Abstract
RNA abundance and DNA copy number are routinely measured in high-throughput using microarray and next-generation sequencing (NGS) technologies, and the attributes of different platforms have been extensively analyzed. Recently, the application of both microarrays and NGS has expanded to include microRNAs (miRNAs), but the relative performance of these methods has not been rigorously characterized. We analyzed three biological samples across six miRNA microarray platforms and compared their hybridization performance. We examined the utility of these platforms, as well as NGS, for the detection of differentially expressed miRNAs. We then validated the results for 89 miRNAs by real-time RT-PCR and challenged the use of this assay as a “gold standard.” Finally, we implemented a novel method to evaluate false-positive and false-negative rates for all methods in the absence of a reference method.
Keywords: miRNA, microRNA, differential expression, microarray, real-time PCR, sequencing, pyrosequencing, miRNA-seq
INTRODUCTION
MicroRNAs (miRNAs) are regulatory noncoding RNA molecules ∼20–23 nucleotides (nt) long, generated by two cleavage events mainly from RNA Pol II primary transcripts (pri-miRNAs) via a ∼70-nt imperfect stem–loop intermediate (pre-miRNA). Over 10,000 miRNAs from 115 species, ranging from vertebrates (Lagos-Quintana et al. 2001) to viruses (Pfeffer et al. 2004), are currently deposited in the miRNA registry (miRBase version 14) (Griffiths-Jones et al. 2008). These include ∼700 (out of up to ∼3400 predicted) (Sheng et al. 2007) human miRNAs.
miRNAs mediate the translational repression, and sometimes degradation, of target mRNAs mostly by directing an RNA-induced silencing protein complex to imperfect complementary sequences in their 3′UTRs (van den Berg et al. 2008). Up to ∼60% of human genes are putative targets of one or more miRNA (Friedman et al. 2009). miRNAs play a role in all major biomolecular processes, including metabolism (Krutzfeldt and Stoffel 2006), cell proliferation (Bueno et al. 2008) and apoptosis (Jovanovic and Hengartner 2006), development and morphogenesis (Stefani and Slack 2008; He et al. 2009), stem cell maintenance, and tissue differentiation (Shi et al. 2006). miRNAs are reported to be involved in 94 human diseases (Jiang et al. 2009), ranging from psychiatric disorders (Barbato et al. 2008) through diabetes (Hennessy and O'Driscoll 2008) to cancer (Medina and Slack 2008).
Three principal methods are used to measure the expression levels of miRNAs: real-time reverse transcription-PCR (qPCR) (Chen et al. 2005; Shi and Chiang 2005), microarray hybridization (Yin et al. 2008; Li and Ruan 2009), and massively parallel/next-generation sequencing (NGS) (Hafner et al. 2008), all of which face unique challenges compared to their use in mRNA profiling. In terms of microarray analysis, the short length of mature miRNA sequences constrains probe design, such that often the entire miRNA sequence must be used as a probe. Consequently, the melting temperatures of miRNA probes may vary by >20°C. qPCR assays, traditionally relying on the specificity provided by a number of contiguous probes, compensate for the compromised sequence specificity by a stringent spatial constraint (3′ terminal or near-terminal sequences). A similar constraint is also imposed by stem–loop microarray probes (Agilent). NGS of miRNAs can be influenced by sequencing errors and often requires search and removal of adaptor sequences before the miRNA sequence itself can be elucidated.
A second challenge in measuring miRNA levels arises from the existence of miRNA families, the largest encompassing nine variants (hsa-let-7a–i), whose members differ by as little as one nucleotide but nevertheless exhibit differential expression patterns (Roush and Slack 2008). The stringency required to differentiate between these closely related miRNA species surpasses that of conventional mRNA microarrays. This challenge is partly addressed by ensuring that hybridization-based assays are performed at high enough temperatures to reject cross-hybridizing transcripts. In addition, microarrays with probes containing locked nucleic acid (LNA) bases (Exiqon) provide higher annealing affinities, potentially allowing the assay to discern between individual miRNA family members and somewhat equalizing the melting temperatures of probe sequences. Finally, advances in sequencing technology have accelerated both the discovery rate of new miRNAs and modifications to existing miRNA entries, reflecting subtle variations in mature miRNA sequences (e.g., post-transcriptional editing or terminal residue addition) (Landgraf et al. 2007). As a result, the continued refinement of miRNA databases necessitates frequent changes to miRNA array probe design and annotation.
The technical merits and drawbacks of qPCR, microarrays, and sequencing of miRNAs are similar to their application for RNA or genomic DNA quantitation. The clear advantage of high-throughput sequencing is the ability to identify novel miRNAs. This technology is not hindered by variability in melting temperatures, coexpression of nearly identical miRNA family members, or post-transcriptional modifications. However, both the RNA ligation (Bissels et al. 2009) and the PCR amplification (see below) steps bear inherent biases, the method is laborious and costly, and associated tools for computational analysis are in their infancy. qPCR is often considered a “gold standard” in the detection and quantitation of gene expression. However, the rapid increase in number of miRNAs renders qPCR inefficient on a genomic scale, and it is probably better used as a validation rather than a discovery tool.
