Abstract
6S RNA is an important noncoding RNA that regulates eubacterial transcription. In Escherichia coli this RNA binds to the σ70 RNA polymerase holoenzyme and is released by the synthesis of a short product RNA. In order to determine how binding and release are controlled by the 6S RNA sequence, we used in vitro selection to screen a high diversity library containing ∼4 × 1012 sequences for functional 6S RNA variants. Residues critical for binding were found to be located in a “-35” region upstream of the 6S RNA transcription bubble mimic structure. Mutating these phylogenetically conserved residues invariably led to decreases in binding and removing them abolished binding, implicating these nucleotides in a biologically important interaction with the Eσ70 complex. Interestingly, mutation of phylogenetically conserved “-10” residues that were also upstream of the site of pRNA synthesis was found to influence 6S RNA release rates in addition to modulating -35 binding. These results indicate how 6S RNA -35 binding to σ70 RNA polymerase holoenzyme can regulate expression from “strong” and “weak” -35 DNA promoters and suggest that 6S RNA release rates have been fine tuned over evolutionary time so as to correctly regulate cellular levels of transcription.
Keywords: 6S RNA, bacterial transcriptional regulation, in vitro selection, σ factors
INTRODUCTION
Escherichia coli 6S RNA was identified in the late 1960's as a highly abundant species in stationary phase cells and was one of the first RNAs to be sequenced (Hindley 1967; Brownlee 1971). This RNA is now known to regulate hundreds of σ70 (housekeeping)-dependent promoters in E. coli (Wassarman and Storz 2000; Trotochaud and Wassarman 2005; Cavanagh et al. 2008). While cells lacking 6S RNA are viable, they are unable to effectively compete with wild-type cells in low nutrient conditions (Trotochaud and Wassarman 2004; Wassarman and Saecker 2006). Critical insight into 6S RNA-dependent transcriptional regulation was obtained from studies demonstrating the ability of the 6S RNA to bind tightly to the RNA polymerase (RNAP) holoenzyme (Eσ70) and to be displaced from the holoenzyme when used as a template to synthesize a short product RNA (pRNA) in the presence of NTPs (Wassarman and Saecker 2006).
The 6S RNA is found across the eubacteria and folds into a well-conserved bulged hairpin structure that resembles an elongating DNA transcription complex (Barrick et al. 2005). A large internal DNA transcription bubble mimic (TBM) structure is intimately associated with the RNAP active site and pRNA synthesis is initiated from template nucleotide U44 (Wassarman and Saecker 2006) within this region. The 6S RNA contains three evolutionary conserved regions that are linked by regions of low sequence conservation: A downstream region that is utilized as a template during pRNA synthesis, and two upstream regions whose biological functions are unknown. These upstream islands of sequence conservation are named the “-35” and “-10” regions after Barrick et al. (2005), who noted by simple geometry that these regions loosely correspond to the location of the -35 and -10 DNA promoter elements in a DNA initiation complex. Consistent with this hypothesis, a series of protein truncation and mutation experiments have since demonstrated the importance of the σ704 subdomain, which has previously been shown to be responsible for -35 DNA promoter recognition in 6S RNA binding (Mooney et al. 2005; Lane and Darst 2006; Cavanagh et al. 2008; Klocko and Wassarman 2009).
Here we explore for the first time how the 6S RNA regulates transcription at the nucleotide level by using in vitro selection to isolate 6S RNA variants that are competent in binding and pRNA-induced release. We find that a tight web of residues located within the -35 region plays an essential role in 6S RNA binding to Eσ70. Residues downstream from this region serve to both modulate the intrinsic binding conferred by the -35 region and, more interestingly, influence pRNA-induced release rate. Significantly, release mutants were clustered in the -10 region and were never found in the -35 region, implying that at least part of the phylogenetic conservation observed in the -10 region must result from a requirement for the regulation of the 6S RNA release rate. Taken together, these results suggest a model where primary 6S RNA binding is achieved by the -35 region in combination with the -10 region, which both modulates this binding and regulates 6S RNA release. This model provides a natural mechanism for the 6S RNA to distinguish “strong” and “weak” -35 elements of σ70-dependent DNA promoters and suggests a process by which the 6S RNA can adjust overall levels of the free RNAP complex by appropriately calibrating the 6S RNA release rate.
