initiate the truncated proteins. The C-terminal RRM3 domain is highlighted in the gray box. The tandem RRGR motifs in the N-terminal domain of S. cerevisiae are denoted by brackets.
TABLE 3.
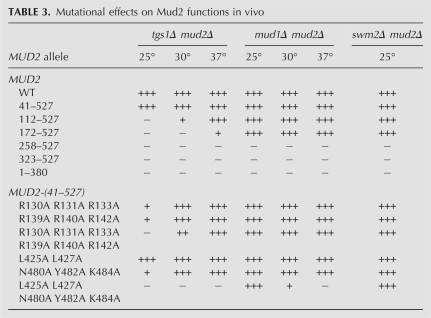
Mutational effects on Mud2 functions in vivo
To gauge whether the requirements for Mud2 function varied with the genetic background in which Mud2 becomes essential for yeast growth, we performed a parallel series of plasmid shuffles in a mud1Δ mud2Δ strain. We found that MUD2-(258–527) and MUD2-(323–527) were again unconditionally lethal (Table 3). However, the effects of shorter N-terminal deletions were strikingly different in the mud1Δ background, where MUD2-(172–527) and MUD2-(112–527) supported +++ growth at 25°C, 30°C, and 37°C (Table 3). These results demonstrate a separation of function, whereby the portion of the N-terminal domain from residues 41 to 171 that is required conditionally or unconditionally for Mud2 function in tgs1Δ cells is inessential in mud1Δ cells.
The lethality of MUD2-(258–527) in two different assays indicates that the segment from 172 to 257 embraces essential components of Mud2, and the C-terminal RRM3 domain does not suffice for Mud2 function in vivo. The necessity of the RRM3 domain for Mud2 function was established by the finding that a C-terminal truncation allele, MUD2-(1–380), that lacks RRM3 was uniformly lethal in tgs1Δ and mud1Δ backgrounds (Table 3). These results agree with the initial report that a nonsense mutation that causes a translation stop after Mud2 residue 448 (which deletes most of the RRM3 domain) elicits the “MUD” phenotype (Abovich et al. 1994).
Effects of alanine-cluster mutations in the Mud2 RRM3 and N domains
The 131-amino acid segment (amino acids 41–171) of the N domain important for Mud2 activity in tgs1Δ cells is strongly hydrophilic (calculated pKa 11.4) and highly enriched for arginine (n = 23) and asparagine (n = 18) side chains, along with serines (n = 13) and glycines (n = 11) (Fig. 3B). The arginine richness raises the prospect that the N-domain might mediate the interaction of Mud2 with pre-mRNA during spliceosome assembly (McPheeters and Muhlenkamp 2003) and thereby be pertinent to Mud2 function in the tgs1Δ strain. Here we tested the effects of triple arginine-to-alanine cluster mutations in two tandem Arg/Gly motifs—130RRGR133 and 139RRGR142—located within the essential segment from amino acids 112 to 171 (denoted by brackets in Fig. 3B). These clustered changes were made in the context of Mud2-(41–527), which is fully active in vivo. We found that the R130A R131A R133A and R139A R140A R142A mutants displayed +++ growth at 37°C and 30°C, but only + at 25°C (Table 3). Combining the two alanine clusters to form a sextuple mutant preserved +++ growth at 37°C, but slowed growth at 30°C and was lethal at 25°C (Table 3). Thus, the double-cluster mutation effacing the two RRGR motifs mimicked to a degree the effects of deleting amino acids 112–171. Note that these N-domain Arg-to-Ala cluster mutations, singly or together, had no effect on Mud2 function in the mud1Δ background (Table 3).
