Abstract
With over 200 million people infected with hepatitis C virus (HCV) worldwide, there is a need for more effective and better-tolerated therapeutic strategies. The HCV genome is a positive-sense; single-stranded RNA encoding a large polyprotein cleaved at multiple sites to produce at least ten proteins, among them an error-prone RNA polymerase that confers a high mutation rate. Despite considerable overall sequence diversity, in the 3′-untranslated region of the HCV genomic RNA there is a 98-nucleotide (nt) sequence named X RNA, the first 55 nt of which (X55 RNA) are 100% conserved among all HCV strains. The X55 region has been suggested to be responsible for in vitro dimerization of the genomic RNA in the presence of the viral core protein, although the mechanism by which this occurs is unknown. In this study, we analyzed the X55 region and characterized the mechanism by which it mediates HCV genomic RNA dimerization. Similar to a mechanism proposed previously for the human immunodeficiency 1 virus (HIV-1) genome, we show that dimerization of the HCV genome involves formation of a kissing complex intermediate, which is converted to a more stable extended duplex conformation in the presence of the core protein. Mutations in the dimer linkage sequence loop sequence that prevent RNA dimerization in vitro significantly reduced but did not completely ablate the ability of HCV RNA to replicate or produce infectious virus in transfected cells.
Keywords: HCV, core protein, X RNA, RNA kissing complex, fluorescence spectroscopy, 2-amino purine
INTRODUCTION
First identified in 1989 (Choo et al. 1989), it is now estimated that hepatitis C virus (HCV), a blood-borne pathogen, persistently infects over 200 million people worldwide (3% of human population) (Lindenbach et al. 2007; Paula et al. 2009). A major cause of liver disease globally and the leading cause of liver transplantation in North America, chronic HCV infection is often associated with significant liver disease, including cirrhosis and liver cancer (Brass et al. 2007; Qureshi 2007). Although as many as 50% of patients with chronic genotype 1 HCV infection may be cured by the current standard-of-care therapy with pegylated interferon-α and ribavirin, many infected patients are either intolerant of this treatment or fail to achieve a sustained viral response (Hadziyannis et al. 2004; Muir et al. 2004).
HCV is an enveloped virus belonging to the Flaviviridae family. Its genome is a 9.6-kb positive-sense RNA molecule containing a long open reading frame (ORF), flanked by highly structured and conserved 5′- and 3′-untranslated regions (UTRs) (Fig. 1A; Lindenbach et al. 2005; Moradpour et al. 2007). This ORF encodes a single polyprotein of ∼3000 amino acids, which is proteolytically cleaved into at least 10 structural and nonstructural viral proteins (Tan et al. 2002). The nonstructural proteins are NS2, NS3, NS4A, NS4B, NS5A, and NS5B, while E1, E2, p7, and the capsid core protein constitute the structural proteins (Penin et al. 2004). The nonstructural NS5B protein is an error-prone RNA-dependent RNA polymerase, which results in a high mutation rate during replication of the virus (Penin et al. 2004; Moradpour et al. 2007).
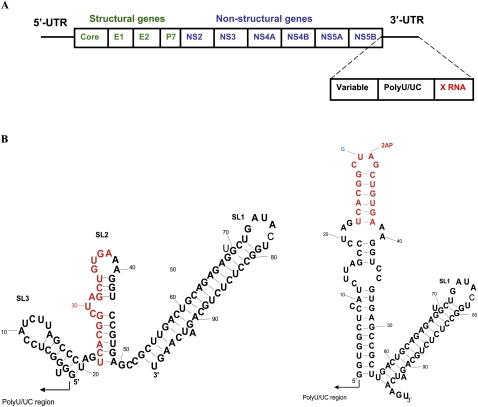
FIGURE 1.
(A). Schematic representation of HCV genome. (B) Alternate predicted conformations of X RNA obtained by Mfold. The DLS region is highlighted in red. The mutations introduced in the X98_G30_31AP are shown in blue and red, respectively.
The 5′- and 3′-UTRs contain cis-acting elements that are crucial for RNA replication and translation (Ito and Lai 1997; Song et al. 2006). The 341-nucleotide (nt) 5′-UTR is a complex, four-domain structure that contains an internal ribosome entry site (IRES), which directs cap-independent translation of the ORF, as well as upstream signals required for viral RNA synthesis (Tsukiyama-Kohara et al. 1992; Wang et al. 1993). The 225-nt 3′-UTR has a tri-domain organization, with a nonconserved 40-nt variable region, a poly (U/UC) stretch of 20–200 nt, and a 3′-terminal, conserved 98-nt sequence named X-RNA (Fig. 1A; Kolykhalov et al. 1996; Blight and Rice 1997).
The 3′ X RNA region is essential for RNA replication and may also facilitate viral protein translation directed by the viral IRES (Song et al. 2006; You and Rice 2008). As shown in Figure 1B, there are two alternate conformations predicted for the 3′-UTR sequence when analyzed by the MFold program (Ivanyi-Nagy et al. 2006). Both predicted conformations include the formation of the extreme 3′ terminal SL1 stem–loop; however, they differ with regard to the folding of a 55-nt-long upstream segment, which we name the X55 RNA region in this study. Two stem–loops (SL2 and SL3) are present within the X55 region in one predicted conformation (“SL2” and “SL3”) (Fig. 1B, left), whereas an extended single stem–loop containing internal bulges is present in the second (Fig. 1B, right). While chemical probing data and mutational analysis both support the existence of the 3′SL1 stem–loop at the extreme 3′-end (Blight and Rice 1997; Yi and Lemon 2003a; Dutkiewicz and Ciesiolka 2005), the structure adopted by the X55 RNA region (which is 100% conserved in its sequence among different HCV strains) remains controversial. No homologous sequences have been found in related viruses, in phylogenetically distinct viruses, or in cellular mRNAs (Dutkiewicz and Ciesiolka 2005). While the reasons underlying the extraordinarily high level of sequence conservation within the unique X55 region is unknown, it is likely due, at least in part, to the need to maintain higher ordered RNA structures required for essential processes in the viral life cycle.
Previous studies suggest that the X55 region is involved in long-range kissing interactions with a conserved stem–loop located in the NS5B coding region, which resides at the 3′ end of the ORF (named 5BSL3.2) (Friebe et al. 2005; Diviney et al. 2008; You and Rice 2008). The biological significance of these interactions has been proven in the context of subgenomic HCV RNA replicons, as well as in a recently developed system allowing for production of infectious HCV in cell cultures (You and Rice 2008); mutations in either of the involved stem–loops (5BSL3.2 or X RNA SL2) that altered the complementarity of the sequences involved in the proposed kissing loop interactions led to a complete block of RNA replication, while complementary mutations restored this function.
Interestingly, the same X55 region contains a 16-nt palindromic sequence, which has been suggested to be responsible for homodimerization of the HCV RNA in vitro in the presence of the HCV core protein (Fig. 1B, highlighted in red; Ivanyi-Nagy et al. 2006). This 16-nt palindromic HCV sequence was named the dimer linkage sequence (DLS), in analogy to a sequence with similar function in human immunodeficiency virus type 1 (HIV-1) (Ivanyi-Nagy et al. 2006). The mechanism by which homodimers of the HCV genome might form is unknown. Moreover, the biological significance of the homodimerization of the genome and its relevance to the HCV life cycle have yet to be established. The finding that the HCV genome dimerizes in vitro is surprising. Dimerization of genomic RNA is common to the Retroviridae, where it allows two copies of the genome to be encapsidated in the virion (Kung et al. 1976; Bender et al. 1978; Maisel et al. 1978; Murti et al. 1981). However, this is not known to occur in the Flaviviridae family, and the HCV virion is believed to contain a single copy of the genomic RNA. Thus, dimerization of the HCV genome, if it occurs within infected cells (as it does in vitro in the presence of the core protein), is likely to play a different role in the viral life cycle.
Here, we have studied the mechanism by which the highly conserved HCV X55 region mediates the dimerization of the HCV genome. We show that this process involves the formation of a homodimeric kissing complex intermediate involving the HCV DLS region, and that this is converted to a more stable extended duplex by the viral core protein. Since the involvement of the DLS region in this process would preclude its ability to engage in the long-range kissing interaction with the NS5B-coding region that is known to be essential for RNA replication, we propose that the DLS region might have dual, mutually exclusive functions in defining alternative conformations of the 3′X RNA. These alternate conformations may constitute a molecular switch that directs plus-strand genomic RNA from RNA synthesis to packaging, and eventually to cellular export.
