Abstract
Years after the discovery that Dicer is a key enzyme in gene silencing, the role of its helicase domain remains enigmatic. Here we show that this domain is critical for accumulation of certain endogenous small interfering RNAs (endo-siRNAs) in Caenorhabditis elegans. The domain is required for the production of the direct products of Dicer, or primary endo-siRNAs, and consequently affects levels of downstream intermediates, the secondary endo-siRNAs. Consistent with the role of endo-siRNAs in silencing, their loss correlates with an increase in cognate mRNA levels. We find that the helicase domain of Dicer is not necessary for microRNA (miRNA) processing, or RNA interference following exposure to exogenous double-stranded RNA. Comparisons of wild-type and helicase-defective strains using deep-sequencing analyses show that the helicase domain is required by a subset of annotated endo-siRNAs, in particular, those associated with the slightly longer 26-nucleotide small RNA species containing a 5′ guanosine.
Keywords: Dicer, helicase, miRNA, RNAi, siRNA
INTRODUCTION
Dicer is a member of the RNase III family of nucleases that target double-stranded RNA (dsRNA) (for review, see MacRae and Doudna 2007). When exogenous dsRNA (exo-dsRNA) triggers an RNA interference (RNAi) response, it is Dicer that cleaves dsRNA into short interfering RNAs (siRNAs) (Bernstein et al. 2001; Grishok et al. 2001; Ketting et al. 2001; Knight and Bass 2001). Furthermore, of the three known classes of small RNAs produced naturally in a cell, Dicer is required for producing micro-RNAs (miRNAs) and endogenous siRNAs (endo-siRNAs) (for review, see Golden et al. 2008), but not piwi-interacting RNAs (piRNAs) (for review, see Carthew and Sontheimer 2009).
Dicer is a large protein (∼150–220 kDa) with multiple domains (Fig. 1A). Biochemical and structural studies have provided good models for how the RNase III nuclease domains cleave dsRNA in the active site of Dicer (Zhang et al. 2004; MacRae et al. 2006). However, as yet, how the helicase domain contributes to Dicer's function is unclear. A surprise from the first biochemical characterization of purified, recombinant human Dicer was that a mutation in the helicase domain did not alter its ability to cleave RNA in vitro (Zhang et al. 2002). A more recent in vitro study shows that full-length human Dicer cleaves a pre-miRNA faster than a short, completely base-paired dsRNA, but if the entire helicase domain is removed, cleavage of these two substrates is equally rapid (Ma et al. 2008). One interpretation of this result is that the helicase domain is not required for pre-miRNA processing but provides a way to regulate cleavage of the completely base-paired dsRNA that gives rise to siRNAs.
FIGURE 1.
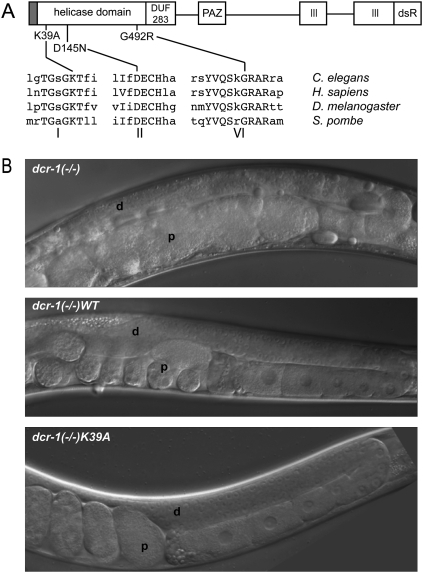
Germline defects of dcr-1(−/−) animals are rescued by transgenes encoding wild-type Dicer and Dicer with point mutations in the helicase domain. (A) The DCR-1 open-reading-frame is depicted as a line with domains as rectangles. Relative positions of domains and point mutations are to scale; sequence surrounding mutations is shown for species indicated. Gray rectangle at N terminus, histidine-FLAG tag; DUF 283, domain of unknown function 283; PAZ, Piwi Argonaute Zwille domain; III, RNase III domains; dsR, dsRNA binding motif. (B) Differential interference contrast micrographs show distal (d) and proximal (p) regions of gonad for dcr-1(−/−) and transgenic rescue strains.
There are other hints that Dicer's helicase domain is required in the siRNA, but not the miRNA, pathway. For example, Drosophila has two Dicer enzymes, one for the miRNA pathway (Dicer-1) and one for the siRNA pathway (Dicer-2), but only the latter has a functional helicase domain. Consistent with a role for Dicer-2 in the siRNA pathway, a mutation in the Walker A motif of the helicase domain (G31R) reduces siRNA production from dsRNA expressed from a transgene in vivo (Lee et al. 2004). In Schizosaccharomyces pombe, which has an siRNA, but not a miRNA, pathway, strains expressing Dicer with a point mutation in the same motif (K38A) of the helicase domain are defective for centromeric silencing and generation of siRNAs (Colmenares et al. 2007).
Here we report studies of DCR-1, the sole Dicer enzyme in Caenorhabditis elegans. We show that a transgene encoding wild-type DCR-1, as well as transgenes encoding three different helicase-mutant forms of DCR-1, all rescue germline defects of homozygote dcr-1(−/−) animals. In addition, all animals are capable of mounting an RNAi response to exo-dsRNA. However, while normal levels of miRNAs exist, animals expressing helicase-mutant forms of Dicer have severe defects in the production of endo-siRNAs. A deep-sequencing analysis of small RNAs from wild-type and helicase-defective strains indicates the defect arises during the production of primary (1°) endo-siRNAs, and also reveals unique features of loci that require Dicer's helicase domain for accumulation of endo-siRNAs.