As with genomic DNA and RNA analysis, microarrays are still the best choice for a standardized genome-wide assay that is amenable to high-throughput applications. Over 400 existing publications have utilized commercial or in-house printed miRNA microarrays. The differences between available platforms range from surface chemistry and printing technology, through probe design and labeling techniques, to cost. Unlike for mRNA gene expression (Shi et al. 2006), comparative genomic hybridization (Baumbusch et al. 2008), or chromatin immunoprecipitation (Johnson et al. 2008) assays, few attempts have been made to establish rigorous parameters for the evaluation of a miRNA microarray platform, especially in light of the specific challenges miRNAs present.
We have undertaken a systematic comparison of six commercially available miRNA microarray platforms representing single- and dual-channel fluorescence technologies, using three well-defined RNA samples (Git et al. 2008), and compared the results with NGS and qPCR. This study represents, to the best of our knowledge, the most comprehensive comparison of the performance of methods to detect differentially expressed miRNAs to date.
RESULTS
Microarray comparison study design
As a preface to this study, we extensively evaluated RNA extraction and quality control (QC) methods to ensure a high standard of quality for the RNA samples used (data not shown). The biological samples were representative of a realistic application of miRNA microarrays in a cancer research institute. Moreover, biological replicates of these samples have been previously profiled by contact-printed and bead-based microarrays (Git et al. 2008), providing a comparative reference for QC during preliminary stages as well as in final analyses (e.g., tumor suppressor [TS] miRNAs) (see Fig. 3B, below).
FIGURE 3.
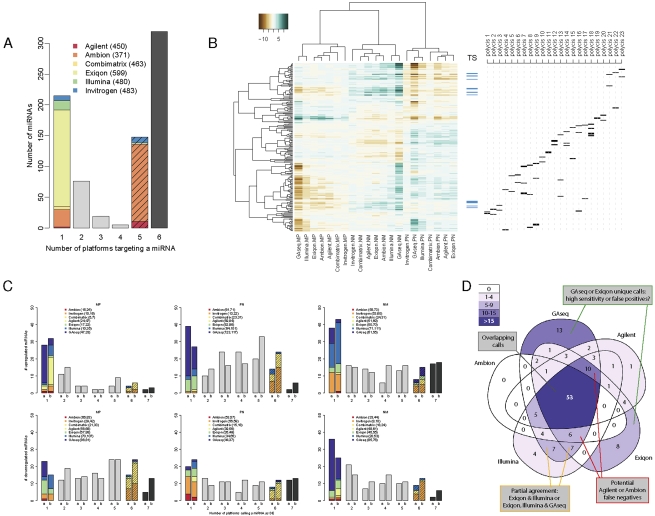
Analysis of differential expression. (A) miRNA targeting by platforms. The number of reannotated miRNAs targeted by varying numbers of platforms was calculated. Solid colors indicate miRNAs found only on the indicated platform; striped colors, miRNAs found on all platforms except the indicated platform. The total number of human miRNAs on each platform is indicated in parenthesis. Black bar indicates 319 miRNAs represented on all microarrays. (B) Clustering of the common probe M-values. M-values of 204 human probes common to all microarray platforms with no predicted cross-hybridization and detectable by GAseq were subjected to unsupervised clustering using Pearson correlation. Ticks indicate the position of potential tumor suppressor (TS) miRNAs (blue) and miRNAs arising from a single genomic location contained in a putative polycistronic pri-miRNA (black). A list of polycistrons is provided in Supplemental file “Polycistrons.” (C) Consistency of DE calls by all platforms. The number of platforms calling each miRNA as DE (up-regulated, top; down-regulated, bottom) in each of the three biological comparisons was recorded. DE calls were derived (1) using a uniform threshold of log2 fold-change>1 or (2) using optimal thresholds calculated for each platform by the iMLE algorithm. The overall number of relevant DE calls made by each platform is indicated in parenthesis. (D) Overlap in DE calls of five platforms. The number of miRNAs called by five platforms as up-regulated in P versus N sample using iMLE-optimized cutoffs was plotted inside a Venn diagram. Areas are shaded according to number of DE calls and their relative sizes bear no meaning.
Three samples were analyzed in this study: a pool of commercial RNAs from normal breast tissue (N), the luminal breast cancer cell line MCF7 (M), and a breast progenitor cancer cell line PMC42 (P), which exhibits many normal-like characteristics. All three samples, extracted in bulk and quality assured, were labeled and hybridized in quadruplicate to six commercially available microarray platforms in strict accordance with the protocols recommended by the manufacturers (see Materials and Methods). The microarray platforms used in this study were the Agilent Human miRNA Microarray 1.0; Exiqon miRCURY LNA microRNA Array, v9.2; Illumina Sentrix Array Matrix 96-well MicroRNA Expression Profiling Assay v1; Ambion mirVana miRNA Bioarrays v2; Combimatrix microRNA 4X2K Microarrays; and Invitrogen NCode Multi-Species miRNA Microarray v2. For simplicity, each platform is referenced throughout after its manufacturer name.