RESULTS AND DISCUSSION
HCV RNA dimerization occurs via a kissing complex intermediate
The HCV X RNA region has been shown to be involved in the in vitro homodimerization of HCV genomic RNA in the presence of the viral core protein (Ivanyi-Nagy et al. 2006). This is an interesting result considering that a role for genomic RNA dimerization has thus far been described only in the life cycle of retroviruses (Kung et al. 1976; Bender et al. 1978; Maisel et al. 1978; Murti et al. 1981). The sequence responsible for HCV RNA dimerization is a 16-nt palindrome within X RNA (Fig. 1B, highlighted in red), named DLS in analogy with the DLS sequence of HIV-1, which is a 23-nt palindrome that has been shown to have a major role in the selection of the copackaged RNA partner in the HIV-1 virion in vivo (Moore et al. 2007). In HIV-1, genomic RNA dimerization initially involves the formation of a kissing complex intermediate via the Watson–Crick pairing of the DLS, which is converted to an extended duplex in the presence of the HIV-1 nucleocapsid (NC) NCp7 protein (Rist and Marino 2002; Mihailescu and Marino 2004). A similar mechanism of RNA genomic dimerization has been demonstrated for the murine leukemia virus, and this has been proposed to be a general feature of other members of the Retroviridae, including viruses such as avian leucosis sarcoma virus and Mason-Pfizer monkey virus (Aagaard et al. 2004).
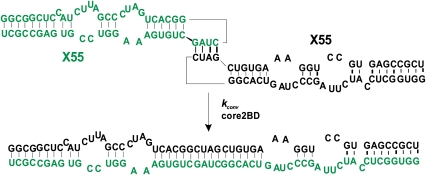
Given the previous report of HCV RNA dimerization (Ivanyi-Nagy et al. 2006), we asked whether a mechanism of dimerization similar to that taking place in Retroviridae might also occur in the case of HCV, a member of the Flaviviridae family. Specifically, we considered the possibility that HCV dimerization might involve the initial formation of a kissing complex intermediate engaging its DLS region, in the absence of the core protein (Fig. 2, top), which in the presence of the core protein is converted to a more stable, extended duplex conformation (Fig. 2, bottom). Since the structure adopted by the first 55 nt of the X RNA segment of HCV (which includes the 16-nt DLS sequence) is uncertain, we designed a series of experiments to test this hypothesis using the entire 55-nt segment, which will be referred to here as X55 RNA.
FIGURE 2.
Model for the kissing complex formed by the X55 RNA (top) and its proposed conversion into the more thermodynamically stable duplex conformation (bottom) in the presence of the core 2BD peptide. The two X55 RNA molecules are identical, but they are illustrated in different colors for clarity purposes.
Native gel electrophoresis studies
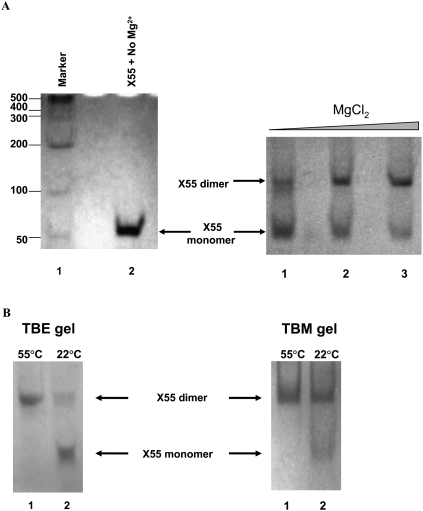
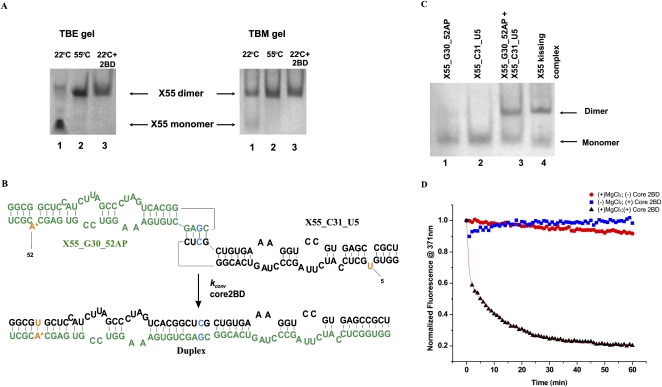
The formation of RNA kissing complexes requires Mg2+ ions (Marquet et al. 1991; Ohuchi and Nakamura 2007). Thus, the first step in testing the above hypothesis was to investigate the conformational status of X55 RNA, whether it exists as a monomer or dimer in the absence of Mg2+ ions, and subsequently to determine if X55 RNA dimerizes in the absence of the core protein and in a MgCl2-dependent manner. In the absence of Mg2+ ions, X55 RNA is monomeric, as evidenced by its migration pattern on a native TBE gel (Fig. 3A, left panel). However, as shown in Figure 3A, at MgCl2 concentrations in the range of 1–10 mM, there are two bands present on a native TBM gel of X55 RNA, with the monomer band that is most intense in the presence of 1 mM MgCl2 (Fig. 3A, right panel, lane 1) decreasing in intensity with a concomitant increase in intensity of the dimer band at 5 mM MgCl2 (Fig. 3A, right panel, lane 2) and 10 mM MgCl2 (Fig. 3A, right panel, lane 3). While this result indicates that the X55 dimer concentration increases with increasing MgCl2 concentrations, it does not prove unambiguously the existence of an X55 kissing complex dimer, as both the kissing complex and the duplex conformations are dimeric (Fig. 2) and should migrate similarly on a native TBM gel. However, since the kissing complex conformation depends on the presence of Mg2+ ions, it should dissociate into monomers while migrating through a TBE gel in which these ions are chelated by EDTA (Mujeeb et al. 2007). In contrast, once formed, the duplex conformation will not be affected, as this conformation is not dependent on the presence of Mg2+ ions. Thus, to distinguish between these two potential X55 RNA dimeric conformations (kissing complex versus duplex), we ran two parallel gels, one in TBM that contained 10 mM MgCl2 and one in TBE in which the Mg2+ ions are chelated (Fig. 3B). X55 RNA was snap-cooled and upon the addition of 10 mM MgCl2, two sets of samples were prepared: the first set was incubated at 22°C (temperature at which the kissing complex conformation is maintained) for 2.5 h, whereas the second set was incubated for 2.5 h at 55°C (temperature that promotes the conversion of any kissing complex to the duplex conformation) (Mujeeb et al. 2007). As seen in Figure 3B, in a TBM gel two bands, monomer and dimer (kissing complex), are present in Figure 3B, right panel, lane 2, which contains the X55 RNA sample incubated at 22°C; whereas, as predicted, a single dimeric band (duplex) is present in the X55 RNA sample incubated at 55°C (Fig. 3B, right panel, lane 1). In contrast, in a TBE gel, in which the Mg2+ ions are chelated by EDTA, the dimer band of the X55 RNA incubated at 22°C is greatly diminished with a concomitant increase in the intensity of the monomer band (Fig. 3B, left panel, lane 2), indicating the dissociation of the kissing complex. As expected, the dimer band corresponding to the X55 RNA incubated at 55°C was not changed during passage through the TBE gel (Fig. 3B, left panel, lane 1), since the duplex conformation is not affected by the absence of the Mg2+ ions. These results strongly support the existence of a kissing complex dimer intermediate formed by the X55 HCV RNA, in the absence of the core protein. These results differ from the previous description of HCV RNA dimerization, which suggested that this does not involve kissing dimer intermediates formed in the absence of the viral core protein (Ivanyi-Nagy et al. 2006). This difference is likely due to the fact that micromolar RNA concentrations were used in our native gels, as opposed to the picomolar concentrations used in previous studies (Ivanyi-Nagy et al. 2006). For a dimer complex with a dissociation constant in the nanomolar range (see below) (Fig. 4), the dimer species is negligible when the total RNA concentration used in the dimerization assay is in the picomolar range.