RESULTS
We previously characterized a C. elegans strain containing a homozygous deletion in the Dicer gene (dcr-1(ok247); hereafter called dcr-1(−/−)), and reported that these animals are sterile, with severe germline defects (Knight and Bass 2001). To study the function of Dicer's helicase domain, we created four transgenes, one encoding the wild-type enzyme and three containing point mutations in the helicase domain. The K39A and D145N mutations lie within the Walker A and B motifs respectively, at positions known to be essential for ATP hydrolysis and helicase activity in many other proteins (Fig. 1A; for reviews, see Singleton et al. 2007; Pyle 2008). While less well studied, the amino acid affected by the G492R mutation has been observed to perturb ATP hydrolysis and reduce helicase function (Kim et al. 1997); this mutation was included to allow comparison to dcr-1(mg375) animals, which were isolated in a genetic screen and found to contain a G492R homozygous mutation in the chromosomal copy of dcr-1 (Pavelec et al. 2009). Each transgene was introduced into dcr-1(−/−) animals and monitored for its ability to rescue germline defects and sterility. All transgenic strains had a normal germline and were fertile when cultivated at normal growth temperatures (16°C–20°C; Fig. 1B; data not shown). At higher temperatures (25°C), while the germline of all animals appeared normal, the fertility of helicase mutant strains remained defective, likely due to a sperm defect as observed in the dcr-1(mg375) strain (Pavelec et al. 2009). We conclude that DCR-1's helicase domain is required for fertility at high-temperatures, but is not required for a normal germline and fertility at normal growth temperatures.
Production of endo-siRNAs is defective in strains containing a mutation in Dicer's helicase domain
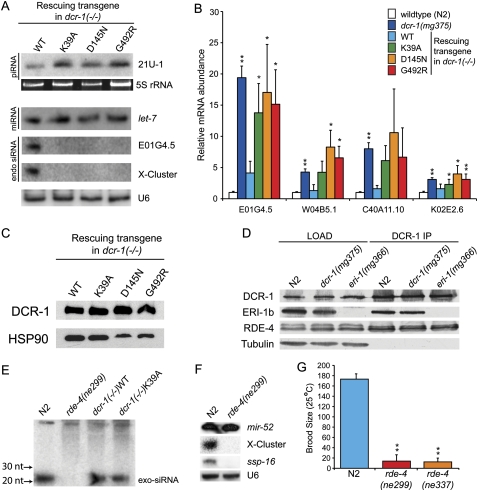
To test for defects in helicase-mutant strains at the molecular level, we monitored small RNAs known to exist in C. elegans by Northern analyses. The 21U-RNAs that are classified as piRNAs (Batista et al. 2008; Das et al. 2008) appeared at similar levels in RNA isolated from dcr-1(−/−)WT and all three helicase-mutant strains (Fig. 2A). Similarly, Northern blots probed for let-7 miRNA showed wild-type levels of mature miRNA in all strains. We next probed for several previously identified endo-siRNAs (Ambros et al. 2003; Ruby et al. 2006). While endo-siRNAs (X-cluster, E01G4.5) were detected in the wild-type (WT) rescue strain, they were absent in strains rescued with transgenes containing mutations in the helicase domain. This trend was observed for four out of five endo-siRNAs monitored (Supplemental Fig. 1A).
FIGURE 2.
C. elegans with mutations in Dicer's helicase domain are defective for production of endo-siRNAs, but not exo-siRNAs or other small RNAs. (A) Total RNA from mixed stage animals of indicated genotypes was subjected to Northern analysis using probes to a piRNA (21U-1), a miRNA (let-7), or endo-siRNAs (E01G4.5, X-cluster). 5S RNA stained with SYBR Gold, or a second hybridization with U6 snRNA probes, provided loading controls. (B) cDNA from total RNA of indicated strains (colors) was subjected to qRT-PCR to assess mRNA levels for indicated genes (x-axis). mRNA levels were normalized to eft-3 mRNA [N2, dcr-1(mg375); n = 6] or act-1 (transgenic rescue lines; n = 3) and plotted as average ratio of mRNA in mutant animals relative to wild type. Error bars, standard deviation; single asterisks, P < 0.05, and double asterisks, P < 0.01. Identical results were obtained using strand-specific primers/probes for reverse transcription or Northern analysis (data not shown), confirming measured mRNA levels derive from sense rather than antisense transcripts. (C) Embryo extracts of indicated strains were analyzed by Western analysis using α-Flag antibody (top panel) or α-HSP90 antibody (bottom panel) to assess levels of transgenic DCR-1. (D) Embryo extracts of indicated genotype (LOAD) were immunoprecipitated (IP) with an α-DCR-1 antibody. Co-precipitating proteins were analyzed by Western blot using the indicated antibodies. eri-1 encodes two isoforms, and the longer (ERI-1b) interacts with DCR-1 (Duchaine et al. 2006). eri-1(mg366) encodes a premature stop codon, so ERI-1 is not detected in extracts from these animals. Bottom panel, tubulin loading control. (E) Small RNAs were isolated from indicated strains (after feeding E. coli expressing dsRNA to the sel-1 gene), and Northern analyses performed with a probe to detect antisense sel-1. Upper bands are unprocessed sel-1 exo-dsRNA that served as an internal loading control; position of exo-siRNAs is indicated. (F) Small RNAs of mixed stage N2 and rde-4(ne299) C. elegans were subjected to Northern analysis using probes as indicated. Re-probing for U6 served as loading control. (G) N2 and rde-4(ne299) worms (n = 7) were grown at 25°C, and progeny counted to obtain average brood size. Asterisks, P < 0.01.
RNAi induced with exo-dsRNA yields small exo-siRNAs that base-pair to the complementary mRNA to mediate its cleavage. By analogy to this pathway, a loss of endo-siRNAs should result in an increased level of the cognate mRNA. We used quantitative real-time PCR (qRT-PCR) to monitor mRNA targets of endo-siRNAs in wild-type (N2), dcr-1(mg375), and all four rescue strains (Fig. 2B). As predicted, levels of complementary mRNAs were increased in all strains that contained a mutation in Dicer's helicase domain. In some cases we observed ∼20-fold increase in mRNA level in the helicase mutant strains compared to wild type.
We verified that differences between strains rescued with WT or helicase-defective transgenes were not due to differential expression of the transgene, by comparing levels of DCR-1 by Western analysis (Fig. 2C). When normalized to HSP90, DCR-1 levels were equivalent in dcr-1(−/−)WT and dcr-1(−/−)K39A strains, and slightly elevated in dcr-1(−/−)D145N and dcr-1(−/−)G492R strains. DCR-1 associates with other proteins in the ERI/DCR complex (Duchaine et al. 2006), and we also considered that the helicase domain might be important for assembly or stability of this complex. However, immunoprecipitation of DCR-1 in extracts of wild-type (N2) and dcr-1(mg375) animals, followed by Western blot analyses to monitor factors of the ERI/DCR complex (Fig. 2D), showed no differences between these strains. For example, DCR-1 immunoprecipitates from wild type and dcr-1(mg375) contained similar levels of ERI-1 and RDE-4 (Fig. 2D) as well as RRF-3 and ERI-3 (data not shown).