Structure–activity relations for the Mud2 RRM3 domain were gauged by introducing alanine-cluster mutations in the RNP2 and RNP1 motifs. The choice of residues to mutate was guided by a PSI-BLAST alignment of the Mud2 RRM3 to the RRM3/UHM domains of fission yeast and human U2AF65 (Supplemental Fig. S2B), and by reference to the atomic structure of the human RRM3 domain (Supplemental Fig. S2A; Selenko et al. 2003). We specifically targeted Leu425 and Leu427 in the β1 (RNP2) strand 424LLLLNC429 and Asn480, Tyr482, and Lys484 in the β3 (RNP1) strand 478AGNIYIKF485; these side chains project outward on the surface of the β-sheet opposite the two α-helices (Supplemental Fig. S2B) and would correspond to residues in canonical RRMs that interact with RNA. The RNP2 cluster mutant L425A L427A was fully active in complementing tgs1Δ mud2Δ (Table 3). The RNP1 cluster mutant N480A Y482A K484A displayed +++ growth at 37°C and 30°C, but only + growth at 25°C (Table 3). The instructive finding was that the combined RNP2/RNP1 penta-alanine cluster mutation was unconditionally lethal, signifying that this face of the β-sheet is functionally essential in the tgs1Δ background (Table 3). In contrast, the same RNP motif mutations had much less impact on complementation of the mud1Δ mud2Δ strain, to wit: (1) the RNP1 triple-alanine mutation per se had no effect on growth; and (2) the RNP2/RNP1 penta-alanine mutant supported +++ growth of mud1Δ cells at 25°C and + at 30°C (Table 3). These results reinforce the conclusions from the deletion analyses that structure–function relations for Mud2 are context-dependent and probably indicative of multiple functions for Mud2 during pre-mRNA splicing.
It is useful to compare our results with the effects of RNP1 mutations reported by Abovich et al. (1994). They found that mutating Tyr482 to aspartic acid had no impact on complementation of mud1Δ mud2Δ, which is consonant with what we see for the Y482A change and attests to tolerance of an ectopic negative charge on the exposed surface of the RRM3 β-sheet, with respect to complementation of mud1Δ. They also noted that a Mud2 RNP1 double mutation in which Tyr482 and Phe485 were both changed to aspartic acid was lethal in a mud1Δ background (Abovich et al. 1994). The Mud2 Phe485 equivalent in human U2AF65 is Phe436, which projects inward to the hydrophobic core of the RRM2 α–β fold (Selenko et al. 2003). We suspect that an ectopic acidic residue would exert a significant local structural effect on the RRM3 module, which makes it difficult to extract a meaningful structure–activity relationship from the lethality of the Y484D F485D change.
Deletion of YNR004w(SWM2) is synthetic with mud2Δ
The relatively higher stringency of Mud2 structural requirements for its function in the tgs1Δ strain prompted us to seek further genetic connections between Mud2 and Tgs1. We focused on the product of the nonessential yeast YNR004w gene (Volckaert et al. 2003), which was identified as a Tgs1-binding protein in two separate genome-wide two-hybrid screens, with reciprocal bait–hit arrangements (Uetz et al. 2000; Yu et al. 2008). YNR004w is a 146-amino acid polypeptide with no recognizable structural or functional motifs and a very narrow phylogenetic distribution, with credible homologs in just three other fungi: V. polyspora, Z. rouxii, and L. thermotolerans (Fig. 4B). Wilmes et al. (2008) reported microarray analysis indicating that splicing of a subset of intron-containing mRNAs is affected when ynr004wΔ cells are shifted from 30°C to 16°C and suggested an overlapping genetic interaction network for YNR004w and Tgs1. Here we performed directed tests for synthetic interactions between YNR004w and TGS1 or MUD2 or NAM8. In the experiment shown in Figure 4A, a haploid YNR004w∷kanMX disruptant was mated to a mud2∷natMX haploid, and the resulting diploids were sporulated and subjected to tetrad dissection. The growth of the haploid progeny of five individual tetrads revealed that either one or two of the four spores germinated and/or grew extremely slowly (Fig. 4A). Testing for G418 and Nourseothricin sensitivity/resistance verified that the ynr004w∷kanMX and mud2∷natMX alleles segregated independently and in a 2:2 pattern with respect to the intact YNR004w and MUD2 genes (data not shown). The slow-growing haploid(s) in each tetrad corresponded to the ynr004w∷kanMX mud2∷natMX double knockout. We conclude that ynr004wΔ and mud2Δ display a strong synthetic sick phenotype. In contrast, we observed no synthetic growth defects in a ynr004wΔ tgs1Δ double knockout compared to the tgs1Δ single mutant, which is cold-sensitive (Supplemental Fig. S3). Moreover, there were no growth defects in a ynr004wΔ nam8Δ double knockout (Supplemental Fig. S4). Consequently, we renamed the YNR004w gene SWM2 (synthetic with mud2Δ).
FIGURE 4.
