Abstract
Viral small interfering RNA (siRNA) accumulation in plants is reported to exhibit a strong strand polarity bias, with plus (+) strand siRNAs dominating over minus (−) strand populations. This is of particular interest, as siRNAs processed from double-stranded RNA would be expected to accumulate equivalent amounts of both species. Here, we show that, as reported, (−) strand viral siRNAs are detected at much lower levels than (+) strand-derived species using standard Northern hybridization approaches. However, when total RNA is spiked with in vitro-transcribed antisense viral genomic RNA, (−) strand viral siRNAs are detected at increased levels equivalent to those of (+) strand siRNA. Our results suggest that (+) and (−) strand viral siRNAs accumulate to equivalent levels; however, a proportion of the (−) strand siRNAs are sequestered from the total detectable small RNA population during gel electrophoresis by hybridizing to the high-molecular-weight sense strand viral genomic RNA. Our findings provide a plausible explanation for the observed strand bias of viral siRNA accumulation, and could have wider implications in the analysis of both viral and nonviral small RNA accumulation.
Keywords: virus, satellite RNA, siRNA, small RNA, RNA silencing
INTRODUCTION
RNA silencing is an adaptive immune response of plants against viruses, with viruses being both an inducer and a target of RNA silencing. Infection with all types of plant viruses and subviral agents, including DNA and RNA viruses, viroids, and viral satellites, is associated with viral-derived small interfering RNA (siRNA) accumulation. Viral siRNAs are 21–25 nucleotides (nt) in length and are processed from viral-derived double-stranded RNA (dsRNA) molecules by Dicer-like (DCL) proteins (Ding and Voinnet 2007). The mature viral siRNAs are bound by an Argonaute (AGO) protein to form the RNA-induced silencing complex (RISC), which in turn uses the viral siRNA to guide the cleavage of complementary viral RNAs. To evade RNA-silencing mechanisms of their host(s), viruses have in turn evolved counterdefense mechanisms, such as expressing RNA-silencing suppressor proteins or adopting stable RNA secondary structures that are resistant to RNA-silencing-mediated degradation (Wang et al. 2004; Ding and Voinnet 2007). The majority of plant viruses and subviral RNAs have a positive-sense, single-stranded, RNA genome. Replication of these positive-strand viral and subviral agents involves the synthesis of complementary minus (−) strand intermediates from the parental RNA template followed by synthesis of nascent plus (+) strands by viral-encoded RNA-dependent RNA polymerases (Buck 1996; Ahlquist 2002). The (+) and (−) strand RNAs accumulate in an asymmetric ratio, ranging from 10:1 to 1000:1 (Buck 1996).
Recent studies based on deep sequencing and gel-blot hybridization analyses have shown that the accumulation of siRNAs from positive-strand RNA viruses in plants also exhibit a strand polarity bias; siRNAs of the (+) strand almost always dominate the detectable siRNA population over those derived from the (−) strand. Plus-strand siRNAs typically account for 60%–80% of the total small RNA (sRNA) population (Ding and Voinnet 2007; Donaire et al. 2008; Qi et al. 2009). This strand bias appears counterintuitive, as siRNAs, if processed from dsRNA, should contain relatively equivalent amounts of both sense and antisense species. To account for this strand bias, a model has been proposed in which regions of single-stranded viral genomic RNA form partially double-stranded secondary structures that are processed by a DCL into siRNAs (Molnár et al. 2005; Ding and Voinnet 2007). This model would account for the more abundant accumulation of (+) strand siRNAs, as (+) strand viral genomic RNAs are present at a much higher level than the (−) strand in virus-infected tissue (Buck 1996). However, recent studies with several positive-strand RNA viruses have shown that viral siRNAs are produced from all regions of the viral genome (e.g., Qi et al. 2009). In addition, these studies demonstrate that there is no clear correlation between siRNA-rich regions (known as “hot spots”) and local secondary structures (e.g., Donaire et al. 2008; Qi et al. 2009), which suggests that secondary structures are not a major determinant of viral siRNA biogenesis. Furthermore, the accumulation of siRNAs from several RNA viruses, and from a viroid, has been shown to be highly dependent on the activity of host-encoded RNA-dependent RNA polymerases (RDRs) (Diaz-Pendon et al. 2007; Donaire et al. 2008; Gómez et al. 2008; Qi et al. 2009). This RDR dependence suggests that dsRNAs synthesized by host RDRs from single-stranded (ss) viral RNA, rather than local secondary structures, are the precursor of viral siRNA biogenesis.
