Abstract
The signal recognition particle (SRP) is a ubiquitous cytoplasmic ribonucleoprotein complex required for the cotranslational targeting of proteins to the endoplasmic reticulum (ER). In eukaryotes, SRP has to arrest the elongation of the nascent chains during targeting to ensure efficient translocation of the preprotein, and this function of SRP is dependent on SRP9/14. Here we present the results of a mutational study on the human protein h9/14 that identified and characterized regions and single residues essential for elongation arrest activity. Effects of the mutations were assessed both in cell-free translation/translocation assays and in cultured mammalian cells. We identified two patches of basic amino acid residues that are essential for activity, whereas the internal loop of SRP14 was found to be dispensable. One patch of important basic residues comprises the previously identified basic pentapetide KRDKK, which can be substituted by four lysines without loss of function. The other patch includes three lysines in the solvent-accessible α2 of h9. All essential residues are located in proximity in SRP9/14 and their basic character suggests that they serve as a positively charged platform for interactions with ribosomal RNA. In addition, they can all be lysines consistent with the hypothesis that they recognize their target(s) via electrostatic contacts, most likely with the phosphate backbone, as opposed to contacts with specific bases.
Keywords: SRP, endoplasmic reticulum, translation, RNA–protein complexes, Alu RNA
INTRODUCTION
The universally conserved signal recognition particle (SRP) and its membrane-associated receptor (SRP receptor [SR], or docking protein) are responsible for the cotranslational targeting of secretory and membrane proteins to the endoplasmic reticulum (ER). SRP-mediated targeting is achieved via a series of ordered steps that are closely coordinated (for review, see Keenan et al. 2001; Pool 2005). The hydrophobic signal sequence, a common hallmark of ER-targeted proteins, is first recognized by the SRP54 subunit of SRP. Upon binding, SRP causes a delay in the elongation of the nascent chain that is termed the elongation arrest activity (Walter and Blobel 1981). The ribosome-nascent-chain–SRP complex (RNC-SRP) is then docked to the ER membrane through an interaction of SRP with SR (Gilmore et al. 1982; Meyer 1985), which is controlled by GTP binding and hydrolysis (Connolly and Gilmore 1986; Connolly et al. 1991). After docking of the ribosome onto the protein-conducting channel (translocon) (Gorlich et al. 1990), SRP and SR dissociate from the ribosome and from each other and translation resumes at its normal speed.
The Alu domain of SRP comprises SRP9/14 and the 5′ and 3′ ends of the mammalian SRP RNA, and it is responsible for the elongation arrest function of SRP (Siegel and Walter 1985) that is also dependent on signal sequence recognition by SRP54. The elongation arrest function is physiologically important (Mason et al. 2000; Lakkaraju et al. 2008). In mammalian cells, the translocation efficiency is significantly reduced, when the elongation arrest function of SRP is abrogated (Lakkaraju et al. 2008). However, efficient translocation can be restored by either slowing down the elongation of nascent chains with antibiotics or by increasing the cellular levels of SR. The latter indicates that limiting cellular concentrations of SR are the reason why nascent chains fail to reach the translocon in a translocation-competent state at normal elongation rates. These results suggest that the elongation arrest function of SRP is part of a mechanism used to control protein influx into the ER.
SRP9 and SRP14 form a heterodimer (Strub and Walter 1990), and the crystal structure of free SRP9/14 revealed structurally homologous polypeptides, despite limited primary sequence homology (Fig. 1; Birse et al. 1997). They both form a three-stranded antiparallel β-sheet stacked against two helices (Birse et al. 1997; Weichenrieder et al. 2000). However, SRP14 differs from SRP9 in possessing an extended loop between β strands β1 and β2. The structures of the internal loop and the basic C-terminal tails in SRP14 and SRP9 (Fig. 1, beyond 14K95 and 9A76) have not been solved, because they are disordered in the crystals obtained so far. The Srp14p homodimer of the yeast Schizosaccharomyces pombe also has a similar structure (Brooks et al. 2009). Within the heterodimeric protein, SRP9 and SRP14 are related by a pseudodyad axis and form a continuous antiparallel six-stranded β-sheet stacked against four helices. The β-sheet formed between the two subunits presents a highly positive charged concave surface due to an abundance of exposed basic residues and forms the major binding site for the Alu domain RNA (Weichenrieder et al. 2000). The SRP14 proteins of primate species have an additional C-terminal extension of variable size. It results from the amplification of a genomic GCA repeat. It codes mostly for alanine and includes 26 additional amino acid residues in human as compared with mouse protein (Fig. 1A; Bovia et al. 1995; Chang et al. 1995). The truncation of the alanine-rich tail did not impair SRP function in vitro (Bovia et al. 1997). Recently, a basic pentapeptide at position 96–100 in human SRP14 (Fig. 1) was shown to be essential for elongation arrest activity in cell-free assays and in mammalian cells (Lakkaraju et al. 2008). The Alu portion of SRP RNAs is present in SRP of eukaryotic and archaeal and in a few eubacterial organisms (Rosenblad et al. 2003). SRP9/14 and the SRP14 homodimeric complex in certain yeast species (Strub et al. 1999; Mason et al. 2000) have so far been identified only in eukaryotic species.
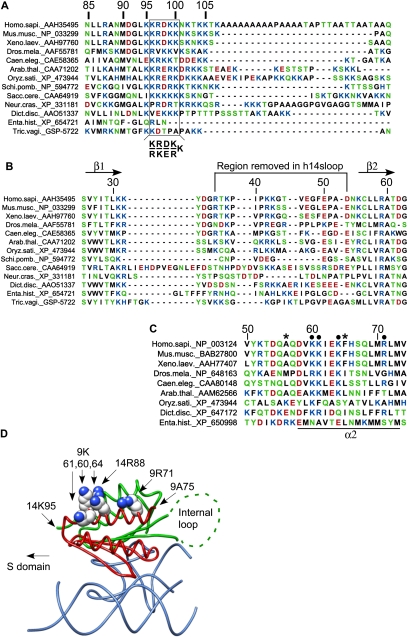
FIGURE 1.
Sequence alignments and structure of SRP9/14. (A) The C-terminal regions of SRP14 proteins from different eukaryotic species. Human 14 contains an alanine-rich tail specific of primate proteins. Box: basic pentapeptide essential for the elongation arrest activity of SRP. (B) Sequences of the internal loop in SRP14 proteins. The internal loop between β1 and β2 is a hallmark of SRP14 proteins and is absent in the structurally homologous SRP9 proteins. (C) Residues 50 to 74 in SRP9 proteins from different eukaryotic species. Black dots, conserved and solvent-exposed residues in SRP9; *, terminal amino acid residues of the truncated proteins SRP14-9-20C and -30C. (D) Structure model of h9/14 bound to Alu RNA. Alu RNA is shown in blue and h9 and h14 are shown in red and green, respectively. Side-chain atoms of K60, K61, K64, and R71 of h9, as well as R88 of h14, are highlighted as spheres. The structure does not include the peptide chains beyond 9A75 and 14K95 as well as the internal loop in h14 (dashed line, see also the text). Dict.disc, Dictyostelium discoideum; Enta.hist: Entamoeba histolytica; Tric.vagi, Trichomonas vaginalis. Color code for amino acids: blue, basic; red, acidic; green, polar; and black, hydrophobic.