RESULTS
Truncation analysis reveals essential binding and release elements
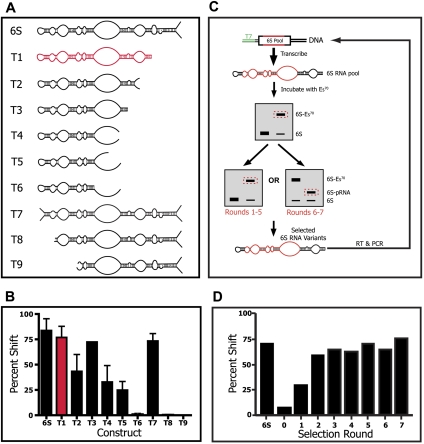
Important sequence elements of the 6S RNA required for binding and release from the holoenzyme complex appeared likely to be revealed by a truncation analysis. As an initial step, we therefore created a range of constructs that systematically truncated the 6S RNA from either end and tested their ability to bind Eσ70. Nine RNA constructs were tested in total. T1–T6 were truncated toward the closing stem of the 6S RNA, with constructs T4–T6 deleting into the central TBM (Fig. 1A). A nearly monotonic decrease in binding, as judged by a native gel shift assay, was observed in this progression, with binding being abolished only after complete removal of the top strand of the TBM (Fig. 1B). This suggested that the sequence downstream from the TBM was not critical for 6S RNA binding. Consistent with this hypothesis, constructs T8 and T9, which partially or completely removed the -35 region from its upstream terminal stem–loop, completely abolished binding, while merely opening the terminal loop structure (construct T7) had no observable effect on binding affinity (Fig. 1B). In all, these data indicated that the bulged stem structure upstream of the TBM plays a fundamental role in 6S RNA binding.
FIGURE 1.
The 6S RNA was truncated by varying amounts (T1–T9) in order to ascertain the minimum motif required for efficient binding to the Eσ70 complex. (A) Structure of the prepared constructs, relative to the 6S RNA sequence. (B) Binding of each construct to Eσ70 as inferred by native gel shift analysis. Highlighted in red is the construct chosen for in vitro selection studies. (C) Schematic of the in vitro selection. A DNA pool having a total diversity of 4 × 1012 sequences was constructed varying residues 42–88 and 104–143 at the level of 10% (residues shown in red, T7 RNAP promoter in green). An excess of RNA pool was incubated with Eσ70. RNA able to form a gel-shifted complex was isolated using two sequential native PAGE each round of selection. For Rounds 6 and 7 the RNA pool was first selected for binding with an excess of Eσ70 and then for its ability to be released from the holoenzyme by the synthesis of pRNA. (D) Fraction of the RNA pool bound to Eσ70 as a function of selective round and compared to the binding of wild-type 6S RNA.
As a second step, we set out to test the ability of these truncated constructs to synthesize pRNA. Truncated 6S RNA variants were briefly incubated with holoenzyme before adding NTPs and supplemental magnesium to induce pRNA synthesis as monitored by the incorporation of [γ-32P] ATP. The T1 and T2 constructs produced pRNA of a length similar to that produced by the 6S RNA control, while the T3 construct, which was truncated upstream of the normal site of pRNA termination, produced a shorter pRNA transcript (Supplemental Fig. S1). All of these constructs appeared to be correctly released from the holoenzyme, as judged by native gel shift analysis. Truncation into the TBM (T4–T6) resulted in a loss of detectable pRNA synthesis, likely due to major destabilization of the template strand. The T7 deletion construct, as expected, gave a pRNA product that was indistinguishable in length from the 6S RNA control. Surprisingly, the T8 construct, which did not bind to the holoenzyme (as judged by native gel analysis), produced a pRNA product that was the same length as the 6S RNA and T1 RNA constructs (Supplemental Fig. S1). The ability of this binding defective RNA to produce a pRNA suggests that the functions of binding and pRNA-induced release are, to a certain extent, independent of each other.
In vitro selection of functional 6S RNA variants
Our truncation experiments strongly implied that the upstream elements of the 6S RNA, as defined by the sequence in the TBM and the terminal loop regions, play a critical role in 6S RNA binding and that the downstream sequence, while essential for correct pRNA synthesis, is of lesser importance. We therefore decided to perform a selection to study how the sequence in the TBM and upstream stem–loop modulates 6S RNA binding and release.
The T1 deletion construct (Fig. 1A), which bound to and released from the holoenzyme in a manner nearly indistinguishable from the 6S RNA, was used as a basis for the pool design. A high diversity DNA pool was synthesized, chemically mutating nucleotide positions 42–88 and 104–143 of the 6S RNA to a level of ∼10% (Zaher and Unrau 2005). A DNA population containing ∼4 × 1012 distinct DNA sequences was then transcribed into RNA resulting in ∼10 RNA copies of each DNA sequence for the first selective round.
