Abstract
Alternative splicing accounts for much of the complexity in higher eukaryotes. Thus, its regulation must allow for flexibility without hampering either its specificity or its fidelity. The mechanisms involved in alternative splicing regulation, especially those acting through coupling with transcription, have not been deeply studied in in vivo models. Much of our knowledge comes from in vitro approaches, where conditions can be precisely controlled at the expense of losing several levels of regulation present in intact cells. Here we studied the relative order of removal of the introns flanking a model alternative cassette exon. We show that there is a preferential removal of the intron downstream from the cassette exon before the upstream intron has been removed. Most importantly, both cis-acting mutations and trans-acting factors that regulate the model alternative splicing event differentially affect the relative order of removal. However, reduction of transcriptional elongation causing higher inclusion of the cassette exon does not change the order of intron removal, suggesting that the assumption, according to the “first come, first served” model, that slow elongation promotes preferential excision of the upstream intron has to be revised. We propose instead that slow elongation favors commitment to exon inclusion during spliceosome assembly. Our results reveal that measuring the order of intron removal may be a straightforward read-out to discriminate among different mechanisms of alternative splice site selection.
Keywords: pol II elongation, alternative splicing, coupling, order of intron removal
INTRODUCTION
Alternative splicing regulation must be strict in order to guarantee it remains flexible and dynamic while still displaying high specificity and fidelity. Many of the approaches used to study the splicing mechanism rely on well-characterized in vitro splicing systems that allow for precisely dissecting regulatory functions (Lim and Hertel 2004; House and Lynch 2006; Kotlajich et al. 2009). While more difficult to control and perform, in vivo experiments addressing mechanistic questions of splicing regulation have shed light on the events that couple splicing to upstream and downstream events. This is of particular importance under the current view of cotranscriptional processing of mRNA where molecular machineries function in a coordinated manner, sharing components and affecting each other's activities (Kornblihtt et al. 2004). In vivo approaches used to study splicing mechanisms, where the timing of events has been taken into account, have been followed in yeast and in mammalian cells. For instance, it has been shown that the recruitment of general splicing factors to the splicing sites in yeast occurs cotranscriptionally in a stepwise manner (Kotovic et al. 2003; Gornemann et al. 2005; Lacadie and Rosbash 2005). Similar approaches in mammalian cells have led to similar conclusions (Listerman et al. 2006). Many reports have demonstrated the importance of transcriptional control on alternative splicing, which together with the action of cis-acting sequences, trans-acting factors, and secondary structures, precisely regulates alternative splicing both spatially and temporally (Kornblihtt et al. 2004).
Despite this progress, in vivo approaches used to study the mechanisms underlying the regulation of alternative splicing are scarce. We have applied a strategy previously used to study the order of constitutive splicing events (i.e., the order of intron removal [Kessler et al. 1993]) as an in vivo approach to study the mechanism of alternative splicing of a prototypical alternative exon cassette: human fibronectin (FN) exon 33 (E33), also known as EDI or EDA. We show that the two introns flanking E33 are removed at different rates, and these relative rates are differentially affected by cis-elements and trans-acting factors regulating E33 alternative splicing. We show that two different cis mutations, both leading to higher E33 exon inclusion, affect the relative rate of intron removal (RRIR) of the flanking introns in opposite ways. Similarly, we demonstrate that several SR proteins that cause an increase in exon inclusion significantly differ in their ability to affect the order of intron removal of the flanking introns. Finally, we show that different treatments that reduce polymerase II (pol II) elongation, and as a consequence promote E33 exon inclusion, do not affect the RRIRs, suggesting that slower elongation promotes commitment of splicing factors that favor E33 exon inclusion rather than increased excision of the upstream intron.
RESULTS
Introns flanking the fibronectin alternative exon 33 are removed at different rates
Previous studies on constitutive splicing events have determined that the relative order of intron removal does not follow a strictly 5′–3′ order (Kessler et al. 1993; Schwarze et al. 1999). Introns can be eliminated according to preferred pathways that differ in different genes. We asked if the introns flanking a prototypical alternative exon cassette (FN E33) were following a preferred pathway of intron removal that could be sensitive to changes in exon inclusion rate.
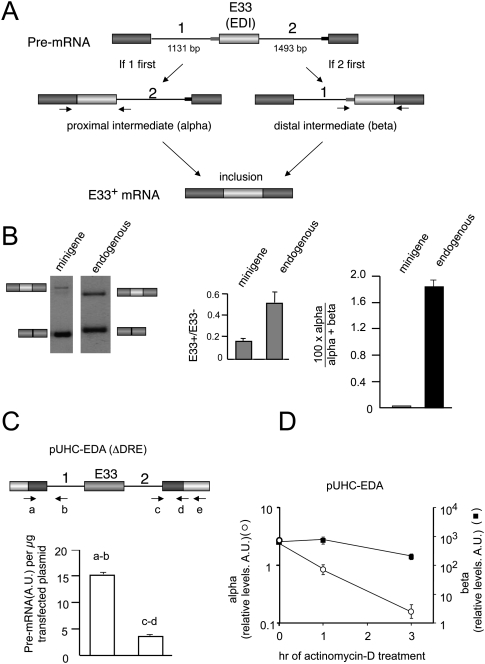
As a first step toward measuring the RRIRs of the introns flanking E33, we set up two real-time RT-PCR reactions, α and β, which amplified the splicing intermediates resulting from either intron 1-first or intron 2-first pathways of intron removal, respectively (Fig. 1A). The PCR strategy was as described before (Kessler et al. 1993), but conditions were adjusted to obtain only one PCR product per primer pair. This was possible because of the very low amplification efficiency of intermediates retaining any of the long introns (Fig. 1A). Human hepatoma Hep3B cells were transfected with an inducible FN E33 minigene (pUHC-EDA), plus the tetracycline-repressible tTA-VP16 transactivator (de la Mata and Kornblihtt 2006), and harvested 48 h later. Total RNA was then isolated, and the two RT-PCR reactions were performed to detect the specific intermediates.