Cryo-electron microscopy (cryo-EM) images of SRP bound to ribosomes containing nascent chains of a defined length showed that the Alu domain of SRP is located in the elongation factor-binding site (Halic et al. 2004). Since slowing elongation overcomes growth and translocation defects caused by low cellular levels of SRP in yeast and in mammalian cells, it appears that SRP interacts with the ribosome during EF2-catalyzed translocation of the tRNA from the A to the P site (Ogg and Walter 1995; Lakkaraju et al. 2007). The Alu domain might then delay the elongation cycle by preventing the binding of EF2. Cross-linking studies during ongoing translation demonstrated that the Alu domain is already in contact with the ribosome in the absence of a signal sequence in the nascent chain. Upon recognizing a signal sequence, the cross-links between SRP14 and the ribosome change, thereby revealing conformational changes at the interface of the Alu domain with the ribosome (Terzi et al. 2004). However, the exact mechanism by which SRP9/14 confers elongation arrest activity to SRP is still unclear.
In these studies, we used in vitro and in vivo experiments to examine the structural requirements in SRP14 and in SRP9 for the elongation arrest function. Our studies demonstrate that along with SRP14, SRP9 is also critical for the elongation arrest function. Overall, one patch of positive charges in each subunit SRP14 and SRP9 are required for function. In addition, when the positive charges in SRP14 are shifted toward the C-terminus in the polypeptide chain by the insertion of as few as two alanine residues, their function is abrogated. Our experiments also define regions in SRP9 and SRP14 that are dispensable for this function. The essential residues are located at the surface of SRP9/14 facing the S domain of SRP and form a positively charged platform that is likely to bind to the ribosome to arrest nascent chain elongation. The three essential basic residues in mammalian SRP9 are absent in the yeast Srp14p homodimer complex, suggesting differences in the exact mechanism by which the SRP Alu domain arrests nascent chain elongation in mammalian and in yeast species.
RESULTS
Four lysines following K95 in human SRP14 are sufficient to preserve elongation arrest activity of SRP
We have previously shown that the basic pentapeptide KRDKK (Fig. 1A) is essential for elongation arrest activity in vitro and in vivo (Lakkaraju et al. 2008). To exclude that sequences beyond K100 contribute to the elongation arrest activity of SRP, we engineered two truncated versions of h14, h14-95 and h14-100 (Fig. 2A). The recombinant h14 proteins were purified as complexes with recombinant human SRP9 (h9) (Huck et al. 2004) and assembled into SRP particles together with all other SRP proteins and synthetic SRP RNA (see Materials and Methods). The reconstitution reactions (RCs) were added directly to wheat germ lysate programmed for translation with synthetic mRNAs encoding the secreted protein preprolactin and a truncated form of the cytosolic cyclin D. The elongation arrest and translocation activities of SRP are determined by measuring the relative inhibition in the accumulation of preprolactin compared with cyclin D and by assessing processing of preprolactin to prolactin in the presence of microsomes, respectively. Particles reconstituted with h9/14 (wild type [WT]) showed maximal elongation arrest and translocation activities confirming that in vitro reconstituted particles were functional (Fig. 2B,C). The lack of elongation arrest activity in reactions with buffer (B) and without h9/14 (−9/14), demonstrated that our assay system was dependent on exogenous SRP and h9/14, respectively. In addition, in the absence of elongation arrest activity (RC-9/14), the translocation efficiency was reduced by about 50%. RC-100 was fully active in both assays, whereas RC-95 completely lacked elongation arrest activity and had a reduced translocation activity. Both particles were able to process preprolaction to prolactin demonstrating that they had intact signal sequence recognition and targeting functions and that the defect in translocation with RC95 was due to the absence of elongation arrest activity.
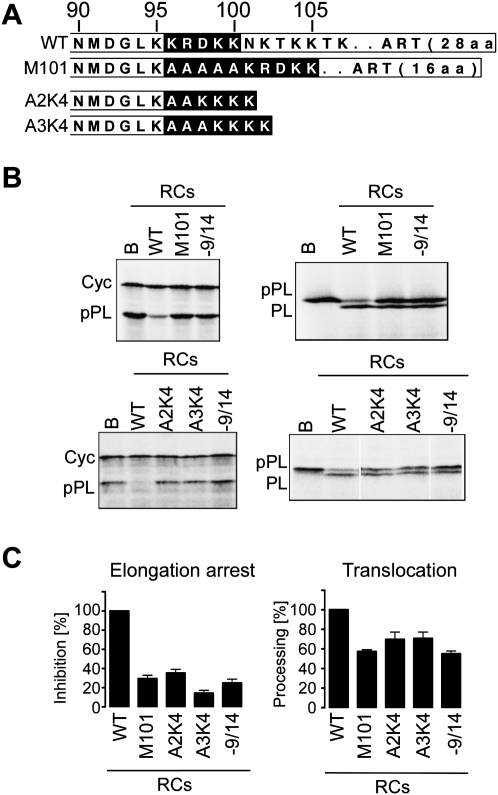
FIGURE 2.
Effects of mutations in the C-terminal region of h14 on elongation arrest activity of SRP. (A) Mutated h14 proteins used for the assembly of SRP in vitro. Amino acids of interest are in black. ART: alanine-rich tail specific for primate h14. (B) Elongation arrest and translocation activities of reconstituted SRPs (RCs). RCs contain recombinant and canine SRP proteins, synthetic SRP RNA, and the h9/14 protein indicated. WT: Wild type. Final concentration of RC: 100 nM; EKRM: 0.02 eq./μL. [35S]methionine-labeled translation products displayed by SDS-PAGE. (Left panels) The elongation arrest activity is monitored as the relative inhibition in the accumulation of preprolactin compared with cyclin. (Right panels) The translocation efficiency is determined by the percentage of total preprolactin processed into prolactin (see Materials and Methods). Cyc, a truncated version of cyclin; pPL, preprolactin; PL, prolactin. (C) Quantification of the results (n ≥ 3). Activities were normalized to RCWT, which was set to 100%. Calculated values are shown in Supplemental Table S1. B, Buffer; −9/14, RC without h9/14.
To show that h14-95 assembled into SRP, we performed equilibrium competition experiments (see Supplemental Material). Both proteins h9/14-95 and h9/14-100 competed well with h9/14 for the binding to the Alu portion of SRP RNA (Supplemental Fig. S1). The relative dissociation constants were increased 2.3- and 3.2-fold, which may be explained by a slight decrease in the affinity for the RNA or a difference in the activities of the proteins. In agreement with the crystal structure (Weichenrieder et al. 2000), these results confirmed that residues beyond 14K95 are not important for RNA binding. Furthermore, the basic pentapeptide KRDKK is the only element required for elongation arrest activity beyond 14K95.
