Abstract
Trans-translation relieves a stalled translation on the bacterial ribosome by transfer-messenger RNA (tmRNA) with the help of SmpB, an essential cofactor of tmRNA. Here, we examined the role of the unstructured C-terminal tail of SmpB using an in vitro trans-translation system. It was found that truncation of the C-terminal tail or substitution of tryptophan residue at 147 in the middle of the C-terminal tail affected the activity in the early stage of trans-translation. Our investigations also revealed that the C-terminal tail is not required for the events until GTP is hydrolyzed by EF-Tu in complex with tmRNA-SmpB. A synthetic peptide corresponding to the C-terminal tail of SmpB inhibited peptidyl-transfer of alanyl-tmRNA and A-site binding of SmpB, but not GTP hydrolysis. These results suggest that the C-terminal tail has a role in the step of accommodation of alanyl-tmRNA-SmpB into the A-site. Directed hydroxyl radical probing indicated that tryptophan residue at 147 is located just downstream of the decoding center in the mRNA path when SmpB is in the A-site.
Keywords: trans-translation, SmpB, tmRNA, ribosome, translation
INTRODUCTION
In bacterial cells, a ribosome stalled on a problematic mRNA, such as that lacking a stop codon, is rescued by a noncanonical translation system called trans-translation, which is mediated by transfer-messenger RNA (tmRNA), also known as 10Sa RNA or SsrA RNA. Initially, tmRNA is aminoacylated with alanine by alanyl-tRNA synthetase (Komine et al. 1994; Ushida et al. 1994). Alanyl-tmRNA enters the A-site of the stalled ribosome, just as aminoacyl-tRNA does but without a codon–anticodon interaction, to receive a nascent polypeptide from peptidyl-tRNA in the P-site (Keiler et al. 1996; Himeno et al. 1997; Muto et al. 1998). Subsequently, tmRNA serves as an mRNA for the tag-peptide. Consequently, the tag-peptide is fused to the C terminus of the nascent peptide as the degradation signal (Gottesman et al. 1998; Moore and Sauer 2007), allowing the ribosome to be recycled back to a new round of translation for various kinds of cellular events (Abo et al. 2000; Huang et al. 2000; Muto et al. 2000; Fujihara et al. 2002; Keiler 2007; Abe et al. 2008; Himeno et al. 2009). tmRNA has two functional domains, a tRNA-like domain (TLD) partially mimicking tRNA (Komine et al. 1994; Ushida et al. 1994; Felden et al. 1998) and an mRNA domain including the coding region for the tag-peptide surrounded by four characteristic pseudoknot structures (Nameki et al. 1999). An elaborate interplay of these two domains should be required for the yet-mysterious trans-translation processes in the ribosome.
Several factors, elongation factor Tu (EF-Tu) (Rudinger-Thirion et al. 1999; Barends et al. 2000; Hanawa-Suetsugu et al. 2001; Stepanov and Nyborg 2003), SmpB (Karzai et al. 1999; Barends et al. 2001; Hanawa-Suetsugu et al. 2002), and ribosomal protein S1 (Wower et al. 2000; Barends et al. 2001; Hanawa-Suetsugu et al. 2002), have been identified as tmRNA-binding proteins. Unlike S1 (Qi et al. 2007; Saguy et al. 2007; Takada et al. 2007), SmpB is essential for trans-translation in vivo and in vitro. It binds to TLD of tmRNA (Barends et al. 2001; Hanawa-Suetsugu et al. 2002; Wower et al. 2002; Gutmann et al. 2003; Nameki et al. 2005) and the ribosome (Karzai et al. 1999) to perform multiple functions in the process of trans-translation, enhancement of aminoacylation efficiency of tmRNA (Barends et al. 2001; Hanawa-Suetsugu et al. 2002; Shimizu and Ueda 2002), protection of tmRNA from degradation in the cell (Hanawa-Suetsugu et al. 2002; Hong et al. 2005), and recruitment of tmRNA to the stalled ribosome (Karzai et al. 1999; Hanawa-Suetsugu et al. 2002). NMR studies have revealed that SmpB is comprised of an antiparallel β barrel core with three α helices and C-terminal basic residues that are disordered in solution (Dong et al. 2002; Someya et al. 2003). In a crystal structure of SmpB lacking an unstructured C-terminal tail in complex with a fragment of TLD (Gutmann et al. 2003; Bessho et al. 2007), SmpB binds the elbow of TLD, in agreement with the results of biochemical studies. SmpB can be superimposed on the anticodon stem and loop of tRNA, if TLD is fixed on the amino acid acceptor stem and the T-arm. It has been shown that SmpB having a mutation or truncation in the C-terminal tail does not facilitate trans-translation in vivo (Jacob et al. 2005; Sundermeier et al. 2005), suggesting that the C-terminal tail plays a crucial role in some step of trans-translation. Although several complexes of SmpB with the ribosome have been visualized by cryo-EM studies (Valle et al. 2003; Gillet et al. 2006; Kaur et al. 2006), the C-terminal tail has not yet been identified in these complexes. In a previous study, we demonstrated the sites and modes of binding of SmpB on the ribosome by directed hydroxyl radical probing (Kurita et al. 2007). It was revealed that there are two SmpB-binding sites on a ribosome, one in the A-site and the other in the P-site, and that the C-terminal tails of both A-site SmpB and P-site SmpB are located along the mRNA path in the ribosome. This led us to propose a novel mechanism of trans-translation, in which SmpB mimics both tRNA and mRNA (Kurita et al. 2007; Himeno et al. 2009). The interaction of SmpB with the vicinity of the decoding center has also been supported by chemical probing and NMR studies (Nonin-Lecomte et al. 2009).
In this study, we examined the effects of truncations or mutations introduced to the C-terminal region of SmpB on the early stage of trans-translation. Our results revealed a novel behavior of the C-terminal tail of SmpB: it interacts with the mRNA path on the stalled ribosome to accommodate tmRNA-SmpB into the A-site after GTP hydrolysis by EF-Tu. Tryptophan residue at 147 in the C-terminal tail would have a crucial role for this interaction.
RESULTS
Effects of mutations in the C-terminal tail of SmpB on peptidyl-transfer of alanyl-tmRNA
In a previous study, we have established an in vitro system to measure the peptidyl-transfer from polyphenylalanyl-tRNAPhe to alanyl-tmRNA (first step) directed by SmpB in trans-translation (Asano et al. 2005; Konno et al. 2007; Takada et al. 2007). First, polyphenylalanine was synthesized from a synthetic mRNA, (UUC)10, using E. coli ribosomes, [14C]phenylalanyl-tRNAPhe, and elongation factors to produce a stalled complex of ribosome (Asano et al. 2005; Konno et al. 2007). This stalled complex of ribosome with [14C]polyphenylalanyl-tRNAPhe at the P-site was incubated with [3H]alanyl-tmRNA and SmpB to evaluate the efficiency of the first step.