FIGURE 1.
Fibronectin E33 (EDI or EDA) is spliced through a pathway that removes its downstream intron prior to the upstream one. (A) Diagrams of the two possible pathways leading to E33 inclusion and the two resulting splicing intermediates. Thin rectangles depict weak (gray) and strong (black) 3′ splice sites. Arrows depict the primers used to detect and quantify the proximal and distal intermediates, hereafter named α and β, respectively. (B) Hep3B cells transfected with the reporter minigene pUHC-EDA were analyzed by RT-PCR for E33 inclusion ratios of both the minigene and the endogenous E33 using specific primers (left, gels; center, quantification, gray bars). (Right) Real-time RT-PCR analysis for the quantification of splicing intermediates calculated as the %α-intermediate (black bars) as explained in Materials and Methods. (C) Real-time RT-PCR of proximal (a,b) and distal (c,d) pre-mRNA regions of Hep3B cells transfected with a ΔDRE mutant of the reporter minigene pUHC-EDA. Proximal region: Primer b (5′-GCATTCAGACACCCAAGAAC-3′) was used for cDNA synthesis; primers a (5′-TTCTCTGCACAGCTCCTAAG-3′) and b were used for PCR. Distal region: Primer e (5′-GGTATTTGGAGG TCAGCA-3′) was used for cDNA synthesis; primers c (5′-TTGGAACTACGTTTATTTTCC-3′) and d (5′-GCGGCCAGGGGTCACGAT-3′) were used for PCR as previously described (de la Mata et al. 2003). A.U. indicates arbitrary units. (D) Quantification of the α intermediates (empty circles) and β intermediates (black squares) of Hep3B cells transfected with pUCH-EDA at different times after treatment with actinomycin D. A.U. indicates arbitrary units. Calculations show mean ± SD.
We first determined that the two RT-PCR reactions had equivalent efficiencies in order to allow for calculation of the percent abundance of intron 1-first and intron 2-first splicing intermediates [100 × α/(α + β) or 100 × β/(α + β)] (Supplemental Fig. 1). We next determined the 100 × α/(α + β) for the transfected E33 minigene, a parameter that will be used in this study as a measure of the RRIR of the E33 flanking introns, which we will also refer to as “%α-intermediate.” The rationale for this assumption is that these intermediates are short-lived due to their fast downstream processing, but their relative abundances reflect the order of intron removal (Kessler et al. 1993). In parallel, to rule out any artificial results resulting from using a minigene, we designed equivalent primers to study the RRIRs of introns flanking the endogenous E33 exon. Figure 1B shows that E33 inclusion levels are higher for the endogenous gene than for the minigene (Fig. 1B, left panel, center panel). Although 100 × α/(α + β) is also significantly higher for the endogenous gene (Fig. 1B, right panel), in both cases the very low %α-intermediate reveals that the intron 2-first pathway is dramatically more efficient than intron 1-first pathway. Note that both splicing intermediates measured correspond to the fraction of pre-mRNAs that will ultimately lead to the mRNA isoform that includes E33. The exon skipping intermediates are not detected with these PCRs and do not interfere in the analysis. The only consequence of a high exon skipping will be a lower amount of splicing intermediates being detected by the α and β PCRs. Two additional experiments support the observation that excision of the downstream intron is more efficient than that of the upstream one. First, when cells were transfected with a reporter minigene that expresses almost exclusively the E33+ mRNA isoform (ΔDRE construct, see below); i.e., where conversion of pre-mRNA into the E33 exclusion isoform is negligible, the steady-state levels of pre-mRNAs harboring intron 2 are four times lower than those of pre-mRNAs harboring intron 1 (Fig. 1C). Second, if transcription is inhibited with actinomycin D, decay of the α intermediate is faster than that of the β intermediate (Fig. 1D). Although it would be virtually impossible to determine experimentally the proportions of this decay that correspond to splicing or to a putative pre-mRNA degradation, based on recent evidence that FN E33 splicing is mainly cotranscriptional (Pandya-Jones and Black 2009) and that cotranscriptional splicing protects pre-mRNAs from entering degradation pathways (Das et al. 2007), we favor the interpretation that this decay mostly reflects splicing.
We conclude that the splicing of pre-mRNAs committed to include the E33 exon progress through a pathway that preferentially eliminates intron 2 in the first place. Our results are in agreement with a recent report demonstrating that introns flanking both FN exon 25 (EDII or EDB) and FN exon 33 (EDI or EDA) follow the same pathway of intron removal as the one described here (Pandya-Jones and Black 2009). Moreover, this work demonstrates that these intermediates are mostly present in a chromatin-associated fraction and are virtually absent from nucleoplasm and cytoplasmic fractions, pointing to the nascent nature of these RNAs. Together with our results, this raises the question of whether changes in splice site selection could alter the relative order of intron removal in a way that reflects the specific mechanism involved in the regulation.