Microarray hybridization performance
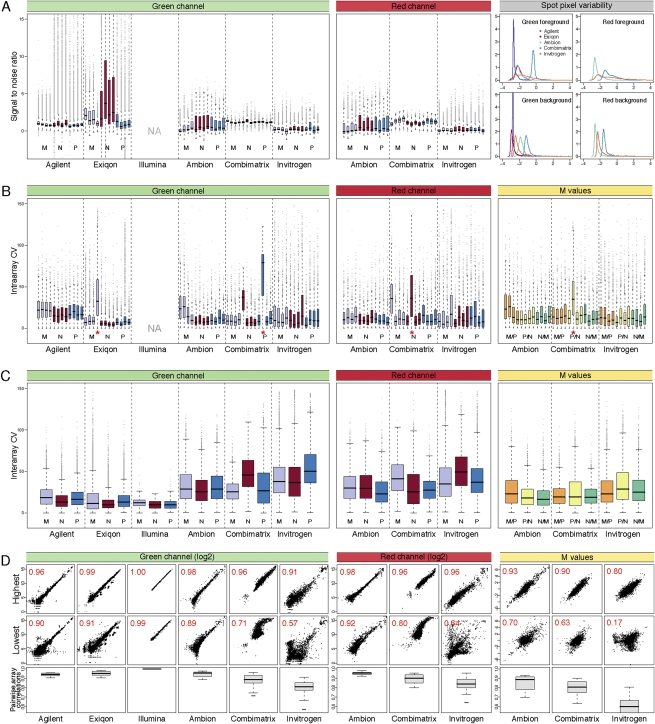
Figure 1 depicts the distributions of several measures of hybridization quality and consistency, such as the signal-to-noise ratio (SNR) (Fig. 1A), the coefficient of variation (CV) between replicate spots and arrays (Fig. 1B,C), and pairwise correlation between arrays (Fig. 1D). Overall, the SNR generated by the Normal samples was the highest, and MCF7 was the lowest for each platform evaluated. This agrees with the observation that overall miRNA content is reduced in cell lines compared to tissue (Lee et al. 2008; C Blenkiron and LD Goldstein, pers. comm.). PMC42, a normal-like cell line, demonstrated intermediate levels of overall miRNA expression. The difference was least pronounced on platforms with a high within-spot pixel variability (Combimatrix, Invitrogen) (Fig. 1A, right panel), since the SNR not only depends on fluorescent signal intensities but is inversely proportional to the standard deviation of both foreground and background pixels, so that high spot uniformity contributes to higher SNR. Some typical spot artifacts leading to low uniformity were indeed observed during feature extraction (“doughnut-shaped” spots for Invitrogen indicate high signal on the outside of spots, low on the inside, and the opposite pattern for Combimatrix) (data not shown).
FIGURE 1.
Analysis of hybridization performance. (A) Signal-to-noise ratio for the raw 532 nm/Cy3 (green banner) and 635 nm/Cy5 (red banner) intensities for all spots on the individual arrays was calculated using the SSDR method. For Illumina arrays, this calculation was impossible as only the foreground intensities were available. Purple indicates arrays with M samples; red, N, and blue, P. For clarity of presentation, the y-axis was truncated at 15, thereby excluding some extreme outliers. The distribution of the log2 standard deviation between pixels within each spot scaled to the median spot intensity is shown on the right (gray banner). (B) Intra-array coefficients of variation across replicated spots on each array were calculated for the unprocessed Cy3 and Cy5 intensities (bar and banner colors as above), and the log2 ratios (M-values, yellow banner; orange bars indicates M/P; yellow; P/N, green, N/M). Arrays with a red asterisk were excluded from subsequent analysis. (C) Interarray coefficients of variation were calculated for arrays hybridized with the same samples (bar and banner colors as above). (D) Pairwise correlations for arrays hybridized with the same samples were calculated (15–18 correlations). Distribution of R2 values are shown in box plots (bottom row), with the highest (top row) and lowest (middle row) correlations shown as examples. The axis for the bottom row was truncated at 0.55 for clarity, excluding some of the values for Invitrogen.
We then examined the variability between replicates spotted on the same array (intra-array CV) (Fig. 1B). For dual-channel arrays, the 532-nm (Cy3, green) and 635-nm (Cy5, red) fluorescence intensities and their log2 ratios (M-values) were treated separately, since localized signal variations occurring in both channels may cancel each other out. We observed no consistent differences between single- and dual-color platforms, and although CV's varied considerably between some platforms, they tended to be consistent within platforms, with the exception of two arrays subsequently excluded from downstream analysis.
The interarray CV's (Fig. 1C) were calculated for each type of probe (several different probe types might target the same miRNA) across all replicate spots on the four replicate arrays. These typically include 12–24 values, although some probes (e.g., controls or empty spots) were present in greater numbers; for example, Agilent arrays contain more than 3000 empty spots. Single-channel hybridization was more consistent across replicates, as evident from the overall lower CV values, but these differences were ameliorated when the M-values, rather than the individual Cy3 and Cy5 intensities, were considered for the dual-color platforms. Reproducibility between hybridizations was also assessed by pairwise comparison of replicate arrays. The distribution of the resulting R2 values as well as the most and least consistent examples from each platform, are illustrated in Figure 1D. Although all platforms demonstrated at least one replicate pair with greater than 0.9 (and usually >0.95) R2 correlation, their distribution was much wider. Notably, unlike the interarray CV values, the dual-channel replicates with low correlation (below an R2 of 0.8) showed poorer agreement when treated as M-values instead of Cy3 and Cy5 intensities. This may be due to the inaccuracy of M-values for low-intensity spots. In particular, negative control or empty replicate spots were considered individually for the pairwise comparison, thus strongly affecting the distribution of M-value correlations, but were condensed into single values across all interarray replicates for the interarray CVs (Fig. 1C).