The sequence alignment of the C-terminal region of SRP14 proteins from different eukaryotic species shows that four positions in the pentapeptide are frequently occupied with basic residues (K or R). The conservation of basic residues at positions 96 and 99 is 85%, whereas it is 70% and 62% at positions 97 and 100, respectively. Arginine is more frequent at position 97, whereas lysine is more frequent at the other positions. Acidic residues (D or E) in the middle position (98) are conserved to 54% (Fig. 1A). To examine the role of D98 and the positive charge, we replaced D98 in WT h14 with an alanine residue (Fig. 2A, h14D98A) and added either two or four lysines to the elongation arrest-defective protein h14-95 (Fig. 2A, h14K2 and h14K4). The three recombinant proteins were purified and assayed as before. The protein h9/14K2 completely lacked the capacity to confer elongation arrest activity to SRP, and consequently, translocation was inefficient by RC14K2. In contrast, SRP particles comprising h9/14K4 and h9/14D98A displayed significant elongation arrest activity and promoted efficient translocation (Fig. 2B,C). From these results we concluded that beyond K95, four lysines are sufficient for elongation arrest activity of SRP. Hence, R97 can be replaced with K and the acidic residue at position 98 is not essential.
Next, we wanted to know whether it was important that the positive charge was immediately following K95 or whether it could be moved C-terminally in the polypeptide chain without loss of function. To this end, the entire basic pentapeptide of five amino acids was shifted toward the C terminus by insertion of five alanine residues (h14M101) into WT SRP9/14. H14M101 was inactive in elongation arrest (Fig. 3B,C). It was conceivable that the flanking stretches of alanines might have had a negative effect on the pentapeptide function. In the wild-type protein, the C-terminal alanine-rich tail is not directly adjacent to the basic pentapeptide. We therefore inserted two and three alanines between K95 and the four-lysine stretch in the h14K4 protein, which is active in elongation arrest (h14A2K4 and h14A3K4). Both mutated proteins completely failed to confer elongation arrest activity to SRP and SRPs comprising these proteins had reduced translocation capacities (Fig. 3B,C). Hence, the positive charge at positions 96 and 97 is very important, since shifting the position of the positively charged patch by two residues completely abrogated its activity in arresting elongation.
FIGURE 3.
Insertion of alanine residues between h14K95 and the basic pentapetide KRDKK or the four-lysine stretch in h14K4 abrogates the elongation arrest function. (A) Mutated h14 proteins used for the assembly of SRP in vitro. Amino acid residues of interest are in black. (B) Elongation arrest and translocation activities of RCs containing mutated h9/14 proteins. WT: wild type. Final concentration of RC: 100 nM; EDTA- and salt-washed membranes (EKRM): 0.02 eq./μL. [35S]methionine-labeled translation products displayed by SDS-PAGE. Cyc, truncated version of cyclin; pPL, preprolactin; PL, prolactin. (C) Quantification of the results (n ≥ 3). Activities were normalized to RCWT, which was set to 100%. Calculated values are shown in Supplemental Table S1. B, Buffer; −9/14, RC without h9/14.
Comparable effects of the mutations were observed with complementation experiments in mammalian cells
Next, we examined the effects of the mutations in h14 on protein translocation in mammalian cells. Using the silencing/complementation assay described previously (Lakkaraju et al. 2008), we replaced the endogenous h14 protein in HEK 293T cells with mutated versions, which were expressed as C-terminal fusions of the GFP protein. The G14K4 and G14A2K4 proteins have the same mutations in the basic peptide as the h14K4 and h14A2K4 proteins studied in the cell-free assay. However, they both have the alanine-rich tail specific for the human protein (Fig. 4A, ART).
FIGURE 4.
Secretion and translocation efficiencies of HEK 293T cells expressing mutated versions of h14. (A) The different h14 proteins expressed as C-terminal fusions with GFP. Amino acid residues of interest are in black. (B) Relative expression levels of 7SL RNA. Cells were depleted of the endogenous h14 protein using shRNA complementary to the 3′ UTR of h14 mRNA (sh14) or mock depleted with shLuc and complemented with the expression of the protein indicated. The expression levels of 7SL RNA were quantified with RT-PCR and normalized to the one of mock-depleted cells expressing G14 (shLuc), which was set to 100% (n = 2). (C) SEAP activity in the medium of cells collected between 144 and 168 h and standardized to the amount of protein present in extracts prepared from the secreting cells. Activities were normalized to that of the mock-depleted cells expressing G14 (n = 2). Calculated values for C and D are shown in Supplemental Table S2. (D) Accumulation of preprolactin (pPL3F), prolactin (PL3F), and phosphorylated prolactin (*PL3F) in cells treated for 8 h with 10 μM MG132. Proteins were revealed with anti flag-tag antibodies. A: anisomycin was added to the medium at 0.03 μg/mL. The accumulation of the translocated protein prolactin in elongation arrest-defective cells is most likely explained by secondary effects on the secretory pathway as a result of inefficient translocation of endogenous proteins.
The proper assembly of the mutated proteins was confirmed by monitoring the levels of 7SL RNA. Normal cellular levels of SRP require the presence of h9/14 (Lakkaraju et al. 2007). In cells depleted of h14 and complemented with GFP, the levels of 7SL RNA are decreased sixfold (Fig. 4B). The levels of 7SL RNA in cells expressing any of the G14 chimeras were restored to 78%–90% of normal levels, confirming that all proteins effectively assembled into SRP. SEAP, a secreted version of alkaline phosphatase, is a sensitive tool to monitor loss of the elongation arrest function (Lakkaraju et al. 2008). As expected, cells with low levels of SRP (sh14/GFP) showed a strong decrease in the secretion of SEAP, whereas the complementation with G14 restored normal levels. The elongation arrest-defective G14A5 characterized previously (Lakkaraju et al. 2008) displayed a 50% loss in secretion efficiency. Compared with these two references, G14K4 restored SEAP secretion to a significant level, whereas G14A2K4 failed to do so (Fig. 4C).
To confirm that the secretion defects were due to translocation defects, we monitored the accumulation of preproteins (proteins with an uncleaved signal sequence) after complementation with G14A2K4 and, as controls, with G14 and G14A5. We chose bovine preprolactin, a well-studied SRP substrate, as a reporter (pPrl3f). To reveal nontranslocated preproteins, which are rapidly degraded by the proteasome, cells were cultured in the presence of proteasome inhibitor for 8 h before harvesting. The specific accumulation of preprolactin in the cells expressing G14A2K4 and G14A5 (Fig. 4D) compared with control cells demonstrated the translocation defects. We have previously shown with pulse-labeling experiments that the accumulation of precursor protein reflects a reduction in translocation efficiency of about 50%. Furthermore, the defects can be rescued by slowing down nascent chain elongation with antibiotics (Lakkaraju et al. 2008). Indeed, treatment of G14A2K4-expressing cells with sublethal doses of anisomycin rescued the translocation defect (Fig. 4D).