Figure 1C summarizes the in vitro selection protocol. During each round a 2.5-fold excess of the RNA pool was incubated with Eσ70 in the presence of heparin, a nonspecific competitor. Following a brief incubation, bound complexes were purified through a native gel, and a band corresponding to the RNA:holoenzyme construct was carefully excised. The resulting RNA was then incubated with a fresh aliquot of Eσ70 and purified for a second time prior to reverse transcription and PCR amplification (Fig. 1C, left selection path). Careful gel purification was critical, as we had previously noticed that random sequence RNA had a propensity to bind to the holoenzyme by an unknown mechanism that resulted in a characteristic gel shift mobility slightly faster than the 6S RNA containing complex. This nonspecific interaction could be observed in our initial RNA pool, but disappeared over the course of the selection (Supplemental Fig. S2).
After five rounds of selection binding activity appeared to plateau (Fig. 1D). At this point the focus of the selection was redirected in order to enrich the RNA pool in 6S RNA variants able to promote fast pRNA-induced release. In Rounds 6 and 7, the RNA population was first selected for binding using only a single native gel. The recovered RNA was then added to fresh Eσ70 as before, but in addition, magnesium and NTPs were added so as to induce pRNA-mediated 6S RNA release (see Materials and Methods). The resulting population was then purified on a second native gel, with a band corresponding to the RNA:pRNA hybrid complex being carefully excised instead of the RNA:holoenzyme complex (Fig. 1C, right selection path). Pool RNA was then reverse transcribed and PCR amplified. The Round 7 population of DNA, when transcribed into RNA, was found to efficiently bind and release from the holoenzyme.
-35 Region further implicated in binding
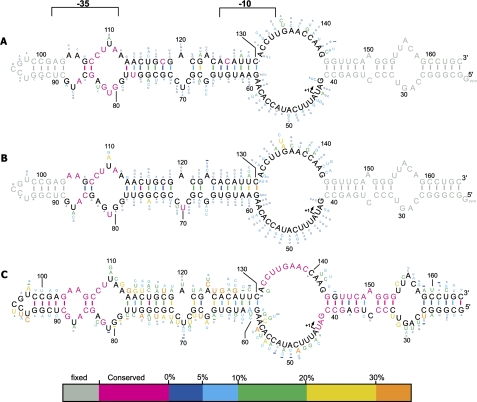
Isolates from Round 5 (binding) and Round 7 (binding and release) of the selection were cloned and sequenced. In total, 60 sequences from each round were aligned and analyzed (Fig. 2A,B; Supplemental Fig. S3). Within the sequenced isolates, absolute conservation was seen for 12 residues in the binding population and 10 residues in the binding and release population. Six of these residues were in common, and many of the conserved residues from each selection were densely clustered in the same immediate region of the terminal bulged loop -35 region (i.e., see residues 78–88 and 104–113), giving this region a very high statistical significance.
FIGURE 2.
6S RNA consensus sequences inferred by in vitro selection for binding and release compared with biological phylogeny. (A) Consensus structure based on 60 sequences from the end of the binding selection stage (Round 5). (B) Consensus structure based on 60 sequences from the end of the binding and release selection stage (Round 7). (C) The 6S RNA biological consensus showing only residues found in the T1 construct (see Fig. 1). This structure was derived from the 6S RNA sequence found in three orders of the gammaproteobacteria namely, the enterobacteriaceae, alteromonadales, and vibrionales (See Supplemental Fig. S3). Two phylogenetically conserved upstream regions are labeled -35 and -10, respectively, for their correspondence to the equivalent regions in a bound DNA promoter complex (Barrick et al. 2005). In all panels, primary sequence conservation and base-pair conservation is summarized by the color bar. Nucleotide numbering corresponds to full-length E. coli 6S RNA.
Examining the -35 region in more detail at the binding stage of the selection, we noticed that a short 2 base pair (bp) helix was absolutely conserved in the sequenced isolates (G84:C107, C85:G106), containing a single C85A:G106U covariation. Downstream, a U78:A113 base pair was absolutely conserved with a single U78C:A113G covariation observed in the 60 isolates sequenced. Extensive sequence conservation was also observed in single-stranded regions upstream of this base pair. In particular, a C108, U109, A111, G80, U81, and G82 constellation, found in the loop between the covarying stem structures and a single A86 residue found in the smaller loop immediately upstream of the 2-bp helix, appeared likely to be important for 6S RNA binding. After two rounds of selection for both binding and pRNA-induced release the previously observed pairing pattern was substantially maintained, but with a low level of U78:G113 wobble pairs and the infrequent formation of A:C pairs in the 2-bp stem. The conserved -35 region constellation of single-stranded residues shifted slightly, with G80 and G82 now having an R80 and R82 conservation pattern and A104 and A105 in the upstream loop now being absolutely conserved within the sequenced isolates. The conservation of these residues, from Round 5 to Round 7 of the selection, strongly implicates the -35 region in an important binding interaction with Eσ70.