Dicer's helicase domain is not required for RNAi by exogenous dsRNA
At present it is unclear how similar the endo-siRNA pathway is to that mediating RNAi in response to exo-dsRNA (e.g., see Lee et al. 2006a). Thus, we also monitored silencing induced by feeding of exo-dsRNA. Strains were grown on Escherichia coli expressing dsRNA corresponding to the unc-22 gene, which in the case of successful RNAi, results in twitching of F1 progeny (Table 1). Wild-type (N2) animals showed a robust RNAi response, with 97% of animals exhibiting twitching; twitching was not observed among F1 progeny of rde-4(ne299) animals, which are defective in RNAi induced by feeding exo-dsRNA (Tabara et al. 1999). Somewhat surprisingly, we observed a wild-type RNAi response for all transgenic rescue strains. Twitching was observed in >90% of F1 progeny of dcr-1(−/−)WT, dcr-1(−/−)K39A, dcr-1(−/−)D145N, and dcr-1(−/−)G492R (data not shown) transgenic rescue strains.
TABLE 1.
RNAi response to dsunc-22
We wondered if helicase mutant strains had defects in processing exo-dsRNA that were not evident when assaying for RNAi by phenotypic outcome. For example, helicase-mutant strains might have lower than normal levels of exo-siRNAs that were still sufficient to trigger a wild-type RNAi response. However, we found that wild-type (N2), dcr-1(−/−)WT, and dcr-1(−/−)K39A stains exhibited similar levels of exo-siRNAs (Fig. 2E) when fed E. coli expressing dsRNA matching the sel-1 gene. Thus, by phenotypic and molecular analysis, we found no difference in the RNAi response to exo-dsRNA between strains expressing wild-type or helicase-mutant DCR-1. This result indicates that Dicer's helicase domain is essential for normal levels of endo-siRNAs, but not exo-siRNAs.
The dsRNA binding proteins (dsRBPs) that facilitate Dicer functions are sometimes specialized for different pathways. In Drosophila, production of most endo-siRNAs requires the dsRBP Loquacious, while production of exo-siRNAs during RNAi depends on the dsRBP R2D2 (Czech et al. 2008). Further, certain dsRBPs, including Loquacious, physically interact with Dicer's helicase domain (Lee et al. 2006b; Ye et al. 2007). We wondered if the requirement for Dicer's helicase domain in the production of endo-siRNAs but not exo-siRNAs reflected a differential requirement for an accessory dsRBP. However, we found that RDE-4, the dsRBP that acts with Dicer in the production of exo-siRNAs (see Fig. 2E), was also required for production of endo-siRNAs (Fig. 2F). Consistent with the requirement of RDE-4 in endo-siRNA accumulation, like the helicase mutants, rde-4-deficient animals exhibited fertility defects at 25°C (Fig. 2G).
Dicer helicase mutants have defects in production of primary endo-siRNAs
Dicer products have a monophosphate at each 5′ terminus and hydroxyl groups at 3′ termini, which overhang the duplex by 2 nucleotides (nt). Here we refer to siRNAs with a 5′ phosphate as primary (1°) siRNAs. In some organisms, including C. elegans and plants, the 1° siRNA signal is amplified by an RNA-dependent RNA polymerase (RdRP) to produce secondary (2°) siRNAs. By analogy to the pathway that leads to silencing with exo-dsRNA (Aoki et al. 2007), and as inferred from deep-sequencing data (Pak and Fire 2007; Sijen et al. 2007), C. elegans 1° endo-siRNAs are proposed to facilitate recruitment of an RdRP to the cognate mRNA, which serves as a template for de novo, primer-independent, synthesis of 2° endo-siRNAs by the RdRP. Secondary siRNAs are distinguished from 1° siRNAs because they are only one strand (antisense), they have triphosphates at their 5′ termini, and they are much greater in abundance.
Except when very high levels of dsRNA are produced from a transgene (e.g., Habig et al. 2008) and certain longer 26-nt endo-siRNAs in C. elegans (Supplemental Fig. 1; Ruby et al. 2006; Han et al. 2009), 1° siRNAs exist at levels that are too low to be detected by Northern analysis. Thus, the endo-siRNAs we detect in animals expressing wild-type DCR-1 (Fig. 2A), as well as the exo-siRNAs produced by feeding dsRNA (Fig. 2E), are largely, if not exclusively, 2° siRNAs. While the absence of 2° endo-siRNAs in Dicer helicase mutant strains indicated a defect in the pathway, from the Northern analysis we could not tell if the helicase domain was required in the production of 2° endo-siRNAs, or an upstream step, such as production of 1° endo-siRNAs.
To address this issue we performed Illumina sequencing on cDNA libraries prepared from small RNAs isolated from dcr-1(−/−)WT and dcr-1(−/−)K39A strains. We prepared libraries using a 5′ ligation-dependent protocol that excludes triphosphorylated RNAs and reports on monophosphorylated RNAs (Ruby et al. 2006; see Materials and Methods). We reasoned that information about levels of monophosphorylated 1° endo-siRNAs, combined with our Northern analyses, would provide information about the step at which Dicer's helicase domain was required.
Sequencing reads from small RNAs of dcr-1(−/−)WT and dcr-1(-/)K39A samples were aligned to the C. elegans genome and selected for further analysis based on a >90% confidence of alignment to the indicated position, thus eliminating reads with significant mismatches and highly repetitive elements. We retained 3,348,593 reads for dcr-1(−/−)WT and 8,254,341 reads for dcr-1(−/−)K39A. Reads for dcr-1(−/−)K39A derived from two Illumina sequencing reactions so as to increase coverage and facilitate our ability to discern statistically significant differences at endo-siRNA loci.
We used the Defined Region Scan Seq algorithm from the USeq software package (Nix et al. 2008) to parse reads into categories that included previously annotated miRNAs, piRNAs, and endo-siRNAs (Fig. 3A). In defining the latter, we curated a published list (Asikainen et al. 2008) based on previous analyses of C. elegans small RNAs (Ambros et al. 2003; Ruby et al. 2006). The curated list included 2179 genes (Supplemental Table 1), and we identified endo-siRNA sequences in at least one of our samples for 1936 of these genes.