Overall, these results corroborated the conclusions drawn from the results obtained in the cell-free assay. The fact that G14K4 restores secretion only partially might be due to the immediate vicinity of the four lysines to the alanine-rich tail. It is conceivable that the long alanine-rich tail with its strong propensity to form a helix may hinder the lysine residues to adopt the conformation required for function.
The internal loop present in all SRP14 proteins is dispensable for the elongation arrest function
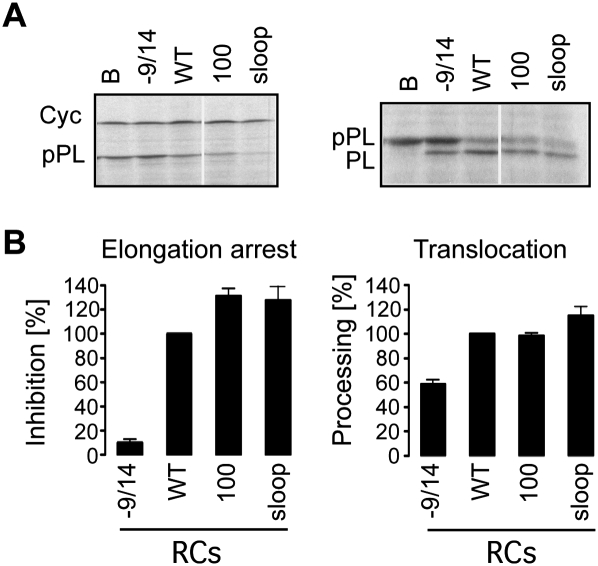
The internal loop between β1 and β2 is present in all SRP14 proteins, although its size varies in yeast species (Fig. 1B). Biochemical experiments suggested a role in RNA-binding (Bui et al. 1997). In the crystal structures of h9/14 alone and with Alu RNA (Birse et al. 1997; Weichenrieder et al. 2000), most of it is disordered and its structure is therefore unknown. The loop is located at the opposite face of the basic pentapeptide in the protein (Fig. 1C) and contains a number of conserved charged amino acid residues. It is solvent-exposed and might therefore make contacts with the ribosome. To examine whether it plays a role in arresting elongation of the nascent chain, we expressed and purified the human protein h14sloop lacking residues 36–53 as complex with h9 (Fig. 1B).
In RNA binding experiments using the Alu portion of 7SL RNA, we found the affinity of the purified h9/14sloop to be about 10-fold decreased as compared with h9/14, which is consistent with our previous results (J Häsler and K Strub, unpubl.; Bui et al. 1997). Since the dissociation constant of h9/14 is in the subnanomolar range, a 10-fold reduction was not expected to interfere with the functional assays in vitro, which are done at concentrations of 100 nM SRP. The h9/14sloop protein was reconstituted into SRP and the elongation arrest and translocation activities were assayed in the cell-free system. Compared with RCWT, RC-100, and RC-9/14, the positive and negative controls, respectively, RC9/14sloop is fully active (Fig. 5A,B). Hence, the internal loop between β1 and β2 of SRP14 is not required for the elongation arrest function.
FIGURE 5.
SRP14 lacking the internal loop between β1 and β2 can confer elongation arrest activity to SRP. (A) Translation products displayed by SDS-PAGE. (B) Quantification of the elongation arrest and translocation activities of RCs (n ≥ 3). Activities were normalized to RCWT (100%). Calculated values are shown in Supplemental Table S1. B, buffer; −9/14, RC without h9/14. [RC]final, 100 nM; EKRM, 0.02 eq/μL. The differences in absolute activities between the three fully reconstituted RCs (RCWT, RC100, RCsloop) are most likely explained by differences in the reconstitution efficiencies.
Mutation of three conserved basic residues in SRP9 abrogates the elongation arrest function of SRP
Preliminary experiments with murine SRP14-9 fusion proteins truncated at positions 66 (SRP14-9-20C) and 56 (SRP14-9-30C) (see also Fig. 1C) revealed a partial and a complete loss of elongation arrest activity (Supplemental Fig. S2). Since removal of the same region did not abolish RNA binding of the heterodimeric protein (Bui et al. 1997), we concluded that amino acid residues 56–66 of SRP9 have a role in arresting elongation. Since activity was only partially lost with SRP14-9-20C, it was conceivable that insufficient amounts of the protein were synthesized in vitro for full reconstitution of SRP. By examining both the primary sequence conservation and the structure of h9/14 (Weichenrieder et al. 2000), we identified four basic residues in α2 of h9 (Fig. 1C,D, K60, K61, K64, R71) that are conserved, solvent-exposed and, for three of them, located in proximity of the basic pentapeptide in the RNA–protein complex. The conservation of basic residues at positions 60, 61, 64, and 71 is 89%, 78%, 67%, and 56%, respectively. The three first positions are primarily occupied by lysines, whereas position 71 is occupied by arginine. We made a mutational study of these residues both in the cell-free system and in mammalian cells. For the in vitro studies, we used two h9 proteins with three and four of the basic residues changed into alanine (Fig. 6A, h9-3A, h9-4A). Purified recombinant proteins were produced as described. Although R71 of h9 makes contact with R80 of h14 (Birse et al. 1997), the mutation did not interfere with the purification of the h9-4A/14 complex.
FIGURE 6.
Mutations of the conserved lysine residues at positions 60, 61, 64, of SRP9 abrogate elongation arrest activity of SRP. (A) Sequence alignment of the C-terminal portions of WT and mutated h9 proteins. Sequences of interest are highlighted in black and mutated amino acid residues are highlighted in gray. (B) Elongation arrest (left panel) and translocation (right panel) activities (n ≥ 2) of particles (RCs) reconstituted with the h9 proteins indicated. B, buffer; −9/14, RC without h9/14. [RC]final, 100 nM; EKRM, 0.02 eq/μL. Activities were normalized to RCWT (100%). Calculated values are shown in Supplemental Table S1. (C) Relative expression levels of 7SL RNA in 293T cells collected at 144 h (left panel). Expression levels of 7SL RNA were normalized to the one of mock-depleted and G9 expressing cells, which was set to 100% (n = 2). (D) SEAP activities in medium of 293T cells collected between 120 and 144 h and standardized to the amount of proteins present in extracts prepared from the secreting cells at 144 h (right panel). Activities were normalized to that of the mock-depleted cells expressing G9 (n = 2). Calculated values for C and D are shown in Supplemental Table S2. (D) Accumulation of preprolactin (pPL3F), prolactin (PL3F), and phosphorylated prolactin (*PL3F) in 293T cells after h9-depletion and complementation with the G9 proteins indicated. Prolactin was revealed with anti flag-tag antibodies.