We then proceeded to analyze the consistency of the detected spots for each platform. First, the frequency of “present/marginally present” or “absent” calls was calculated for each spot on the arrays based on the intensity of negative controls and empty spots (see Materials and Methods) (Fig. 2A). The platforms varied significantly in the consistency of associated present/absent calls, visually represented by the thickness of the “belt” region separating the red (consistently “present”) and blue (consistently “absent”) zones of the bars: whereas the “belt” values comprised fewer than 20% of the probes in Agilent, they accounted for over half the probes in Invitrogen arrays. This variation stems from interarray variability and the availability of spots to evaluate the background distribution. For example, despite the very similar interarray CV of the M-values in Ambion and Combimatrix assays (Fig. 1C), the consistency of calls on the Ambion array platform was higher.
FIGURE 2.
Analysis of detected probes. (A) Consistency of present/absent calls among human miRNAs. (Top) For each human probe, the percentage of replicates detected (called present) by the platform was calculated and summarized (bars). The numbers above the bars indicate number of probe replicates. (Bottom) Intensity distribution of human miRNAs (black) and the empty and negative spots used to calculate the nonspecific binding (red), with the number of probes of each type listed below the plot. Illumina array data are missing from panels A and B, as information regarding negative or empty spots was not available. (B) Detected spot types. Probes have been categorized based on their target miRNAs (see Materials and Methods). The number of unique spots from each category being detected as “present” in >90% of its replicates across all arrays was calculated for each of the three samples types. For categories with 10 or more present probes, the count is shown next to the figure, with the proportion of the “present” calls out of the total probes in that category (%). The radius of each chart is proportional to the total number of present spots, indicated above. The legend is shared with panel C. PosControl and NegControl are positive and negative controls, respectively; MM_human, mismatched human. (C) Intensity range of the different spot types. For each of the spot types of panel B, the distribution of intensities of background-corrected and normalized green or red log2 values across all arrays was calculated.
Microarray probe mapping and hybridization specificity
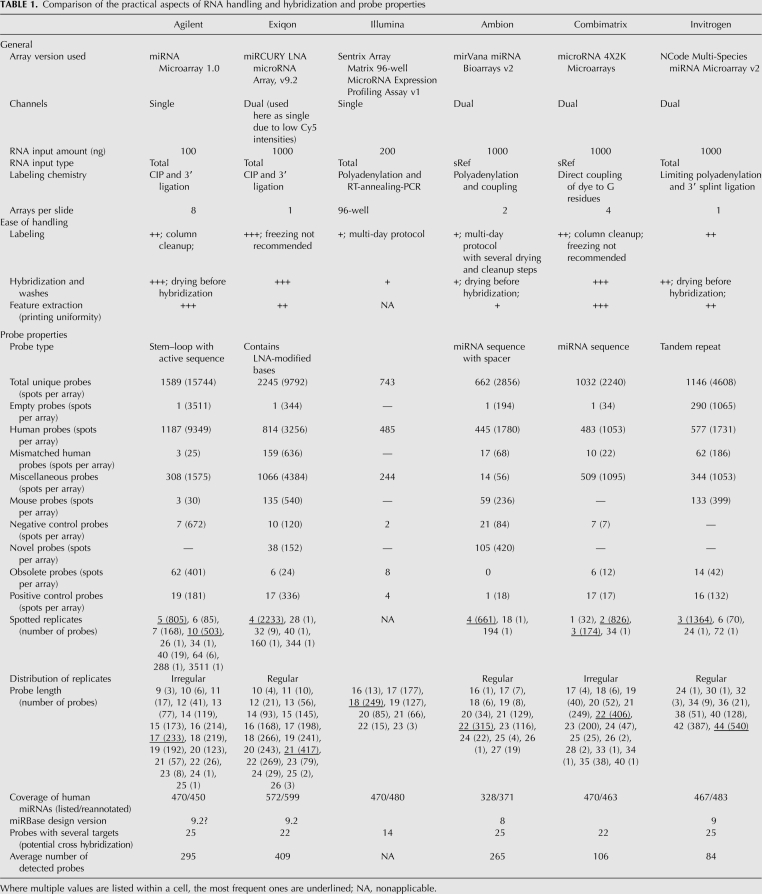
Due to the inherent difficulties associated with miRNA probe design outlined in the introduction, the complements of miRNAs targeted by each platform are difficult to compare. To allow an accurate comparison between the platforms, we reannotated all the probes against miRBase version 12 using uniform criteria (see Materials and Methods). Although the total number of probes varied significantly across platforms, the number of human miRNAs represented on the array was fairly constant and depended mainly on the miRBase version at the time of array design. The overall characteristics of probes represented in each platform and the effect of reannotation are summarized in the “probe properties” section of Table 1.