FIGURE 3.
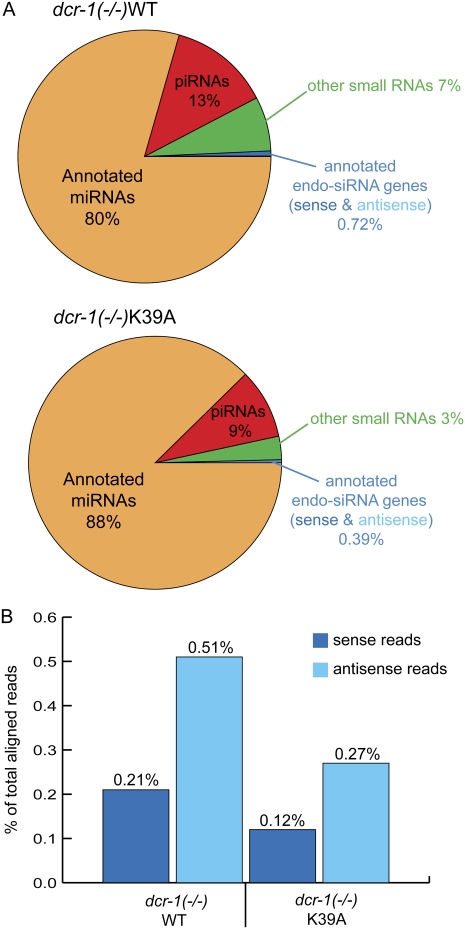
endo-siRNAs are a smaller fraction of reads for dcr-1(−/−)K39A animals compared to dcr-1(−/−)WT animals. (A) Pie charts show percentage of reads in classes defined by previous annotations (see Materials and Methods). Binomial P-value for differences in piRNA, miRNA, and endo-siRNA fractions, <4.9e−324. (B) The plot shows percentage of reads corresponding to sense or antisense annotated endo-siRNAs (see Materials and Methods).
When compared to the dcr-1(−/−)WT sample, the dcr-1(−/−)K39A sample was slightly enriched for annotated miRNAs (88% versus 80%) and slightly depleted for annotated piRNAs (9% versus 13%). Studies using protocols that clone both 1° and 2° endo-siRNAs indicate that, together, these small RNAs are comparable in abundance to miRNAs (Pak and Fire 2007). However, as in other studies that focused on 1° endo-siRNAs (Ruby et al. 2006), we found the fraction of endo-siRNAs to be far less than the fraction of miRNAs [∼1:100 for dcr-1(−/−)WT and ∼1:200 for dcr-1(−/−)K39A]. Importantly, while endo-siRNAs were a small fraction of both samples, the fraction of dcr-1(−/−)K39A reads matching endo-siRNA loci was about half that of the dcr-1(−/−)WT sample (0.39% versus 0.72%). The lower fraction of endo-siRNAs in dcr-1(−/−)K39A reads reflected a reduction of both sense and antisense strands (Fig. 3B), consistent with the idea that these reads derive from cleavage of dsRNA by Dicer. That said, because it is difficult to conclusively determine if a sense siRNA is a Dicer product or an intermediate of mRNA degradation, in many analyses that follow we focused on antisense reads.
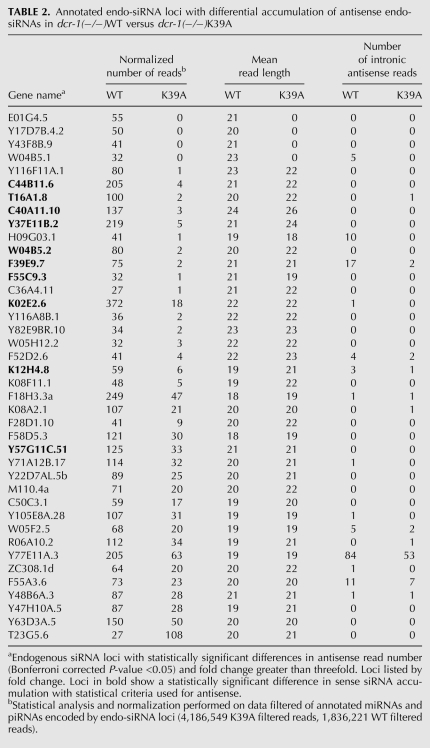
One explanation for the lower fraction of endo-siRNAs in dcr-1(−/−)K39A animals (Fig. 3A,B) was that there was a global reduction of endo-siRNAs across all annotated endo-siRNA-producing genes. Alternatively, large differences in a subset of endo-siRNA-producing genes could account for the lower fraction. Our analysis supported the latter, since we identified only 80 genes with a statistically significant difference in antisense endo-siRNAs between samples (Bonferroni corrected P-value <0.05). Of these genes, 79 showed reduced endo-siRNA reads in dcr-1(−/−)K39A, while only one gene (T23G5.6) had increased reads in the helicase mutant. Half of these genes (40/80) displayed greater than a threefold change (Table 2).
TABLE 2.
Annotated endo-siRNA loci with differential accumulation of antisense endo-siRNAs in dcr-1(−/−)WT versus dcr-1(−/−)K39A
Using the same statistical criteria as in the antisense analysis, we found 21 genes with differences in sense reads greater than threefold (Supplemental Table 2), all showing reduced reads in dcr-1(−/−)K39A. Ten of these genes overlapped with genes showing differences in antisense reads greater than threefold (Table 2, bold), and this overlap represented a 23-fold enrichment over what is expected by chance (P < 0.001). While we cannot completely rule out the possibility that some sense reads are a result of nonspecific mRNA degradation, the strong overlap between genes that show reduction in both sense and antisense reads suggests that these small RNAs arose during cleavage of dsRNA by Dicer.