Particles reconstituted with h9-4A/14 and h9-3A/14 lacked elongation arrest activity and displayed decreased translocation efficiencies as expected of elongation arrest-defective SRP (Fig. 6B). These results indicated that both reconstituted particles possessed intact signal recognition and targeting activities and demonstrated that the SRP9 mutations specifically abrogated elongation arrest activity. As expected from the structure and as shown in the in vivo experiments (see below), the mutated proteins assembled well into SRP. Hence, changing the three residues 60, 61, and 64 from lysine into alanine is sufficient to abolish elongation arrest activity.
To confirm the functional role these basic residues in the homologous in vivo system, we established a silencing/complementation assay similar to the one used for h14. To reduce h9 levels in mammalian cells, we expressed a short hairpin RNA (sh9) with complementarities to the 3′ UTR of the h9 mRNA. The level of the endogenous h9 was efficiently reduced about eightfold at 120 h after transfection (Supplemental Fig. S3). As for h14, we expressed the exogenous h9 as a fusion protein with GFP. The protein chimeras, GFP-h9 (G9) and h9-GFP (9G) worked with similar efficiency in complementation assays (Supplemental Table S2). We first examined the effects of the mutations on SRP assembly. Like in cells with low levels of h14, without complementation the amount of 7SL RNA decreased upon sh9 treatment by about sixfold (Fig. 6C, GFP lane). Expression of the wild-type and mutated G9 chimeras restored 7SL RNA levels to ∼80%–90% compared with the level of control cells (Fig. 6C), indicating that all mutated h9 proteins were well expressed and assembled into SRP in vivo.
We next examined the effects of the mutations on SEAP secretion. Without complementation, SEAP secretion was decreased to about 20%, whereas complementation with any of the two protein chimera G9 (Fig. 6C) or chimera 9G (Supplemental Table S2) restored normal secretion levels. G9-3A and G9-4A both had a secretion phenotype typical for elongation arrest-defective SRP. As in the cell-free assay, simultaneous mutation of K60, K61, and K64 was sufficient to abrogate function. To identify more specifically the residues required for elongation arrest activity, we examined three additional G9 mutants (Fig. 6A). In G9R71A and G9K64A, basic amino acid residues were replaced individually with alanine. In G9-2A, K60 and K61 were simultaneously changed into alanine. Mutation of R71 had no effect on secretion, demonstrating that it is not essential for activity. Complementation with G9-2A and G9K64A led to an intermediate secretion defect demonstrating that all three lysine residues or a combination of K64 and K60 or K61 are required for full activity (Fig. 6C). We again confirmed that the secretion defects reflected a defect in translocation using preprolactin as a reporter. We see specific accumulation of preprolactin in cells complemented with G9-3A and the translocation defect disappears upon treatment with anisomycin (Fig. 6D).
We noted that R88 in α2 of h14 is in very close proximity of the three essential lysines in h9 and a basic residue at this position is very frequent in SRP14 proteins of different species (76%) (Fig. 1A,D) suggesting that it might also contribute to elongation arrest activity. We therefore mutated R88 into alanine and tested the activities of the mutated protein in HEK 293T cells. R88 in h14 makes a hydrogen bond with E63 in h9, whereas R71 in h9 makes a hydrogen bond with Q80 of h14. Since G9R71A expression restored normal SRP levels (Fig. 6C), we did not expect G14R88A to interfere with SRP assembly. However, G14R88A expression restored SRP levels only partially. On average, it was about 50%, but the values were quite variable between experiments. Possibly, the assembly efficiency was dependent on the expression level of h14R88A (Fig. 4B). Invariably, expression of G14R88A resulted in the same secretion-defective phenotype as the expression of other h9 and h14 proteins that fail to confer elongation arrest activity to SRP (Fig. 4C). This defect is unlikely to be explained solely by the twofold reduction in SRP levels. Small reductions in SRP levels were found to decrease protein translocation/secretion only moderately or not at all (Ren et al. 2004; Lakkaraju et al. 2007). Hence, R88 is a very important amino acid for SRP assembly and might possibly also be involved in arresting the elongation cycle of the ribosome.
DISCUSSION
In this study we identified two positively charged patches one in each polypeptide SRP9 and SRP14 that are essential for elongation arrest activity. One patch is in the α2 helix of SRP9 and the other follows immediately K95 is SRP14 (Fig. 1D). All of the essential residues are located in proximity on the same side of SRP9/14 facing the S-domain of SRP (Fig. 1D) and most likely provide a platform for interactions with the ribosome. 14R88 might also be part of the functional platform. However, we have not been able to dissociate unambiguously its role in assembly from its putative role in the elongation arrest function. The positive patch in SRP14 has to be directly adjacent to 14K95, indicating that the positive charges at positions 96 and 97 are very important. One plausible explanation for this observation is that the positive patch in SRP14 has to be properly positioned with regard to the positive patch in SRP9 for function. The fact that amino acid residues 93–95 are firmly held in place by a network of interactions with SRP9 (Birse et al. 1997), supports this interpretation. Alternatively, we cannot exclude that the insertion of two alanines in 14A2K4 abrogates function per se. Notably, although conserved to 54%, the acidic residue at position 98 in the basic pentapeptide can be substituted by lysine without loss of function.
The other dispensable elements for activity include the conserved internal loop of SRP14, 9R71, and the C-terminus of SRP9 (Bovia et al. 1994). These nonessential regions are located on the same side of the complex as the 5′ and 3′ ends of the RNA (Fig. 1D). The internal loop is present in all SRP14 proteins and it is directed toward the kink in the SRP RNA that folds it back by almost 180°C (L Bousset, M Brooks, and S Cusack, pers. comm.; Weichenrieder et al. 2000). Together with the decreased affinity of 9/14sloop for the RNA, it appears that it plays primarily a role in SRP assembly.
The hypothesis that the basic patches contact ribosomal RNA is consistent with the images of the RNC-SRP complex obtained by cryo-EM (Halic et al. 2004). In these images, the SRP Alu domain is located in the elongation factor-binding site and the region of SRP9 comprising the key residues is in proximity of h5 and h15 of 18S ribosomal RNA. Lysine usually makes nonspecific electrostatic interactions with the phosphate backbone of nucleic acids, whereas arginine has the additional ability to engage in diverse ionic, weakly polar, and van der Waals interactions as well as in hydrogen bonding with the phosphate backbone and the base (Bayer et al. 2005). Since the critical residues in SRP9/14 can all be lysines, we hypothesize that the putative interactions with the ribosomal RNA are mediated by nonbase-specific electrostatic contacts.