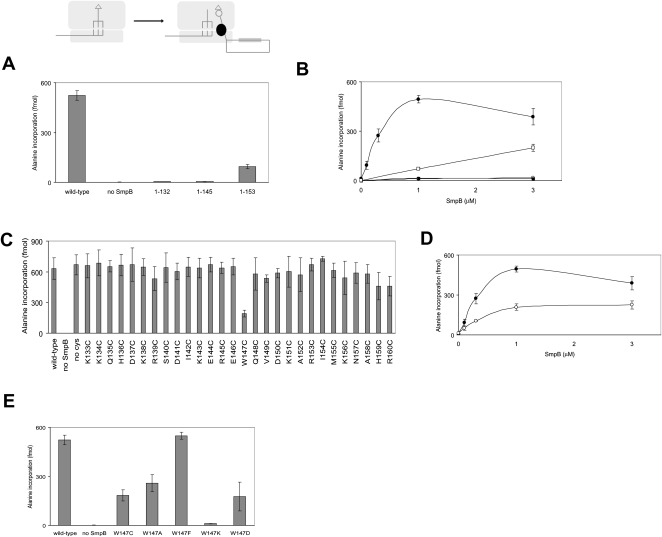
In the present study, we focused on the function of the C-terminal tail of SmpB encompassing residues 133–160. Using our in vitro system, the effects of truncation of the C-terminal tail of SmpB were examined. An entire truncation of the C-terminal tail (1–132) led to an almost complete loss of promotion of the first step as shown in a previous study (Konno et al. 2007). We also prepared mutants 1–145 and 1–153, in which the C-terminal tail was partially truncated. When 1–145 was used, almost no activity of the first step was detected (Fig. 1A). In contrast, 1–153 retained about 20% of the first step activity of wild-type SmpB (Fig. 1A). Sundermeier et al. (2005) have reported that this truncated mutant (1–153) is deficient in tagging activity in vivo. Our results suggest that the latter half of the C-terminal tail has a moderate effect on the activity of the first step.
FIGURE 1.
Effects of mutation of the C-terminal tail of SmpB on the first step of trans-translation. The effects of truncation (A,B) and substitution of SmpB mutants (C–E) on the first step of trans-translation were examined. (B) The level of incorporation of [3H]alanine into the polypeptide fraction after 10 min of incubation was monitored to evaluate the first step of trans-translation mediated by various concentrations of wild-type SmpB (•), 1–132 (○), 1–145 (■), and 1–153 (□). (D) The level of incorporation of [3H]alanine into the polypeptide fraction was monitored to evaluate the first step of trans-translation mediated by various concentrations of wild-type SmpB (•) and W147C mutant (○). The values obtained from experiments without SmpB were subtracted as backgrounds.
Next, we examined the activity of the first step with various concentrations of SmpB (Fig. 1B). When wild-type SmpB was used, the first step activity increased with increase of SmpB up to 1 μM, and an excess of SmpB apparently inhibited the activity. This inhibition might be the result of competition of free SmpB binding to the A-site and the complex of Ala-tmRNA-SmpB binding to the site of initial binding to the ribosome (see Discussion). On the other hand, as the concentration of 1–153 increased, the activity increased linearly.
Positively charged amino acid residues, lysine and arginine, are abundant in the C-terminal tail of SmpB, and many of them are highly conserved among bacterial species (Jacob et al. 2005). We selected several sets of two positively charged residues, K133/K134, K138/R139, K143/R145, and K151/R153, to substitute each of them with Ala/Ala (K133A/K134A, K138A/R139A, K143A/R145A, and K151A/R153A, respectively). Among these positively charged residues, K138 and R139 are of especially high conservation. Sundermeier et al. (2005) have shown that introduction of alanine at any of these sites leads to a defect in tagging activity in vivo. Here, we found that K138/R139 had a critical role in the first step, although the three other sets did not (Supplemental Fig. 1). They have also shown that a double point mutation of I154/M155 to D154/E155 (I154D/M155E) seriously affects the tagging activity in vivo (Sundermeier et al. 2005). We confirmed that the same mutation led to a failure of promotion of the first step in our in vitro system (Supplemental Fig. 1).
In a previous study, we have prepared a series of SmpB mutants, each having a single cysteine residue everywhere in the C-terminal tail for directed hydroxyl radical probing (Kurita et al. 2007). In this study, we used these SmpB mutants to examine the effect of point mutations in the C-terminal tail on the activity of the first step. In these mutants, two naturally occurring cysteine residues at 82 and 123 have been replaced by alanines. We confirmed that the SmpB mutant free of a cysteine residue, designated SmpB (no Cys), had activity of the first step comparable to that of wild-type SmpB (Fig. 1C).
Although most of the mutations in the C-terminal tail, including those at 138, 139, 154, and 155, had only moderate effects on the first step, a pronounced decrease in the first step activity was detected in the W147C mutant (Fig. 1C). In our previous study, this mutant had only a weak activity of poly(U)-dependent alanine incorporation directed by tmRNA in vitro using crude cell extracts of E. coli (Kurita et al. 2007). We confirmed that this mutant has activities of tmRNA binding (data not shown) and enhancement of aminoacylation efficiency comparable to those for wild-type SmpB (Supplemental Fig. 2). We also compared the first step activity of wild-type SmpB with that of W147C mutant at various concentrations (Fig. 1D), and it was found that the activity of W147C reached a plateau around 1 μM, like that of wild-type SmpB. However, apparent inhibition by a higher concentration of SmpB was not as significant as that in the case of wild-type SmpB (see Discussion).
To further study the functional importance of Trp147, it was replaced by other amino acids, alanine (W147A), phenylalanine (W147F), lysine (W147K), and aspartic acid (W147D). The activities of the first step in W147A, W147F, W147K, and W147D were about 50%, 105%, 2.9%, and 35%, respectively, of that in the wild-type SmpB (Fig. 1E). Interestingly, W147F mutant had activity of the first step comparable to that of wild-type SmpB.