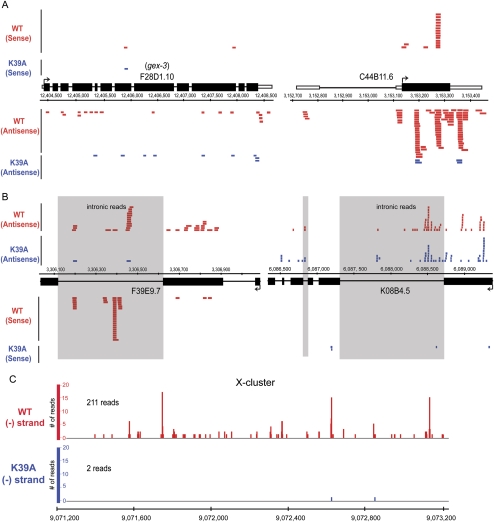
A subset of endo-siRNA loci were completely devoid of reads in helicase mutant samples (Table 2), while others were dramatically reduced in number (Fig. 4). Consistent with previous reports (Ambros et al. 2003; Ruby et al. 2006), most endo-siRNAs mapped to exons (Fig. 4A), but some loci had reads mapping to sense and antisense strands of introns (Fig. 4B; Supplemental Fig. 1B; Supplemental Table 3). For the 2179 endo-siRNA loci, ∼95% of antisense reads mapped to exons and 5% to introns.
FIGURE 4.
Screen shots of representative loci show endo-siRNAs mostly in exons. (A,B) Sense and antisense reads are shown for dcr-1(−/−)WT (red) and dcr-1(−/−)K39A (blue) with gene structures for plus strand loci above coordinates and those for minus below. Each bar is one read (size to scale, except F28D1.10 [gex-3]), and here data were not normalized to account for the ∼2.5-fold more reads of dcr-1(−/−)K39A samples. In A reads map almost exclusively to exons (black rectangles), while those in B map to introns (lines, reads in shaded boxes) and exons. (C) Bar height is number of reads at each genomic position of X-cluster for dcr-1(−/−)WT (red) and dcr-1(−/−)K39A (blue) samples.
Among loci for which endo-siRNA accumulation was dependent on Dicer's helicase domain was an intergenic region of chromosome X, termed the X-cluster (Ambros et al. 2003). We parsed reads mapping to the X-cluster and found 211 reads for dcr-1(−/−)WT, and only two reads for dcr-1(−/−)K39A (Fig. 4C). Similar to previous reports (Ruby et al. 2006) we observed a strong strand bias for small RNAs of the X-cluster, and despite cloning 213 total antisense reads, only obtained two sense reads, both in dcr-1(−/−)WT samples.
Characteristics of endo-siRNAs that depend on Dicer's helicase domain
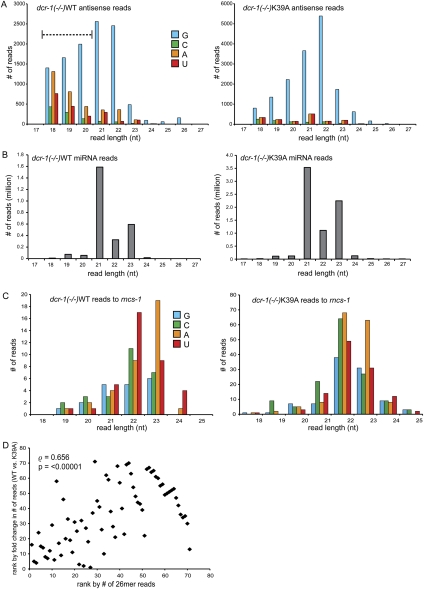
Previous deep-sequencing experiments performed to enrich for monophosphorylated, and exclude triphosphorylated, small RNAs reveal endo-siRNAs that usually have a guanosine (G) at their 5′ terminus, with lengths enriched for species of 21, 22, and 26 nt (Ruby et al. 2006). For both dcr-1(−/−)WT and dcr-1(−/−)K39A endo-siRNAs we observed a preference for a G at 5′ termini and lengths of 21–22 nt (Fig. 5A).
FIGURE 5.
Features of dcr-1(−/−)K39A small RNAs compared to those of dcr-1(−/−)WT. Bar height is number of reads of a particular length and 5′ nt (colors as indicated) for: (A) all 2179 annotated endo-siRNA-producing genes for which we obtained reads (1936 genes); (B) annotated miRNAs; (C) sense and antisense reads mapping to rncs-1. (D) Spearman's rank correlation analysis (ρ, rho) indicates a positive correlation between number of 26 nt reads of a particular gene and the degree to which reads are reduced in dcr-1(−/−)K39A at that loci (fold change = (normalized reads in dcr-1(−/−)WT + 1)/(reads in dcr-1(−/−)K39A + 1). The P-value was estimated based on randomizing ranked lists and computationally determining ρ 1e5 times; under no circumstances was ρ > 0.656.
Recent studies indicate that 26-nt endo-siRNAs that begin with G (26G RNAs) have 5′ monophosphates and require an RdRP (RRF-3), an exonuclease (ERI-1), and likely DCR-1 for their accumulation (Han et al. 2009). All of these data are consistent with 26G RNAs being 1° endo-siRNAs, and accordingly 26G RNAs do not depend on RRF-1, the RdRP believed to be involved in 2° siRNA production (Han et al. 2009). While 26G RNAs were observed in the dcr-1(−/−)WT sample, they were almost nonexistent in dcr-1(−/−)K39A samples, again consistent with the idea that the helicase domain of DCR-1 is required for the production of certain 1° endo-siRNAs (Fig. 5A). Even in the dcr-1(−/−)WT samples, 26G RNA reads were fewer than in a previous analysis of wild-type C. elegans (Ruby et al. 2006). Since cosuppression of transgenes in the germline is common (Kelly et al. 1997), the Dicer transgenes in our study are likely expressed at low levels in the germline. This, combined with the fact that loci giving rise to 26G RNAs are enriched for germline functions (Ruby et al. 2006; Han et al. 2009), may explain the low levels of 26G RNAs in our dcr-1(−/−)WT sample. Regardless, by comparison, 26G RNAs in dcr-1(−/−)K39A were dramatically reduced, suggesting a correlation between number of 26G RNAs from a locus and its dependence on the helicase domain for endo-siRNA accumulation. Because the number of 26G RNA reads in our samples was low, to increase statistical significance, we tested this possibility using 26G RNA reads from a previous study of C. elegans small RNAs (Ruby et al. 2006). We created a list of genes with three or more 26G RNA reads (71 genes), ranked according to their number of 26G RNA reads. For these 71 genes, we also created a list ranked according to the fold change in their endo-siRNA reads observed in our dcr-1(−/−)WT and dcr-1(−/−)K39A samples. Using these two lists, we found a positive correlation between the number of 26G RNAs expressed from a given gene and the extent to which endo-siRNAs are reduced in dcr-1(−/−)K39A samples at that gene (Fig. 5D; Spearman's rank correlation coefficient, ρ = 0.656; P-value < 0.00001). This analysis suggested that loci enriched for 26G RNA reads are more likely to depend on a functional helicase domain for the accumulation of endo-siRNAs.