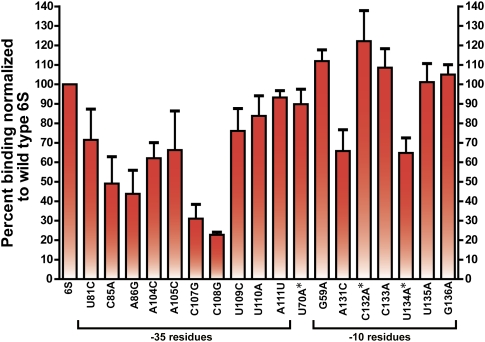
To test explicitly the importance of these -35 region residues in binding, individual 6S RNA point mutants were constructed for a range of residues and their binding efficiency was judged relative to the 6S RNA wild-type sequence. Notably, disrupting the 2-bp internal helix by either a C85A or a C107G mutation lowered binding efficiency by more than a factor of 2 in either case, implying that this short helix is important for correct docking to the protein complex (Fig. 3). Mutation of single-stranded residues found within the conserved constellation also resulted in marked decreases in binding. In particular C108G significantly lowered binding efficiency, while U81C, A86G, A104C, A105C, and U109C all had notable declines (Fig. 3). These point mutant experiments are entirely consistent with our in vitro selection results and truncation analysis, where complete loss of binding was observed after deletion through this region (Fig. 1A,B).
FIGURE 3.
Binding of full-length 6S RNA point mutants to Eσ70 as judged by native gel shift. Experiments were performed in triplicate and normalized to 6S RNA binding. Asterisks highlight release mutants. Residue numbers are as labeled in Figure 2 and correspond to E. coli 6S RNA numbering.
It was striking how well the sequence conservation observed in the -35 region during our selection agrees with the phylogenetic pattern of sequence conservation found in eubacteria closely related to E. coli (Fig. 2C; Supplemental Fig. S3). In this regard, we note that, in addition to the pattern of nucleotide conservation, there are also patterns of sequence variability that are preserved between the biological and in vitro systems. For example, U87 and U110 have very similar mutational patterns when biological variation is compared to our in vitro selection results (Fig. 2B,C). This detailed and shared pattern of conservation between biological and in vitro selected 6S RNA variants combined with the importance of residues in this region for in vitro binding (Figs. 1–3) implies that the -35 region has been biologically selected for optimal binding toward its protein target, just as in our in vitro selection. This protein target appears likely to involve the positively charged σ704.2 subdomain as deletion or mutation of this domain is known to ablate 6S RNA binding (Cavanagh et al. 2008). It is notable that the σ704 subdomain is highly conserved (Supplemental Fig. S4) consistent with the evolution of a single optimal RNA binding motif in the -35 region. It will be of considerable interest to map the interactions between the RNA residues in the -35 region and specific amino acids in the σ704.2 subdomain.
Sequence downstream from the -35 region modulates binding and 6S RNA release rate
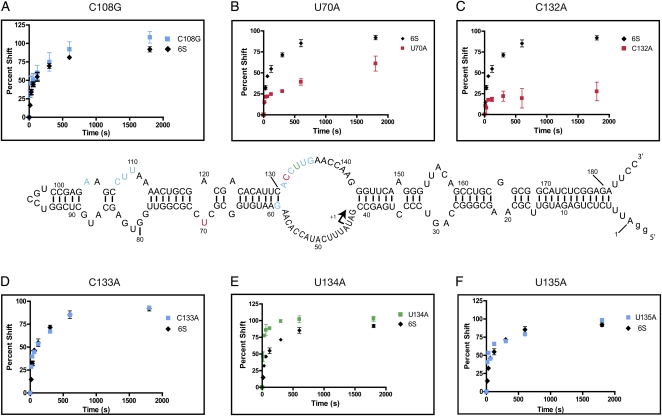
The remaining sequence element variations resulting from the selection were initially difficult to interpret, as nearly all residues outside of the -35 binding region appeared to have been under relatively modest selective pressure, with few positions being highly conserved in our in vitro selection. To make progress we initially focused on the U70A mutant, which was only slightly defective in binding (Fig. 3). This mutation was present in Round 5, but was sufficiently disfavored during the selection for rapid pRNA-induced release that by Round 7 all of the U70 variants were lost (i.e., absolute conservation was observed in the 60 analyzed sequences) (Fig. 2A,B). We reasoned therefore that this residue was likely to influence pRNA release rate. To this end, we developed a pRNA release assay where formation of 6S RNA:pRNA hybrids was tracked as a function of time by gel shift analysis (see Materials and Methods). We found that the U70A mutant showed a marked decrease in release rate relative to its parent (Fig. 4). As a slower release rate would not be favored in our selection, a simple explanation for the extinction of U70A emerged.