Here, we have examined whether the observed strand bias is, in fact, due to unequal detection of (+) and (−) strand viral siRNAs during the routinely used polyacrylamide gel electrophoresis step of cloning or hybridization-based detection of small RNAs. We provide strong evidence that the under-representation of (−) strand viral siRNAs is due to the sequestration of these small RNAs to complementary high-molecular-weight (+) strand viral RNAs during gel electrophoresis. Our findings could have wider implications in the interpretation of results generated in future studies of both viral and nonviral small RNA accumulation.
RESULTS
The presence of complementary high-molecular-weight RNA reduces the detection of sRNAs
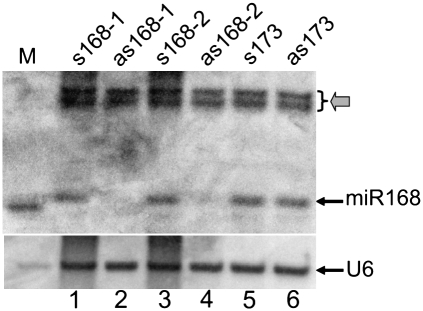
To examine whether sRNA detection, involving standard denaturing polyacrylamide gel electrophoresis (PAGE) and nucleic acid hybridization analyses, would be affected by the presence of complementary high-molecular-weight (HMW) RNAs that have the potential to hybridize to the sRNA species, we performed PAGE and subsequent sRNA Northern blot hybridization assays to detect the presence of an endogenous microRNA (miRNA), namely, miR168. Total RNA was precipitated from Arabidopsis thaliana plant extracts that were spiked with sense or antisense in vitro-transcribed primary transcripts of miR168 (pri-miR168). The RNA was resuspended in 100% formamide and then size separated on 17% polyacrylamide gels. As shown in Figure 1, miR168 was readily detectable in samples spiked with sense pri-miR168 transcripts (s168-1 is a full-length pri-miR168 transcript, whereas s168-2 is a truncated version that also contained the miR168 sequence) (see Fig. 1, lanes 1,3). In contrast, miR168 was almost undetectable in the presence of the antisense version of the pri-miR168 transcript (Fig. 1, lanes 2,4). The intensities of the two larger nonspecific bands (indicated by the gray arrow) were not affected by the addition of antisense transcripts, indicating that the reduced miR168 signals in Figure 1, lanes 2 and 4, were not caused by RNA degradation. Furthermore, the detection of miR168 was not affected by the presence of a sense or an antisense primary transcript of an unrelated miRNA, namely miR173 (Fig. 1, lanes 5,6). This result indicated that the mature miR168 may have become bound to complementary antisense pri-miR168 transcripts during PAGE, thus sequestering this sequence from the detectable miRNA signal. This result also suggested that the standard urea-based denaturing conditions used in this assay were insufficient to fully separate 21-nt small RNAs from their complementary sequences.
FIGURE 1.
Presence of in vitro antisense RNA of the miR168 primary transcript interferes with detection of mature miR168. Twenty micrograms of total RNA were loaded per sample. The membrane was hybridized with a 21-nt DNA oligonucleotide probe complementary to the mature Arabidopsis miR168. (M) A labeled 21-nt RNA oligonucleotide size maker. The “s” and “as” prefixes denote in vitro sense and antisense transcripts of either the miR168 or miR173 primary transcript, respectively. The gray arrow indicates two nonspecific bands that do not vary in intensity among the samples.