FIGURE 3.
(A, right) TBM native gel of X55 RNA (20 μM) at different MgCl2 concentrations: 1 mM (lane 1), 5 mM (lane 2), and 10 mM (lane 3). Both the gel and the running buffer contained 1 mM MgCl2. (Left) TBE native gel with no magnesium: Abnova RNA marker R0001 (lane 1) and wild-type X55 RNA (lane 2). (B, right) TBM gel containing 10 mM MgCl2. Both lanes contain X55 RNA annealed, treated with 10 mM MgCl2 and incubated for 2.5 h at 55°C (lane 1) and 22°C (lane 2), respectively. (Left) TBE native gel with no magnesium. Both lanes contain X55 RNA annealed, treated with 10 mM MgCl2 and incubated for 2.5 h at 55°C (lane 1) and 22°C (lane 2), respectively.
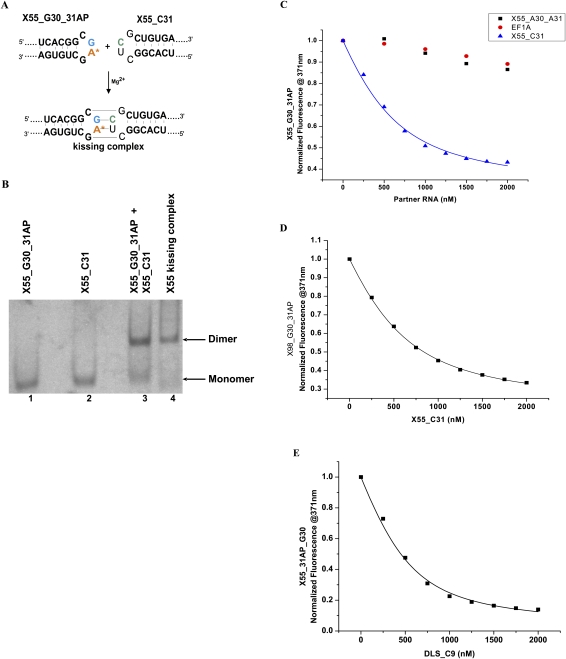
FIGURE 4.
(A) RNA samples used in the fluorescence spectroscopy assay to detect kissing complex formation (the samples were 55 nt, but only the DLS region with the indicated mutations highlighted in red, blue, and green is shown [top]) and their proposed kissing interactions (bottom). (B) TBM native gel showing that neither X55_G30_31AP RNA (lane 1) nor X55_C31 RNA (lane 2) is able to homodimerize in the presence of 10 mM MgCl2, but they heterodimerize when mixed (lane 3), similar to the wild-type X55 RNA (lane 4). (C) Plots of the steady-state fluorescence of X55_G30_31AP (500 nM) in 1 mM cacodylic acid (pH 6.5) and 10 mM MgCl2, titrated with nanomolar increments of X55_C31 RNA (blue triangles), with a mutant RNA X55_A30_A31, which contains a CAAG loop as opposed to the wild-type CUAG loop (black squares), or with a 21-nt fragment derived from EF1A mRNA (red circles). (D) Plot of the steady-state fluorescence of the full-length X98_G30_31AP RNA (500 nM) in 1 mM cacodylic acid (pH 6.5) and 10 mM MgCl2, titrated with nanomolar increments of X55_C31. (E) Plot of the steady-state fluorescence of X55_G30_31AP (500 nM) in 1 mM cacodylic acid (pH 6.5) and 10 mM MgCl2, titrated with nanomolar increments of DLS_C9 RNA.
Fluorescence spectroscopy studies
To obtain quantitative information about the kissing complex intermediate formed in the HCV RNA dimerization process, we used a fluorescence spectroscopy assay initially developed to characterize HIV-1 RNA dimerization (Rist and Marino 2002; Mihailescu and Marino 2004). In this assay, the highly fluorescent purine analog 2-amino purine (2AP) replaces the adenine in the CUAG DLS hairpin loop (at position 31 in X55 RNA) (shown in red in Fig. 4A). The 2-AP steady-state fluorescence is very sensitive to its microenvironment, being quenched by stacking interactions (Heppell et al. 2009). Thus, if a kissing complex is formed by X55 RNA in the presence of MgCl2, the steady-state fluorescence of the 2AP reporter will be quenched due to the change in its microenvironment from a single-stranded loop nucleotide (Fig. 4A, top) to a base-paired stacked nucleotide in the kissing complex (Fig. 4A, bottom). To ensure that a single 2AP reporter is present in the HCV X55 kissing complex, we designed an X55 RNA construct in which the A at position 31 in the CUAG DLS loop sequence was replaced by C (named X55_C31) (Fig. 4A). To maintain the complementary base pairs in the DLS loop, we replaced the U at position 30 in the CUAG DLS loop with G, thus generating X55_G30_31AP (Fig. 4A). This type of loop sequence alteration to construct heterodimers has been previously employed in the studies of the HIV-1 dimerization for both, the Lai variant (Paillart et al. 1996, Takahashi et al. 2000), and Mal variant (Rist and Marino 2002; Mihailescu and Marino 2004). We used TBM native gel electrophoresis to demonstrate that neither X55_G30_31AP, nor X55_C31 RNA is able to form homodimers due to their mismatched loop sequences (Fig. 4B, lanes 1,2), whereas they form dimers when mixed in equimolar ratios (Fig. 4B, lane 3), similar to those formed by the wild-type X55 RNA (Fig. 4B, lane 4).
The titration of the labeled X55_G30_31AP RNA (500 nM in 1 mM cacodylic acid at pH 6.5 and 10 mM MgCl2) with nanomolar increments of the complementary unlabeled construct, X55_C31 RNA, resulted in a decrease in the fluorescence emission at 371 nm (Fig. 4C, blue triangles), due to the base stacking interactions arising when the 2AP in the labeled RNA (X55_G30_31AP) and the uracil in the unlabeled complementary RNA construct (X55_C31) become involved in base pairs (Fig. 4A). This interpretation is supported by the results of a control experiment that demonstrated that when a mutated X55 RNA with the sequence CAAG in the DLS loop (X55_A30_A31) was titrated into X55_G30_31AP, the 2AP fluorescence of X55_G30_31AP did not change significantly (Fig. 4C, black squares), as the noncomplementarity of the DLS loops of these two RNAs prevents the formation of a kissing complex. Similarly, no significant decrease in the 2AP reporter signal was observed in a second control experiment in which an unrelated single-stranded RNA derived from the EF1A mRNA was titrated into X55_G30_31AP (Fig. 4C, red circles).
The binding curve of X55_C31 was fitted with Equation 1 (Materials and Methods) to determine the dissociation constant for the heterodimeric kissing complex formed by X55_G30_31AP and X55_C31 as 318 ± 58 nM (Fig. 4C, blue triangles). The mutated terminal loop HCV DLS sequences used in the determination of this dissociation constant potentially could create RNA structure interactions that differ subtly from those of the native sequence. Nonetheless, this value determined by fluorescence spectroscopy provides an upper limit for the dissociation constant of the kissing complex formed by the wild-type X55 RNA, as the wild-type RNA appears to form tighter kissing homodimers than the heterodimeric kissing dimers formed by X55_G30_31AP and X55_C31 (Fig. 4B, cf. relative intensities of monomer/dimer bands in lanes 3 and 4 of the native TBM gel Fig. 4B). It is noteworthy that this value is in the same range as the dissociation constant of 200 nM determined for the HIV-1 DLS kissing complex (Zhao and Marino 2007) and for other RNA kissing complexes (Argaman and Altuvia 2000; Di Primo 2008). With respect to the biological relevance of these results, it is important to consider that the replication of HCV RNA, as with other positive-strand RNA viruses, is directed by membrane-associated replication complexes. In the case of HCV, these appear as “membranous webs,” composed of vesicles 80–180 nm in diameter (Egger et al. 2002; Gosert et al. 2003; Wolk et al. 2008). The local concentration of the genomic RNA within this microenvironment, while unknown, may be quite high, potentially reaching levels sufficient for the formation of a homodimeric kissing complex with a dissociation constant in the nanomolar range.