Another difference between the characteristics of 1° endo-siRNAs in the two samples was the greater proportion of reads of 18–20 nt in the dcr-1(−/−)WT sample compared to the dcr-1(−/−)K39A sample (Fig. 5A; dashed over-bar). One explanation for enrichment of 18–20 nt reads was that RNA of the dcr-1(−/−)WT sample was subject to more degradation by nonspecific ribonucleases during purification. However, this seemed unlikely since miRNA reads between strains showed no difference in length distribution, with the majority of reads being 21–23 nt with very few shorter or longer species (Fig. 5B).
For contrast, we also analyzed a locus where production of endo-siRNAs did not depend on a functional helicase domain in Dicer. Other studies in our laboratory identified a noncoding RNA called rncs-1 that folds into a double-stranded structure of ∼300 nearly contiguous base pairs (Hellwig and Bass 2008). In young adults, steady-state levels of rncs-1 are not different between wild-type and dcr-1(−/−) animals. We observed numerous reads matching the rncs-1 locus, with no significant difference in number of normalized reads between dcr-1(−/−)WT and dcr-1(−/−)K39A animals. The majority of reads were 22–23 nt, and these reads did not have a bias for G at the 5′ position. Interestingly, reads mapping to rncs-1 were devoid of the 26G RNAs, again consistent with the idea that loci that encode 26G RNAs are highly dependent on Dicer's helicase domain for endo-siRNA accumulation. Despite many attempts, we were unable to detect small RNAs for rncs-1 by Northern analyses (data not shown), possibly because rncs-1 small RNAs are not amplified to make 2° endo-siRNAs.
miRNA levels are similar in dcr-1(−/−)WT and dcr-1(−/−)K39A animals
Northern analysis (Fig. 2A) and morphology of dcr-1(−/−)K39A, D145N, and G492R animals (Fig. 1B) suggested there were unlikely to be gross defects in miRNA processing in helicase mutants. Indeed, we found no obvious differences in miRNA length distribution between dcr-1(−/−)WT and dcr-1(−/−)K39A samples, with most reads being 21 nt (Fig. 5B). Reads were obtained for 138/155 annotated miRNAs (Supplemental Table 2) including 77 reads to lsy-6 miRNA, which is expressed in only a few cells (Johnston and Hobert 2003). Most differences between samples were modest (less than twofold change), but still 78 miRNA loci showed a small but statistically significant difference (Bonferroni corrected P-value <0.05); in most cases, miRNA reads were slightly increased in dcr-1(−/−)K39A. Of the miRNAs with reduced expression in dcr-1(−/−)K39A, many were previously suggested to be misannotated, and better classified as endo-siRNAs (Ruby et al. 2006). We conclude that Dicer's helicase domain plays a minor role in processing C. elegans miRNA precursors.
DISCUSSION
We compared C. elegans strains expressing wild-type Dicer with those containing mutations in Dicer's helicase domain, using phenotypic, molecular, and high-throughput sequencing analyses. We find that Dicer's helicase domain is required for the production of a subset of endo-siRNAs but is dispensable for the production of miRNAs and RNAi triggered by exo-dsRNA. For the subset of endo-siRNAs that require a functional helicase domain, high-throughput sequencing experiments indicate the helicase domain contributes to the production of the direct products of Dicer, the 1° endo-siRNAs, and accordingly, Northern analyses suggest a loss of the downstream intermediates, the triphosphorylated 2° endo-siRNAs. Loci whose endo-siRNAs depend on Dicer's helicase domain are enriched for 26G RNAs, suggesting a correlation between this small RNA species and the mechanistic function of the helicase domain.
What is the mechanistic function of Dicer's helicase domain?
The diverse group of proteins originally classified as helicases are now recognized as proteins that couple NTP hydrolysis to many activities, including, but not limited to, unwinding of nucleic acids (for review, see Lohman et al. 2008). Conserved sequences in Dicer's helicase domain place it in the Superfamily 2 (SF2) group of helicases, many of which couple NTP hydrolysis to movement along polynucleotides, or translocation. Our data are consistent with the idea that Dicer's helicase domain also functions as a translocase, allowing the enzyme to catalyze multiple, processive, cleavage events before dissociation from a dsRNA substrate. According to this model, the helicase domain would be required for processive cleavage of certain long dsRNA substrates that give rise to endo-siRNAs. As we observe, the helicase domain would not be required for processing miRNAs, which requires only one double-stranded cleavage event, and is thus optimal without processivity. Dicer enzymes with mutations in the helicase domain would only be capable of a nonprocessive mode, where each cleavage event would be followed by dissociation of the enzyme, and rebinding in preparation for another cleavage event. Especially in the presence of high concentrations of dsRNA, such as occurs during feeding of exogenous dsRNA, a nonprocessive mode of cleavage may produce enough 1° siRNAs to trigger production of 2° siRNAs that can then elicit silencing, and this could explain why helicase defective strains are capable of a wild-type RNAi response.
We observed similar levels of exo-siRNAs corresponding to the sel-1 gene following feeding of dsRNA to sel-1 (Fig. 2E). Since the assay used to detect these siRNAs mainly reports on 2° siRNAs, we cannot rule out the possibility that helicase mutants still have a defect in the production of 1° siRNAs in response to exogenous dsRNA. The true structure of the exogenous dsRNA used in RNAi feeding experiments remains unclear. This dsRNA is typically delivered by feeding worms bacteria that are expressing the dsRNA, and by the time it reaches Dicer for cleavage it has likely been exposed to various ribonucleases, phosphatases, and kinases. Perhaps the helicase domain of Dicer is only required for processing dsRNA with specific features, while other dsRNA precursors are processed in a helicase-independent manner.
What is the precursor of small RNAs that depend on Dicer's helicase domain?