The (−) strand siRNAs from cucumber mosaic virus Y satellite (Y-Sat) are sequestered away by the (+) strand genomic RNAs during PAGE
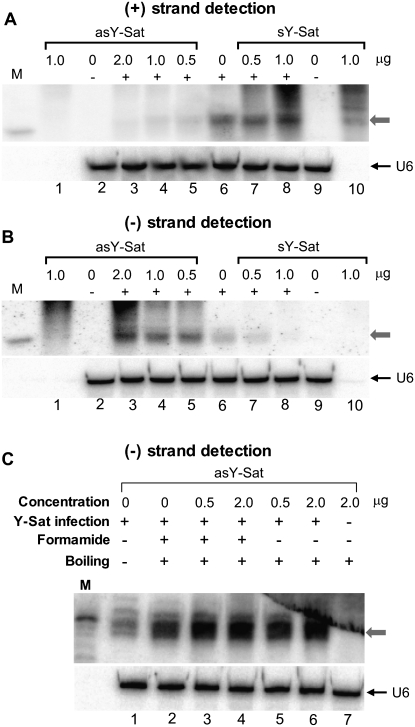
To examine whether underrepresentation of detectable (−) strand siRNAs was due to their sequestering by HMW (+) strand genomic RNAs, we spiked total RNA samples isolated from Y-Sat-infected tobacco plants with either sense (sY-Sat) or antisense (asY-Sat) in vitro Y-Sat transcripts. Total RNA samples were boiled in water, then mixed with formamide and immediately separated on 17% denaturing PAGE gels. As shown in Figure 2A, the addition of sense Y-Sat (sY-Sat) in vitro transcripts caused no dramatic increases in the levels of detectable (+) strand siRNAs (Fig. 2A, lanes 6–8); the stronger siRNA signal in Figure 2A, lane 8, is likely to result from the residual degradation product of the sY-Sat transcript (see Fig. 2A, lane 10). This result indicated that the (+) strand Y-Sat siRNAs were not significantly sequestered, presumably because the (−) strand HMW Y-Sat RNAs were bound to the highly abundant (+) strand HMW Y-Sat RNAs and therefore not available to hybridize with the (+) strand siRNAs. However, when the samples were spiked with 0.5–2 μg of in vitro antisense Y-Sat (asY-Sat) transcripts, (+) strand YSat siRNAs became almost undetectable (Fig. 2A, lanes 3–5), indicating that (+) strand viral siRNAs can also be sequestered when (−) strand HMV viral RNAs are present in excessive amounts.
FIGURE 2.
Presence of in vitro antisense Y-Sat transcript improves the detection of (−) strand Y-Sat siRNAs. (A,B) Detection of (+) strand (A) or (−) strand (B) Y-Sat siRNAs in the presence or absence of an in vitro–transcribed antisense (asY-Sat) or sense (sY-Sat) Y-Sat transcript. The amount of in vitro transcript added to each sample is given at the top of each lane. The “+” symbol indicates that RNA samples were extracted from Y-Sat-infected tobacco, while the “−” symbol indicates RNA from uninfected tobacco plants. (Lanes 1,10) In vitro transcripts only. The gray arrows indicate the position of siRNAs. (C) Detection of (−) strand Y-Sat siRNAs in the presence or absence of in vitro antisense Y-Sat transcript. The samples were treated differently prior to loading: (lane 1) total RNA dissolved in water and mixed with an equal volume of formamide loading buffer with no heating; (lanes 2–4) total RNA samples were dissolved in formamide, heated in boiling water for 5 min, and chilled on ice for 5 min; (lanes 5,6) total RNA samples were dissolved in water, boiled for 5 min, then mixed with an equal volume of 100% formamide loading buffer. Five micrograms of total RNA was loaded for each sample, except lanes 1 and 10 in A and B, which contain only the in vitro transcripts. (M) 21-nt radiolabeled RNA size marker.