FIGURE 4.
The release kinetics of a subset of the 6S RNA point mutants tested for binding (Fig. 3). The central schematic summarizes the 12 point mutants tested. Blue indicates no significant change, red indicates a decreased release rate, and green indicates increased release rate relative to wild-type 6S RNA. The time courses of six point mutants are shown explicitly as insets: (A) C108G, (B) U70A, (C) C132A, note apparent failure to completely release for this mutant, (D) C133A, (E) U134A, and (F) U135A. Percent shift refers to the fraction of 6S:pRNA complex formed.
To explore the importance of RNA release rate further, we tested additional 6S RNA point mutants that were suggested by mutational frequency trends observed in the Round 5 and Round 7 sequence data (Fig. 2A,B). Mutants in the -35 binding region, such as A104C (mildly binding defective), C108G (strongly binding defective), and U109C (mildly binding defective), all released from the holoenzyme with normalized rate profiles indistinguishable from wild type (Fig. 4, blue residues; Supplemental Fig. S5), further reinforcing the idea that natural selection has selected for optimal binding alone in the -35 region and that mutational frequencies in this region were determined by their binding ability and not by their release phenotype.
Puzzled by the lack of in vitro sequence conservation in the -10 region relative to biological phylogeny (Fig. 2), we found a set of point mutants within this region that could either increase or decrease pRNA-induced release rate and also stabilize or destabilize binding. A U134A mutation that was not observed in Round 5 had accumulated significantly by Round 7, implicating this point mutant in fast release and/or a tight binding phenotype (Fig. 2A,B). This mutant, which was actually moderately defective in binding (Fig. 3), would have been disfavored during the binding selection and it could only have been enriched upon selection for its fast pRNA release phenotype (Fig. 4) during the last two rounds of selection. Likewise, a C132A mutation was found that increased only slowly in frequency from Round 5 to Round 7 (Fig. 2A,B), despite having a higher binding affinity than the 6S RNA wild-type sequence (Fig. 3). As a mutant with such tight binding would have been expected to rapidly accumulate, we hypothesized that this mutant must have a slow rate of release to offset this advantage. Consistent with our expectations, this mutant, when tested, exhibited a defective relative release profile—where even after prolonged incubation, less than 30% of the RNA had been released (Fig. 4). Other point mutants in this region had no detectable release phenotype (G59A, A131C, C133A, U135A, and G136A: Fig. 4, blue residues) and appear to have been enriched or depleted during the selection based largely on their binding phenotypes that were either superior or inferior, respectively, to wild-type binding (Figs. 2, 3). Thus, in contrast to the -35 region, which by all indications has been optimized for binding alone, downstream residues located in the -10 region have not been biologically selected to maximize or minimize either binding or release as demonstrated by our in vitro data. This basic finding has a number of important biological implications that will now be discussed.
DISCUSSION
It is a striking observation that the pattern of sequence conservation found in nature and in our in vitro selection match nearly exactly in the -35 region. As our in vitro selection optimized for binding affinity and as our truncation and mutation experiments demonstrate, this region is essential for 6S RNA binding. Together, these results implicate the phylogenetic pattern of conservation in the -35 region as being derived from a local optimization in binding affinity. If, as it seems likely, the -35 region interacts specifically with positively charged amino acid residues found in the σ704.2 subdomain, which are known to play an important role in 6S RNA binding and -35 DNA promoter recognition (Klocko and Wassarman 2009; Cavanagh et al. 2008), then this would naturally provide a mechanism for competitive binding (Cavanagh et al. 2008) between -35 DNA promoter elements and the -35 region of the 6S RNA for the σ704 domain. Thus, from this framework strong and weak -35 DNA promoter elements can, to a large extent, be defined by their binding affinities relative to that generated by the -35 binding domain.
An intriguing aspect of this regulatory system is that, so long as the -35 region binds optimally to the σ704.2 subdomain, the relative concept of strong and weak -35 promoter binding is unambiguously and stably defined by the 6S RNA and cannot easily drift with evolutionary time. This would not be the case if -35 binding was suboptimal. Such a situation would allow quick and presumably undesirable fluctuations in the regulation of weak promoters by the 6S RNA system and so is biologically selected against. Nevertheless, this primary binding interaction can be modulated by downstream residues, in particular those in the -10 region, that we have demonstrated can influence binding. This secondary pattern of binding might be expected to play an important role in regulating transcription based on -10 promoter affinity. This would be consistent with previous studies by Wassarman and Storz (2000), who demonstrated that extended -10 promoters can be suppressed by the 6S RNA regulatory system (Cavanagh et al. 2008).