To exclude any possibility of dimerization artifacts due to truncation of the 3′ X RNA sequence in the X55 constructs, we also analyzed the dimerization properties of the full-length 98-nt X RNA. To accomplish this, we constructed a 98-nt X RNA that, like X55, had the A at position 31 replaced by 2AP and the U at position 30 replaced by G (X98_G30_31AP) (indicated in Fig. 1B, right), and analyzed its dimerization with X55_C31 RNA. By fitting the binding curve of X98_G30_31AP to Equation 1 (Fig. 4D), we determined a dissociation constant of 323 ± 16 nM. The fact that the dissociation constants determined for X55_G30_31AP and full-length X RNA X98_G30_31AP are the same (within experimental error) suggests that the truncated X55 RNA mimics the behavior of the full-length X RNA, and validates the selection of X55 RNA as a useful model for the analysis of the X RNA homodimerization process.
We also asked whether X55 RNA is able to dimerize with the isolated 16-nt DLS sequence, which due to its size, is expected to fold as a simple stem–loop–hairpin (Fig. 1B, right, highlighted red sequence). For this, we constructed a 16-nt DLS that had the A at position 9 (equivalent to position 31 in X55 RNA and full-length X RNA) in the CUAG loop mutated to C (DLS_C9) to maintain loop complementarity with X55_G30_31AP. We determined the dissociation constant for the kissing complex formed between X55_G30_31AP and the 16-nt DLS_C9 to be 188 ± 33 nM (Fig. 4E). This value is in the same range as the dissociation constants determined for the X55_G30_31AP: X55_C31 and X98_G30_31AP: X55_C31 kissing complexes, corresponding to a difference in free energy of kissing complex formation of only 0.3 kcal/mol. This result suggests that, when participating in the formation of homodimeric kissing complex interactions between two X RNA molecules, the DLS region adopts a stem–loop conformation as depicted in Figure 1B, right. However, it should be noted that this finding does not rule out the existence of alternative conformations of X55 RNA, which would make possible its long-range interactions with the 5BSL3.2 hairpin.
The HCV core protein 2BD domain converts the X55 kissing complex intermediate to a more stable duplex conformation
The HCV core protein is a 191 amino acid, highly basic, RNA binding protein with a hydrophilic N terminus (containing three basic domains) and a hydrophobic C terminus (Cristofari et al. 2004). The core protein acts as a building block of the viral NC and together with the envelope glycoproteins forms the structure of the infectious virion (Ivanyi-Nagy et al. 2006; Brass et al. 2007; Qureshi 2007). The core protein has RNA-binding and nucleic acid chaperone properties (Ivanyi-Nagy et al. 2006), similar to those found in retroviral NC proteins (Tsuchihashi and Brown 1994; Lapadat-Tapolsky et al. 1995; Rein et al. 1998; Bampi et al. 2004; Levin et al. 2005). Since it has been shown that the full-length HCV core protein and its isolated basic domains (either alone or in combination) have similar nucleic acid chaperoning ability, we used a synthetic peptide (2BD) representing a fusion of the first two basic domains of core, comprising residues 2–23 and 38–74 (Ivanyi-Nagy et al. 2006) to examine the role of this protein in X RNA dimerization.
As described above, previous studies with HIV-1 RNA suggest by analogy that the kissing complex intermediate formed by the HCV X55 RNA might be converted to a more thermodynamically stable extended duplex conformation by the core 2BD peptide (Fig. 2). To test this hypothesis, we initially used native TBM gel electrophoresis: The core 2BD peptide was directly added to the preformed wild-type X55-X55 kissing complex, and following incubation for 0.5 h, proteinase K was added to the samples to degrade the 2BD core peptide. As shown in Figure 5A, the addition of the 2BD core peptide caused X55 RNA, which in the absence of the peptide exists as a mixture of monomer and dimer (Fig. 5A, right panel, lane 1), to exist only in the dimer conformation (Fig. 5A, right panel, lane 3). This dimer migrated at the same position as the control X55 duplex obtained by incubating the X55 RNA in the presence of 10 mM MgCl2 for 2.5 h at 55°C (Fig. 5A, right panel, lane 2). To determine whether the X55 conformation obtained after addition of the 2BD core peptide was indeed the duplex conformation, we used TBE native gel electrophoresis. As seen in Figure 5A, in the presence of EDTA, which chelates Mg2+ ions, X55 RNA incubated in the absence of the 2BD peptide at 22°C reverts to the monomeric state (Fig. 5A, left panel, lane 1). In contrast, neither the X55 sample incubated in the presence of the 2BD core peptide (Fig. 5A, left panel, lane 3) nor the control duplex X55 RNA obtained by incubation at 55°C for 2.5 h (Fig. 5A, left panel, lane 2) was affected by EDTA. This result indicates that the core 2BD peptide is able to act on the X55 RNA to promote its structural isomerization to the duplex conformation.
FIGURE 5.
(A, right) TBM native gel: X55 RNA samples (20 μM) were annealed and treated with 10 mM MgCl2, followed by incubation at 22°C (lane 1), incubation at 55°C (lane 2), and incubation at 22°C and treatment with 1:2 RNA: core 2BD peptide (lane 3). (Left) TBE native gel: X55 RNA samples (20 μM) were annealed and treated with 10 mM MgCl2, followed by incubation at 22°C (lane 1), incubation at 22°C, and treatment with 1:2 RNA: core 2BD peptide (lane 2) and incubation at 55°C (lane 3). (B) RNA samples used in the fluorescence spectroscopy assay used to detect the kissing complex to duplex structural isomerization. The inserted 2AP and uracil are highlighted in red, and the loop mutations are highlighted in blue. (C) TBM native gel showing that neither X55_G30_52AP RNA (lane 1) nor X55_C31_U5 RNA (lane 2) is able to homodimerize, but they heterodimerize when mixed in equimolar ratio (lane 3), similar to the wild-type X55 RNA (lane 4). (D) Conversion of the kissing complex conformation to duplex by the 2BD core peptide (black triangles). The curve was fit with equation 2 (Materials and Methods) to determine the structural isomerization rate. Blue squares indicate that the addition of the 2BD core peptide in the absence of MgCl2 does not quench the 2AP fluorescence. Red circles indicate that no structural isomerization occurs in the absence of the core 2BD peptide.
To elucidate the mechanism of this structural isomerization, we used a fluorescence spectroscopy assay similar to that previously employed for the study of the HIV-1 dimer initiation site (Rist and Marino 2002; Mihailescu and Marino 2004). We constructed two additional X55 RNA molecules: X55_G30_52AP, which retains the (U→G) at position 30 (Fig. 5B, shown in blue) and has 2AP inserted in the lower stem region at position 52 (Fig. 5B, shown in red) to form a single nucleotide bulge; and X55_C31_U5, which retains the point mutation (A→C) at position 31 (Fig. 5B, shown in blue) and has a uracil inserted in the lower stem at position 5 (Fig. 5B, shown in red). Due to the G30 and C31 mutations, these two X55 molecules have their DLS loops mismatched and are not able to homodimerize, but they can heterodimerize when mixed in an equimolar ratio, as shown when analyzed in a native TBM gel (Fig. 5C). The steady-state fluorescence of the 2AP in X55_G30_52AP is expected to be high when this RNA is involved in a kissing complex with X55_C31_U5 due to its internal bulge conformation (Fig. 5B, top). If the core protein converts the kissing complex to the stable duplex conformation by a strand exchange mechanism, the 2AP in X55_G30_52AP will base pair with the uracil at position 5 of X55_C31_U5, and since this newly formed 2AP-U base pair (Fig. 5B, shown in red) will become part of the extended duplex, the steady-state fluorescence of the 2AP in X55_G30_52AP will be quenched over time due to stacking interactions.
As shown in Figure 5D, this was indeed the case, as an 80% decrease in the 2AP steady-state fluorescence was observed upon the addition of an excess of 2BD core peptide to the preformed kissing complex between X55_G30_52AP and X55_C31_U5 in the presence of 10 mM MgCl2 (Fig. 5D, black triangles). This result indicates that the 2BD core peptide assists in the conversion of the kissing complex intermediate to the duplex conformation by a strand-exchange mechanism, similar to the mechanism reported for the NCp7-chaperoned structural isomerization of the HIV-1 RNA genome (Rist and Marino 2002; Mihailescu and Marino 2004). To rule out the possibility that the observed quenching could be due to the direct interactions of the core 2BD peptide with the 2AP reporter and not the strand-exchange mechanism, we carried out a control experiment in which the same excess of the core 2BD peptide was added to a mixture of X55_G30_52AP and X55_C31_U5, but in the absence of MgCl2. As shown in Figure 5D (blue squares), this led to an initial 10% drop in the 2AP steady-state fluorescence, which over time returned to the 100% intensity value, in contrast to the drop in intensity associated with the conversion of the kissing complex to the duplex conformation (Fig. 5D, black triangles).