In contrast to the (+) strand siRNAs, the detection of (−) strand Y-Sat siRNAs were markedly improved in the presence of increasing amounts of the asY-Sat in vitro transcript (Fig. 2B, lanes 3–6). This indicated that a large proportion of (−) strand Y-Sat siRNAs are normally sequestered to HMW (+) strand Y-Sat RNAs, but are released in the presence of the antisense in vitro transcript.
Different treatments of RNA preparations prior to their loading onto PAGE gels could affect the level of siRNA sequestration. To investigate this, the extracted RNA was subjected to the following treatments prior to loading: (1) boiling in 100% formamide for 5 min, then immediately chilling on ice for an additional 5 min, (2) boiling in 100% formamide for 5 min in the presence of the asY-Sat in vitro transcript, then chilling on ice for 5min, and (3) boiling in water for 5 min in the presence of asY-Sat in vitro transcript, then mixing with an equal volume of formamide loading buffer. These individual treatments demonstrated that boiling of the extracted RNA sample in formamide prior to loading increased the detection of the (−) strand Y-Sat siRNAs (Fig. 2C, cf. lane 2 and lane 1). However, boiling the RNA in the presence of the asY-Sat in vitro transcript further improved the detection of (−) strand Y-Sat siRNAs (Fig. 2C, cf. lanes 3–6 and lane 2), indicating that siRNA sequestration by HMW viral RNAs was not prevented by the boiling pretreatment. Boiling the RNA extract in water, as opposed to 100% formamide, in the presence of asY-Sat in vitro transcript, resulted in a similar level of detection of (−) strand Y-Sat siRNAs (Fig. 2C, cf. lanes 5,6 and lanes 3,4).
Minus (−) strand Y-Sat siRNAs are present at similar levels to (+) strand Y-Sat siRNAs in infected tobacco
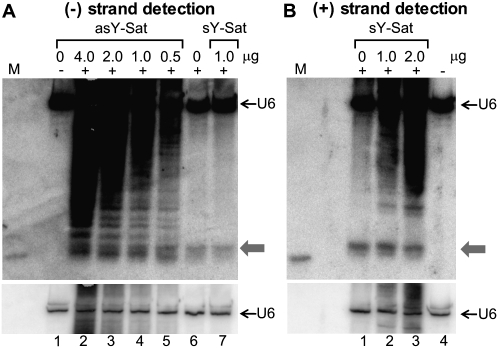
The experimental data presented in Figure 2 indicated that the (−) strand Y-Sat siRNAs are underrepresented using standard PAGE-based detection procedures, which is consistent with a recent report showing that 65% of Y-Sat siRNAs are (+) strand derived (Ebhardt and Unrau 2009). To compare the abundance of (+) and (−) strand Y-Sat-derived siRNAs in the presence of in vitro Y-Sat transcripts, we designed two chimeric oligonucleotide probes containing a 31-nt sequence complementary to the U6 small nuclear RNA fused with a 40-nt sequence of either the sense or antisense strand of the Y-Sat genome (Supplemental Data 1). These oligonucleotide probes were predicted to hybridize to both U6 RNA and Y-Sat-derived siRNAs on the same Northern blot filter, allowing for comparison of (+) to (−) strand Y-Sat siRNA abundance across two individual gels, by using the U6 signal as a normalization reference. The selected 40-nt Y-Sat sequence corresponds to a region (nucleotides 111–150) previously shown to have strong siRNA strand bias, with 21 (+) strand siRNAs compared with three (−) strand siRNAs (Ebhardt and Unrau 2009).