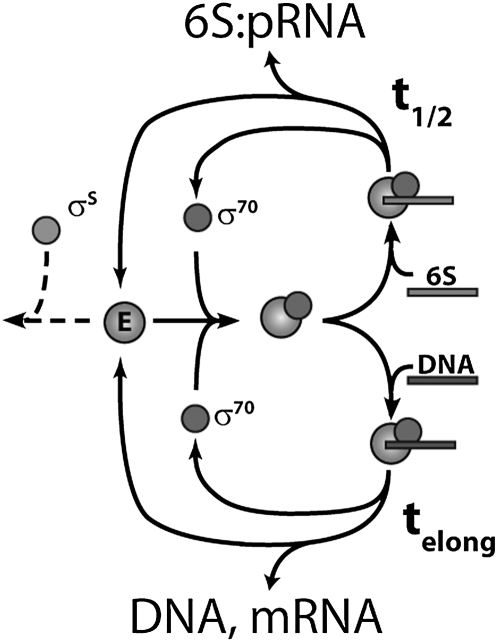
While competitive binding of the 6S RNA plays a large role in defining promoter strength in stationary phase, our findings suggest that pRNA-induced 6S RNA release rate may contribute an equally important function in dynamically regulating overall levels of transcription. In any given cellular condition the transcriptional state can be defined by specifying the concentrations of the various transcriptional components summarized in Figure 5. Competition between DNA promoters and 6S RNA results in free Eσ70 being committed either to DNA-dependent transcription (Fig. 5, lower cycle) or to pRNA synthesis using the 6S RNA as a template (Fig. 5, upper cycle). If we presume that elongated DNA and 6S RNA complexes have increased stability due to additional interactions with the polymerase catalytic site that are required for polymerization, then these complexes can serve to sequester core enzyme and potentially σ70, making these factors temporarily unavailable for transcription. This sequestration can be characterized by the time required for pRNA-induced release and DNA-dependent transcription termination to occur (t1/2, and telong respectively; Fig. 5). While telong should be highly variable and depend on the transcript length, t1/2 should have a well-defined value for any particular cellular condition.
FIGURE 5.
A model summarizing the two modes of 6S RNA-dependent regulation: 6S RNA binding affinity to Eσ70 and pRNA-induced release rate. Levels of free core enzyme (E, large circle) and free sigma factors (σ70 and σS shown) determine the formation rate of Eσ70 complex (superimposed circles). This complex is depleted by the formation of either a bound 6S RNA:Eσ70 complex (top path) or DNA:Eσ70 complex (bottom path). This competitive binding provides a 6S RNA-dependent mechanism to distinguish between strong and weak DNA promoters that depend on 6S RNA intrinsic binding affinity and cellular concentration. The characteristic time for pRNA-induced 6S RNA release (t1/2), which is sensitive to free magnesium and weakly to NTP levels, provides a mechanism to sequester core polymerase from use, effectively lowering the concentration of free polymerase (E) in the cell.
Motivated by this simple model we tried to find in vitro conditions (varying NTP and magnesium concentration), whereby 6S RNA release could be precluded, while still allowing DNA-dependent transcription to occur as suggested by Wassarman and Saecker (2006) and failed. This, together with our discovery of release mutants that enhance or compromise pRNA-induced release, indicated to us that 6S RNA release rate could play an important role in transcriptional regulation over a broad range of cellular conditions. Interestingly, 6S RNA release rate is relatively insensitive to NTP concentration, but is very sensitive to free magnesium levels, with rates of release varying over at least two orders of magnitude (Supplemental Fig. S6). If even a small fraction of this free magnesium range is biologically relevant, then transient contacts between nucleotides in the -10 region could well make important contacts with the β′ rudder and σ702 and σ703 subdomains (Mooney et al. 2005). As such protein–RNA contacts would be sensitive to RNA folding time scales, the dynamics of such interactions might be expected to change as a function of magnesium concentration. If so, then -10 residues, in addition to the ones found in our screen, remain to be found that serve to define pRNA release for a variety of physiologically relevant conditions not tested in our study. Encouragingly, an ongoing screen for upstream 6S RNA mutants that fail to release in the presence of NTPs and 4 mM free magnesium has found a sizable population of mutants that are defective in release, suggesting further that the -10 sequence plays a key role in defining the dynamics of biologically “optimal” pRNA-induced release (data not shown). Biologically then, optimal 6S RNA release rate can be seen as a mechanism to adjust the levels of cellular transcription in any particular cellular environment by sequestering core enzyme in a 6S RNA:Eσ70 complex for a characteristic time (t1/2) that is defined by cellular state and the exact sequence of the 6S RNA (Fig. 5).