Requirement of SmpB for GTP hydrolysis of EF-Tu
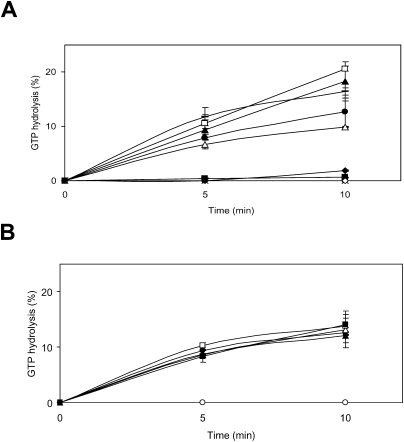
The first step defined here might include several events, initial binding of alanyl-tmRNA–SmpB–EF-Tu–GTP, GTP hydrolysis by EF-Tu to the stalled ribosome, accommodation of alanyl-tmRNA into the A-site, and peptidyl-transfer to alanyl-tmRNA. It has been believed that when a ternary complex (aminoacyl-tRNA–EF-Tu–GTP) binds to the ribosomal A-site for canonical translation, the codon–anticodon interaction triggers GTP hydrolysis, leading to a conformational change of EF-Tu and dissociation of EF-Tu–GDP (for review, see Rodnina and Wintermeyer 2001; Ogle and Ramakrishnan 2005). However, it is unclear what mechanistic event triggers GTP hydrolysis by EF-Tu in the absence of a codon–anticodon interaction when the alanyl-tmRNA–SmpB–EF-Tu–GTP complex binds to the ribosome for trans-translation. We therefore measured alanyl-tmRNA–SmpB-dependent GTP hydrolysis by EF-Tu on a model substrate as the stalled ribosome, in which the P-site is occupied by the AUG codon of a synthetic mRNA and tRNAfMet, but with a vacant A-site. As shown in Figure 2A, in the absence of either SmpB or alanyl-tmRNA, little activity of GTP hydrolysis by EF-Tu was detected, in agreement with a previous report (Shimizu and Ueda 2006). To our surprise, all of the SmpB mutants tested, including those that were deficient in the activity of the first step or even that of complete truncation of the C-terminal tail, had GTPase activities comparable to that of wild-type SmpB. These results suggest that the C-terminal tail of SmpB is not required for the events before GTP hydrolysis. We also prepared a synthetic peptide corresponding to the C-terminal tail of SmpB encompassing 133 to 160 (133–160). As expected, it did not promote GTP hydrolysis by EF-Tu (Fig. 2A).
FIGURE 2.
GTPase activity by EF-Tu in the stalled ribosome. (A) The effects of truncation and substitution of SmpB mutants on GTPase activity of EF-Tu were examined. The values obtained from experiments without SmpB (○) were subtracted as backgrounds. There was no detectable activity in the absence of EF-Tu (crosses) or alanyl-tmRNA (⋄). Little activity was detected when deacyl-tmRNA (◆) was added instead of alanyl-tmRNA. EF-Tu, [α-32P]GTP, alanyl-tmRNA, and wild-type (•), 1–132 (△), 1–145 (▲), 1-153 (□) or W147C (bar) of SmpB were added to the stalled ribosome. Little activity was detected when the synthetic peptide corresponding to the C-terminal tail of SmpB (133–160) was added (■). (B) The effects of the synthetic peptide (133–160) on alanyl-tmRNA–SmpB-dependent GTPase activity of EF-Tu were examined. EF-Tu, [α-32P]GTP, alanyl-tmRNA, wild-type SmpB (1 μM), and 0 (•), 0.1 (△), 1 (▲), 10 (□), or 100 (■) μM of synthetic peptide were incubated with the stalled ribosome.
Crucial role of tryptophan residue at 147 for binding to the A-site
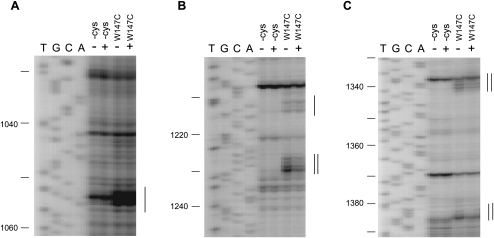
In a previous study, we have demonstrated the sites and modes of binding of SmpB on the ribosome by directed hydroxyl radical probing with Fe(II)-BABE. Fe(II)-BABE is a specific modifier of the cysteine residue of a protein, which generates hydroxyl radicals to cleave the RNA chain, allowing mapping of amino acid residues of a binding protein on an RNA-based macromolecule such as the ribosome. It revealed that there are two SmpB-binding sites on a ribosome, one in the A-site and the other in the P-site, presumably reflecting the pre- and post-translocation steps of trans-translation, respectively (Kurita et al. 2007). In this study, to examine the effect of mutation at residue 147 on the binding of SmpB to the A-site and P-site, we compared the patterns of cleavage from Fe(II) tethered to SmpB mutants having only one cysteine residue at 20, in which position 147 is occupied by tryptophan (A20C) or alanine (A20C/W147A). Note that W147A mutant had reduced activity of the first step as well as W147C mutant (Fig. 1E). A previous study has shown that A20C mutant has activity of tag-peptide synthesis comparable to that of wild-type SmpB and that Fe(II) tethered to residue 20 cleaves both the A-site and P-site of 16S rRNA (Kurita et al. 2007). As shown in Figure 3A–D, Fe(II) tethered to A20C/W147A mutant provided only weak cleavages in the A-site even at a high concentration compared with A20C mutant. In contrast, there was no drastic change in cleavage at the P-site at any concentration tested. These results confirm that tryptophan at 147 is critical for binding of SmpB to the A-site but not to the P-site. The binding site of A-site SmpB revealed by a previous directed hydroxyl radical probing study (Kurita et al. 2007) may reflect the accommodated state rather than the preaccommodated state in consideration of the absence of alanyl-tmRNA and EF-Tu in this complex.
FIGURE 3.
Role of Trp147 in binding to the A-site and P-site. (A–D) The sites of cleavage in 16S rRNA were detected by primer extension at various concentrations (1.2, 6, and 30 μM) of SmpB mutant (A20C or A20C/W147A). The cleavages are designated by bars. The lanes of DNA sequencing with the same primer are designated by T, G, C, and A. The bands due to A-site SmpB and P-site SmpB are designated by a single line and a double line, respectively.
Sundermeier et al. (2005) have shown that functional binding of SmpB to the ribosome can specifically be detected under the condition of high concentrations of NH4Cl both in vivo and in vitro. We performed Western blotting analysis at various concentrations of NH4Cl to compare the ribosome binding of W147C mutant with that of wild-type SmpB (Supplemental Fig. 3). To prepare a model substrate as the stalled ribosome in which the A-site is vacant but the P-site is occupied, a synthetic mRNA encompassing the SD sequence to the P-site AUG codon together with tRNAfMet corresponding to the AUG codon was added to the ribosomes prior to the addition of SmpB. Compared with wild-type SmpB, reduction in ribosome binding of W147C mutant was obvious under high-stringency conditions (300 or 600 mM NH4Cl).
Effect of synthetic peptide corresponding to the C-terminal tail of SmpB
Next, we examined the effects of the synthetic peptide (133–160) on trans-translation. This synthetic peptide as well as wild-type SmpB had little effect on (UUC)10-dependent polyphenylalanine synthesis (data not shown). In contrast, the first step was significantly inhibited by adding the synthetic peptide (Fig. 4A). The activity of the first step was reduced to 73% and 49% by the addition of the synthetic peptide at 10- and 100-fold molar excesses of wild-type SmpB, respectively. Notably, this inhibition was almost entirely abolished when tryptophan at 147 in the synthetic peptide was replaced with cysteine (133–160/W147C).