Figure 3 shows that (−) strand Y-Sat siRNAs were detected at much lower levels than (+) strand-derived siRNAs without the introduction of in vitro transcripts (Fig. 3, cf. panel A, lane 6, and panel B, lane 1). However, in the presence of greater than 1 μg of in vitro asY-Sat transcript, (−) strand siRNAs were detected at levels equivalent to that of (+) strand siRNAs (Fig. 3, cf. panel A, lanes 2–4, and panel B, lanes 1–3). This result is in sharp contrast to the previously reported sequence data suggesting a 7:1 ratio between (+) and (−) siRNAs derived from this region (Ebhardt and Unrau 2009). Again, the detection of (+) strand siRNAs was not significantly affected by adding the sY-Sat transcript (Fig. 3B, cf. lane 1 and lanes 2,3). Taken together, these results strongly suggested that (−) strand Y-Sat siRNAs accumulate at a similar abundance to (+) strand siRNAs; however, a large proportion of these (−) strand siRNAs are sequestered to the HMW (+) strand Y-Sat RNAs during PAGE separation.
FIGURE 3.
(−) Strand Y-Sat siRNAs are detected at similar levels as (+) strand siRNAs in the presence of in vitro antisense Y-Sat transcript. (A) Detection of (−) strand Y-Sat siRNAs in the presence (lanes 2–5) or absence (lanes 6,7) of in vitro antisense Y-Sat transcript. (B) Detection of (+) strand Y-Sat siRNAs in the presence (lanes 2,3) or absence (lane 1) of an in vitro–transcribed sense Y-Sat transcript. Blotted membranes were hybridized with an oligonucleotide probe containing a 31-nt U6 RNA sequence fused to either a 40-nt sense (A) or antisense (B) Y-Sat sequence (Supplemental Data 1). (Bottom) Shorter exposure of the U6 RNA signal in the top panels to indicate even loading. Ten micrograms of total RNA were loaded for each sample. The gray arrows indicate the position of the 21-nt siRNA species.
Minus (−) strand siRNAs from cucumber mosaic virus (CMV) are also sequestered by (+) strand CMV genomic RNA
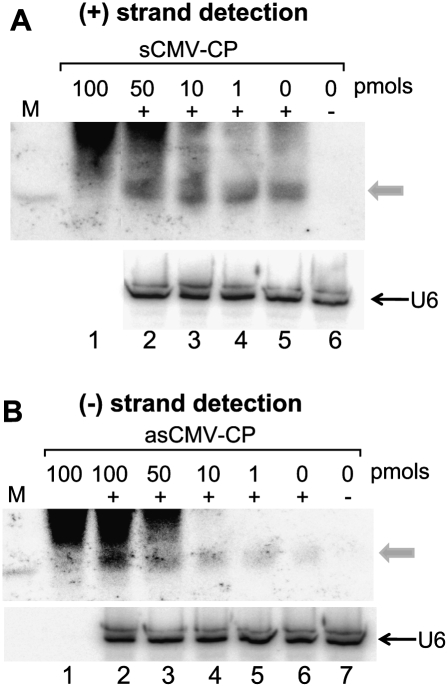
The abundance of genomic RNAs from an infecting virus is generally lower than that of a satellite RNA (e.g., Takanami 1981), so the sequestering of (−) strand siRNAs might not be as significant as that observed for Y-Sat siRNAs. To investigate whether the detection of (−) strand siRNAs from RNA viruses could also be reduced by sequestration, we isolated total RNA from tobacco plants infected with a satellite RNA-free CMV isolate, and performed Northern blot hybridization using a RNA probe specific for the coat protein (CP) sequence of CMV. In this experiment we spiked total RNA samples with five phased sense or antisense DNA oligonucleotides covering a 392-nt segment of the CMV CP sequence (Supplemental Data 2; Supplemental Data 3). As expected, CMV-derived siRNAs were more difficult to detect than Y-Sat siRNAs, presumably due to the relatively low abundance of viral RNAs in infected tissue. Nevertheless, as with Y-Sat siRNAs, the detection of (+) strand CMV siRNAs was not significantly affected by the presence or absence of the five sense CMV CP oligos (Fig. 4A). In contrast, however, the detection of (−) strand CMV siRNAs was enhanced in the presence of increasing amounts of the five antisense CMV CP oligos (Fig. 4B, cf. lane 6 and lanes 2–5). This result indicated that a proportion of the (−) strand siRNAs from CMV remained bound to HMW (+) strand viral RNAs during standard PAGE separation.