Different cis-acting mutations enhance E33 inclusion through different intron removal pathways
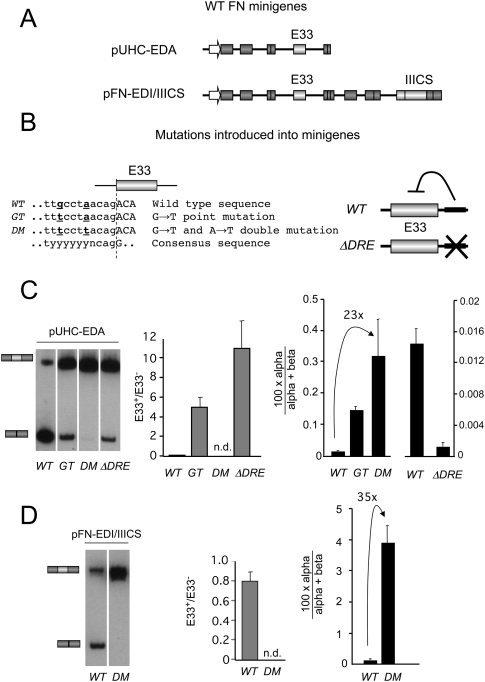
We next asked whether different cis mutations known to increase E33 inclusion by different mechanisms could differentially affect the RRIRs of E33 flanking introns. For this we introduced into the inducible E33 minigene (Fig. 2A, top) a set of previously described simple (GT) and double (DM) mutations (Nogues et al. 2003), or a deletion that eliminates an intronic downstream regulatory element (ΔDRE) (Fig. 2B; Gromak et al. 2008). All three mutations highly increase E33 inclusion levels by means of different mechanisms: GT and DM mutants strengthen the polypyrimidine tract (PPT) of the 3′ splice site (3′ss) upstream of E33 (Nogues et al. 2003) and ΔDRE mutant removes an intronic splicing silencer (Gromak et al. 2008).
FIGURE 2.
Different cis-acting mutations enhance E33 inclusion through different intron removal pathways. (A) Diagrams representing the two basic minigenes used in this study. (B) Mutations introduced in the minigenes shown in A. WT indicates wild-type sequence; GT and DM, G→T and G→T+A→T point mutations, respectively, at the polypyrimidine tract of the 3′ss upstream of E33 (Nogues et al. 2003); ΔDRE, deletion of the intronic downstream regulatory element (Gromak et al. 2008). The minigenes containing two alternative splicing regions, WT (pFN-EDIWT/IIICS) and DM (pFN-EDIC/IIICS), were previously described (Fededa et al. 2005). (C,D) Hep3B cells were transfected with the WT and mutant versions of pUHC-EDA (C) and pFN-EDI/IIICS (D), and analyzed by RT-PCR for E33 inclusion ratios (left), the quantification of E33+/E33− ratios (middle), and the quantification of splicing intermediates (right, black bars) calculated as the %α-intermediate. Calculations show mean ± SD.
Hep3B cells were transiently transfected with different wild-type (WT) and mutant minigenes and assessed for both E33 inclusion and percentage of abundance of α. As previously described, all three mutations highly increased E33 inclusion on the transfected minigene (Fig. 2C, left panel, center panel). However the %α-intermediate differed between them (Fig. 2C, right panel). Both the GT and DM mutations significantly increased the %α-intermediate to extents that were proportional to their effect on exon inclusion; i.e., the higher the increase in E33 inclusion the higher the increase in the 100 × α/(α + β). This is consistent with these mutations acting by strengthening the PPT of the 3′ss upstream of E33 to different extents, and thus preferentially favoring the excision of the upstream intron (Nogues et al. 2003). On the contrary, the ΔDRE mutant significantly decreased the percentage of α, which is consistent with a different mechanism of action of this mutant to promote E33 inclusion.
Together these results demonstrate that different cis-acting mutations that enhance E33 exon inclusion into the mature mRNA can act by enhancing opposite intron removal pathways for introns flanking the alternative exon.
The order of intron removal does not depend on the relative gene position of E33
FN E33 is placed as the penultimate exon within the reporter minigene, an architecture that is clearly different from the endogenous FN gene, where E33 has more than 10 exons downstream. To rule out that the measured %α-intermediate was a consequence of E33 being the penultimate exon within the minigene, an additional control was performed. We used an alternative splicing reporter minigene (pFN-EDIWT/IIICS) (Fededa et al. 2005) in which E33 is placed in a position upstream of subsequent constitutive and alternative exons, including the alternative IIICS alternative region (Fig. 2A, bottom). We then transfected Hep3B cells with pFN-EDIWT/IIICS or with a variant bearing a DM E33 (pFN-EDIC/IIICS). Virtually the same result was obtained with these minigenes compared with the regular minigenes; i.e., the %α was lower with pFN-EDIWT/IIICS than with the DM version (pFN-EDIC/IIICS) used as reporters (Fig. 2D). This points to a regulation that is independent of the exon being placed at the penultimate position, and it suggests that the effect is not mediated by a direct coupling with the 3′end processing of the mRNA.