TABLE 1.
Comparison of the practical aspects of RNA handling and hybridization and probe properties
Reannotated probes were divided into categories based on information from the manufacturers and our remapping of probe sequences (see Materials and Methods). The categories are listed and color coded in the legend to Figure 2, B and C, according to our approximate expectation regarding their intensity (high/red to low/blue). Of particular interest were human miRNAs and potential cross-hybridizing probes (mouse miRNAs and probes with mismatches to human miRNAs; MM_human). We counted the number of probes called as “present” in each category (Fig. 2B) and examined the distribution of their normalized signal intensities (Fig. 2C). Platforms varied both in overall signal intensity and number of probes called “present.” The former property is affected by a combination of labeling chemistry, input RNA concentration, and hybridization efficiency, with Combimatrix arrays producing the brightest signal. However, the low numbers of “present” calls (127, 85, 105) on this platform are similar to those produced by the low-intensity Invitrogen arrays (49, 103, 100), underscoring the importance of distinguishing between the two metrics.
Within most platforms the signal and “present” rate also varied extensively depending on spot category. As expected, positive controls were usually among the brightest spots, and probes targeting human miRNAs had the broadest range representing varying levels of expression or tissue specificity of miRNAs. Probes matching mouse miRNAs or MM_human miRNAs were clearly “present” in some platforms and not others, indicating a degree of cross-hybridization between similar probes. For example, the intensities of mouse probes or mismatched probes in Agilent did not differ greatly from the negative or empty probes, and indeed, less than 10 probes were called “present” in each category. In contrast, the spread of Exiqon intensities in the mouse and human_MM categories was large, and in the Normal sample, “present” calls were made for 68 mismatched probes and 43 mouse probes, representing 28% and 36% of the total number of probes in the respective category. We note that most of the mismatched probes identified by our uniform reannotation are classified by Exiqon as “obsolete” or “not_designed_for_hsa,” so they may not exhibit the same LNA spiking pattern as their perfect-match counterparts. The distribution of signals upon removal of these probes is available in Supplemental file “Exiqon annotation.” Probe specificity is evaluated by Exiqon using synthetic RNA spike-ins in a relatively low complexity background (yeast tRNA). The biological relevance of the two analyses (mismatched probes versus spike-ins) remains to be elucidated.
Worthy of mention is the considerable number of “present” calls made by most platforms in other categories, such as miscellaneous or obsolete, and in particular the novel category in Ambion containing proprietary “Ambi-miRs.” These results emphasize the limited information offered by overall signal intensities or total number of detected features, often quoted as measures of hybridization performance and platform sensitivity.
Correlation of microarray and NGS data
We proceeded to sequence the mature miRNAs from each of the samples using a Genome Analyzer II platform (Illumina; hereafter abbreviated as GAseq). On average, 12 million reads were obtained in each sequencing run, and after filtering, 32%, 35%, or 63% (M, P, and N, respectively) could be mapped to known miRNAs. Overall, 733 miRNAs were detectable (501 in M, 588 in P, and 608 in N), and 472 of those had at least 10 cumulative counts across the three samples. The number of reads obtained for each miRNA was well-correlated to the respective microarray hybridization intensity (Pearson correlation 0.66 ± 0.12, ranging between 0.42 and 0.87; see Supplemental file “Intensity Correlations”). The 45 miRNAs that were not identified in the sequencing data set, but for which expression levels were in the detection range of at least one microarray platform, were typically called “marginally present” in the latter, suggesting low cross-hybridization of the corresponding array probes.
To allow a direct comparison of the platforms' performances, we focused on the intersection of miRNAs represented on all platforms (Fig. 3A). A total of 215 miRNA probes were included on only one microarray platform (most often Exiqon), while 148 miRNAs were represented on five out of the six microarrays. This was predominantly due to their absence from the Ambion arrays, which were designed against an earlier version of miRBase. Three hundred nineteen miRNAs were targeted by all six microarray platforms; of these, 204 had no predicted cross-hybridization and at least 10 GA sequencing reads mapped to mature sequences.
For these 204 miRNAs, the log2 ratios were calculated for the M/P, P/N, and N/M comparisons and clustered based on Pearson correlation (Fig. 3B). Importantly, as this analysis is limited to a subset of miRNAs, it should not be considered as a complete comparison of the three biological samples. All M-values clustered according to the biological comparison rather than platform type. Data obtained from the two PCR-based methods (GAseq sequencing and Illumina microarrays) consistently clustered together, as did the data from the three microarray platforms exhibiting greater reproducibility (Exiqon, Ambion, and Agilent). The clustering of Invitrogen and Combimatrix data was inconsistent.