FIGURE 4.
Presence of antisense CMV-CP oligonucleotides improves the detection of (−) strand CMV-CP siRNAs. (A) Total RNA (20 μg) mixed with increasing amounts of five phased 72- or 80-nt sense CMV-CP oligonucleotides (Supplemental Data 2) and hybridized to antisense CMV-CP RNA. (B) Total RNA (20 μg) mixed with increasing amounts of five phased 72- or 80-nt antisense CMV-CP oligonucleotides (Supplemental Data 2) and hybridized to sense CMV-CP RNA. The gray arrows indicate the position of siRNAs.
DISCUSSION
In this study we demonstrate that (−) strand Y-Sat and CMV siRNAs are sequestered from the total detectable sRNA population during polyacrylamide gel electrophoresis. These (−) strand siRNAs were released in the presence of an in vitro–transcribed antisense viral transcript or a synthetic antisense DNA oligonucleotide. This demonstrates that (−) strand siRNAs are sequestered via their binding to complementary HMW (+) strand RNAs, but this species of sRNA can be displaced through the addition of in vitro antisense (−) molecules. Through the addition of complementary HMW in vitro transcripts, we demonstrate that (−) strand Y-Sat siRNAs could be detected at levels equivalent to (+) strand siRNAs. Taken together, these results suggest that (+) and (−) strand viral-derived siRNAs accumulate to equivalent ratios in infected plants, and that the under-representation of (−) strand siRNAs reported in previous studies is due to the sequestering of this siRNA population by complementary HMW viral genomic RNAs.
Our finding provides a plausible alternative to the previously proposed, local secondary structure-based, siRNA biogenesis model (Molnár et al. 2005; Ding and Voinnet 2007), to explain the strand polarity bias of viral siRNA accumulation. The results presented in this study would also have implications for the development of improved viral siRNA detection and sequencing strategies, as well as inviting a reinterpretation of the existing viral siRNA detection data from previous studies. Our results may also have implications for the analysis of nonviral sRNAs in plants as well as in other organisms. The majority of methods currently used for cloning and sequencing of sRNA involve separation of this population from HMW RNAs using denaturing PAGE (Lu et al. 2007). Our results indicate that the denaturing conditions adopted in standard PAGE are not sufficient to fully separate small RNAs from their complementary HMW RNAs. Furthermore, in some protocols, sRNAs are first enriched by removal of HMW RNA through the preferential precipitation of this larger nucleic acid species prior to further purification using PAGE (e.g., Lu et al. 2007). This HMW RNA precipitation is carried out at low temperatures that are favorable for RNA–RNA hybridization, and thereby could potentially result in the loss of many sRNA species that are sequestered by these HMW molecules. Thus, sequestering of sRNAs by complementary HMW RNA could occur for many endogenous sRNA species, leading to the generation of biased sequencing results. Consequently, sRNAs with relatively abundant target transcripts could be underrepresented in sequenced populations, resulting in misinterpretation of sRNA expression in vivo.
MATERIALS AND METHODS
Isolation of RNA
Tobacco plants (Nicotiana tobacum Wisconsin 38) were grown in the glasshouse (25°C) with natural light for 2 wk. These plants were inoculated with leaf extracts from tobacco plants infected with a Cucumber mosaic virus (CMV) isolate with or without the Y-satellite (Y-Sat) RNA (Wang et al. 2004). Total RNA was isolated from 100 mg of tobacco leaves 4 wk post-inoculation using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Isopropanol precipitation of the total RNA was carried out overnight at −20°C to maximize recovery of the small RNA fraction.