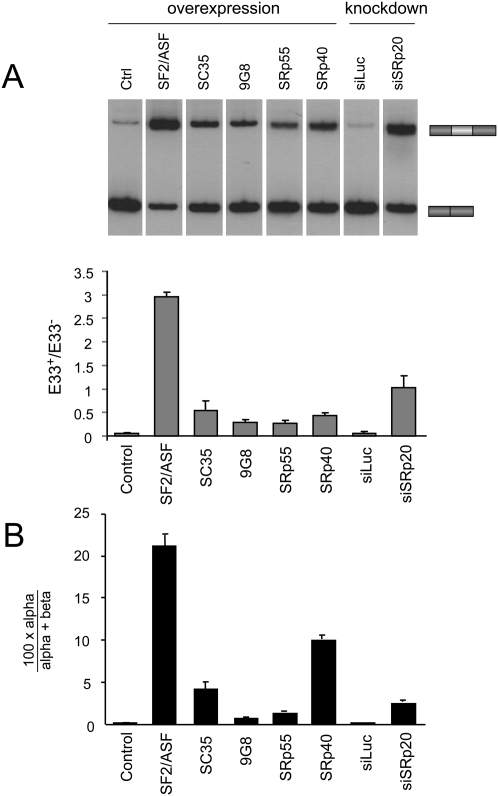
SR proteins regulate E33 inclusion through different order of intron removal pathways
We next asked whether the effect of different SR proteins on E33 inclusion was correlated with the RRIRs of E33 flanking introns. A clear correlation between the effect on the RRIRs and E33 inclusion could suggest a similar mechanism for the different SR proteins. Hep3B cells were transfected with the inducible WT minigene and identical amounts of expression vectors for different SR proteins (SF2/ASF, 9G8, SC35, SRp40, and SRp55). Since these constructs share the same plasmid backbone and promoter, and transfection efficiencies were identical, we assumed similar expression levels as done before (Cramer et al. 1999). Figure 3A shows that E33 inclusion is stimulated by all tested SR proteins to different extents, with SF2/ASF having the strongest effect, as previously reported (Cramer et al. 1999). We then determined the respective effects on the RRIRs by measuring the %α-intermediate for E33 inclusion. As shown in Figure 3B, SF2/ASF displayed the largest effect in increasing the %α-intermediate; i.e., favoring the removal of the upstream intron, with the other SR proteins showing smaller effects yet in the same direction as SF2/ASF. This result is consistent with previous results demonstrating that SF2/ASF binds to an exonic splicing enhancer within E33 and stimulates the use of the suboptimal 3′ss upstream of E33 (Lavigueur et al. 1993). The correlation of E33 inclusion ratios with the %α-intermediate for the different SR proteins was significantly high (Pearson correlation coefficient 0.95), suggesting that all of them may act through a similar mechanism; i.e., favoring the removal of the upstream exon. However, the correlation is not perfect, and the magnitudes of the two effects differ among the different SR protein members. In contrast to the above studied SR proteins, SRp20 has a known negative effect on E33 inclusion (Cramer et al. 1999; de la Mata and Kornblihtt 2006). In order to more effectively determine its effect on both E33 inclusion and on the RRIR, we performed a siRNA-mediated knockdown of SRp20. As previously shown, knocking down SRp20 by transfecting a specific siRNA (siSRp20) greatly enhanced E33 inclusion compared with the nonspecific siRNA control (siLuc) (Fig. 3A). The %α-intermediate for E33 inclusion was also significantly increased when transfecting with siSRp20 (Fig. 3B), allowing us to infer that the presence of SRp20 favors the elimination of the downstream intron in the first place.
FIGURE 3.
SR proteins regulate E33 inclusion through different order of intron removal pathways. Hep3B cells were cotransfected with the reporter minigene pUHC-EDA and either vectors expressing different SR proteins or SR protein-specific siRNAs. (A) Radioactive RT-PCR analysis of E33 isoforms and quantification of E33+/E33− ratios (gray bars). (B) Real-time RT-PCR analysis for the quantification of splicing intermediates calculated as the %α-intermediate (black bars). Calculations show mean ± SD.
We conclude that SR proteins affect the RRIRs of introns flanking FN E33 in a way that is consistent with their proposed mechanism of action on E33 inclusion. These results provide additional evidence supporting the use of the RRIR as an indicator of the underlying mechanisms of splice site selection.
Inhibition of pol II elongation does not affect the relative rate of intron removal
Up-to-date in vivo approaches used to explain the mechanisms responsible for the regulation of alternative splicing are scarce or are somehow descriptive. They usually lack time considerations necessary for addressing the problem at its real dimension; i.e., taking into account the dynamic nature of the spliceosome assembly and its coupling with transcription and other pre-mRNA processing reactions. For instance, most of the approaches rely on steady-state levels of pre-RNAs and mRNAs. We hypothesized that the RRIR could be used as a useful parameter to dissect the mechanisms of action of several treatments affecting E33 inclusion through pol II elongation. Our approach also relies on measuring steady-state levels of splicing intermediates; however, given these are short-lived species due to their fast downstream processing, we speculated that they could reflect early events happening in close connection with transcription. This is consistent with these splicing intermediates being intimately associated with the chromatin nuclear fraction (Pandya-Jones and Black 2009). Previous reports have also highlighted this by showing that splicing can take place rapidly while transcription is still ongoing (Beyer and Osheim 1988; Lacadie et al. 2006).