A subset of the loci with the most significant differences in number of antisense endo-siRNAs for dcr-1(−/−)WT and dcr-1(−/−)K39A samples were also those with the most differences in sense endo-siRNAs (Table 2). This suggests that the endo-siRNAs arise from a dsRNA precursor, as expected of Dicer products. In other organisms, loci that give rise to endo-siRNAs often have features suggesting a dsRNA precursor, such as hairpin structures, or evidence of sense-antisense pairs arising from converging genes or bidirectional transcription (for review, see Golden et al. 2008; Okamura et al. 2008). In hopes of revealing such features, we compared our lists of endo-siRNA producing loci to published lists of loci known to give rise to sense-antisense pairs (Zhang et al. 2006). In addition, since endo-siRNA loci of flies and mammals are often pseudogenes or transposons (as cited in Golden et al. 2008), we performed comparisons with lists of C. elegans transposons and pseudogenes (see Materials and Methods). There was no significant overlap of these data sets with C. elegans endo-siRNA loci, and such loci were not enriched in the helicase-dependent endo-siRNA loci (Table 2).
One explanation for the lack of features in our endo-siRNA loci suggestive of a dsRNA precursor is that dsRNA arises not from transcription, but from an RdRP that copies an mRNA template. This would be consistent with the fact that the vast majority of endo-siRNAs we characterized match exonic, rather than intronic, sequences. Analyses of DCR-1-containing complexes reveal a tight association of the RdRP, RRF-3 (Duchaine et al. 2006), and, similar to the defects of dcr-1 helicase mutants, rrf-3 mutants show defects in production of certain endo-siRNAs (Lee et al. 2006a; Han et al. 2009) but are capable of an RNAi response to exogenous dsRNA (Simmer et al. 2002).
The idea that RdRP activity is coupled to cleavage by Dicer is consistent with experiments performed in S. pombe (Colmenares et al. 2007) and Tetrahymena thermophila (Lee and Collins 2007), which utilize an RdRP in the production of siRNAs. The C terminus of S. pombe Dicer physically interacts with an RdRP, and reconstitution experiments that include Dicer, the RdRP, and a ssRNA template show production of double-stranded siRNAs over time. If the experiment is performed with S. pombe Dicer containing mutations that disrupt RNase III activity, the product is a dsRNA corresponding to the full-length ssRNA template. The role of Dicer's helicase domain in S. pombe RdRP activity is unclear, but interestingly, addition of Dicer harboring RNase III mutations stimulates RdRP activity.
endo-siRNAs that require Dicer's helicase domain have unique features
Of ∼2000 annotated endo-siRNA loci, we observed only 80, or 4%, that showed a significant difference in abundance when comparing dcr-1(−/−)WT and dcr-1(−/−)K39A animals. We imposed a rigorous statistical cutoff, and it seems probable that additional read coverage would identify other small RNAs that depend on Dicer's helicase domain. However, certainly the majority of annotated endo-siRNAs are not dependent on Dicer's helicase domain, and possibly these small RNAs arise by a mechanism that is independent of Dicer cleavage. In fact, this is entirely consistent with a recent report showing that a large fraction of C. elegans endo-siRNAs, in particular those characterized by a length of 22 nt and a 5′ G (22G RNAs), depend on a Dicer-related helicase, DRH-3, but not Dicer (Gu et al. 2009). Synthesis of 22G RNAs is thought to involve de novo synthesis by an associated RdRP, without the requirement for production of a 1° endo-siRNA by Dicer. These results raise the possibility that all Dicer-dependent endo-siRNAs require its helicase domain, and that the Dicer-helicase dependent endo-siRNAs we describe are the only endo-siRNAs that require Dicer.
Our studies set the stage for experiments designed to understand the role of Dicer's helicase domain in endo-siRNA production, and those aimed at determining the biologic function of these small RNAs. Given the strong correlation of the 26G RNAs with the dependence on the helicase domain (Fig. 5D), it seems likely this small RNA species relates to the helicase function. A Gene Ontology (GO) analysis (Supplemental Table 4; see Materials and Methods) of the 80 genes whose endo-siRNAs depend on Dicer's helicase domain shows a greater than fivefold enrichment in terms related to cytokinesis/cell division, as well as those for cytoskeleton organization/biogenesis (false discovery rate < 0.01), perhaps hinting at specialized functions of these endo-siRNAs. Further, many 26G RNAs map to sperm enriched genes (Ruby et al. 2006; Han et al. 2009), consistent with the idea that the infertility of helicase mutant animals at high temperatures results from sperm defects.
MATERIALS AND METHODS
Plasmids and transgenic strains
Strains are detailed in Supplemental Data. Rescue strains were made by injecting dcr-1(ok247); III/ht2[bli-4(e937) let-?(q782) qIs48] heterozygotes with constructs of 2.5 kb of the putative dcr-1 promoter, a TEV-cleavable affinity tag (10 histidines followed by three copies of FLAG), the genomic region encoding DCR-1 (including exons and introns), and 1.5 kb 3′ of the stop codon. Point mutations were made by PCR mutagenesis. Injection mixes were 5 ng/μL rescue construct, 2 ng/μL dpy-30∷mCherry marker, and 95 ng/μL nonspecific DNA (Invitrogen 1 kb ladder). Homozygotes positive for transformation were singled and screened for viability.
RNAi experiments
Bacteria expressing dsRNA, either unc-22 (Fraser et al. 2000; Kamath et al. 2003) or sel-1 (pJP12.2+), were fed to strains as described (Timmons et al. 2001).
RNA analysis
Total RNA was prepared from mixed-stages or embryos by douncing in TRIzol (Invitrogen) followed by precipitation with isopropanol. For qRT-PCR, cDNA was made with iScript cDNA synthesize kit (Bio-Rad), and qPCR performed on an iCycler (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad). Primers are in Supplemental Data.
For Northern analyses, RNA was further purified using a mirVana miRNA Isolation Kit (Ambion). MirVana-enriched RNAs (3 μg) were separated on a 15% denaturing polyacrylamide gel, transferred to hybond-N+ membrane (GE-Amersham), cross-linked with EDC (30 min, 60°C; Pall et al. 2007), and blotted in Ultrahyb-Oligo hybridization buffer (Ambion). Oligonucleotide probes were 5′ end labeled with γ-[32]p-(dATP) using T4 PNK. Membranes were washed three times in 2 × SSC + 0.5% SDS, and exposed on a PhosphorImager screen (Molecular Dynamics). Probe sequences are in Supplemental Data.