Recent reports have demonstrated the effect of normalization on the interpretation of miRNA expression data (Hua et al. 2008; Pradervand et al. 2009), which is certainly magnified by combining data from several platforms. We therefore tested whether our clustered normalized data reflected the coregulation of biologically meaningful groups, in particular potential TS miRNAs frequently lost in cancer (Git et al. 2008, and references therein), and groups of miRNAs residing in close genomic proximity and potentially cotranscribed as a polycistronic pri-miRNA. Figure 3B (right) indicates the relative positions of these miRNAs in the overall clustering, clearly demonstrating correlated levels of both potential TS miRNAs and the miRNA products of many putative polycistronic transcripts. Among the latter category, those groups that do not demonstrate coregulation may not in fact be polycistronic or may be individually regulated by post-transcriptional mechanisms.
We identified the differentially expressed genes on each platform using a uniform arbitrary fold-change threshold of 2 and corrected P-values of <0.05 (Fig. 3C, bars coded “a”) and examined the agreement between platforms. Surprisingly, the actual overlap between the differentially expressed (DE) calls of the platforms was very low. Consistent with the low rate of “present” calls, Invitrogen and Combimatrix results were most frequently in disaccord with the other microarray platforms, while GAseq, Illumina, and Exiqon assays produced the highest numbers of unsupported DE calls.
To eliminate the possibility that the low degree of overlap between platforms resulted from applying an arbitrary uniform cutoff, we developed a novel iterative maximal likelihood estimate (iMLE) algorithm to establish the optimal cutoff for each platform in view of the combined data of all platforms. The overlap of the resulting DE calls is presented in bars “b” in Figure 3C. Although the optimized cutoffs increased the number of the fully overlapping DE calls in all six sample comparisons, the vast majority of the DE calls were still not unanimous across platforms. Whether this disagreement ensues from nonspecific contributions, varying degrees of cross-hybridization of miRNA family members or reduced discrimination between unprocessed and mature forms of the miRNAs (only Agilent's probes are mature-specific) is at present unknown and will necessitate the use of specific synthetic spike-in oligonucleotides.
The difference in DE calls for each comparison is the net result of sensitivity and specificity characteristics inherent to each platform, and those exhibiting the highest sensitivity are expected to make some unsupported DE calls and to generate increasingly large overlaps with platforms of lower sensitivity, evident as DE calls made by two to four platforms (Fig. 3C, gray bars). We therefore examined the nature of the overlap in DE calls. Figure 3D shows an example of the overlap between GAseq and four of the six microarray platforms tested (Invitrogen and Combimatrix were excluded for ease of plotting) in identifying miRNAs up-regulated in the P/N comparison. Here, 53 miRNAs were called up-regulated by all platforms, and both GAseq and Exiqon yielded a large number of unique DE calls (13 and eight, respectively), suggesting that at least one of the platforms exhibits high false-positive (FP) calls (i.e., reduced specificity). Similarly, 10 and six miRNAs were called significantly up-regulated by all platforms except Ambion and Agilent, respectively (false negatives [FNs]), indicative of lower sensitivity. In more complex overlap patterns, the same number (seven) of Exiqon and Illumina's overlapping DE calls was supported or rejected by GAseq. Since all platforms were given equal status, such data could not easily be translated into specificity (true negative [TN]) and sensitivity (true positive [TP]) values.
Correlation with qPCR results
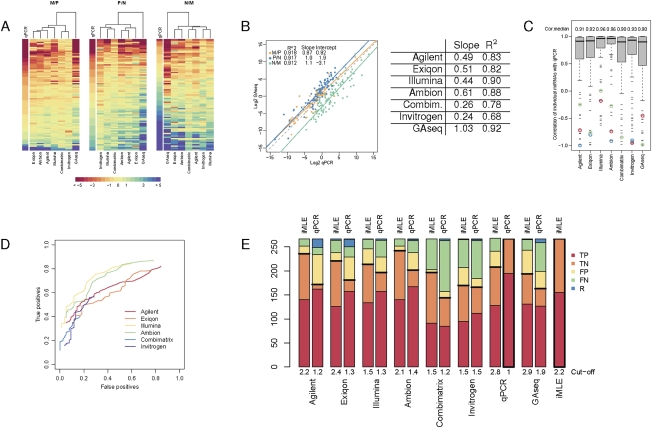
Microarray and NGS data are regularly validated by qPCR. We analyzed the expression of 89 miRNAs from multiple overlap categories using either TaqMan or SYBR Green assays. The log2 ratios of this miRNA subset for all platforms were sorted according to the corresponding qPCR values (Fig. 4A). Although the trend of M-values follows that of the qPCR data, the magnitude of the M-value is clearly different between platforms (ratio compression). Occasional spurious values in single platforms are noticeable as red or blue “islands.”
FIGURE 4.