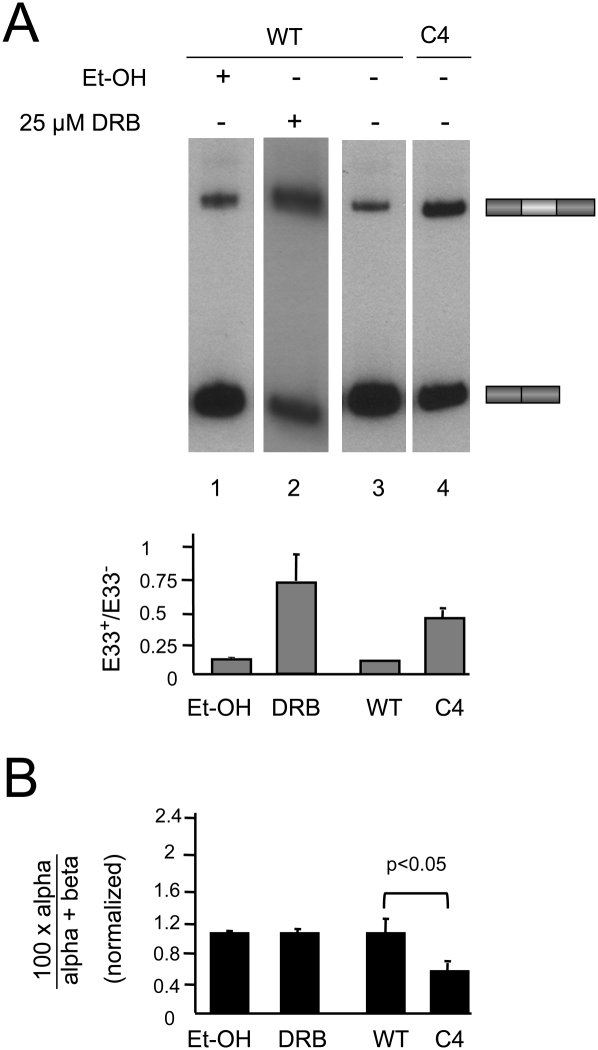
For this purpose, we transfected Hep3B cells with the inducible E33 minigene in the presence of tetracycline to keep its transcription off. Twenty-four hours later, tetracycline was washed out and cells were treated with 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) or camptothecin, two drugs reported to affect elongation through different mechanisms. DRB is known to inhibit pTEFb and in turn decrease pol II phosphorylation at Ser 2 within the C-terminal domain (CTD) heptads of pol II (Saunders et al. 2006), resulting in inhibition of transcriptional elongation as demonstrated in vivo and in real time by imaging methodologies (Darzacq et al. 2007). In these studies, DRB concentrations as high as 150 μM were shown to only inhibit the elongation phase of transcription. However, in a more recent study (Singh and Padgett 2009), similar concentrations of DRB were used to fully inhibit transcription. In order to minimize the risk of full inhibition, we treated the cells with lower doses of DRB (25 μM) and harvested them 8 h after drug addition. Upon this treatment, we observed an enhancement of E33 inclusion (Fig. 4A, lanes 1,2) as previously reported (Nogues et al. 2002). Notably, this was not accompanied by a change in the RRIRs of E33 flanking introns (Fig. 4B). We next tested camptothecin, which is known to inhibit topoisomerase I in a template-dependent manner, thus slowing down pol II elongation on some target genes (Listerman et al. 2006). Similar to DRB, treatment of cells with 1 μM camptothecin for 16 h did not affect the RRIRs of E33 flanking introns but increased E33 inclusion (data not shown). Due to the long time needed to see effects of camptothecin on E33 splicing, we cannot rule out indirect effects of this drug.
FIGURE 4.
Inhibition of pol II elongation does not affect the relative rates of intron removal. Hep3B cells were transfected with the reporter minigene pUHC-EDA and treated with the ethanol vehicle (lane 1) or 25 μM DRB for 8 h (lane 2) following induction by withdrawal of tetracycline. (Lanes 3,4) Cells were cotransfected in the presence of tetracycline with the reporter minigene pUHC-EDA and expression vectors for human α-amanitin resistant pol II large subunits (hRpb1), WT (lane 3) and slow mutant (C4, lane 4). Minigene transcription was induced 18 h post-transfection by washing out tet and adding α-amanitin (10 μg mL−1) to inhibit endogenous pol II transcription. Cells were harvested 20 h post-induction and used for subsequent assays. (A) Results of radioactive RT-PCR analysis of E33 isoforms and quantification of E33+/E33− ratios (gray bars). (B) Real-time RT-PCR analysis for the quantification of splicing intermediates calculated as %α-intermediate (black bars). Calculations show mean ± SD.
To investigate the effects of other means of inhibiting pol II elongation, we cotransfected the E33 inducible minigene together with a vector expressing the large subunit of pol II bearing mutations (C4 mutant) that confers both a reduced elongation rate and α-amanitin resistance (de la Mata et al. 2003; Boireau et al. 2007). The human C4 pol II mutant was shown to inhibit elongation in vivo and in real-time experiments (Boireau et al. 2007). Twenty-four hours post-transfection, cells were treated with 10 μg/mL α-amanitin and harvested 16 h later. Whereas the slow pol II increased E33 inclusion by fourfold (Fig. 4A, lanes 3,4), it did not increase the %α-intermediate (it even showed a small but statistically significant decrease) (Fig. 4B). Together with the drug treatment experiments, this shows that reducing pol II elongation affects E33 alternative splicing without significantly altering the RRIR, whereas other means of regulating E33 inclusion, such as cis mutations on splice sites or SR proteins, do change the RRIR, revealing different underlying mechanisms of action.