This result also indicates that the kissing complex conformation is required for the core 2BD peptide activity, as in the absence of Mg2+ ions (conditions in which the kissing complex does not form), the peptide cannot convert X55_G30_52AP and X55_C31_U5 to the dimer conformation. This result was confirmed by native gel electrophoresis of the wild-type X55 RNA, in which no dimer band was observed in the absence of Mg2+ ions despite an excess of the 2BD peptide (data not shown). Thus, it is likely that when assisting in the structural isomerization of the HCV DLS, the core protein does not employ a “zipper” pathway involving nucleation at the 3′-/5′-terminal ends, such as the HIV-1 NCp7 protein has been suggested to use in annealing of the full-length transactivation response element (TAR) (Vo et al. 2009). Also shown in Figure 5C is the result of a second control experiment, in which we confirmed that in the absence of the core 2BD protein, the steady-state fluorescence of X55_G30_52AP, which is involved in a kissing complex with X55_C31_U5 in the presence of 10 mM MgCl2, remains almost constant over time (Fig. 5D, 5% drop in intensity, red circles). This indicates that within the timeline of the experiment, conversion to the duplex conformation is minimal without the assistance of the 2BD core peptide. The rate of conversion from the kissing complex to the duplex conformation, kconv, was obtained by fitting the conversion curve (Fig. 5D, black triangles) with Equation 2 (Materials and Methods). The value of kconv = 1.7 ± 0.2 min−1 is very similar to that of kconv = 2.2 ± 0.2 min−1, reported for the conversion rate of the HIV-1 DLS kissing dimer to the mature extended duplex by the NCp7 protein (present in a 5:1 ratio, as in our experiments).
Taken together, these results indicate that the mechanism of HCV RNA dimerization involves the formation of a kissing complex intermediate, which in the presence of the core peptide undergoes isomerization through the melting of intramolecular stem base pairs and reformation of intermolecular base pairing to form a more thermodynamically stable duplex conformation. These results thus demonstrate a remarkable similarity in the mechanisms of genomic RNA dimerization of HCV and HIV-1.
Possible biological role of HCV genomic RNA dimerization
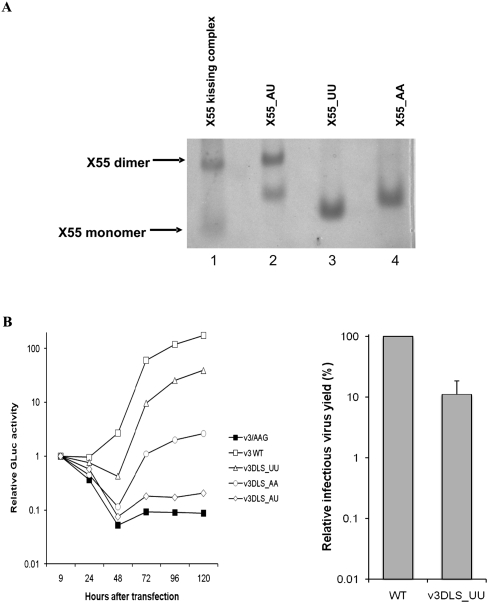
We have shown unambiguously that the mechanism by which genomic HCV dimerization occurs in vitro is very similar to that of HIV-1, involving the formation of a kissing complex intermediate that is converted to a duplex conformation by the basic core protein. Despite these similarities, the finding that the HCV RNA genome dimerizes in vitro is very surprising. Unlike HIV-1, HCV genomic RNA is generally believed to be packaged in viral particles as a monomer (although this has never been formally demonstrated). This suggests that dimerization of the HCV genome may play a completely different role in the life cycle of the virus. To explore a possible role for HCV genomic RNA dimerization in virus replication, we determined whether mutations in the DLS loop sequence that prevent the RNA dimerization in vitro have any effect upon the replication capacity of HCV RNA in transfected cells (Yi and Lemon 2003b). For these studies, we transfected cells with a modified, genome-length, genotype 1a HCV RNA in which the sequence encoding Gaussia luciferase (GLuc) was inserted in-frame into the viral polyprotein-coding region of an infectious molecular clone (H77Sv3; see Materials and Methods). Three different mutations were introduced into the 3′-UTR sequence of this modified viral RNA, replacing the CUAG wild-type DLS loop sequence with CUUG (DLS_UU), CAAG (DLS_AA), and CAUG (DLS_AU). It was expected that the mutations in the DLS_UU and DLS_AA would prevent RNA dimerization due to their mismatched loops, whereas although not wild type, the DLS_AU sequence would retain the ability to dimerize since its mutations are compensatory. To test this hypothesis, related X55 sequences containing identical mutations in the DLS loops were prepared in vitro by transcription reactions off synthetic DNA templates (X55_UU, X55_AA and X55_AU). As showed by native TBM gel electrophoresis (Fig. 6A), the X55_UU and X55_AA were not able to dimerize (Fig. 6A, lanes 3,4), whereas X55_AU formed dimers (Fig. 6A, lane 2). It is interesting to note that although these mutated X55 RNAs behaved as expected with respect to the formation of the kissing complex dimer, the migration pattern of their monomeric form differed from that of the wild-type X55 (Fig. 6A, lanes 1,2), suggesting that they might adopt different conformations (whereas this was not the case with the mutated X55 samples used in the fluorescence spectroscopy studies).
FIGURE 6.
(A) Native TBM gel showing that wild-type X55 RNA (lane 1) and its DLS loop mutated constructs: the double mutant (DLS_AU) with compensatory mutations can homodimerize (lane 2), whereas the single mutants—DLS_UU (lane 3) and DLS_AA (lane 4) —are not able to homodimerize due to their mismatched loops. (B, left) Normalized GLuc activity in culture supernatants collected at different times after transfection of cells with replication competent genome-length HCV RNA with a wild-type DLS (v3 WT, □), a negative-control RNA with defective RNA-dependent RNA polymerase activity (v3/AAG, ■), and related DLS-mutant RNAs: DLS_AA (○), DLS_UU (△), and DLS_AU (⋄). (Right) Yield of infectious virus released into supernatant fluids of cell cultures transfected with the H77Sv3DLS-UU viral RNA. Virus titers were determined by an infectious focus assay and are reported relative to that released by cells transfected in parallel with the wild-type H77Sv3 RNA (100%). Error bars, range of results obtained in replicate independent experiments.
The impact of the DLS mutations upon HCV RNA replication was assessed by measuring GLuc activity in cell culture media following transfection with genome-length RNAs containing the wild-type DLS (v3 WT), a negative-control RNA with a replication-lethal mutation in the active site of the NS5B RNA-dependent RNA polymerase (v3/AAG), or RNAs with mutated DLS sequences: DLS_AA, DLS_UU and DLS_AU. The GLuc activities were normalized to that present at 9 h following transfection, to control for variations in transfection efficiency, with the relative GLuc activity at 24, 48, 72, 96, and 120 h taken as a measure of genome amplification. As shown in Figure 6B, left panel, RNA containing the wild-type DLS (v3 WT) demonstrated robust replication, with a 175-fold increase in secreted GLuc activity by 120 h after transfection, whereas the negative-control v3/AAG RNA containing a lethal polymerase mutation demonstrated a >90% decline in GLuc activity consistent with a failure to replicate and express the polyprotein. The DLS_AA and DLS_UU mutations both partially inhibited but did not ablate replication of the viral RNA. The DLS_UU mutant replicated with moderate efficiency, demonstrating an approximately 40-fold increase in GLuc activity between 9 and 120 h after transfection, while the DLS_AA mutant was severely impaired for replication, generating only a 2.5-fold increase in GLuc activity. Both mutants demonstrated substantial delays in the replication kinetic, with increases in GLuc not observed until after 48 h, ∼24 h later than wild type (Fig. 6B). Importantly, the double compensatory mutations in DLS_AU failed to rescue RNA replication, as might have been expected if the RNA dimerization were essential for RNA replication. In contrast, the impact of the double mutations appeared to be additive, giving rise to a near-lethal RNA replication phenotype and GLuc activities minimally above background at 120 h.