FIGURE 4.
Effects of the addition of synthetic peptide corresponding to the C-terminal tail of SmpB on the canonical translation and the first step of trans-translation. (A) The level of incorporation of [3H]alanine into the polypeptide fraction after 10 min of incubation was monitored to evaluate the first step of trans-translation in the presence of 1 μM wild-type SmpB and various concentrations of synthetic peptide (133–160; •) or mutational synthetic peptide (133–160/W147C; ○). (B–D) The sites of cleavage in 16S rRNA by Fe(II) tethered to 153 (B,C) and 159 (D) were detected by primer extension in the presence of various concentrations (0, 1, 10, and 100 μM) of synthetic peptide (133–160) or mutational synthetic peptide (133–160/W147C). The bands of cleavages are designated by bars. The lanes of DNA sequencing with the same primer are designated by T, G, C, and A. The bands due to A-site SmpB and P-site SmpB are designated by a single line and a double line, respectively.
We compared the patterns of cleavage from hydroxyl radicals to evaluate the effect of the synthetic peptide. Fe(II) tethered to the cysteine residue at 153 of A-site SmpB cleaved nucleotides around 422, 513, and 533–535 of the 16S rRNA, while that tethered to 159 of P-site SmpB cleaved nucleotides around 1381–1384 of 16S rRNA (Fig. 4B–D). When the synthetic peptide was added, cleavages from A-site SmpB disappeared (Fig. 4B,C), while a cleavage from P-site SmpB remained (Fig. 4D). In contrast, when the mutational synthetic peptide (133–160/W147C) was used, no inhibition of cleavages from A-site SmpB was detected, even at high concentrations (Fig. 4B,C), while a cleavage from P-site SmpB still remained (Fig. 4D). We further confirmed that Fe(II) tethered to residue 153 of the synthetic peptide conferred a typical hydroxyl radical probing pattern of A-site SmpB, but the mutational synthetic peptide did not (Supplemental Fig. 4). Further study using the single-molecule-fluorescence system indicated that the wild-type peptide bound to the ribosome with a dissociation constant (Kd) ∼125 nM (data not shown). In contrast, the mutant peptide (W147C) showed no detectable binding with Kd less than 1250 nM. These results suggest that the ribosome had a single binding site of the synthetic peptide in which the C-terminal tail of A-site SmpB binds, confirming the importance of Trp residue at 147 for binding.
We also measured GTP hydrolysis by EF-Tu on the stalled ribosome in the presence of the synthetic peptide (133–160). If the initial binding of alanyl-tmRNA/SmpB to the stalled ribosome requires interaction of the C-terminal tail with the mRNA path, the synthetic peptide should inhibit this event, consequently inhibiting subsequent GTP hydrolysis by EF-Tu. However, the synthetic peptide (133–160), even at the concentration of 100-fold molar excess of wild-type SmpB, had no inhibitory effect on GTPase activity (Fig. 2B), suggesting that the C-terminal tail of SmpB is located out of the mRNA path until GTP hydrolysis.
Location of the tryptophan residue at 147 in the ribosome
To identify the location of tryptophan residue at 147 in the ribosome, we performed directed hydroxyl radical probing toward W147C mutant. Fe(II) tethered to W147C provided cleavages of helix 28 (1382–1384), helix 29 (1339–1340), helix 30 (1229–1230), helix 32 (1212–1214), and helix 34 (1052–1054) of 16S rRNA (Fig. 5A–C). The cleavages of helices 28, 29, and 30 disappeared when a synthetic mRNA encompassing the SD sequence to the P-site AUG codon together with tRNAfMet was added to the ribosome prior to the complex formation with Fe(II) tethered to W147C to distinguish the cleavages by A-site SmpB from those by P-site SmpB. The cleavages by A-site SmpB were detectable only at high concentration (30 μM) of Fe(II)-tethered SmpB, while the cleavages by P-site SmpB were obvious even at a low concentration (6 μM) of Fe(II)-tethered SmpB (data not shown). These results suggest that W147C mutant has lower affinity to the A-site than wild-type SmpB, which is consistent with the results shown in Figure 4.
FIGURE 5.
Directed hydroxyl radical probing of 16S rRNA by Fe(II) tethered to 147 of SmpB. (A–C) The sites of cleavage in 16S rRNA were detected by primer extension. 70S ribosome was probed with Fe(II) tethered to 147 of SmpB in the presence (+) or absence (−) of the set of mRNA and tRNAfMet. The bands of cleavages are designated by bars. The lanes of DNA sequencing with the same primer are designated by T, G, C, and A. The bands due to A-site SmpB and P-site SmpB are designated by a single line and a double line, respectively.
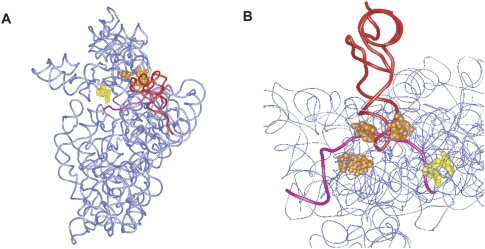
Based on a crystal structure of the 30S subunit from T. thermophilus in complex with tRNA and mRNA (Yusupova et al. 2006), we built a model of the complex of ribosome and SmpB, in which the cleavage sites of Trp147 of A-site SmpB are located on the mRNA path downstream of the decoding center, while those of P-site SmpB are located around the region of the codon–anticodon interaction of the P-site (Fig. 6). A site of cleavage by Trp147 of A-site SmpB is located in helix 34, very close to the decoding center.
FIGURE 6.
Mapping the cleavage sites in the tertiary structure of the 30S subunit. (A,B) Nucleotides of cleavages from the A-site and P-site SmpB W147C are colored yellow and orange, respectively. P-site tRNA and mRNA are colored red and pink, respectively, in a crystal structure of the 30S subunit from T. thermophilus (PDB ID 2HGR).
DISCUSSION
Our previous study has shown that the C-terminal tail of SmpB is located on the mRNA path, suggesting a new concept of “molecular mimicry of mRNA” or “a protein mimicking mRNA” during trans-translation. Results of earlier in vivo studies suggest that the C-terminal tail of SmpB has a critical role in trans-translation (Jacob et al. 2005; Sundermeier et al. 2005). However, it has remained unclear which residues are involved and which event they are involved in. The C-terminal tail has not been identified in any of the complexes of SmpB with the ribosome that have been visualized by cryo-EM studies (Valle et al. 2003; Gillet et al. 2006; Kaur et al. 2006). In this study, we examined the significance of the C-terminal tail of SmpB for trans-translation by using an in vitro system to evaluate the early stage of trans-translation.