DISCUSSION
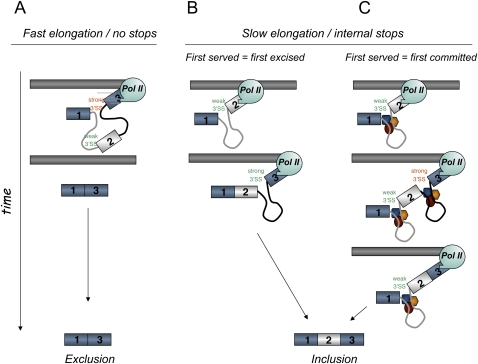
Our results indicate that different cis-acting mutations and trans-acting factors that affect the inclusion levels of the FN E33 cassette exon into mature mRNA use mechanisms that differ in the preferential order of intron removal. However, we found that three treatments reported to inhibit pol II elongation using different mechanisms coincide in promoting E33 inclusion, but none of them affects the RRIR, as could be predicted by the most parsimonious interpretation of the “first come, first served” model of splice site selection (Aebi and Weissmann 1987), where “first served” implies “first excised” (Fig. 5A,B). E33 skipping occurs because the 3′ss of the upstream intron is suboptimal compared with the 3′ss of the downstream intron. In fact, in our previous interpretations we have speculated that if the polymerase paused anywhere between these two sites, elimination of the upstream intron would be favored. Once the pause was passed or the polymerase proceeded, there would be no option for the splicing machinery but to excise the downstream intron, which would lead to higher exon inclusion. Alternatively, highly elongating pol II would favor the simultaneous presentation of both introns to the splicing machinery (Fig. 5A), a situation in which the stronger 3′ss of the downstream intron would outcompete the weaker 3′ss of the upstream intron, resulting in exon skipping (de la Mata et al. 2003; Howe et al. 2003). In view of the results shown in Figure 4, we must now abandon this interpretation, at least for FN E33. This does not mean we should abandon the “first come, first served” model, but that we should rethink it. Indeed, one could argue that reducing elongation may act on the overall recruitment of splicing factors to the splice sites without affecting the kinetics of intron removal. Keeping this view in mind, slow elongation would favor recruitment of splicing factors to the pre-mRNA that would favor E33 recognition and commitment to subsequent inclusion. Once commitment is achieved, the order of intron removal becomes irrelevant to guarantee inclusion. According to this interpretation, which we used before for the role of elongation on E1a 5′ splice selection (de la Mata et al. 2003), “first served” would not be equivalent to “first excised” but to “first committed” (Fig. 5C).
FIGURE 5.
Alternative models for the “first come, first served” mechanism of splice site selection. (A) Fast elongation promotes usage of the stronger downstream 3′ splice site. (B) Slow elongation causes preferential excision of the upstream intron (first served equals first excised). (C) Slow elongation causes commitment to E33 inclusion via recruitment of splicing factors (first served equals first committed). Both introns are excised individually and in an order that is independent of elongation.
Recent in vitro studies have discriminated between two levels of commitment during spliceosome assembly: exon/intron definition (or early commitment to a splicing pathway) and juxtaposition of exons within the spliceosome (or splicing pairing) (Lim and Hertel 2004; Kotlajich et al. 2009). These two levels of commitment do not necessarily coincide with the formation of the same spliceosome complex and, in principle, splicing regulation could impact on different commitment levels depending on the underlying mechanism involved. An overall increase in exon definition of a given alternative exon due to enhanced early recruitment of general splicing factors would not necessarily be accompanied by a preferred subsequent order of exon juxtaposition to its flanking exons. However, regulatory mechanisms acting at a later stage, such as A complex formation, might impact on the juxtaposition to its flanking exons, thereby affecting the RRIRs of introns flanking the alternative exon.
We can thereby speculate that the effect of slowing down transcription elongation might impact mostly on early exon definition events without affecting the subsequent juxtaposition of exons, as evidenced by the fact that the RRIR is unaffected after elongation reduction induced by different treatments. This situation clearly differs from the effect of SR protein or cis-acting mutations on regulatory regions, which might mostly impact the juxtaposition of exons after exon definition, thereby affecting the RRIR.
A recent report shows results that complement our findings (Pandya-Jones and Black 2009). In that study, the investigators show that introns flanking FN E33 (EDI or EDA) and FN E25 (EDII or EDB) follow the same order of intron removal as described in this work, with the downstream intron being removed prior to the upstream one. Most interestingly, they show that these intermediates almost exclusively purify with a chromatin-associated RNA fraction (by over 85%), meaning these are most probably nascent transcripts, and confirming the cotranscriptional nature of E33 splicing. They also show that not all alternative exons follow the same splicing pathway. In fact, c-Src exon N1 follows the opposite pathway, with the upstream intron being removed prior to the downstream one. It is known that different mechanisms govern the regulation of these alternative exons (Cramer et al. 1999; de la Mata et al. 2003; Underwood et al. 2005; de la Mata and Kornblihtt 2006), and it will be interesting to study whether they respond in a similar way to changes in transcriptional elongation.
In summary, our results suggest that measuring the order of intron removal may be a useful parameter to consider when studying exon skipping regulation in vivo, serving as a straightforward read-out discriminating among different mechanisms of alternative splice site selection.
MATERIALS AND METHODS
Plasmids
Alternative splicing reporter minigenes
pUHC-EDA has been previously described (de la Mata and Kornblihtt 2006). pUHC-EDA mutants GT and DM were generated by replacing the fragment HindIII-NcoI into the pUHC-EDA minigene. ΔDRE is based on pUHC-EDA but bears a deletion between sites NcoI-NdeI according to a previous report (Gromak et al. 2008). Tandem reporter minigenes bearing a WT EDI exon (pFN-EDIWT/IIICS) or DM EDI exon (pFN-EDIC/IIICS) were previously described (Fededa et al. 2005).