In summary, we find evidence that the -10 region can influence two distinct regulatory roles of the 6S RNA. The first role is to modulate the intrinsic binding of the -35 region and consequently regulate promoters with extended -10 elements, while the second fine tunes overall levels of transcription by controlling 6S RNA release rates from the Eσ70 complex. The combination of these two complex biological functions defines an overall selective pressure that results in conservation of sequence in the -10 region. In addition to these two regulatory functions, we cannot preclude that the conserved -10 domain may serve additional biological roles. For example, phylogenetic similarities between the σS and σ70 might result in the natural selection of a -10 6S RNA sequence that specifically precludes EσS binding, but not Eσ70 (Fig. 5). We note that, in principle, any sequence constraints imposed by the presence of the σS can be directly examined by future in vitro experiments. Such discrimination, if found, would have a number of important implications for bacterial transcriptional control, which is under the control of a diverse set of evolutionarily related sigma factors.
Our findings have a number of important biological and medical implications. If, as we speculate, the 6S RNA has been selected to differentially regulate σ70 promoters, while modulating overall levels of transcription in a kinetic fashion, then it should be possible to adjust each of these regulatory parameters independently by expressing more than one 6S RNA. Notably some eubacteria, such as B. subtilis and Prochlorococcus, contain two distinct 6S RNAs (Barrick et al. 2005; Axmann et al. 2007). We would predict, based on our in vitro results, that each paralog should have distinct binding or release properties so as to allow transcriptional regulation to be accurately controlled in a highly contextual fashion. Potentially these RNAs might target more than one σ factor, but could equally well regulate a single σ in a highly contextual fashion. Medically, the in vitro discovery that 6S RNA point mutants can significantly alter pRNA-induced release rate implies that small molecule drugs might be able to achieve the same effect with wild-type 6S RNA in pathogenic bacteria. A screen for such molecules has the potential to discover new antibiotics that act by interfering with the release of this important master regulator of eubacterial transcriptional regulation.
MATERIALS AND METHODS
6S RNA and truncations
6S DNA was amplified directly from E. coli K12 cells using 6S specific primers (Supplemental Table 1), confirmed by sequencing and transcribed in vitro (40 mM Tris at pH 7.9, 5 mM DTT, 2.5 mM spermidine, 26 mM MgCl2, 0.01% Triton X-100, 8 mM GTP, 5 mM ATP, 5 mM CTP, 2 mM UTP, template DNA, 5 U/μL T7 RNA polymerase, with or without 10.0 μCi/μL [α32P]-UTP, 1 h at 37°C). Reactions were quenched by the addition of one volume of: 90% formamide, 50 mM EDTA at pH 8.0, 0.025% xylene cyanol, 0.025% bromophenol blue, and the RNA separated by 8% PAGE. RNA was excised, eluted overnight (300 mM NaCl, 4°C), and recovered by ethanol precipitation. Truncations of 6S RNA were generated by PCR amplification of the 6S template, using appropriate primers (Supplemental Fig. S7), and RNAs were generated as described.
Library synthesis
DNA having the sequence: 5′-GTAACCCTTGAACCcttggttcaaggtgaatgtgtcgtcgcagttttaaggcttCTCGGACGGACCGAGcatgctcaccaaccgcggagcgccacattcttgtggtatgaaatatcGGCTCAGGGGACTG was synthesized on an ABI 392 DNA synthesizer using standard phosphoramidite chemistry and 2000 Å CPG (upper case residues were fixed, lower case residues were mutagenized at the ∼10% level). The pool was amplified by PCR using Library Primers (Supplemental Fig. S7) and mutagenesis rate confirmed by sequencing.