Our results demonstrated that several kinds of truncations of the C-terminal tail significantly inhibited the activity of the first step. In addition, single-point mutations at Trp147 in the middle of the C-terminal tail resulted in drastic reduction in the first step in the present in vitro system. The following question then arose: Which process in the first step is defective in these mutants? Our results demonstrated that the level of GTPase activity of EF-Tu directed by these mutants is comparable to that of wild-type SmpB, suggesting that W147 is involved in a process after GTP hydrolysis by EF-Tu rather than the initial binding of alanyl-tmRNA–SmpB–EF-Tu–GTP to the ribosome. Our directed hydroxyl radical probing results revealed that the interaction of Trp147 with the mRNA path downstream of the decoding center is required for the binding of SmpB to the ribosomal A-site. We also found that the synthetic peptide corresponding to the C-terminal tail inhibits the binding of SmpB to the A-site and that this inhibition requires Trp147. Taken together, our results suggest that interaction of the C-terminal tail of SmpB with the mRNA path is required for accommodation of Ala-tmRNA–SmpB into the A-site.
Given the requirements of the C-terminal tail for A-site binding, we can rationalize the present results showing that the first step activity was inhibited by an excess of wild-type SmpB but not a truncated mutant (1–153) or W147C mutant (Fig. 1B,D). At a high concentration of SmpB, free SmpB would preferentially bind to the A-site, which would inhibit the initial binding of the quarternary complex (alanyl-tmRNA–SmpB–EF-Tu–GTP) to the ribosome. Inhibition by W147C mutant and that by a truncated mutant (1–153) were weak and undetectable, respectively, even at high concentrations of SmpB, which may be a result of the difference in affinity of SmpB to the A-site relative to that of the quarternary complex to the initial binding site.
Our previous study has shown that SmpB interacts with the upstream region of the resume codon of tmRNA, although the C-terminal tail of SmpB is not involved in this interaction (Konno et al. 2007). Thus, Trp147 may contribute to the interaction with the ribosome rather than tmRNA. Interestingly, W147F mutant had activity of the first step comparable to that of wild-type SmpB, suggesting that the aromatic side chain of Trp147 is required for the first step. It is possible that the aromatic ring stacks on a base of rRNA or an aromatic amino acid of a ribosomal protein. No aromatic amino acids in ribosomal proteins were found around the sites of cleavage in 16S rRNA by hydroxyl radicals from Fe(II) tethered to 147 of A-site SmpB. Thus, an aromatic ring of Trp147 may be recognized by a base of rRNA rather than a ribosomal protein or tmRNA. The aromatic side chain of tryptophan residue often stacks on the base of an RNA chain such as in a complex of a transcription antiterminator N-peptide with a stem–loop RNA hairpin (boxB) (Legault et al. 1998; Scharpf et al. 2000).
A cryo-EM study has shown that two molecules of SmpB (SmpB-1 and SmpB-2) bind to the ribosome in complex with Ala-tmRNA/SmpB/EF-Tu/GDP/kirromycin, putatively reflecting the preaccommodation step, whereas SmpB-1 is missing in a complex without kirromycin, presumably reflecting the step after accommodation of Ala-tmRNA into the A-site (Kaur et al. 2006). SmpB-2 is bound to the decoding center of the 30S subunit and SmpB-1 is bound to the GTPase-associated center (GAC) of the 50S subunit. SmpB-2 is in agreement with a previous directed hydroxyl radical probing, a chemical modification, and X-ray crystallographic studies (Gutmann et al. 2003; Kurita et al. 2007; Nonin-Lecomte et al. 2009). These have raised the possibility that two molecules of SmpB are involved in the initial binding of Ala-tmRNA to the ribosome for trans-translation and that SmpB-1 is released from the ribosome upon GTP hydrolysis by EF-Tu. The affinity of SmpB to the 50S subunit presumably reflecting the binding of SmpB-1 to the ribosome has also been shown by biochemical studies (Ivanova et al. 2005; Hallier et al. 2006), although another biochemical study has argued against the involvement of the second SmpB molecule (Sundermeier and Karzai 2007). Even if SmpB-1 is actually involved in the mechanism of trans-translation, however, neither of the C-terminal tails of the two SmpB molecules would be involved in its initial binding to the ribosome and in subsequent GTP hydrolysis by EF-Tu.
NMR studies have shown that the C-terminal tail of SmpB has a random-coil-like structure in solution (Dong et al. 2002; Someya et al. 2003), while it has been predicted as an α-helical structure from the periodical occurrence of positively charged residues (Jacob et al. 2005). An α-helical structure of the latter half of the C-terminal tail of A-site SmpB is also suggested by the results of the present and previous studies showing that the signals of directed hydroxyl radical probing appear at intervals of three residues (147, 150, and 153 for A-site SmpB). Although our results showed that K138/R139 is important for the activity of the first step, no signals of hydroxyl radical probing have been found from the former half of the C-terminal tail spanning 134–146 of A-site SmpB. In addition, cleavage sites from 133 and 147 of A-site SmpB are both in the vicinity of the decoding region (Kurita et al. 2007; this study), raising the possibility that this span is out of the decoding center.
On the basis of the present results, we propose a model of the early stage of trans-translation in light of the behavior of the C-terminal tail of SmpB (Fig. 7). First, a quaternary complex of alanyl-tmRNA–SmpB–EF-Tu–GTP forms an initial binding complex with the stalled ribosome. In this state (preaccommodated state), the aminoacyl end of tmRNA, like that of tRNA in the canonical translation, is still in contact with the binding pocket in EF-Tu, thus preventing it from entering the peptidyl transferase center. The main body of SmpB is located in the decoding center of the 30S subunit, which triggers GTP hydrolysis by EF-Tu. The present results suggest that the C-terminal tail of SmpB is not involved in this process. After GTP hydrolysis, EF-Tu-GDP is released from the ribosome, allowing alanyl-tmRNA–SmpB to be accommodated into the A-site and its aminoacyl end to reach the peptidyl-transferase center. Upon accommodation, the C-terminal tail of SmpB is set to the mRNA path, and this process requires Trp147, the conserved residues K138/R139 and the latter half of the C-terminal tail (154–160). It has been shown that alanyl-tmRNA–SmpB enters the stalled ribosome in the absence of EF-Tu (Shimizu and Ueda 2006), suggesting the binding of alanyl-tmRNA–SmpB to the stalled ribosome with affinity higher than that of aminoacyl-tRNA to the elongating ribosome.
FIGURE 7.