Pol II expression vectors
The expression vectors for human α-amanitin resistant pol II large subunits (hRpb1), WT and slow mutant (C4), have been previously described (de la Mata et al. 2003). hRpb1 C4 mutant (pAT7Rpb1αAmrR749H) expresses a hRpb1 with an amino acid change (R749H) that is equivalent to the Drosophila C4 mutation.
SR expression vectors
Plasmids expressing SF2/ASF, SC35, 9G8, SRp55, SRp40, and SRp20 have been described elsewhere (Cramer et al. 1999).
Transfections
Conditions for transfection of human hepatoma Hep3B cells were repeated from our previous report (de la Mata and Kornblihtt 2006). Briefly, Hep3B cells were cotransfected with lipofectamine following the manufacturer's instructions with a tetracycline (tet)-controlled transactivator (tTA) and a tet-inducible minigene. Cells were harvested 48 h post-transfection and total RNA extracted. When cotransfecting with vectors for human α-amanitin resistant pol II large subunits (hRpb1), tet (1 μg mL−1) was added shortly after transfection, and 18 h post-transfection, cells were induced by washing out tet and adding tet-free medium with α-amanitin (10 μg mL−1) to shut down endogenous pol II transcription. Cells were harvested 20 h post-induction and used for subsequent assays.
For RNAi functional knock-down experiments, the following siRNAs duplexes were used (only sense sequence shown): SRp20 (Stealth siRNA) 5′-GGACUGUAAGGUUUAUGUAGGCAAU-3′; Luc (siRNA against GL3 Luciferase) 5′-CUUACGCUGAGUACUUCGAdTdT-3′. DRB and camptothecin treatments were done simultaneously with minigene induction at final concentrations of 25 μM and 1 μM, respectively.
RNA analysis
RNA preparation and reverse transcriptase reactions were previously described (Kadener et al. 2001). RT was performed using the following oligo specific to the 3′ region of the EDI minigene: pSVcDNA (5′-GGTATTTGGAGGTCAGCA-3′). Radioactive PCR amplification of cDNA splicing isoforms was performed using reported specific primers for E33 (EDI or EDA) (Cramer et al. 1999). Quantification of splicing intermediates was carried out by real-time RT-PCR using a Mastercycler realplex real-time PCR system. The following primers were used in the analysis: pSVcDNA (5′-GGTATTTGGAGGTCAGCA-3′) was used for cDNA synthesis; 5′J (5′-CACTGCCTGCTGGTGACTCGA-3′) and Donor_Rev (5′-CCACCTGCTGCTGCAATTAAC-3′) were used for PCRα; and Py_For (5′-TCTTACCATTAATTTGCCTAAC-3′) and 3′J (5′-GCGGCCAGGGGTCACGAT-3′) were used for PCRβ, except as shown in Figure 2 and for the minigene in Figure 1, where Py_For 2 (5′-TCACAACTTGCTTTCACATAAC-3′) and 3′J were used instead.
For measuring endogenous α and β splicing intermediates, we used the following primers: AMPL5 (5′-GCAGCCCACAGTGGAGTATG-3′) and Donor_Rev were used for PCRα; Py_For and FN E34_Rev were used for PCRβ.
Endogenous E33 splicing isoforms were detected by radioactive RT-PCR using the same conditions as for the minigene but with primer FN E34-Rev (5′-CTGTCAGGAGCAAGGTTGATTTC-3′) for reverse transcription and primers hFN For (5′-AGCCCCGCAAGCAGCAAGCC-3′) and FN E34_Rev for the radioactive PCR.
Quantification of α and β intermediates
Splicing intermediates were quantified using real-time RT-PCR and following the calculations described below. For both PCR reactions (α and β), a calibration curve was performed from serial dilutions of the same cDNA preparation obtained by reverse transcription with the oligo pSVcDNA as a primer. α and β PCRs were performed on each sample, and the threshold cycle (Ct) values were used to calculate the relative amounts of each intermediate by interpolating them into the corresponding calibration curve. The resulting relative amounts are arbitrary values arising from the dilution factors of the standards and are dimensionless quantities (they lack units). Since the absolute abundances of α and β intermediates differ to a large extent by an unknown factor (β intermediates being much more abundant), and because the same set of arbitrary dilution values was used for the standards of the two calibration curves, we needed to rescale the values of the α intermediate relative to those of the β intermediate in order to compare them directly. This was possible given that the two PCRs have virtually identical efficiencies (0.99 and 0.97) (see Supplemental Fig. 1). For the rescaling, we interpolated the α Ct values into both α and β calibration curves, generating two relative amount values, Rα and Rβ, respectively (see Supplemental Table 1). A ratio was then calculated by dividing them (Rα/Rβ), and a rescaling factor (RF) was generated by taking the average Rα/Rβ ratio. α Relative amounts were finally rescaled (relative to β values) by dividing the initial α relative amounts (obtained by interpolation into the α calibration curve) by the RF. This strategy allows rescaling the PCR data while keeping the original internal relationship between values, and thus not introducing further distortions within each PCR.