As little as one and two amino acid mutations in SRP9 and SRP14 were sufficient to abrogate elongation arrest activity. Such small changes are unlikely to modify the overall structure of SRP and therefore its signal sequence-dependent binding to the ribosome as confirmed by the result that mutant SRPs promoted the translocation of nascent chains, albeit inefficiently. We therefore believe that these strong phenotypes cannot be explained by a simple spatial competition between the elongation factor and SRP for ribosome binding. Rather, the mechanism of the elongation arrest function appears to require few, specific interactions between SRP9/14 and the ribosome. In addition, these contacts do not occur in the absence of a signal sequence in the nascent chain, although the Alu domain is already in close proximity to the ribosome as shown by cross-linking experiments (Terzi et al. 2004). Hence, signal sequence recognition by SRP54 is transmitted via SRP or via the ribosome to the Alu domain, resulting in putative specific contacts that then interfere with the elongation cycle.
At this moment the exact molecular target(s) in the ribosome of the essential basic residues in SRP9/14 remain to be identified. They are likely conserved, since the effects of the mutations in SRP9/14 were essentially the same with wheat germ and with human ribosomes. As mentioned before, cryo-EM images show the essential region of SRP9 in close proximity of helices 5 and 15 of 18S RNA, whereas the basic pentapeptide cannot be traced. The addition of a bivalent cross-linker during translation of preprolactin in reticulocyte lysate revealed three covalent links between SRP14 and proteins of the large subunit and only one link to a protein of the small subunit (Terzi et al. 2004). Three of the four cross-links are exclusively observed during synthesis of a protein bearing a signal sequence.
The essential residues in SRP9 and in SRP14 are not highly conserved in protozoan species and in the plant species Oryza sativa (Fig. 1). Some of the protozoan species lack SRP9/14 and display major structural changes in the Alu portion of SRP RNA (Rosenblad et al. 2004). Trypanosomal SRP comprises a tRNA-like molecule that might substitute SRP9/14 for activity (Lustig et al. 2005, 2010). In yeast, the homodimeric complex of SRP14 has replaced the heterodimer complex in yeast species (Strub et al. 1999; Mason et al. 2000). K60, 61, and 64 in h9 are not conserved in the homologous positions of yeast Srp14p (Birse et al. 1997; Brooks et al. 2009) and the pentapeptide is also less conserved in the yeast Neurospora crassa (Fig. 1). In addition, SRP RNAs of yeast species have simple stem–loop structures instead of the typical two-stem–loop structure of the metazoan Alu domain. The divergence may therefore reflect differences in the exact molecular mechanism of the SRP-mediated arrest in different species.
Basic oligopeptides related to the pentapeptide of SRP14 are found in several proteins involved in translation and ER translocation (Dudek et al. 2002) such as translation initiation factors, EF2, the NAC β subunit, ERj1p, and Sec62. ERj1p is a membrane protein of the ER, with a cytosolic domain that interacts with translating ribosomes on the ER (Chevalier et al. 2000; Kroczynska et al. 2004). In functional assays, the basic oligopeptide of ERj1p inhibits translation initiation through binding to 28S ribosomal RNA (Dudek et al. 2002). Cryo-EM images of the ERj1p–ribosome complex show the protein bound to both sides of the nascent chain exit tunnel forming a bridge (Blau et al. 2005). This position does not straightforwardly explain the mechanism of the translational inhibition, and further studies are therefore required. Notably, a similar basic oligopeptide is present at the C terminus of EF2, which is located close to the GTPase activating center when bound to ribosomes (Gomez-Lorenzo et al. 2000; Spahn et al. 2004). The conservation of short, lysine-rich regions in several ribosomal ligands suggests that their interactions with specific structures of ribosomal RNA could be a common feature of some positive and negative effectors of translation. The recognition of different ribosomal structures by the various lysine-rich motifs is reminiscent of the arginine-rich peptide motifs capable of binding unique RNA structures (ARMs) (for review, see Chen and Varani 2005). They are very short in length and adopt a variety of different structures upon association with their specific RNA targets.
Finally, our results underline the requirement of the protein subunit for the elongation arrest function in species where it is present. It is therefore not surprising that these proteins are conserved in sequence (Gorodkin et al. 2001; Andersen et al. 2006). In contrast, the sequence and structure of the Alu portion of SRP RNA are less conserved. Specifically, the solvent accessible loops L1 and L1.2 and their stems are not conserved in size and shape. Furthermore, all the mutations in the Alu portion of SRP RNA including the L1.2 and L2 loops interfered primarily with the assembly of SRP (Andreazzoli and Gerbi 1991; Chang et al. 1997; Huck et al. 2004) and affected elongation arrest activity only indirectly. It is therefore conceivable that the recruitment of the subunit SRP9/14 into SRP allowed eukaryotic organisms to acquire the elongation arrest function that ultimately served to control protein influx into the ER by limiting receptor concentrations (Lakkaraju et al. 2008).
MATERIALS AND METHODS
Plasmid constructions
Plasmids coding for the mutated versions of the h14 protein were obtained using the QuickChange method (Stratagene). Primers containing the desired mutations were flanked by 15 complementary nucleotides on both sides (Microsynth). Clones pBh14K95 and pBh14K100 were obtained by introducing two stop codons after the K95 and the K100 codons, respectively. pBh14K2 and pBh14K4 were obtained by inserting codons for two and four lysine residues before the first stop codon of the cDNA in pBh14K95. Introducing two or three alanine codons after the K95 codon in pBh14K4, respectively, generated clones pBh14A2K4 and pBh14A3K4. pEth14M101 was generated from the h14 cDNA in pEth14 (Bovia et al. 1997). pGh14K4 was engineered by introducing four lysine codons after the K95 codon in pGh14A12 (Lakkaraju et al. 2008). pGh14A2K4 was obtained from pGh14K4 by introducing two lysine codons after K95 in the cDNA region. pGFH9 or pH9G were engineered by cloning the cDNA of the human SRP9 from pEth9 (Bovia et al. 1997) into pEGFP-C1, pEGFP-N1. Clones pEth9-4A, pEth9-3A, pGFh9-3A pGFh9-4A, pGh9-2A, pGh9R71A, and pGh9K64A were obtained from pEth9 and pGFH9 by changing the nucleotide sequence encoding K60, K61, K64, and R71 into alanine codons in the h9 cDNA. shRNAs were produced from the expression vectors pSUPER.retro.puro (Oligoengine) containing the appropriate inserts (pSR14 and pSR9) complementary to the following target sequences: 14 sense: 5′-AGGGCATACATTTCCTGCT-3′, 9 sense: 5′-GAAATGAACAGGAGTCAAT-3′.
Protein synthesis and purification
Proteins h14, h14M101, and h9 were expressed in BL21 LysS Escherichia coli (Novagen) and h14-95, h14-100, h1414K2, h14K4, h14A2K4, and h14A3K4 in Tuner (DE3) pLacI cells (Novagen), respectively. Purification of the proteins was described earlier (Huck et al. 2004; Terzi et al. 2004). All proteins were stored in 20 mM HEPES (pH 7.5), 500 mM KOAc (pH 7.5), 0.1 mM EDTA (pH 7.5), 0.01% NIKKOL, 10 mM DTT, and 10% glycerol. Their purity was >95% and their concentrations were calculated from the absorbance at 280 nm using the specific molar extinction coefficient calculated for each protein (Gill and von Hippel 1989). All the purified h9/14 proteins had an equimolar composition of h9 and h14 proteins based on Coomassie staining and on the biochemical purification protocol, which removed excess of h9 in one of the purification steps.