A model of the early stages of trans-translation. As described in the Discussion section, the C-terminal tail of SmpB is not located on the mRNA path in the processes before accommodation. Note the dynamic behavior of the C-terminal tail of SmpB in the course of trans-translation.
This study has revealed the molecular mechanism of the early stage of trans-translation. The molecular mechanism of later steps will be focused on in the future.
MATERIALS AND METHODS
Preparation of tmRNA
E. coli tmRNA was induced from an overproducing strain by the addition of 0.5 mM isopropyl-β-thiogalactopyranoside (IPTG) (Himeno et al. 1997) and was purified as described previously (Ushida et al. 1994). The nucleic acid fraction was extracted with phenol from the cells and then subjected to ethanol precipitation. The resulting fraction was subjected to differential isopropylalcohol precipitations to roughly remove DNA, followed by incubation with RNase-free DNase I (Pharmacia). tmRNA was purified by electrophoresis on a 5% polyacrylamide gel containing 7 M urea, heated for 3 min at 75°C, and slowly cooled for 1 h at room temperature in a refolding buffer containing 10 mM HEPES-KOH (pH 7.5), 5 mM magnesium chloride, and 20 mM ammonium chloride. Spectrophotometric measurements were made to determine the concentration of RNA.
Preparation of SmpB variants
Plasmids encoding truncation variants (1–132, 1–145, and 1–153) were constructed from pGEMEX-2 (Promega) carrying the gene for N-terminally His6-tagged SmpB (Hanawa-Suetsugu et al. 2002). Double-point mutants (K133A/K134A, K138A/R139A, K143A/R145A, K151A/R153A, and I154D/M155E) and single-point mutants at residue 147 (W147C, W147A, W147F, W147K, and W147D) were constructed from pGEMEX-2 (Promega) carrying the gene for C-terminally His6-tagged SmpB (Hanawa-Suetsugu et al. 2002). Plasmids encoding cysteine variants, having a single cysteine residue anywhere on the C-terminal tail (positions 133–160), were constructed from the plasmid in which two naturally occurring cysteines at positions 82 and 123 were replaced by alanines (Kurita et al. 2007). SmpB mutants were purified as described previously (Hanawa-Suetsugu et al. 2002).
In vitro translation and trans-translation
Poly (Phe) synthesis was performed by incubating 10 pmol ribosomes, 100 pmol (UUC)10 as an mRNA, 100 pmol EF-Tu, 3 pmol EF-G, and 10 pmol [14C]phenylalanyl-tRNAPhe in 40-μL of TMND buffer containing 80 mM Tris-HCl (pH 7.5), 150 mM ammonium chloride, 7 mM magnesium chloride, 2.5 mM dithiothreitol, 2 mM spermidine, 1 mM ATP, 0.2 mM GTP, and 5 mM phospho(enol)pyruvate at 37°C. A 40-μL aliquot of the mixture was withdrawn, diluted in 4 mL of 5% trichloroacetic acid, and incubated for 10 min at 90°C. The polypeptide fraction precipitated was recovered by filtration with a mixed cellulose membrane (Advantec). The filter was dried, and the radioactivity on the filter was counted by a liquid scintillation counter (Aloka).
The stalled ribosome was prepared from the reaction mixture for (UUC)10-dependent poly (Phe) synthesis as described above. The trans-translation reaction was initiated by adding a 10-μL aliquot of a mixture containing 5 pmol of [3H]alanyl-tmRNA, 50 pmol of SmpB and 0, 5, 50, 500, or 5000 pmol of synthetic peptide corresponding to the C-terminal tail (133–160) of SmpB in TMND buffer to 40 μL of stalled ribosomes. After incubation at 37°C for 10 min, 40 μL of the reaction mixture was withdrawn. The product was detected by the same procedure as that used for the detection of poly (Phe) described above.
GTPase assay
The reaction mixtures (50 μL) contained 80 mM Tris-HCl (pH 7.5), 150 mM ammonium chloride, 7 mM magnesium chloride, and 2.5 mM dithiothreitol. 70S ribosomes (20 pmol) were incubated with a synthetic mRNA having the sequence of 5′-AAGGAGGUAAAAAUG-3′ (100 pmol) and E. coli tRNAfMet (100 pmol) at 37°C for 10 min to form the stalled ribosomes. They were subsequently incubated with a mixture (10 μL) containing 5 pmol of alanyl-tmRNA, 100 pmol of EF-Tu, 0.7 pmol of [α-32P]GTP, and 50 pmol of SmpB at 37°C. At each time point, a 10-μL aliquot was withdrawn and 10 μL of 20 mM EDTA was added to stop the reaction. It was then spotted on a PEI cellulose sheet for thin-layer chromatography. After development using 0.75 M KH2PO4 as a solvent, the radioactivities of the spots of GTP and GDP were monitored using a Bio-Image-Analyzer FLA3000 (Fuji Film) (Himeno et al. 2004).
Directed hydroxyl radical probing
Preparation of Fe(II)-tethered SmpB was performed according to Kurita et al. (2007). 70S ribosomes (20 pmol) were incubated with or without a synthetic mRNA with the sequence of 5′-AAGGAGGUAAAAAUG-3′ (100 pmol) and E. coli tRNAfMet (100 pmol) at 37°C for 10 min. They were subsequently incubated with Fe(II)-tethered SmpB mutant (60, 300, or 900 pmol) in 30 μL of 50 mM MOPS (pH 8.0), 120 mM NaCl, 0.1 mM EDTA, 10 mM MgCl2, and 10% glycerol at 37°C for 10 min. Hydroxyl radical formation and primer extension were performed as previously described (Kurita et al. 2007). Inhibition by synthetic peptide was performed under the conditions described below. 70S ribosomes (20 pmol) were incubated with Fe(II)-tethered SmpB mutant (300 pmol) and synthetic peptide (133–160) (0, 50, 500, or 5000 pmol) at 37°C for 20 min in 50 μL of reaction mixture for hydroxyl radical probing described above.
Ribosome binding assay
70S ribosomes (20 pmol) were incubated with synthetic mRNA with the sequence of 5′-AAGGAGGUAAAAAUG-3′ (100 pmol) and E. coli tRNAfMet (100 pmol) at 37°C for 10 min in 30 μL of 10 mM Tris (pH 7.5), 10 mM MgCl2, 0, 30, 60, 150, 300, or 600 mM NH4Cl and 1 mM DTT. After incubation, SmpB (100 pmol) was added to the reaction mixture and incubated at 37°C for 10 min. Free SmpB was removed by ultrafiltration (Microcon YM-100) and washed twice with the buffer used upon binding. Complexes were electrophoresed on 15% SDS-polyacrylamide gel and subjected to Western blotting using a polyclonal antibody raised against SmpB.
tmRNA binding assay
tmRNA (20 pmol) was incubated with SmpB (2, 6, 20, 60, or 200 pmol) in 20 μL of 5 mM HEPES (pH 7.5), 10 mM MgCl2, 40 mM NH4Cl, 10 mM KCl, and 1 mM DTT at 37°C for 10 min. Free SmpB was removed by ultrafiltration (Microcon YM-100) and washed twice with the above buffer. Complexes were electrophoresed on a 15% SDS–polyacrylamide gel. The gel was stained with silver.