Synthetic interactions of yeast Mud2 and Swm2. (A) MUD2 mud2∷natMX SWM2 swm2∷kanMX diploids were sporulated, five asci (1, 2, 3, 4, and 5) were dissected, and the haploid spores (A, B, C, and D) were germinated on YPD agar for 4 d at 30°C. All of the tiny colonies seen were ynr004w∷kanMX mud2∷natMX double-knockout progeny. (B) The primary structure of S. cerevisiae Swm2(YNR004w) is aligned to the homologous polypeptides of other fungi: V. polyspora, Z. rouxii, and L. thermotolerans. (•) Positions of side chain identity/similarity in all four proteins; (—) gaps in the alignment.
swm2Δ mud2Δ cells displayed + growth on YPD agar; +++ growth was restored by transformation with a CEN MUD2 plasmid (Table 3). A plasmid shuffle assay for complementation of swm2Δ mud2Δ p(CEN URA3 SWM2) cells by the collection of mutated CEN LEU2 MUD2 plasmids provided a “third” test of mutational impact on Mud2 function. Those MUD2 alleles that restored normal growth were scored as +++; those that offered no improvement over the basal + growth of swm2Δ mud2Δ were scored as − (Table 3). We found that the activity of the various MUD2 alleles in the swm2Δ mud2Δ background mirrored that in rescuing the lethality of the mud1Δ mud2Δ strain. We surmise that the structural requirements for Mud2 function are more demanding when yeast cells lack TMG caps than when they lack Mud1 or Swm2.
An issue arises as to whether the losses of function attendant on Mud2 mutations might reflect gross deficiencies in the steady-state levels or folding of the mutated proteins in yeast. Although we have not assayed directly the steady-state levels of the Mud2 polypeptides, we can surmise that most of the mutants analyzed were produced in sufficient quantity in vivo, insofar as they retained activity in at least two of the three genetic assays employed (Table 3). This applies to N-terminal deletions of up to 171 amino acids and to the various alanine-cluster mutations in the N- and C-domains of Mud2. The situation is unclear for the large domain deletions that were lethal in all three genetic backgrounds. The Mud2-(323–527) fragment includes the RRM3 module that suffices for interaction with BBP (Rain et al. 1998; Selenko et al. 2003). To gauge if Mud2-(323–527) is produced in vivo at levels capable of eliciting a phenotype, we placed the MUD2 and MUD2-(323–527) alleles on CEN plasmids under the control of a GAL promoter and introduced the plasmids into mud2Δ yeast cells. The pertinent findings were that: (1) induced expression of wild-type Mud2 had no apparent effect on cell growth on galactose-containing medium, for example, compared with isogenic cells bearing the empty vector; and (2) induced expression of Mud2-(323–527) suppressed cell growth on galactose medium at 18°C and 25°C, while no effects of the MUD2-(323–527) plasmid on growth were seen on repressive glucose-containing plates (Supplemental Fig. S5). Thus, even though Mud2 is inessential, the isolated Mud2-(323–527) fragment is deleterious at ≤25°C (although not at ≥30°C) (data not shown). We take this as evidence that the C-domain fragment of Mud2 is made in vivo in a form that has some bioactivity. It is conceivable that galactose-induced Mud2-(323–527) sequesters the essential BBP protein in an inactive state.
Concluding remarks on TMG caps and Mud2
The genetic connections between TMG caps and proteins involved in early steps of yeast pre-mRNA splicing are deep and broad (Hausmann et al. 2008). The tightest mutational synergy is seen in cells that simultaneously lack TMG caps and either Mud2 or Nam8, both of which are nonessential early-stage splicing factors that contain one or more RRM domains. We have shown here that Mud2 function in the absence of TMG caps requires its C-terminal RRM3 domain as well as the arginine-rich segment of the N-terminal domain. Mud2 activity is more acutely dependent on the outwardly projecting residues of the RRM3 RNP2 and RNP1 motifs in tgs1Δ cells than in two other genetic backgrounds (mud1Δ and swm2Δ) where Mud2 is essential or important for growth. We suggest the following scenario, admittedly speculative at this stage, as a potential explanation for our results. We envision that spliceosome assembly on some of the essential yeast pre-mRNAs