The DLS mutations were also placed directly into the background of the parental genotype 1a infectious molecular clone (H77Sv3) that was used for construction of the GLuc RNAs described above. H77Sv3 RNA is capable of producing fully infectious virus particles when transfected into cells (Yi et al. 2006), allowing an assessment of the effect of DLS mutations on viral assembly and release. Supernatant fluids were collected from RNA-transfected cells 96 h after transfection, and the yield of virus was determined by an infectious focus assay following inoculation of naïve cells, as described previously (Yi et al. 2006). In replicate experiments, the presence of the DLS_UU mutation resulted in an 89% ± 7% reduction in infectious virus yield relative to the wild-type RNA. This reduction is consistent with the impairment in RNA replication (77%) demonstrated by the DLS_UU mutant in Figure 6B. The severely attenuated replication phenotypes of the DLS_AA and double DLS_AU mutants precluded testing of their ability to produce infectious particles.
The fact that the DLS_UU and DLS-AA mutants are incapable of dimerization in vitro, yet capable of at least limited replication, suggests that dimerization of the genomic RNA is not absolutely essential for viral RNA replication. While rescue of the replication defects in DLS_AA and DLS_UU by the double compensatory mutations in DLS_AU would have suggested that dimerization facilitates efficient RNA replication, the absence of such a result cannot be considered to exclude this possibility. Given the high degree of conservation of the DLS sequence, it is possible that the loss of RNA replication competence in the mutated DLS constructs could have reflected perturbation of essential interactions other than dimerization of the RNA (including, for example, the kissing-loop interaction with the upstream NS5B sequence) (Friebe et al. 2005; Diviney et al. 2008; You and Rice 2008). Previous studies have shown that very few mutations introduced into the first 55 nt of X55 RNA are tolerated, and with a single exception, those that do not ablate replication of the RNA are located outside the DLS region (Yi and Lemon 2003b). The results of the infectious RNA experiments also suggest that dimerization of the genome is not absolutely essential for other aspects of the viral life cycle, including packaging of the viral RNA during virion assembly.
As mentioned earlier, it has been proposed that a fragment of the HCV DLS sequence (Fig. 1B, nucleotides 32–38) is involved in long-range kissing interactions with a conserved cis-acting element located in the coding sequence of the NS5B protein (named 5BSL3.2) (Friebe et al. 2005; Diviney et al. 2008; You and Rice 2008), and it has been shown that these interactions are absolutely required for RNA replication (You and Rice 2008). Since both types of kissing interactions (homodimeric, between two HCV RNA genomes, or heterodimeric, a cis-interaction with the 5BSL3.2 stem–loop within a single RNA genome) involve the same DLS region, they are likely to be mutually exclusive. It is possible that the DLS region might thus function as a molecular switch, influencing processes such as replication, translation, and/or packaging, all of which involve the same plus-strand genomic RNA. As infection progresses within a cell, the abundance of both genomic RNA and HCV core protein increase, thereby potentially favoring formation of the homodimeric RNA conformation over the monomolecular interaction of the DLS with the 5BSL3.2 sequence. We envision a model in which the sequestration of the DLS region in the duplex dimer with another genomic RNA molecule (promoted through the action of the core protein) excludes the DLS sequence from the long-range kissing interaction with the 5BSL3.2 CRE required for RNA replication, thus leading to inhibition of RNA replication and perhaps promoting packaging of the viral RNA. It is also possible that the homodimeric interaction has no positive role in the viral life cycle but rather may have evolved as a “brake” on viral replication, so as to maintain a relatively low overall abundance of viral RNA and viral proteins within infected hepatocytes, which might in turn favor escape from immunologic surveillance and long-term persistence of the virus.
It is interesting to note that the genome of GB virus B (GBV-B), the only other member of the Hepacivirus genus within which HCV is classified, is also capable of dimeric conformation in the presence of its core protein (Ivanyi-Nagy et al. 2006, 2008). This suggests that dimerization of genomic RNA is a common feature of the Hepacivirus genus. Since the core proteins of other related viruses (such as West Nile virus, a member of the Flavivirus genus, and bovine viral diarrhea virus, a member of the Pestivirus genus) also exhibit RNA chaperone functions (Ivanyi-Nagy et al. 2008), it is interesting to speculate that genomic dimerization might be a feature shared more broadly among other genera within the family Flaviviridae.
Conclusions
In this study we analyzed the HCV 3′-UTR and its involvement in the genomic RNA dimerization. Our results indicate that the mechanism of dimerization involves the formation of a kissing complex intermediate, which is converted to a more stable duplex conformation by the core protein. We show that dimerization is not absolutely essential for viral RNA replication or infectious virus production, and propose a model according to which the HCV DLS region might function as a molecular switch operated through kissing interactions to regulate the timing of different HCV life cycle events, such as replication, packaging, and/or viral protein translation.
MATERIALS AND METHODS
RNA samples
A series of RNA oligonucleotides (the 55-nt wild-type X55 RNA, related mutated forms used in the fluorescence spectroscopy studies, X55_C31 and X55_C31_U5 RNAs, mutated 16-nt DLS_C9 RNA, and 55-nt DLS_AA, DLS_UU and DLS_AU) were transcribed as previously described (Milligan et al. 1987; Milligan and Uhlenbeck 1989) from synthetic DNA templates in vitro using the T7 RNA polymerase. RNAs were purified by 10% polyacrylamide, 8 M urea gel electrophoresis, recovered by electrophoretic elution, and dialyzed against either 1 mM cacodylic acid (pH 6.5) or 10 mM Tris (pH 7.5). Unless otherwise specified, all RNA samples were annealed prior to use by heating for 5 min at 95°C, followed by snap-cooling on ice.
HCV core peptide
The full-length 191 amino acid HCV core protein contains three basic domains (Cristofari et al. 2004; Ivanyi-Nagy et al. 2006). The first two basic domains, comprising amino acid residues 2–23 and 38–74, were combined to form a 58 amino acid sequence core peptide, which was named the 2BD core peptide (Ivanyi-Nagy et al. 2006). The core 2BD peptide was chemically synthesized and purified by the Peptide Synthesis Unit at the University of Pittsburgh, Center for Biotechnology and Bioengineering. The peptide was reconstituted in 1 mM cacodylic acid (pH 6.5) at a final concentration of 618 μM.
Native gel electrophoresis
Individual wild-type RNAs, or mixtures of RNAs (such as labeled and unlabeled samples used in fluorescence spectroscopy assays), were diluted to 20 μM concentrations. Following annealing, the RNA samples were incubated either in the absence of MgCl2 or in the presence of 10 mM MgCl2, at 22°C (temperature at which a kissing complex conformation is maintained) or at 55°C (temperature that promotes the formation of the duplex conformation; see Results) for 2.5 h. The samples were electrophoresed on a 10% native gel in either TBE (Tris Borate EDTA) buffer or TBM (Tris Borate Magnesium) buffer at 4°C. To study the effect of the 2BD core peptide on the magnesium treated samples, the peptide was directly added to the RNA, followed by incubation for 30 min at room temperature. These samples were subsequently treated with proteinase K for peptide digestion (concentration of 190 μg/mL) for 1 h prior to being run on native gels. The native gels were run at 4°C, 100 V for 1.5 h and were visualized by ultraviolet shadowing at 254 nm using an AlphaImager HP (AlphaInnotech).
Fluorescence spectroscopy
Experiments were performed on a J.Y. Horiba Fluoromax-3 fluorimeter equipped with variable temperature control. The excitation wavelength was set at 310 nm, and the emission spectra were recorded from 330–450 nm. All experiments were performed at 22°C.