Validation by real-time RT-PCR. (A) M-values of miRNAs tested by qPCR. Eighty-nine miRNAs validated by qPCR (rows) are sorted by their qPCR M-values. Platforms (columns) are clustered by Euclidean distance. (B) Overall correlation between GAseq and qPCR data. For each biological comparison, the ratios of miRNA expression calculated from GAseq were plotted against those derived from qPCR. Best linear regression fit (solid lines; R^2 values, intercept with y-axis and slope indicated in legend); Y = X (dotted line). Average correlations and slopes across the three comparisons are listed for each platform compared to qPCR. (C) Correlation between microarray/NGS and qPCR data. Boxes depict the distribution of correlation for the M-values generated by qPCR and indicated platforms for each miRNA in all three comparisons (MP, PN, NM), and the median value (Cor.median) is indicated above. Examples of consistent outliers are circled; hsa-miR-484 (red), hsa-miR-15a (green), and hsa-miR-215 (blue). (D) Effect of DE cutoff on the TP and FP rate of each platform. The number of TP and FP DE calls, compared with qPCR calls at fold-change >2 was calculated across a range of thresholds (0–5 in 0.1 increments). Only miRNAs with P-value <0.05 were included for each platform; hence, the ROC curves do not cover the entire range of TP and FP rates. (E) True and false call rates of each platform at optimal cutoffs. The number of TP and FP and FN DE calls was calculated at the optimal log2 cutoffs calculated based on a qPCR reference or on the iMLE algorithm with qPCR as an unknown platform. The number of DE (equivalent to TP) and non-DE (equivalent to TN) calls made by these references is shown with a thick frame. A horizontal black thick line separates true calls (below) from false calls (above). Abbreviations as in panel C.
The ratio compression can also be visualized by the slope of the concordance between each platform and qPCR data for each of the three biological comparisons, exemplified for GAseq data in Figure 4B (average slope ∼1; i.e., no compression). The average slopes for the microarray platforms are listed in Figure 4B and range between 0.24 (Invitrogen) and 0.61 (Ambion). Also evident in this plot is the shift in the y-axis intercept, representing a consistent drift in the measured ratio, also evident in microarray/qPCR plots (data not shown). This shift arises from the inherently different ratio of the overall miRNA population and external reference genes used in qPCR normalization (e.g., 5S rRNA). It has been repeatedly observed by ourselves and others (Lee et al. 2008; C Blenkiron and LD Goldstein, pers. comm.) that cell lines have a lower miRNA content per total RNA (>85% of which is rRNA species) than tissue samples. This trend is supported by the fact that despite similar quality and quantity of RNA input the overall hybridization signal in MCF7 arrays is lower than in normal breast tissue arrays (Fig. 1A) with the normal-like cell line PMC42 showing intermediate values. NGS and microarrays are for the most part blind to such fluctuations as they employ normalization techniques within the miRNA population. As a result, every miRNA appears to be better-expressed in M samples when measured by GAseq compared with equivalent qPCR measurements, where its levels are normalized to high 5S content, thus consistently shifting the GAseq N/M ratios down (intercept = −3.1). Similarly, M/P and P/N correlations are shifted by +0.92 and +1.9, respectively.
The concordance of each platform with qPCR data was measured as either Pearson correlation of all array M-values against the matching qPCR M-values (comparing columns in a traditional table layout) (Fig. 4B, R^2 values) or the distribution of correlations of the M-values of individual miRNAs in the three comparisons (comparing rows in a traditional table layout) (Fig. 4C, box plots). The two measures do not necessarily agree (e.g., Invitrogen's median correlation is 0.93, although the overall average correlation is only 0.68). Discrepancies could arise due to a relatively small number of poorly correlating outliers (counted once for box plots but strongly skewing an overall linear fit) or as a result of differences in the correlation slopes of individual probes (i.e., rows), which—while possibly scoring well in a box plot analysis—reduce the quality of the overall (i.e., columns) linear fit.
We then extended our analysis from continuous M-values to discrete DE calls. Using the calls generated by qPCR as a standard reference (195 DE/positive and 72 non-DE/negative calls across all three comparisons), we counted the number of TN and TP calls made by each platform at multiple threshold values. The resulting ROC curves (Fig. 4D; Supplemental file “4D-Detailed”) exemplify the effect of a chosen cutoff on the perceived sensitivity and specificity of each microarray platform. The threshold values generating the highest overall number of TP calls for each platform was determined to be optimal and is consistent with the ratio compression of each platform such that the platforms exhibiting greater compression (e.g., Combimatrix) perform better at lower cutoffs than those with lower compression (e.g., Ambion). The number of TP and FP DE and non-DE calls made by each platform is presented in Figure 4E (qPCR bars).
Unexpectedly, some outliers in Figure 4C are miRNAs that correlate poorly with qPCR across all platforms (colored circles), suggesting that the FPs were generated by qPCR (similarly to a recent observation by Ach et al. 2008), rather than consistent errors across platforms incorporating different probe design, hybridization conditions, and labeling chemistries. We therefore repeated the DE analysis with the qPCR data incorporated into the iMLE algorithm. Figure 4E contrasts the number of true and false calls made by each platform at the optimal cutoffs calculated using qPCR either as a reference or integrated into iMLE. Consistently across all platforms, the number of true calls calculated under the iMLE algorithm was greater than those calculated using qPCR as a gold standard. The iMLE (TP/TN) rates are as follows: Agilent, 0.90/0.86; Exiqon, 0.82/0.85; Illumina, 0.87/0.71; Ambion, 0.91/0.91; Combimatrix, 0.59/0.95; Invitrogen, 0.61/0.67; qPCR, 0.83/0.71; and GAseq, 0.85/0.56. Omission of “obsolete” or “not_designed_for_hsa” Exiqon probes resulted in minimal changes to these numbers (±0.2 in optimal fold-change cutoff and ±0.04 in TP/TN rates; data not shown). The low sensitivity (TP) of GAseq contradicts the commonly expressed expectation of digital miRNA profiling and was also recently reported in a comparative study using a pool of synthetic RNAs (Willenbrock et al. 2009).