In vitro selection
RNA from pool DNA was transcribed and gel purified. Pool RNA at 2.5 μM was preannealed in RNA buffer (20 mM HEPES at pH 7.5, 5 mM MgCl2 at 80°C for 2 min followed by 50°C for 5 min). This RNA was then diluted 10-fold into binding mix (20 mM HEPES at pH 7.5, 120 mM KCl, 1 mM DTT, 100 μg/mL heparin, and ∼100 nM or 0.1 U/μL Eσ70 [Epicenter] for 30 min at 37°C). One volume of native loading buffer (50% glycerol, 0.025% xylene cyanol, 0.025% bromophenol blue) was added, and complexes were resolved by native 8% (37.5:1 acrylamide:bis) PAGE run at 4°C. The shifted band corresponding to the T1:Eσ70 complex was carefully excised, and RNA recovered. The resulting RNA was subjected to this procedure a second time prior to RT-PCR. In Rounds 6 and 7 the RNA recovered from the first gel was preincubated as before, but using 500 nM preannealed RNA and 200 μg/mL heparin, and the reactions were supplemented with 225 μM of each NTP and 4 mM MgCl2, and incubated for a further 30 min at 37°C. Following native gel separation, RNA with a mobility corresponding to the T1:pRNA complex was excised and recovered.
RT-PCR
The final RNA recovered from each round was reverse transcribed. After treatment with 100 mM KOH for 10 min at 90°C, the resulting cDNA neutralized with Tris-HCl and PCR amplified for the next round of selection.
Truncation and point mutant binding assay
Body-labeled RNA in the amount of 500 nM was preannealed. The RNA mixture was then diluted 10-fold into the binding reaction using ∼160 nM Eσ70 and incubated for 30 min at 37°C. Gel shifts were resolved on 5% native PAGE gels, containing 5% glycerol.
pRNA Synthesis/Release Assay:
Body-labeled RNA in the amount of 250 nM was preannealed and diluted 10-fold into a binding reaction using 150 nM Eσ70 and incubated at 37°C for 30 min. This premix was then supplemented to achieve the final reaction concentrations that were incubated at 37°C: 18.5 nM body-labeled RNA, 15 mM HEPES at pH 7.5, 1 mM MgCl2, 225 μM each NTP, 90 mM KCl, 0.75 mM DTT, 75 μg/mL heparin, and 110 nM Eσ70. Time points were quenched by addition of one volume of native loading dye supplemented with 10 mM EDTA and were resolved by 8% native PAGE.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Seth Darst, Mariana Oviedo, and Lena Dolgosheina for important discussions and comments on the manuscript. This project was funded by an NSERC grant to P.J.U., a MSFHR fellowship to P.J.U., and a NSERC undergraduate fellowship to L.S.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2036210.
REFERENCES
- Axmann IM, Holzendorff J, Voss B, Kensche P, Hess WR 2007. Two distinct types of 6S RNA in Prochlorococcus. Gene 406: 69–78 [DOI] [PubMed] [Google Scholar]
- Barrick JE, Sudarsan N, Weinberg Z, Ruzzo WL, Breaker RR 2005. 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA 11: 774–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee GG 1971. Sequence of 6S RNA of E coli. Nat New Biol 229: 147–149 [DOI] [PubMed] [Google Scholar]
- Cavanagh AT, Klocko AD, Liu X, Wassarman KM 2008. Promoter specificity for 6S RNA regulation of transcription is determined by core promoter sequences and competition for region 4.2 of σ70. Mol Microbiol 67: 1242–1256 [DOI] [PubMed] [Google Scholar]
- Hindley J 1967. Fractionation of 32P-labeled ribonucleic acids on polyacrylamide gels and their characterization by fingerprinting. J Mol Biol 30: 125–136 [DOI] [PubMed] [Google Scholar]
- Klocko AD, Wassarman KM 2009. 6S RNA binding to Eσ70 requires a positively charged surface of σ70 region 4.2. Mol Microbiol 73: 152–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane WJ, Darst SA 2006. The structural basis for promoter -35 element recognition by the group IV σ factors. PLoS Biol 4: e269 doi: 10.1371/journal.pbio.0040269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney RA, Darst SA, Landick R 2005. Sigma and RNA polymerase: an on-again, off-again relationship. Mol Cell 20: 335–345 [DOI] [PubMed] [Google Scholar]
- Trotochaud AE, Wassarman KM 2004. 6S RNA function enhances long-term cell survival. J Bacteriol 186: 4978–4985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud AE, Wassarman KM 2005. A highly conserved 6S RNA structure is required for regulation of transcription. Nat Struct Mol Biol 12: 313–319 [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Saecker RM 2006. Synthesis-mediated release of a small RNA inhibitor of RNA polymerase. Science 314: 1601–1603 [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Storz G 2000. 6S RNA regulates E. coli RNA polymerase activity. Cell 101: 613–623 [DOI] [PubMed] [Google Scholar]
- Zaher HS, Unrau PJ 2005. Nucleic acid library construction using synthetic DNA constructs. Methods Mol Biol 288: 359–378 [DOI] [PubMed] [Google Scholar]