RNA kissing complex formation
Two distinct RNAs were constructed for the fluorescence spectroscopy assay that analyzes the dimerization of X55 RNA: X55_G30_31AP, which contained a point mutation (U→G) at position 30, and in which the highly fluorescent purine analog 2 amino purine (2AP) replaced the adenine at position 31, and X55_C31, which contained a point mutation (A→C) at position 31 (Fig. 4A). These point mutations prevented the homodimerization of the RNA constructs but allowed their heterodimerization. Similarly, to analyze the interactions of the full-length X RNA, X98_G30_31AP was constructed, which contained a point mutation (U→G) at position 30 and in which 2AP replaced the adenine at position 31. For the analysis of the interactions between X55 and the isolated DLS region, DLS_C9 was constructed, which contains a point mutation (A→C) at position 9 (equivalent to position 31 in X55 of full-length X RNA). The 2AP labeled samples (55-nt X55_G30_31AP and X55_C31_52AP, as well as the full-length 98-nt X98_C31_31AP) were chemically synthesized and purified by Dharmacon.
The dissociation constant of the kissing complex formed by X55 RNA was determined by titrating a fixed concentration of the fluorescent X55_G30_31AP (500 nM) in 1 mM cacodylic acid (pH 6.5) containing 10 mM MgCl2 with increasing concentrations of X55_C31. The kissing complex dissociation constant was determined by fitting the binding curve to Equation 1:
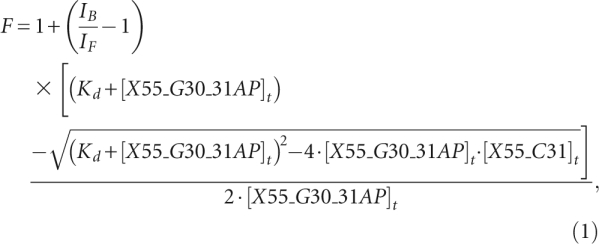 |
in which IB and IF are the steady-state fluorescence intensities of the free and bound X55_G30_31AP, [X55_G30_31AP]t is the total concentration of X55_G30_31AP RNA, and [X55_C31]t is the total concentration of X55_C31 RNA.
X55 RNA kissing complex to duplex conversion
To analyze the formation of the X55 duplex conformation in the presence of the HCV core protein by fluorescence spectroscopy, two additional RNAs were constructed: X55_G30_52AP, which retained the point mutation (U→G) at position 30 and in which 2AP was introduced in the stem region as a single bulged nucleotide at position 52, and its complementary RNA construct X55_C31_U5, which retained the point mutation (A→C) at position 31 and had a uracil inserted at position 5 (Fig. 5B). These mutated constructs are not able to homodimerize but can heterodimerize. The structural isomerization of X55 RNA was monitored by preforming the kissing complex between X55_G30_52AP and X55_C31_U5 (100 nM each) in the presence of 10 mM MgCl2 and 140 mM KCl, and monitoring the decrease of the 2AP steady-state fluorescence upon the addition of an excess (500 nM) of the 2BD core peptide. All conversion curves were corrected by subtracting the contribution of the peptide background. The kissing complex to duplex conversion rate was determined using Equation 2:
where kconv is the observed isomerization rate for conversion of the DLS kissing complex to extended mature duplex dimer, and karr is attributed to the rate of rearrangement of the 2-AP probe subsequent to its stacking in the duplex conformation (Rist and Marino 2002; Mihailescu and Marino 2004).
HCV replication analysis
pH77Sv3 is an improved version of pH77S, a plasmid containing the genome-length sequence of the genotype 1a H77 strain of HCV with five cell culture-adaptive mutations that promote its replication in Huh7 hepatoma cells (Yi and Lemon 2004; Yi et al. 2006). pH77Sv3GLuc2A is a related construct in which GLuc sequence, fused to the 2A auto-catalytic protease of foot-and-mouth virus RNA (Yi et al. 2002), was inserted in-frame between p7 and NS2 (Shimakami et al. 2009). RNAs transcribed from both plasmids contain authentic 5′- and 3′-terminal UTR sequences of genotype 1a virus and produce infectious virus when transfected into Huh7 hepatoma-derived cells. Mutations were made within the DLS sequence in both parental constructs using the QuickChange mutagenesis kit in combination with conventional DNA cloning methods (Stratagene). Wild-type and mutant plasmid DNAs were linearized by XbaI digestion followed by in vitro transcription using the MEGAscript kit (Ambion). Following digestion with RNase-free DNase to remove template DNA, RNA was purified using the RNAeasy Mini Kit (Qiagen), and RNA transfection was carried out using the Trans-IT mRNA transfection kit (Mirus) according to the manufacturer's suggested protocol. Briefly, 250 ng RNA was transfected to 8 × 104 cells/well in 24-well plates. Beginning 9 h after transfection, and at 24-h intervals thereafter, the medium of cultures transfected with the GLuc RNA constructs was replaced with fresh medium. Secreted GLuc activity was measured in 25-μL aliquots of these cell culture supernatant fluids using the Gaussia Luciferase Assay Kit (New England Biolabs). The luminescent signal was measured on a Synergy 2 (Bio-Tek) microplate reader. GLuc activity was normalized to that present at 9 h to control for transfection efficiency.
Infectious virus titers were determined by enumerating foci of infected cells in naïve Huh-7.5 cells inoculated with dilutions of supernatant fluids from H77Sv3 RNA-transfected cultures. The cells were examined for virus-specific immunofluorescence after staining for core antigen expression using an indirect labeling method, as described (Yi et al. 2006).
ACKNOWLEDGMENTS
We thank Dr. MinKyung Yi (University of Texas Medical Branch at Galveston) for the pH77Sv3 plasmid. This work was supported by a Pennsylvania State Health Formula Research Grant (to M.-R.M.) and a grant from the National Institute of Allergy and Infectious Diseases (U19-AI40035, to S.M.L.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1960410.
REFERENCES
- Aagaard L, Rasmussen SV, Mikkelsen JG, Pedersen FS 2004. Efficient replication of full-length murine leukemia viruses modified at the dimer initiation site regions. Virology 318: 360–370 [DOI] [PubMed] [Google Scholar]
- Argaman L, Altuvia S 2000. fhlA repression by OxyS RNA: Kissing complex formation at two sites results in a stable antisense-target RNA complex. J Mol Biol 300: 1101–1112 [DOI] [PubMed] [Google Scholar]
- Bampi C, Jacquenet S, Lener D, Decimo D, Darlix JL 2004. The chaperoning and assistance roles of the HIV-1 nucleocapsid protein in proviral DNA synthesis and maintenance. Curr HIV Res 2: 79–92 [DOI] [PubMed] [Google Scholar]
- Bender W, Chien YH, Chattopadhyay S, Vogt PK, Gardner MB, Davidson N 1978. High-molecular-weight RNAs of AKR, NZB, and wild mouse viruses and avian reticuloendotheliosis virus all have similar dimer structures. J Virol 25: 888–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight KJ, Rice CM 1997. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J Virol 71: 7345–7352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass V, Moradpour D, Blum HE 2007. Hepatitis C virus infection: In vivo and in vitro models. J Viral Hepat 14: 64–67 [DOI] [PubMed] [Google Scholar]
- Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244: 359–362 [DOI] [PubMed] [Google Scholar]
- Cristofari G, Ivanyi-Nagy R, Gabus C, Boulant S, Lavergne JP, Penin F, Darlix JL 2004. The hepatitis C virus Core protein is a potent nucleic acid chaperone that directs dimerization of the viral (+) strand RNA in vitro. Nucleic Acids Res 32: 2623–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Primo C 2008. Real time analysis of the RNAI-RNAII-Rop complex by surface plasmon resonance: From a decaying surface to a standard kinetic analysis. J Mol Recognit 21: 37–45 [DOI] [PubMed] [Google Scholar]
- Diviney S, Tuplin A, Struthers M, Armstrong V, Elliott RM, Simmonds P, Evans DJ 2008. A hepatitis C virus cis-acting replication element forms a long-range RNA-RNA interaction with upstream RNA sequences in NS5B. J Virol 82: 9008–9022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutkiewicz M, Ciesiolka J 2005. Structural characterization of the highly conserved 98-base sequence at the 3′ end of HCV RNA genome and the complementary sequence located at the 5′ end of the replicative viral strand. Nucleic Acids Res 33: 693–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger D, Wolk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K 2002. Expression of hepatitis C virus protein induces distinct membrane alterations including a candidate viral replication complex. J Virol 76: 5974–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe P, Boudet J, Simorre JP, Bartenschlager R 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J Virol 79: 380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol 77: 5487–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadziyannis SJ, Sette H Jr, Morgan TR, Balan V, Diago M, Marcellin P, Ramadori G, Bodenheimer H Jr, Bernstein D, Rizzetto M, et al. 2004. Peginterferon-α2a and ribavirin combination therapy in chronic hepatitis C: A randomized study of treatment duration and ribavirin dose. Ann Intern Med 140: 346–355 [DOI] [PubMed] [Google Scholar]