Aminoacylation assay
Aminoacylation was performed according to Hanawa-Suetsugu et al. (2002). A total of 1 μM tmRNA was incubated with 0, 0.3, 1, or 3 μM SmpB (wild-type or W147C mutant) in 50 μL of TMKD buffer containing 60 mM Tris (pH 7.5), 10 mM MgCl2, 30 mM KCl, 10 mM DTT, 2.5 mM ATP, 14 μM L-[U-14C]alanine (6.07GBq/mmol), and 8.5 × 10−2 U of alanyl-tRNA synthetase at 37°C. At each time point, a 12-μL aliquot was withdrawn and spotted on Whatman 3MM filter paper, and radioactivity in the trichloroacetic acid-insoluble fraction was measured by a liquid scintillation counter.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank the staff of the Gene Research Center of Hirosaki University for the use of the facility. We also thank Natsumi Sugai for technical assistance. This work was supported by a Research Fellowship of Hirosaki University (D.K.), a Grant-in-Aid for Young Scientists Start-up from the Japan Society for the Promotion of Science (D.K., no. 21880009), a Grant-in-Aids for Scientific Research from the Japan Society for the Promotion of Science (A.M., no. 20580068; H.H., no. 20380057), a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology (H.H., no. 20059001), and a Grant for Priority Research Designated by the President of Hirosaki University (H.H.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1916610.
REFERENCES
- Abe T, Sakaki K, Fujihara A, Ujiie H, Ushida C, Himeno H, Sato T, Muto A 2008. tmRNA-dependent trans-translation is required for sporulation in Bacillus subtilis. Mol Microbiol 69: 1491–1498 [DOI] [PubMed] [Google Scholar]
- Abo T, Inada T, Ogawa K, Aiba H 2000. SsrA-mediated tagging and proteolysis of LacI and its role in the regulation of lac operon. EMBO J 19: 3762–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Kurita D, Takada K, Konno T, Muto A, Himeno H 2005. Competition between trans-translation and termination or elongation of translation. Nucleic Acids Res 33: 5544–5552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barends S, Wower J, Kraal B 2000. Kinetic parameters for tmRNA binding to alanyl-tRNA synthetase and elongation factor Tu from Escherichia coli. Biochemistry 39: 2652–2658 [DOI] [PubMed] [Google Scholar]
- Barends S, Karzai AW, Sauer RT, Wower J, Kraal B 2001. Simultaneous and functional binding of SmpB and EF-Tu-TP to the alanyl acceptor arm of tmRNA. J Mol Biol 314: 9–21 [DOI] [PubMed] [Google Scholar]
- Bessho Y, Shibata R, Sekine S, Murayama K, Higashijima K, Hori-Takemoto C, Shirouzu M, Kuramitsu S, Yokoyama S 2007. Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA. Proc Natl Acad Sci 104: 8293–8298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong G, Nowakowski J, Hoffman DW 2002. Structure of small protein B: The protein component of the tmRNA-SmpB system for ribosome rescue. EMBO J 21: 1845–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felden B, Hanawa K, Atkins JF, Himeno H, Muto A, Gesteland RF, McCloskey JA, Crain PF 1998. Presence and location of modified nucleotides in Escherichia coli tmRNA: Structural mimicry with tRNA acceptor branches. EMBO J 17: 3188–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujihara A, Tomatsu H, Inagaki S, Tadaki T, Ushida C, Himeno H, Muto A 2002. Detection of tmRNA-mediated trans-translation products in Bacillus subtilis. Genes Cells 7: 343–350 [DOI] [PubMed] [Google Scholar]
- Gillet R, Kaur S, Li W, Hallier M, Felden B, Frank J 2006. Scaffolding as an organizing principle in trans-translation. The roles of small protein B and ribosomal protein S1. J Biol Chem 282: 6356–6363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Roche E, Zhou Y, Sauer RT 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes & Dev 12: 1338–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann S, Haebel PW, Metzinger L, Sutter M, Felden B, Ban N 2003. Crystal structure of the transfer-RNA domain of transfer-messenger RNA in complex with SmpB. Nature 424: 699–703 [DOI] [PubMed] [Google Scholar]
- Hallier M, Desreac J, Felden B 2006. Small protein B interacts with the large and the small subunits of a stalled ribosome during trans-translation. Nucleic Acids Res 34: 1935–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawa-Suetsugu K, Bordeau V, Himeno H, Muto A, Felden B 2001. Importance of the conserved nucleotides around the tRNA-like structure of Escherichia coli transfer-messenger RNA for protein tagging. Nucleic Acids Res 29: 4663–4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawa-Suetsugu K, Takagi M, Inokuchi H, Himeno H, Muto A 2002. SmpB functions in various steps of trans-translation. Nucleic Acids Res 30: 1620–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno H, Sato M, Tadaki T, Fukushima M, Ushida C, Muto A 1997. In vitro trans translation mediated by alanine-charged 10Sa RNA. J Mol Biol 268: 803–808 [DOI] [PubMed] [Google Scholar]
- Himeno H, Hanawa-Suetsugu K, Kimura T, Takagi K, Sugiyama W, Shirata S, Mikami T, Odagiri F, Osanai Y, Watanabe D, et al. 2004. A novel GTPase activated by the small subunit of ribosome. Nucleic Acids Res 32: 5303–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno H, Kurita D, Muto A 2009. Trans-translation by tmRNA and a protein mimicking tRNA and mRNA. In Protein biosynthesis (eds. Esterhouse TE and Petrinos LB), pp. 69–107 Nova Science Publishers Inc, New York [Google Scholar]
- Hong SJ, Tran QA, Keiler KC 2005. Cell cycle-regulated degradation of tmRNA is controlled by RNase R and SmpB. Mol Microbiol 57: 565–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wolfgang MC, Withey J, Koomey M, Friedman DI 2000. Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability. EMBO J 19: 1098–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Pavlov MY, Bouakaz E, Ehrenberg M, Schiavone LH 2005. Mapping the interaction of SmpB with ribosomes by footprinting of ribosomal RNA. Nucleic Acids Res 33: 3529–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y, Sharkady SM, Bhardwaj K, Sanda A, Williams KP 2005. Function of the SmpB tail in transfer-messenger RNA translation revealed by a nucleus-encoded form. J Biol Chem 280: 5503–5509 [DOI] [PubMed] [Google Scholar]