Reconstitution of SRP and cell-free translation/translocation assays
SRP reconstitutions, activity assays and quantifications were done as described previously (Huck et al. 2004). In short, all SRP proteins and synthetic SRP RNA (7SLC) were combined at 0.5 and 0.6 μM, respectively. The reconstitution reactions and EDTA and salt-washed membranes (EKRM) were added to translation reactions at final concentrations of 100 nM and of 0.02 eq/μL, respectively, as described in Thomas et al. (1997). 7SLC represents a circular permutation of 7SL RNA (5′ and 3′ ends at positions 280 and 277 in 7SL RNA, respectively) that reconstitutes more efficiently into SRP than the previously used construct (A Scherrer and K Strub, unpubl.). The relative inhibition of preprolactin accumulation was calculated as follows:  . Ps and Cs are the amounts of preprolactin and cyclin quantified in the sample and Po and Co the amounts present in the negative control (buffer). The translocation efficiency was calculated as follows: T = 100* PL/(PL + pPL), where PL is prolactin and pPL preprolactin. The translocation efficiencies of the samples in each experiment were normalized to RCWT, which was set to 100%.
. Ps and Cs are the amounts of preprolactin and cyclin quantified in the sample and Po and Co the amounts present in the negative control (buffer). The translocation efficiency was calculated as follows: T = 100* PL/(PL + pPL), where PL is prolactin and pPL preprolactin. The translocation efficiencies of the samples in each experiment were normalized to RCWT, which was set to 100%.
Cell culture, reporter protein assays, and 7SL RNA quantification
HEK293T cells were grown at 37°C in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (both from Sigma). For complementation experiments, equal numbers of 293T cells were initially transfected with shRNA(14) and shRNA(9) and, for mock depletion, with shRNA(Luc) expressing plasmids. After 24 h, cells were selected for 24 h with puromycin (3 μg/mL). At 72 h, cells were plated at equal numbers into 6-cm dishes and transfected with GFP-h14 or GFP-h9 chimeras (or as a control with GFP) and, if applicable, with a plasmid expressing a reporter protein.
The SEAP assay was performed in 96-well plates using 15 μL of cell culture supernatant collected at the times indicated (Lakkaraju et al. 2007) . For the analysis of preproteins, cells were treated with MG132 (10 mM, SIGMA) for 8 h at 136 h and 160 h after the initial transfection for h9 and h14 experiments, respectively. Cells were lysed with 200 μL of 1% SDS in 0.1 M Tris (pH 8) directly in the plate. The lysates were transferred into microtubes and heated to 70°C with occasional vortexing until the lysates became less viscous. The sample was diluted (threefold) in a buffer containg 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS 50 mM Tris-HCl (pH 8), 1 mM EDTA, and 1× protease inhibitor cocktail. In the experiments with anisomycin, cells were first treated for 2 h with anisomycin followed by 8 h of anisomycin and MG132 treatment. Quantification of 7SL RNA was done with qRT-PCR as described in Lakkaraju et al. (2007) using 10 ng total RNA.
Statistical methods
The data in the figures represents the standard error of the mean (SEM).  , where X is the individual data point, M is the mean, and n is the number of samples.
, where X is the individual data point, M is the mean, and n is the number of samples.
Sequence alignment and structure, statistical methods
Molecular graphics images were produced using the UCSF Chimera package from Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (Pettersen et al. 2004). Alignments were performed using the CLC free workbench from CLC Bio.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Dr. L. Terzi for providing purified 9/14-100 and 9/14-95 and Drs. O. Weichenrieder and S. Cusack for the coordinates of the structure model of the Alu RNA–h9/14 complex. We thank A. Chassot, Y. Miao, and Y. Shao for technical assistance. This work was supported by grants from the Swiss National Science Foundation, the Canton of Geneva, and the MEDIC Foundation. Support from NIH Grants R01GM26494 and the Robert A. Welch Foundation (Chair grant BE-0017) are also acknowledged (to A.E.J.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2040410.
REFERENCES
- Andersen ES, Rosenblad MA, Larsen N, Westergaard JC, Burks J, Wower IK, Wower J, Gorodkin J, Samuelsson T, Zwieb C 2006. The tmRDB and SRPDB resources. Nucleic Acids Res 34: D163–D168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreazzoli M, Gerbi SA 1991. Changes in 7SL RNA conformation during the signal recognition particle cycle. EMBO J 10: 767–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer TS, Booth LN, Knudsen SM, Ellington AD 2005. Arginine-rich motifs present multiple interfaces for specific binding by RNA. RNA 11: 1848–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse DE, Kapp U, Strub K, Cusack S, Aberg A 1997. The crystal structure of the signal recognition particle Alu RNA binding heterodimer, SRP9/14. EMBO J 16: 3757–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau M, Mullapudi S, Becker T, Dudek J, Zimmermann R, Penczek PA, Beckmann R 2005. ERj1p uses a universal ribosomal adaptor site to coordinate the 80S ribosome at the membrane. Nat Struct Mol Biol 12: 1015–1016 [DOI] [PubMed] [Google Scholar]
- Bovia F, Bui N, Strub K 1994. The heterodimeric subunit SRP9/14 of the signal recognition particle functions as permuted single polypeptide chain. Nucleic Acids Res 22: 2028–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovia F, Fornallaz M, Leffers H, Strub K 1995. The SRP9/14 subunit of the signal recognition particle (SRP) is present in more than 20-fold excess over SRP in primate cells and exists primarily free but also in complex with small cytoplasmic Alu RNAs. Mol Biol Cell 6: 471–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovia F, Wolff N, Ryser S, Strub K 1997. The SRP9/14 subunit of the human signal recognition particle binds to a variety of Alu-like RNAs and with higher affinity than its mouse homolog. Nucleic Acids Res 25: 318–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks MA, Ravelli RB, McCarthy AA, Strub K, Cusack S 2009. Structure of SRP14 from the Schizosaccharomyces pombe signal recognition particle. Acta Crystallogr D Biol Crystallogr 65: 421–433 [DOI] [PubMed] [Google Scholar]
- Bui N, Wolff N, Cusack S, Strub K 1997. Mutational analysis of the protein subunits of the signal recognition particle Alu-domain. RNA 3: 748–763 [PMC free article] [PubMed] [Google Scholar]
- Chang DY, Sasaki-Tozawa N, Green LK, Maraia RJ 1995. A trinucleotide repeat-associated increase in the level of Alu RNA-binding protein occurred during the same period as the major Alu amplification that accompanied anthropoid evolution. Mol Cell Biol 15: 2109–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DY, Newitt JA, Hsu K, Bernstein HD, Maraia RJ 1997. A highly conserved nucleotide in the Alu domain of SRP RNA mediates translation arrest through high affinity binding to SRP9/14. Nucleic Acids Res 25: 1117–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Varani G 2005. Protein families and RNA recognition. FEBS J 272: 2088–2097 [DOI] [PubMed] [Google Scholar]