- Heppell B, Mulhbacher J, Penedo JC, Lafontaine DA 2009. Application of fluorescent measurements for characterization of riboswitch-ligand interactions. Methods Mol Biol 540: 25–37 [DOI] [PubMed] [Google Scholar]
- Ito T, Lai MM 1997. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J Virol 71: 8698–8706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanyi-Nagy R, Kanevsky I, Gabus C, Lavergne JP, Ficheux D, Penin F, Fosse P, Darlix JL 2006. Analysis of hepatitis C virus RNA dimerization and core-RNA interactions. Nucleic Acids Res 34: 2618–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanyi-Nagy R, Lavergne JP, Gabus C, Ficheux D, Darlix JL 2008. RNA chaperoning and intrinsic disorder in the core proteins of Flaviviridae. Nucleic Acids Res 36: 712–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolykhalov AA, Feinstone SM, Rice CM 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol 70: 3363–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung HJ, Hu S, Bender W, Bailey JM, Davidson N, Nicolson MO, McAllister RM 1976. RD-114, baboon, and woolly monkey viral RNA's compared in size and structure. Cell 7: 609–620 [DOI] [PubMed] [Google Scholar]
- Lapadat-Tapolsky M, Pernelle C, Borie C, Darlix JL 1995. Analysis of the nucleic acid annealing activities of nucleocapsid protein from HIV-1. Nucleic Acids Res 23: 2434–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin JG, Guo J, Rouzina I, Musier-Forsyth K 2005. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: Critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol 80: 217–286 [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309: 623–626 [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Thiel H, Rice CM 2007. Flaviviridae: The viruses and their replication. fields virology, 5th ed Lippincott-Raven Publishers, Philadelphia [Google Scholar]
- Maisel J, Bender W, Hu S, Duesberg PH, Davidson N 1978. Structure of 50 to 70S RNA from Moloney sarcoma viruses. J Virol 25: 384–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquet R, Baudin F, Gabus C, Darlix JL, Mougel M, Ehresmann C, Ehresmann B 1991. Dimerization of human immunodeficiency virus (type 1) RNA: Stimulation by cations and possible mechanism. Nucleic Acids Res 19: 2349–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihailescu MR, Marino JP 2004. A proton-coupled dynamic conformational switch in the HIV-1 dimerization initiation site kissing complex. Proc Natl Acad Sci 101: 1189–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan JF, Uhlenbeck OC 1989. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol 180: 51–62 [DOI] [PubMed] [Google Scholar]
- Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res 15: 8783–8798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DM, Fu W, Nikolaitchik O, Chen J, Ptak RG, Hu W-S 2007. Dimer initiation signal of human immunodeficiency virus type 1: Its role in partner selection during RNA copackaging and its effects on recombination. J Virol 81: 4002–4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradpour D, Penin F, Rice CM 2007. Replication of hepatitis C virus. Nat Rev Microbiol 5: 453–463 [DOI] [PubMed] [Google Scholar]
- Muir AJ, Bornstein JD, Killenberg PG 2004. Peginterferon α-2b and ribavirin for the treatment of chronic hepatitis C in blacks and non-Hispanic whites. N Engl J Med 350: 2265–2271 [DOI] [PubMed] [Google Scholar]
- Mujeeb A, Ulyanov NB, Georgantis S, Smirnov I, Chung J, Parslow TG, James TL 2007. Nucleocapsid protein-mediated maturation of dimer initiation complex of full-length SL1 stem–loop of HIV-1: Sequence effects and mechanism of RNA refolding. Nucleic Acids Res 35: 2026–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murti KG, Bondurant M, Tereba A 1981. Secondary structural features in the 70S RNAs of Moloney murine leukemia and Rous sarcoma viruses as observed by electron microscopy. J Virol 37: 411–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi SP, Nakamura Y 2007. Slight sequence modifications unexpectedly alter the metal-dependency of a kissing-loop interaction. Nucleic Acids Symp Ser 51: 395–396 [DOI] [PubMed] [Google Scholar]
- Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R 1996. A loop–loop ‘kissing’ complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc Natl Acad Sci 93: 5572–5577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paula T, Pablo R, Eugenia V, Pablo B, Sabino P, Jose M, Antonio M, Dolores HM, Pablo L, Javier GS, et al. 2009. New drug targets for hepatitis C and other Flaviviridae viruses. Infect Disord Drug Targets 9: 133–147 [DOI] [PubMed] [Google Scholar]
- Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky J 2004. Structural biology of hepatitis C virus. Hepatology 39: 5–19 [DOI] [PubMed] [Google Scholar]
- Qureshi SA 2007. Hepatitis C virus—biology, host evasion strategies, and promising new therapies on the horizon. Med Res Rev 27: 353–373 [DOI] [PubMed] [Google Scholar]
- Rein A, Henderson LE, Levin JG 1998. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: Significance for viral replication. Trends Biochem Sci 23: 297–301 [DOI] [PubMed] [Google Scholar]
- Rist MJ, Marino JP 2002. Mechanism of nucleocapsid protein catalyzed structural isomerization of the dimerization initiation site of HIV-1. Biochemistry 41: 14762–14770 [DOI] [PubMed] [Google Scholar]
- Shimakami T, Lanford RE, Lemon SM 2009. Hepatitis C: Recent successes and continuing challenges in the development of improved treatment modalities. Curr Opin Pharmacol 9: 537–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Friebe P, Tzima E, Junemann C, Bartenschlager R, Niepmann M 2006. The hepatitis C virus RNA 3′-untranslated region strongly enhances translation directed by the internal ribosome entry site. J Virol 80: 11579–11588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi KI, Baba S, Chattopadhyay P, Koyanagi Y, Yamamoto N, Takaku H, Kawai G 2000. Structural requirement for the two-step dimerization of human immunodeficiency virus type 1 genome. RNA 6: 96–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SL, Pause A, Shi Y, Sonenberg N 2002. Hepatitis C therapeutics: Current status and emerging strategies. Nat Rev Drug Discov 1: 867–881 [DOI] [PubMed] [Google Scholar]
- Tsuchihashi Z, Brown PO 1994. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J Virol 68: 5863–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A 1992. Internal ribosome entry site within hepatitis C virus RNA. J Virol 66: 1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo MN, Barany G, Rouzina I, Musier-Forsyth K 2009. HIV-1 nucleocapsid protein switches the pathway of transactivation response element RNA/DNA annealing from loop–loop ‘kissing’ to ‘zipper’. J Mol Biol 386: 789–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Sarnow P, Siddiqui A 1993. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J Virol 67: 3338–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk B, Buchele B, Moradpour D, Rice CM 2008. A dynamic view of hepatitis C virus replication complexes. J Virol 82: 10519–10531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M, Lemon SM 2003a. Structure–function analysis of the 3′ stem–loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA 9: 331–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M, Lemon SM 2003b. 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J Virol 77: 3557–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M, Lemon SM 2004. Adaptive mutations producing efficient replication of genotype 1a hepatitis C virus RNA in normal Huh7 cells. J Virol 78: 7904–7915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M, Bodola F, Lemon SM 2002. Subgenomic hepatitis C virus replicons inducing expression of a secreted enzymatic reporter protein. Virology 304: 197–210 [DOI] [PubMed] [Google Scholar]
- Yi M, Villanueva RA, Thomas DL, Wakita T, Lemon SM 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc Natl Acad Sci 103: 2310–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You S, Rice CM 2008. 3′ RNA elements in hepatitis C virus replication: Kissing partners and long poly(U). J Virol 82: 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Marino JP 2007. Synthesis of HIV-1 Psi-site RNA sequences with site specific incorporation of the fluorescent base analog 2-amino purine. Tetrahedron 63: 3575–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]