The RRIR was then plotted as the %α-intermediate relative to the total intermediate levels following the formula: 100 × α/(α + β).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Valeria Buggiano, Ezequiel Petrillo, and Juan Pablo Fededa for their invaluable help, and the members of the Srebrow and Kornblihtt laboratories for their support and useful discussions. This work was supported by grants to A.R.K. from the Agencia Nacional de Promoción de Ciencia y Tecnología of Argentina (ANPCYT), the University of Buenos Aires, and the European Alternative Splicing Network (EURASNET). C.L. is recipient of a fellowship from ANPCYT, and A.R.K. is a career investigator from the Consejo Nacional de Investigaciones Científicas y Técnicas of Argentina (CONICET) and an international research scholar of the Howard Hughes Medical Institute. M.d.l.M. designed, performed, and interpreted the results of most experiments, and co-wrote the manuscript draft. C.L. performed some experiments. A.R.K. coordinated the work, designed and interpreted experiments, and co-wrote the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1993510.
REFERENCES
- Aebi M, Weissmann C 1987. Precision and orderliness in splicing. Trends Genet 4: 102–107 [Google Scholar]
- Beyer AL, Osheim YN 1988. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes & Dev 2: 754–765 [DOI] [PubMed] [Google Scholar]
- Boireau S, Maiuri P, Basyuk E, de la Mata M, Knezevich A, Pradet-Balade B, Backer V, Kornblihtt A, Marcello A, Bertrand E 2007. The transcriptional cycle of HIV-1 in real-time and live cells. J Cell Biol 179: 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Caceres JF, Cazalla D, Kadener S, Muro AF, Baralle FE, Kornblihtt AR 1999. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol Cell 4: 251–258 [DOI] [PubMed] [Google Scholar]
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH 2007. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol 14: 796–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R 2007. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell 26: 867–881 [DOI] [PubMed] [Google Scholar]
- de la Mata M, Kornblihtt AR 2006. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat Struct Mol Biol 13: 973–980 [DOI] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell 12: 525–532 [DOI] [PubMed] [Google Scholar]
- Fededa JP, Petrillo E, Gelfand MS, Neverov AD, Kadener S, Nogues G, Pelisch F, Baralle FE, Muro AF, Kornblihtt AR 2005. A polar mechanism coordinates different regions of alternative splicing within a single gene. Mol Cell 19: 393–404 [DOI] [PubMed] [Google Scholar]
- Gornemann J, Kotovic KM, Hujer K, Neugebauer KM 2005. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell 19: 53–63 [DOI] [PubMed] [Google Scholar]
- Gromak N, Talotti G, Proudfoot NJ, Pagani F 2008. Modulating alternative splicing by cotranscriptional cleavage of nascent intronic RNA. RNA 14: 359–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- House AE, Lynch KW 2006. An exonic splicing silencer represses spliceosome assembly after ATP-dependent exon recognition. Nat Struct Mol Biol 13: 937–944 [DOI] [PubMed] [Google Scholar]
- Howe KJ, Kane CM, Ares M Jr 2003. Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. RNA 9: 993–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Cramer P, Nogues G, Cazalla D, de la Mata M, Fededa JP, Werbajh SE, Srebrow A, Kornblihtt AR 2001. Antagonistic effects of T-Ag and VP16 reveal a role for RNA pol II elongation on alternative splicing. EMBO J 20: 5759–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler O, Jiang Y, Chasin LA 1993. Order of intron removal during splicing of endogenous adenine phosphoribosyltransferase and dihydrofolate reductase pre-mRNA. Mol Cell Biol 13: 6211–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G 2004. Multiple links between transcription and splicing. RNA 10: 1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlajich MV, Crabb TL, Hertel KJ 2009. Spliceosome assembly pathways for different types of alternative splicing converge during commitment to splice site pairing in the A complex. Mol Cell Biol 29: 1072–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotovic KM, Lockshon D, Boric L, Neugebauer KM 2003. Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol Cell Biol 23: 5768–5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacadie SA, Rosbash M 2005. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Mol Cell 19: 65–75 [DOI] [PubMed] [Google Scholar]
- Lacadie SA, Tardiff DF, Kadener S, Rosbash M 2006. In vivo commitment to yeast cotranscriptional splicing is sensitive to transcription elongation mutants. Genes & Dev 20: 2055–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavigueur A, La Branche H, Kornblihtt AR, Chabot B 1993. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes & Dev 7: 2405–2417 [DOI] [PubMed] [Google Scholar]
- Lim SR, Hertel KJ 2004. Commitment to splice site pairing coincides with A complex formation. Mol Cell 15: 477–483 [DOI] [PubMed] [Google Scholar]
- Listerman I, Sapra AK, Neugebauer KM 2006. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol 13: 815–822 [DOI] [PubMed] [Google Scholar]
- Nogues G, Kadener S, Cramer P, Bentley D, Kornblihtt AR 2002. Transcriptional activators differ in their abilities to control alternative splicing. J Biol Chem 277: 43110–43114 [DOI] [PubMed] [Google Scholar]
- Nogues G, Munoz MJ, Kornblihtt AR 2003. Influence of polymerase II processivity on alternative splicing depends on splice site strength. J Biol Chem 278: 52166–52171 [DOI] [PubMed] [Google Scholar]
- Pandya-Jones A, Black DL 2009. Co-transcriptional splicing of constitutive and alternative exons. RNA 15: 1896–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT 2006. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol 7: 557–567 [DOI] [PubMed] [Google Scholar]
- Schwarze U, Starman BJ, Byers PH 1999. Redefinition of exon 7 in the COL1A1 gene of type I collagen by an intron 8 splice-donor-site mutation in a form of osteogenesis imperfecta: influence of intron splice order on outcome of splice-site mutation. Am J Hum Genet 65: 336–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Padgett RA 2009. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol 16: 1128–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL 2005. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol 25: 10005–10016 [DOI] [PMC free article] [PubMed] [Google Scholar]