depends on recognition of the TMG caps present on the U1 and/or U2 snRNA, by an as yet undefined yeast TMG-binding protein(s). When Tgs1 is deleted or its methyltransferase disabled, the spliceosomal snRNAs will, by default, have the m7G caps that are formed during transcription by RNA polymerase II. In this case, the ability of the m7G-capped U1 and/or U2 snRNAs to function in splicing relies on a backup system entailing different protein interactions with the residual m7G snRNA 5′ termini. We speculate that Mud2 provides this backup by binding to the m7G cap of either the U2 or U1 snRNP. Consequently, cells lacking TMG caps are strictly reliant on Mud2 for viability. (By extension, we can rationalize the synthetic lethal interactions of tgs1Δ with both mud2Δ and nam8Δ by positing that Nam8 binds the m7G cap of whichever snRNP is not taken by Mud2.) There is ample precedent to invoke m7G cap recognition by an RRM protein. For example: (1) the CBP20 subunit of the nuclear cap binding protein comprises an embellished RRM domain that binds the m7G cap nucleotide on the outward face of its β-sheet (away from the α-helices) via contacts with the side chains of the RNP2 and RNP1 motifs (among others) (Calero et al. 2002; Mazza et al. 2002); and (2) poly(A)-specific ribonuclease exploits an entirely different site on its RRM domain for specific binding to an m7G cap nucleotide (Monecke et al. 2008; Nagata et al. 2008; Wu et al. 2009). We considered the possibility that the yeast nuclear cap binding protein might be a good candidate to mediate snRNA function in the absence of TMG caps, but rejected this idea upon finding no mutational synergy between tgs1Δ and cbc2Δ (a deletion of the gene encoding the cap binding CBP20 subunit of yeast CBC) (Supplemental Fig. S6). The cbc2Δ single mutant per se is slow-growing at warmer temperatures and displays a much stronger cold-sensitive defect than does tgs1Δ, yet there is no significant growth defect in tgs1Δ cbc2Δ cells at 34°C or 37°C versus cbc2Δ cells (Supplemental Fig. S6).
MATERIALS AND METHODS
Assaying mutational effects on hTgs1 function in yeast
Mutations were introduced into the hTGS1-(631–853) gene encoding the C-terminal catalytic domain by two-stage overlap extension PCR as described previously (Hausmann et al. 2008). The mutated genes were inserted into the yeast expression vector pRS425 (2μ LEU2) that contains a 2.2-kb PvuII fragment from pYX132 (Novagen) bearing a yeast TPI1 promoter that drives transcription of hTGS1-(631–853). The plasmid inserts were sequenced completely to ensure that no unwanted mutations were introduced during amplification and cloning. The pRS-hTGS1-(631–853) plasmids were transformed into S. cerevisiae tgs1Δ (tgs1∷natMX) mud2Δ (mud2∷kanMX) p360-TGS1 (CEN URA3 TGS1) cells (Hausmann et al. 2008). Individual Leu+ transformants were streaked on agar medium containing 1.0 mg/mL 5-fluoroorotic acid (FOA). Growth was scored after 7 d of incubation at 18°C, 25°C, 30°C, and 37°C. Lethal mutants were those that failed to form colonies at any temperature. Individual FOA-resistant colonies with viable hTGS1-(631–853) alleles were transferred to YPD agar medium. Two isolates of each mutant were tested for growth on YPD agar at 18°C, 25°C, 30°C, and 37°C. Growth was assessed as follows: (+++) colony size was indistinguishable from strains bearing wild-type hTGS1-(631–853); (++) slightly reduced colony size; (+) only pinpoint colonies were formed; (−) no growth.
Recombinant hTgs1 proteins
The T7 RNA polymerase-based bacterial expression plasmid pET28b-His10Smt3-hTGS1-(631–853) encodes the C-terminal catalytic domain of hTgs1 fused to an N-terminal His10Smt3 tag (Hausmann et al. 2008). Mutated hTGS1-(631–853) genes were excised from their respective yeast pRS vectors (see above) by digestion with BamHI and XhoI and then inserted into pET28b-His10Smt3. The wild-type and mutated pET28-His10Smt3-hTgs1(631–853) plasmids were transformed into E. coli BL21(DE3) cells. Cultures (500 mL) derived from single transformants were grown at 37°C in LB medium containing 50 μg/mL kanamycin until the A600 reached ∼0.6. The cultures were adjusted to 0.2 mM IPTG and 2% (v/v) ethanol, after which incubation was continued for 20 h at 17°C with