- Karzai AW, Susskind MM, Sauer RT 1999. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J 18: 3793–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Gillet R, Li W, Gursky R, Frank J 2006. Cryo-EM visualization of transfer messenger RNA with two SmpBs in a stalled ribosome. Proc Natl Acad Sci 103: 16484–16489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC 2007. Physiology of tmRNA: What gets tagged and why? Curr Opin Microbiol 10: 169–175 [DOI] [PubMed] [Google Scholar]
- Keiler KC, Waller PR, Sauer RT 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271: 990–993 [DOI] [PubMed] [Google Scholar]
- Komine Y, Kitabatake M, Yokogawa T, Nishikawa K, Inokuchi H 1994. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc Natl Acad Sci 91: 9223–9227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno T, Kurita D, Takada K, Muto A, Himeno H 2007. A functional interaction of SmpB with tmRNA for determination of the resuming point of trans-translation. RNA 13: 1723–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita D, Sasaki R, Muto A, Himeno H 2007. Interaction of SmpB with ribosome from directed hydroxyl radical probing. Nucleic Acids Res 35: 7248–7255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legault P, Li J, Mogridge J, Kay LE, Greenblatt J 1998. NMR structure of the bacteriophage lambda N peptide/boxB RNA complex: Recognition of a GNRA fold by an arginine-rich motif. Cell 93: 289–299 [DOI] [PubMed] [Google Scholar]
- Moore SD, Sauer RT 2007. The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem 76: 101–124 [DOI] [PubMed] [Google Scholar]
- Muto A, Ushida C, Himeno H 1998. A bacterial RNA that functions as both a tRNA and an mRNA. Trends Biochem Sci 23: 25–29 [DOI] [PubMed] [Google Scholar]
- Muto A, Fujihara A, Ito K, Matsuno J, Ushida C, Himeno H 2000. Requirement of transfer-messenger RNA for the growth of Bacillus subtilis under stresses. Genes Cells 5: 627–635 [DOI] [PubMed] [Google Scholar]
- Nameki N, Felden B, Atkins JF, Gesteland RF, Himeno H, Muto A 1999. Functional and structural analysis of a pseudoknot upstream of the tag-encoded sequence in E. coli tmRNA. J Mol Biol 286: 733–744 [DOI] [PubMed] [Google Scholar]
- Nameki N, Someya T, Okano S, Suemasa R, Kimoto M, Hanawa-Suetsugu K, Terada T, Shirouzu M, Hirao I, Takaku H, et al. 2005. Interaction analysis between tmRNA and SmpB from Thermus thermophilus. J Biochem 138: 729–739 [DOI] [PubMed] [Google Scholar]
- Nonin-Lecomte S, Germain-Amiot N, Gillet R, Hallier M, Ponchon L, Dardel F, Felden B 2009. Ribosome hijacking: A role for small protein B during trans-translation. EMBO Rep 10: 160–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V 2005. Structural insights into translational fidelity. Annu Rev Biochem 74: 129–177 [DOI] [PubMed] [Google Scholar]
- Qi H, Shimizu Y, Ueda T 2007. Ribosomal protein S1 is not essential for the trans-translation machinery. J Mol Biol 368: 845–852 [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Wintermeyer W 2001. Fidelity of aminoacyl-tRNA selection on the ribosome: Kinetic and structural mechanisms. Annu Rev Biochem 70: 415–435 [DOI] [PubMed] [Google Scholar]
- Rudinger-Thirion J, Giegé R, Felden B 1999. Aminoacylated tmRNA from Escherichia coli interacts with prokaryotic elongation factor Tu. RNA 5: 989–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saguy M, Gillet R, Skorski P, Hermann-Le Denmat S, Felden B 2007. Ribosomal protein S1 influences trans-translation in vitro and in vivo. Nucleic Acids Res 35: 2368–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharpf M, Sticht H, Schweimer K, Boehm M, Hoffmann S, Rosch P 2000. Antitermination in bacteriophage λ. The structure of the N36 peptide-boxB RNA complex. Eur J Biochem 267: 2397–2408 [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Ueda T 2002. The role of SmpB protein in trans-translation. FEBS Lett 514: 74–77 [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Ueda T 2006. SmpB triggers GTP hydrolysis of elongation factor Tu on ribosomes by compensating for the lack of codon-anticodon interaction during trans-translation initiation. J Biol Chem 281: 15987–15996 [DOI] [PubMed] [Google Scholar]
- Someya T, Nameki N, Hosoi H, Suzuki S, Hatanaka H, Fujii M, Terada T, Shirouzu M, Inoue Y, Shibata T, et al. 2003. Solution structure of a tmRNA-binding protein, SmpB, from Thermus thermophilus. FEBS Lett 535: 94–100 [DOI] [PubMed] [Google Scholar]
- Stepanov VG, Nyborg J 2003. tmRNA from Thermus thermophilus. Interaction with alanyl-tRNA synthetase and elongation factor Tu. Eur J Biochem 270: 463–475 [DOI] [PubMed] [Google Scholar]
- Sundermeier TR, Karzai AW 2007. Functional SmpB-ribosome interactions require tmRNA. J Biol Chem 282: 34779–34786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundermeier TR, Dulebohn DP, Cho HJ, Karzai AW 2005. A previously uncharacterized role for small protein B (SmpB) in transfer messenger RNA-mediated trans-translation. Proc Natl Acad Sci 102: 2316–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada K, Takemoto C, Kawazoe M, Konno T, Hanawa-Suetsugu K, Lee S, Shirouzu M, Yokoyama S, Muto A, Himeno H 2007. In vitro trans-translation of Thermus thermophilus: Ribosomal protein S1 is not required for the early stage of trans-translation. RNA 13: 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushida C, Himeno H, Watanabe T, Muto A 1994. tRNA-like structures in 10Sa RNAs of Mycoplasma capricolum and Bacillus subtilis. Nucleic Acids Res 22: 3392–3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Gillet R, Kaur S, Henne A, Ramakrishnan V, Frank J 2003. Visualizing tmRNA entry into a stalled ribosome. Science 300: 127–130 [DOI] [PubMed] [Google Scholar]
- Wower IK, Zwieb CW, Guven SA, Wower J 2000. Binding and cross-linking of tmRNA to ribosomal protein S1, on and off the Escherichia coli ribosome. EMBO J 19: 6612–6621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wower J, Zwieb CW, Hoffman DW, Wower IK 2002. SmpB: A protein that binds to double-stranded segments in tmRNA and tRNA. Biochemistry 41: 8826–8836 [DOI] [PubMed] [Google Scholar]
- Yusupova G, Jenner L, Rees B, Moras D, Yusupov M 2006. Structural basis for messenger RNA movement on the ribosome. Nature 444: 391–394 [DOI] [PubMed] [Google Scholar]