Illumina sequencing
Total RNA (10 μg) was subjected to 5′ ligation-dependent sequencing (Illumina small RNA sequencing protocol). Reads were aligned to the C. elegans genome (May 2008, ce6 assembly), and, using Novoalign software (http://www.novocraft.com), adapter sequence trimmed and a gapped alignment performed. Alignments with a posterior probability of <0.1 were used in downstream analyses. Aligned data sets were compared to reference data sets with USeq software (Nix et al. 2008) using coordinates for miRNA genes (miRBase version 12.0), piRNA genes (Ruby et al. 2006; Batista et al. 2008), or a list of 2179 endo-siRNA genes (Asikainen et al. 2008), curated to remove duplicate genes, ambiguous annotations, and small RNAs with a read length <18 nt. Reads were visualized using the Integrated Genome Browser (IGB-Affymetrix). GO analysis was performed with GoMiner (Zeeberg et al. 2003).
Accession numbers
Raw Illumina data and aligned reads (Novoalign) are available at the NCBI Gene Expression Omnibus (GEO), http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse20336.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank B. Milash and B. Dalley of the University of Utah (UU) Bioinformatics Core Facility, and W.E. Johnson, for advice and technical support; the UU DNA/Peptide Core Facility for DNA/RNA synthesis (partially funded by NCI CA04214); M. Hammerlund, formerly of the Jorgensen laboratory, for dpy-30::mCherry injection marker; and J. Pak and A. Fire for pJP12.2+. This work was supported by funds to B.L.B. from NIH (GM067106); S.K. from NIH (GM076619), the Pew Scholars Program, and the Shaw Scientist Program; T.F.D. from CIHR, NSERC, and a Chercheur-Boursier Junior 1 Award.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2122010.
REFERENCES
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D 2003. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol 13: 807–818 [DOI] [PubMed] [Google Scholar]
- Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H 2007. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J 26: 5007–5019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asikainen S, Heikkinen L, Wong G, Storvik M 2008. Functional characterization of endogenous siRNA target genes in Caenorhabditis elegans. BMC Genomics 9: 270. doi: 10.1186/1471-2164-9-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. 2008. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell 31: 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenares SU, Buker SM, Buhler M, Dlakic M, Moazed D 2007. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell 27: 449–461 [DOI] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. 2008. An endogenous small interfering RNA pathway in Drosophila. Nature 453: 798–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, et al. 2008. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell 31: 79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D Jr, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, et al. 2006. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124: 343–354 [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408: 325–330 [DOI] [PubMed] [Google Scholar]
- Golden DE, Gerbasi VR, Sontheimer EJ 2008. An inside job for siRNAs. Mol Cell 31: 309–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34 [DOI] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. 2009. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell 36: 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habig JW, Aruscavage PJ, Bass BL 2008. In C. elegans, high levels of dsRNA allow RNAi in the absence of RDE-4. PLoS One 3: e4052. doi: 10.1371/journal.pone.0004052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, Thierry-Mieg D, Thierry-Mieg J, Kim JK 2009. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc Natl Acad Sci 106: 18674–18679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwig S, Bass BL 2008. A starvation-induced noncoding RNA modulates expression of Dicer-regulated genes. Proc Natl Acad Sci 105: 12897–12902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Hobert O 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426: 845–849 [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237 [DOI] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, Fire A 1997. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146: 227–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev 15: 2654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Kim J, Gwack Y, Han JH, Choe J 1997. Mutational analysis of the hepatitis C virus RNA helicase. J Virol 71: 9400–9409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Bass BL 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293: 2269–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Collins K 2007. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat Struct Mol Biol 14: 604–610 [DOI] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81 [DOI] [PubMed] [Google Scholar]
- Lee RC, Hammell CM, Ambros V 2006a. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA 12: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN 2006b. The role of PACT in the RNA silencing pathway. EMBO J 25: 522–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman TM, Tomko EJ, Wu CG 2008. Nonhexameric DNA helicases and translocases: Mechanisms and regulation. Nat Rev Mol Cell Biol 9: 391–401 [DOI] [PubMed] [Google Scholar]
- Ma E, MacRae IJ, Kirsch JF, Doudna JA 2008. Autoinhibition of human dicer by its internal helicase domain. J Mol Biol 380: 237–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Doudna JA 2007. Ribonuclease revisited: Structural insights into ribonuclease III family enzymes. Curr Opin Struct Biol 17: 138–145 [DOI] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA 2006. Structural basis for double-stranded RNA processing by Dicer. Science 311: 195–198 [DOI] [PubMed] [Google Scholar]
- Nix DA, Courdy SJ, Boucher KM 2008. Empirical methods for controlling false positives and estimating confidence in chIP-seq peaks. BMC Bioinformatics 9: 523. doi: 10.1186/1471-2105-9-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Balla S, Martin R, Liu N, Lai EC 2008. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat Struct Mol Biol 15: 581–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J, Fire A 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315: 241–244 [DOI] [PubMed] [Google Scholar]
- Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A 2007. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res 35: e60. doi: 10.1093/nar/gkm112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelec DM, Lachowiec J, Duchaine TF, Smith HE, Kennedy S 2009. Requirement for the ERI/DICER complex in endogenous RNA interference and sperm development in Caenorhabditis elegans. Genetics 183: 1283–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle AM 2008. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys 37: 317–336 [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127: 1193–1207 [DOI] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, Plasterk RH 2007. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315: 244–247 [DOI] [PubMed] [Google Scholar]
- Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH 2002. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol 12: 1317–1319 [DOI] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Wigley DB 2007. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem 76: 23–50 [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99: 123–132 [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263: 103–112 [DOI] [PubMed] [Google Scholar]
- Ye X, Paroo Z, Liu Q 2007. Functional anatomy of the Drosophila microRNA-generating enzyme. J Biol Chem 282: 28373–28378 [DOI] [PubMed] [Google Scholar]
- Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, Narasimhan S, Kane DW, Reinhold WC, Lababidi S, et al. 2003. GoMiner: A resource for biological interpretation of genomic and proteomic data. Genome Biol 4: R28. doi: 10.1186/gb-2003-4-4-r28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W 2002. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J 21: 5875–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W 2004. Single processing center models for human Dicer and bacterial RNase III. Cell 118: 57–68 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu XS, Liu QR, Wei L 2006. Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res 34: 3465–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]