constant shaking. Cells were harvested by centrifugation and stored at −80°C. All subsequent procedures were performed at 4°C. Thawed bacteria were resuspended in 25 mL of buffer A (50 mM Tris-HCl at pH 8.0, 200 mM NaCl, 10% glycerol) and supplemented with 1 protease inhibitor cocktail tablet (Roche Applied Science). The suspensions were adjusted to 0.1 mg/mL lysozyme and incubated for 30 min. Imidazole was added to a final concentration of 5 mM, and the lysates were sonicated to reduce viscosity. Insoluble material was removed by centrifugation. The soluble extracts were mixed for 30 min with 1.6 mL of Ni2+-nitrilotriacetic acid-agarose (QIAGEN) that had been equilibrated with buffer A containing 5 mM imidazole. The resins were recovered by centrifugation, resuspended in buffer A with 5 mM imidazole, and then poured into columns. The columns were washed with 8-mL aliquots of 10 and 20 mM imidazole in buffer A and then eluted stepwise with 2.5 mL aliquots of buffer A containing 50, 100, 250, and 500 mM imidazole. The elution profiles were monitored by SDS-PAGE. The 250 mM imidazole eluates containing the His10Smt3-hTgs1-(631–853) polypeptides were dialyzed against buffer containing 50 mM Tris-HCl (pH 8.0), 200 mM NaCl, 2 mM dithiothreitol, 1 mM EDTA, and 10% glycerol and then stored at −80°C. The protein concentrations were determined initially by using the Bio-Rad dye reagent with BSA as the standard. The concentrations of the His10Smt3-hTgs1-(631–853) polypeptides were then compared by SDS-PAGE and staining with Coomassie Blue dye. The staining intensities of the His10Smt3-hTgs1-(631–853) polypeptides were quantified using the Bio-Rad Molecular Imager ChemiDoc gel densitometry system, and each mutant was normalized to the wild-type polypeptide. A subsequent SDS-PAGE analysis of aliquots of the Ni-agarose preparations containing ∼3 μg of wild-type or mutant His10Smt3-hTgs1-(631–853) polypeptide is shown in Supplemental Figure S1.
Methyltransferase assay
Reaction mixtures (10 μL) containing 50 mM Tris-HCl (pH 8.0), 5 mM DTT, 50 mM NaCl, 100 μM [3H-CH3]AdoMet, 1 mM m7GDP or m2,7GDP, and 0.1 μg of wild-type or mutant hTgs1-(631–853) were incubated for 30 min at 37°C. Aliquots (6 μL) were spotted on polyethyleneimine cellulose TLC plates, in parallel with AdoMet and m2,7GDP or m2,2,7GDP standards. The TLC plates were developed with 50 mM (NH4)2SO4. The markers were identified by UV illumination and the portions of the lanes containing the AdoMet substrate, m2,7GDP or m2,2,7GDP products were cut out, and the radioactivity in each was quantified by liquid scintillation counting.
Yeast gene disruptions
We constructed new haploid strains in which MUD2 or NAM8 was replaced by the natMX gene conferring resistance to Nourseothricin. This was accomplished by transforming the mud2∷kanMX and nam8∷kanMX knockouts described previously (Hausmann et al. 2008) with the natMX-containing plasmid p4339 (Tong et al. 2001) that had been restricted with EcoRI to permit substitution of the natMX cassette for kanMX. Nourseothricin-resistant (natR) transformants were selected on YPD agar containing 100 μg/mL clonNAT (Werner BioAgents). We also constructed haploid strains in which the entire YNR004w(SWM2) or CBC2 open reading frames were replaced with the kanMX gene conferring resistance to geneticin. Plasmid pUC19-cbc25′-kanMX-cbc23′ harbors kanMX flanked by the 480-base-pair (bp) genomic DNA segment located immediately 5′ of the CBC2 start codon and the 404-bp segment 3′ of the CBC2 stop codon. In the plasmid pUC19-ynr004w5′-kanMX-ynr004w3′, the kanMX gene is flanked by the 510-bp genomic DNA segment immediately 5′ of the YNR004w start codon and the 313-bp segment immediately downstream from the YNR004w stop codon. DNA restriction fragments encompassing the disruption cassettes were excised and transformed into the diploid yeast strain BY4743 (a/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/met15Δ0 ura3Δ0/ura3Δ0). Geneticin-resistant (kanR) transformants were selected, and the targeted insertions were confirmed by diagnostic Southern blotting. Heterozygous YNR004w ynr007wΔ and CBC2 cbc2Δ diploids were then sporulated, and tetrads were dissected to obtain ynr004w∷kanMX (swm2Δ) and cbc2∷kanMX (cbc2Δ) haploids.