- Chevalier M, Rhee H, Elguindi EC, Blond SY 2000. Interaction of murine BiP/GRP78 with the DnaJ homolog MTJ1. J Biol Chem 275: 19620–19627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly T, Gilmore R 1986. Formation of a functional ribosome–membrane junction during translocation requires the participation of a GTP-binding protein. J Cell Biol 103: 2253–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly T, Rapiejko PJ, Gilmore R 1991. Requirement of GTP hydrolysis for dissociation of the signal recognition particle from its receptor. Science 252: 1171–1173 [DOI] [PubMed] [Google Scholar]
- Dudek J, Volkmer J, Bies C, Guth S, Muller A, Lerner M, Feick P, Schafer KH, Morgenstern E, Hennessy F, et al. 2002. A novel type of co-chaperone mediates transmembrane recruitment of DnaK-like chaperones to ribosomes. EMBO J 21: 2958–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SC, von Hippel PH 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem 182: 319–326 [DOI] [PubMed] [Google Scholar]
- Gilmore R, Blobel G, Walter P 1982. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J Cell Biol 95: 463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Lorenzo MG, Spahn CM, Agrawal RK, Grassucci RA, Penczek P, Chakraburtty K, Ballesta JP, Lavandera JL, Garcia-Bustos JF, Frank J 2000. Three-dimensional cryo-electron microscopy localization of EF2 in the Saccharomyces cerevisiae 80S ribosome at 17.5 Å resolution. EMBO J 19: 2710–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Prehn S, Hartmann E, Herz J, Otto A, Kraft R, Wiedmann M, Knespel S, Dobberstein B, Rapoport TA 1990. The signal sequence receptor has a second subunit and is part of a translocation complex in the endoplasmic reticulum as probed by bifunctional reagents. J Cell Biol 111: 2283–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorodkin J, Knudsen B, Zwieb C, Samuelsson T 2001. SRPDB (Signal Recognition Particle Database). Nucleic Acids Res 29: 169–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Becker T, Pool MR, Spahn CM, Grassucci RA, Frank J, Beckmann R 2004. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427: 808–814 [DOI] [PubMed] [Google Scholar]
- Huck L, Scherrer A, Terzi L, Johnson AE, Bernstein HD, Cusack S, Weichenrieder O, Strub K 2004. Conserved tertiary base pairing ensures proper RNA folding and efficient assembly of the signal recognition particle Alu domain. Nucleic Acids Res 32: 4915–4924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan RJ, Freymann DM, Stroud RM, Walter P 2001. The signal recognition particle. Annu Rev Biochem 70: 755–775 [DOI] [PubMed] [Google Scholar]
- Kroczynska B, Evangelista CM, Samant SS, Elguindi EC, Blond SY 2004. The SANT2 domain of the murine tumor cell DnaJ-like protein 1 human homologue interacts with α1-antichymotrypsin and kinetically interferes with its serpin inhibitory activity. J Biol Chem 279: 11432–11443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakkaraju AK, Luyet PP, Parone P, Falguieres T, Strub K 2007. Inefficient targeting to the endoplasmic reticulum by the signal recognition particle elicits selective defects in post-ER membrane trafficking. Exp Cell Res 313: 834–847 [DOI] [PubMed] [Google Scholar]
- Lakkaraju AK, Mary C, Scherrer A, Johnson AE, Strub K 2008. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell 133: 440–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig Y, Goldshmidt H, Uliel S, Michaeli S 2005. The Trypanosoma brucei signal recognition particle lacks the Alu-domain-binding proteins: Purification and functional analysis of its binding proteins by RNAi. J Cell Sci 118: 4551–4562 [DOI] [PubMed] [Google Scholar]
- Lustig Y, Wachtel C, Safro M, Liu L, Michaeli S 2010. ‘RNA walk’—a novel approach to study RNA–RNA interactions between a small RNA and its target. Nucleic Acids Res 38: e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason N, Ciufo LF, Brown JD 2000. Elongation arrest is a physiologically important function of signal recognition particle. EMBO J 19: 4164–4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer DI 1985. Signal recognition particle (SRP) does not mediate a translational arrest of nascent secretory proteins in mammalian cell-free systems. EMBO J 4: 2031–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg SC, Walter P 1995. SRP samples nascent chains for the presence of signal sequences by interacting with ribosomes at a discrete step during translation elongation. Cell 81: 1075–1084 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Pool MR 2005. Signal recognition particles in chloroplasts, bacteria, yeast, and mammals. Mol Membr Biol 22: 3–15 [DOI] [PubMed] [Google Scholar]
- Ren YG, Wagner KW, Knee DA, Aza-Blanc P, Nasoff M, Deveraux QL 2004. Differential regulation of the TRAIL death receptors DR4 and DR5 by the signal recognition particle. Mol Biol Cell 15: 5064–5074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblad MA, Gorodkin J, Knudsen B, Zwieb C, Samuelsson T 2003. SRPDB: Signal recognition particle database. Nucleic Acids Res 31: 363–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblad MA, Zwieb C, Samuelsson T 2004. Identification and comparative analysis of components from the signal recognition particle in protozoa and fungi. BMC Genomics 5: 5 doi: 10.1186/1471-2164-5-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel V, Walter P 1985. Elongation arrest is not a prerequisite for secretory protein translocation across the microsomal membrane. J Cell Biol 100: 1913–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J 2004. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J 23: 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub K, Walter P 1990. Assembly of the Alu domain of the signal recognition particle (SRP): Dimerization of the two protein components is required for efficient binding to SRP RNA. Mol Cell Biol 10: 777–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub K, Fornallaz M, Bui N 1999. The Alu domain homolog of the yeast signal recognition particle consists of an Srp14p homodimer and a yeast-specific RNA structure. RNA 5: 1333–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzi L, Pool MR, Dobberstein B, Strub K 2004. Signal recognition particle Alu domain occupies a defined site at the ribosomal subunit interface upon signal sequence recognition. Biochemistry 43: 107–117 [DOI] [PubMed] [Google Scholar]
- Thomas Y, Bui N, Strub K 1997. A truncation in the 14 kDa protein of the signal recognition particle leads to tertiary structure changes in the RNA and abolishes the elongation arrest activity of the particle. Nucleic Acids Res 25: 1920–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Blobel G 1981. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J Cell Biol 91: 557–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichenrieder O, Wild K, Strub K, Cusack S 2000. Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature 408: 167–173 [DOI] [PubMed] [Google Scholar]