Sample treatments, polyacrylamide gel electrophoresis (PAGE), and Northern blot hybridization
Total RNA (5–20 μg) was mixed with in vitro-transcribed sense or antisense transcripts of Y-Sat (sY-Sat and asY-Sat, respectively), or with synthetic sense or antisense oligonucleotides corresponding to the CMV CP coding sequence (Supplemental Data 2), in a total volume of 15 μL (nuclease-free water or 100% formamide). Two micrograms of in vitro pri-miR168 transcript were added to plant extracts from 12 mg of leaf tissue before phenol-chloroform extraction and total RNA precipitation (Fig. 1). As specified in the figure legends, samples were boiled for 5 min, then immediately chilled on ice for 5 min, or cooled at room temperature. For samples dissolved in water, an equal volume of formamide loading buffer (98% formamide, 40 mM EDTA, 0.05 mg/mL bromophenol blue, 0.05 mg/mL xylene cyanol FF) was added prior to loading. Treated samples were loaded onto denaturing 17% polyacrylamide gels and run at 40 volts. RNA was electro blotted onto Hybond-N+ membrane (Amersham) in 0.5× TBE buffer, immobilized onto the membrane by UV cross-linking, and hybridized overnight at 42°C (Curtin et al. 2008) with either a [γ-32P]ATP end-labeled U6-YSat DNA oligonucleotide probe (Supplemental Data 1) or with an [α-32P]UTP-labeled RNA transcribed from linearized plasmids containing either the CMV Y-satellite (Wang et al. 2004) or CMV coat protein sequence (Supplemental Data 3) with the SP6 or T7 RNA polymerase. The hybridized membranes were washed three times with 2× SSC and 0.2% SDS for 20 min per wash at 42°C, and visualized on a PhosphorImager.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Chikara Masuta for providing the Y-Sat infectious clone used to start the original Y-Sat culture, and Peter Unrau, Tony Miller, and Liz Dennis for helpful discussions.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2049510.
REFERENCES
- Ahlquist P 2002. RNA-dependent RNA polymerase, viruses, and RNA silencing. Science 296: 1270–1273 [DOI] [PubMed] [Google Scholar]
- Buck KW 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv Virus Res 47: 159–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin SJ, Watson JM, Smith NA, Eamens AL, Blanchard CL, Waterhouse PM 2008. The roles of plant dsRNA-binding proteins in RNAi-like pathways. FEBS Lett 582: 2753–2760 [DOI] [PubMed] [Google Scholar]
- Diaz-Pendon JA, Li F, Li WX, Ding SW 2007. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 19: 2053–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SW, Voinnet O 2007. Antiviral immunity directed by small RNAs. Cell 130: 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaire L, Barajas D, Martínez-García B, Martínez-Priego L, Pagán I, Llave C 2008. Structural and genetic requirements for the biogenesis of Tobacco Rattle Virus-derived small interfering RNAs. J Virol 82: 5167–5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebhardt HA, Unrau PJ 2009. Characterizing multiple exogenous and endogenous small RNA populations in parallel with subfemtomolar sensitivity using a streptavidin gel-shift assay. RNA 15: 724–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez G, Martínez G, Pallás V 2008. Viroid-induced symptoms in Nicotiana benthamiana plants are dependent on RDR6 activity. Plant Physiol 148: 414–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Meyers BC, Green PJ 2007. Construction of small RNA cDNA libraries for deep sequencing. Methods 43: 110–117 [DOI] [PubMed] [Google Scholar]
- Molnár A, Csorba T, Lakatos L, Várallyay E, Lacomme C, Burgyán J 2005. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J Virol 79: 7812–7818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Bao FS, Xie Z 2009. Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA-dependent RNA polymerases in viral siRNA biogenesis. PLoS One 4: e4971. doi: 10.1371/journal.pone.0004971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanami Y 1981. A striking change in symptoms on cucumber mosaic-virus-infected tobacco plants induced by a satellite RNA. Virol 109: 120–126 [DOI] [PubMed] [Google Scholar]
- Wang MB, Bian XY, Wu LM, Liu LX, Smith NA, Isenegger D, Wu RM, Masuta C, Vance VB, Watson JM, et al. 2004. On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc Natl Acad Sci 101: 3275–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]