Assaying Mud2 function in yeast
MUD2 is essential for viability of haploid yeast tgs1Δ and mud1Δ cells (Abovich et al. 1994; Hausmann et al. 2008). Plasmid shuffle assays were employed to test MUD2 activity in mud2Δ tgs1Δ and mud2Δ mud1Δ strains that rely for viability on maintenance of one of the missing essential genes on a URA3 plasmid. tgs1Δ (tgs1∷natMX) mud2Δ (mud2∷kanMX) p360-TGS1 (CEN URA3 TGS1) cells were used to assay complementation in the tgs1Δ background (Hausmann et al. 2008). To establish the complementation assay in the mud1Δ background, we prepared a heterozygous MUD2 mud2∷natMX MUD1 mud1∷kanMX diploid and transformed it with p360-MUD1, a CEN URA3 plasmid bearing a 1.59-kb genomic segment that includes the MUD1 gene plus 420 bp of DNA 5′ of the start codon and 185 bp of DNA 3′ of the stop codon. The diploid strain was sporulated, asci were dissected, and the desired haploid mud1Δ mud2Δ Ura+ progeny were recovered that were: (1) resistant to geneticin and clonNAT and (2) unable to grow on medium containing 5-FOA.
A series of CEN LEU2 plasmids bearing wild-type or mutated MUD2 genes under the control of the native MUD2 promoter was constructed as follows. First, we made plasmid pRS-MUD2(5′3′), a derivative of the yeast shuttle vector pRS415 (CEN LEU2) with a genomic DNA segment spanning 558 bp upstream of the MUD2 ORF cloned between the XhoI and BamHI sites and a genomic DNA segment spanning a 241-bp segment downstream from the MUD2 ORF cloned between the flanking NotI and SacI sites. The wild-type MUD2 ORF was PCR-amplified with primers that introduced a BamHI site preceding the start codon and a NotI site following the stop codon. N-terminal truncation mutants MUD2-(41–527), MUD2-(112–527), MUD2-(172–527), MUD2-(258–527), and MUD2-(323–527) were PCR-amplified with sense strand primers that introduced a BamHI site and a methionine codon in place of the codons for Ile40, Ser111, Leu171, Gly257, or Asn322 and an antisense strand primer that introduced a NotI site 3′ of the stop codon. The C-terminal deletion mutant MUD2-(1–380) was PCR-amplified with an antisense primer that introduced a stop codon in place of the codon for Leu381 and a NotI site 3′ of the new stop codon. Alanine mutations were introduced into MUD2-(41–527) by two-stage PCR overlap extension using mutagenic primers. The MUD2 PCR products were digested with BamHI and NotI and then inserted between the BamHI and NotI sites of pMUD2(5′3′). The inserts of the resulting pRS-MUD2 clones were sequenced completely to exclude the presence of unwanted mutations.
The pRS-MUD2 plasmids were transformed into tgs1Δ mud2Δ p360-TGS1 cells and mud1Δ mud2Δ p360-MUD1 cells. Individual Leu+ transformants were streaked on agar medium containing 1.0 mg/mL FOA. Growth was scored after 7 d of incubation at 25°C, 30°C, or 37°C. Lethal mutants were those that failed to form colonies at any temperature. Individual FOA-resistant colonies with viable MUD2 alleles were transferred to YPD agar medium. Two isolates of each mutant were tested for growth on YPD agar at 25°C, 30°C, or 37°C. Growth was assessed as follows: (+++) colony size was indistinguishable from strains bearing wild-type MUD2; (++) slightly reduced colony size; (+) only pinpoint colonies were formed; (−) no growth (Table 3).
Tests of mutational synergy with tgs1Δ
Haploid tgs1∷natMX cells were mixed with swm2∷kanMX or cbc2∷kanMX strains of opposite mating type, and diploids were selected on YPD agar containing clonNAT and geneticin. The heterozygous diploids were sporulated, and tetrads were dissected to obtain viable haploid tgs1Δ swm2Δ and tgs1Δ cbc2Δ double mutants. The genotypes of the haploid progeny were determined by replica-plating to agar plates containing geneticin, clonNAT, or both.
Tests of mutational synergy with ynr004wΔ(swm2Δ)
Haploid ynr004w∷kanMX (swm2Δ) cells were mixed with mud2∷natMX or nam8∷natMX strains of opposite mating type, and diploids were selected on YPD agar containing clonNAT and geneticin. The heterozygous diploids were sporulated; asci were dissected and germinated on YPD agar. The genotypes of the haploid progeny were determined by replica-plating to agar plates containing geneticin, clonNAT, or both.
To establish a plasmid shuffle assay for complementation of the mud2Δ swm2Δ synthetic growth defect, we transformed the MUD2 mud2∷natMX SWM2 swm2∷kanMX diploid and with p360-SWM2, a CEN URA3 plasmid with SWM2 under the control of the TPI1 promoter. The diploids were sporulated, asci were dissected, and the desired haploid swm2Δ mud2Δ Ura+ progeny were recovered that: (1) were resistant to clonNAT and geneticin; and (2) formed tiny colonies on medium containing FOA.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM52470. S.S. is an American Cancer Society Research Professor.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2082610.
REFERENCES
- Abovich N, Rosbash M 1997. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89: 403–412 [DOI] [PubMed] [Google Scholar]
- Abovich N, Liao XC, Rosbash M 1994. The yeast MUD2 protein: An interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes & Dev 8: 843–854 [DOI] [PubMed] [Google Scholar]
- Benarroch D, Qiu ZR, Schwer B, Shuman S 2009. Characterization of a mimivirus RNA cap guanine-N2 methyltransferase. RNA 15: 666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benarroch D, Jankowska-Anyszka M, Stepinski J, Darzynkiewicz E, Shuman S 2010. Cap analog substrates reveal three clades of cap guanine-N2 methyltransferases with distinct methyl acceptor specificities. RNA 16: 211–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund JA, Abovich N, Rosbash M 1998. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes & Dev 12: 858–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch H, Reddy R, Rothblum L, Choi YC 1982. SnRNAs, SnRNPs, and RNA processing. Annu Rev Biochem 51: 617–654 [DOI] [PubMed] [Google Scholar]
- Calero G, Wilson KF, Ly T, Rios-Steiner JL, Clardy JC, Cerione RA 2002. Structural basis of m7GpppG binding to the nuclear cap-binding complex. Nat Struct Biol 9: 912–917 [DOI] [PubMed] [Google Scholar]
- Gottschalk A, Tang J, Puig O, Salgado J, Neubauer G, Colot HV, Mann M, Séraphin B, Rosbash M, Lührmann R, et al. 1998. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA 4: 374–393 [PMC free article] [PubMed] [Google Scholar]
- Hausmann S, Shuman S 2005a. Specificity and mechanism of RNA cap guanine-N2 methyltransferase (Tgs1). J Biol Chem 280: 4021–4024 [DOI] [PubMed] [Google Scholar]
- Hausmann S, Shuman S 2005b. Giardia lamblia RNA cap guanine-N2 methyltransferase (Tgs2). J Biol Chem 280: 32101–32106 [DOI] [PubMed] [Google Scholar]
- Hausmann S, Ramirez A, Schneider S, Schwer B, Shuman S 2007. Biochemical and genetic analysis of RNA cap guanine-N2 methyltransferases from Giardia lamblia and Schizosaccharomyces pombe. Nucleic Acids Res 35: 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann S, Zheng S, Costanzo M, Brost RL, Garcin D, Boone C, Shuman S, Schwer B 2008. Genetic and biochemical analysis of yeast and human cap trimethylguanosine synthase: Functional overlap of TMG caps, snRNP components, pre-mRNA splicing factors, and RNA decay pathways. J Biol Chem 283: 31706–31718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PC, Hodel MR, Thomas JW, Taylor LJ, Hagedorn CH, Hodel AE 2000. Structural requirements for the specific recognition of an m7G mRNA cap. Biochemistry 39: 13730–13736 [DOI] [PubMed] [Google Scholar]
- Hu G, Oguro A, Li C, Gershon PD, Quiocho FA 2002. The ‘cap-binding’ slot of an mRNA cap-binding protein: Quantitative effects of aromatic side chain choice in the double-stacking sandwich with cap. Biochemistry 41: 7677–7687 [DOI] [PubMed] [Google Scholar]
- Kielkopf CL, Lücke S, Green MR 2004. U2AF homology motifs: Protein recognition in the RRM world. Genes & Dev 18: 1513–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komonyi O, Papai G, Enunlu I, Muratoglu S, Pankotai T, Kopitova D, Maróy P, Udvarty A, Boros I 2005. DTL, the Drosophila homolog of PIMT/Tgs1 nuclear receptor coactivator-interacting protein/RNA methyltransferase, has an essential role in development. J Biol Chem 280: 12397–12404 [DOI] [PubMed] [Google Scholar]
- Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonné R, Lührmann R 2006. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol Biol Cell 17: 3221–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao XC, Tang J, Rosbash M 1993. An enhancer screen identifies a gene that encodes the yeast U1 snRNP A protein: Implications for snRNP protein function in pre-mRNA splicing. Genes & Dev 7: 419–428 [DOI] [PubMed] [Google Scholar]
- Mattaj IW 1986. Cap trimethylation of U snRNA is cytoplasmic and dependent of U snRNP protein binding. Cell 46: 905–911 [DOI] [PubMed] [Google Scholar]
- Mazza C, Segref A, Mattaj IW, Cusack S 2002. Large-scale induced fit recognition of an m7GpppG cap analogue by the human nuclear cap-binding complex. EMBO J 21: 5548–5557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters DS, Muhlenkamp P 2003. Spatial organization of protein–RNA interactions in the branch site-3′ splice site region during pre-mRNA splicing in yeast. Mol Cell Biol 23: 4174–4186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecozzi S, West AP, Dougherty DA 1996. Cation-π interactions in aromatics of biological and medicinal interest: Electrostatic potential surfaces as a useful qualitative guide. Proc Natl Acad Sci 93: 10566–10571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monecke T, Schell S, Dickmanns, Ficner R 2008. Crystal structure of the RRM domain of poly(A)-specific ribonuclease reveals a novel m7G-cap-binding mode. J Mol Biol 382: 827–834 [DOI] [PubMed] [Google Scholar]
- Monecke T, Dickmanns, Ficner R 2009. Structural basis for m7G-cap hypermethylation of small nuclear, small nucleolar and telomerase RNA by the dimethyltransferase TGS1. Nucleic Acids Res 37: 3865–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouaikel J, Verheggen C, Bertrand E, Tazi J, Bordonné R 2002. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol Cell 9: 891–901 [DOI] [PubMed] [Google Scholar]
- Mouaikel J, Bujnicki JM, Tazi J, Bordonné R 2003. Sequence-structure-function relationships of Tgs1, the yeast snRNA/snoRNA cap hypermethylase. Nucleic Acids Res 31: 4899–4909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Suzuki S, Enod R, Shirouzu M, Terada T, Inoue M, Kigawa T, Kobayashi N, Günert P, Tanaka A, et al. 2008. The RRM domain of poly(A)-specific ribonuclease has a noncanonical binding site of mRNA cap analog recognition. Nucleic Acids Res 36: 4754–4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessel G, Fischer U, Lührmann R 1994. m3G cap hypermethylation of U1 small nuclear ribonucleoprotein (snRNP) in vitro: Evidence that the U1 small nuclear RNA-(guanosine-N2)-methyltransferase is a non-snRNP protein that requires a binding site on the Sm core domain. Mol Cell Biol 14: 4160–4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rain JC, Rafi Z, Legrain P, Krämer A 1998. Conservation of functional domains involved in RNA binding and protein–protein interactions in human and Saccharomyces cerevisiae pre-mRNA splicing factor SF1. RNA 4: 551–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz B, Seraphin B 1999. Transient interaction of BBP/ScSF1 and Mud2 with the splicing machinery affects the kinetics of spliceosome assembly. RNA 5: 819–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selenko P, Gregorovic G, Sprangers R, Stier G, Rhani Z, Krämer A, Sattler M 2003. Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/BBP. Mol Cell 11: 965–976 [DOI] [PubMed] [Google Scholar]
- Sickmier EA, Frato KE, Shen H, Paranawithana SR, Green MR, Kielkopf CL 2006. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol Cell 23: 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Barbosa A, Louly C, Franco OL, Rubio MA, Alfonzo JD, Johnson PJ 2008. The divergent eukaryote Trichomonas vaginalis has an m7G cap methyltransferase capable of a single N2 methylation. Nucleic Acids Res 36: 6848–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, et al. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368 [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. 2000. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627 [DOI] [PubMed] [Google Scholar]
- Volckaert G, Voet M, Van der Schueren J, Robben J, Vanstreels E 2003. Disruption of 12 ORFs located on chromosomes IV, VIIA, and XIV of Saccharomyces cerevisiae reveals two essential genes. Yeast 20: 79–88 [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhang L, Lynn B, Rymond BC 2008. A BBP-Mud2p heterodimer mediates branchpoint recognition and influences splicing substrate abundance in budding yeast. Nucleic Acids Res 36: 2787–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, Collins SR, Whitworth GB, Kress TL, Weissman JS, et al. 2008. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol Cell 32: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Nilsson P, Henriksson N, Niedzwiecka A, Lim MK, Cheng Z, Kokkoris K, Virtanen A, Song H 2009. Structural basis of m7GpppG binding to poly(A)-specific ribonuclease. Structure 17: 276–286 [DOI] [PubMed] [Google Scholar]
- Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, et al. 2008. High-quality binary protein interaction map of the yeast interactome network. Science 322: 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]