DISCUSSION
We present a comparison of the suitability of six microarray platforms and one NGS technology to detect differential expression of miRNAs. In our hands, Ambion, Agilent, and Exiqon microarrays ranked highest in the rate of true DE calls. During the course of this study, several changes occurred in the handling protocols and microarray design, some of which are summarized by the manufacturers in the Supplemental file “Manufacturers Comments.” Moreover, NGS and miRNA microarrays are now available from several additional manufacturers (e.g., Affymetrix microarrays, whose performance in comparison to Agilent and Exiqon is currently under evaluation by ABRF) (Web-report 2009). We therefore delineate generic key criteria for the evaluation of current miRNA platforms, including common aspects of microarray technology, such as reproducibility, and aspects particular to miRNAs, such as probe annotation and the utility of qPCR for validation.
Several practical considerations are worthy of mention in miRNA microarray platform selection (Table 1). The choice of single- or dual-channel platform depends on the nature of the biological question investigated, and reliable data were generated by all three single channel platforms and the Ambion dual-channel platform. We found that despite the overall lower signal intensity of cell lines, all platforms were equally applicable to cell line and tissue samples (to be corroborated in additional tissues). The platforms vary widely in their input sample requirement, ranging from 100 ng of total RNA (Agilent) to small RNA-enriched fractions equivalent to ∼10 μg total RNA (Ambion and Combimatrix). Thus, despite Ambion's excellent TP and TN rates, the platform is not suitable for studies where input material is limited. Similarly, Ambion's performance in detection of DE may be secondary to ease of handling or slide layout in studies with large numbers of samples, or in a high-throughput core facility, for which the labeling and hybridization protocols of Agilent and Combimatrix would be better suited. Platforms also varied in the reproducibility of hybridization, enumerated as CV across replicates (Fig. 1B,C) and consistency of present/absent calls (Fig. 2A). Lower reproducibility might prescribe a larger number of replicate arrays, affecting the experimental design, computational analysis and costing. Cross-hybridization can be estimated by the signal distribution and present calls from mouse and mismatched human probes as a surrogate measure (Fig. 2B,C). Surprisingly, the LNA probes used by Exiqon were among the poorest in discriminating the groups of probes classified using our uniform reannotation, although the contribution of suboptimal LNA spike patterns could not be evaluated. Finally, unique features such as the ability to customize the microarray probe sets for specific applications (Agilent and Combimatrix), or supported array stripping and reuse procedures (Combimatrix), come into play for particular experimental needs.
Periodic changes to miRBase necessitate a reannotation of microarray and qPCR probes prior to analysis. For example, 35 novel miRNAs of each Ambion and Exiqon match recent additions to miRBase. Our arrays, although acquired within a few weeks of each other, were designed and annotated against different versions of miRBase, resulting in a low number of initially overlapping miRNA identifiers. A substantial fraction of the discrepancies resulting from changes in miRNA nomenclature can be resolved by consulting the tracking files available on miRBase without further computational manipulation. However, changes to the actual sequences of miRBase entries expose potential cross-hybridization between previously unrelated probes and therefore must be identified computationally. Unfortunately, the sequence information provided by the manufacturers is often partial (e.g., miRNA target rather than probe, or probe without proprietary linker). At the two extremes, Combimatrix provides all probe sequences whereas Exiqon offers only proprietary reannotation against miRBase updates, reserving probe sequence information for users bound by confidentiality agreements. This model restricts the inclusion of sequence information in published research studies. Laboratories with no access to fully exploratory methods (such as deep sequencing) may benefit from microarray platforms that include novel miRNAs (Ambion, Exiqon; annotated by the manufacturers), provided that the underlying probe sequences are disclosed.
High-throughput sequencing of miRNAs is coming into wider use and is unmatched for the discovery and experimental validation of novel or predicted miRNAs. However, library preparation methods seem to have systematic preferential representation of the miRNA complement, resulting in different DE calls (Linsen et al. 2009) and the approach awaits rigorous evaluation. We therefore focused on the differential expression of 204 miRNAs represented by all six microarray platforms as well as detected by sequencing. We observed a low degree of overlap in the DE miRNAs (consistent with Sato et al. 2009), not easily attributable to the strength or weakness of singular platforms. We implemented a novel algorithm (iMLE) integrating partial overlaps of DE calls in the calculation of TP and TN rates. Furthermore, we show that qPCR is not an infallible validation method of miRNA microarray data, especially where the array technology itself incorporates PCR-based amplification (e.g., Illumina). The question of an “industry standard” in miRNA expression awaits further advances in both technology (e.g., deep sequencing) and computation (normalization and DE algorithms). iMLE-based assignment of true values can also potentially help amalgamate other binary datasets, such as peak-calling or miRNA target predictions by different algorithms with no need for a standard reference.
We illustrate the effect of using non-miRNA reference genes for qPCR normalization on the perceived differential expression of tested miRNAs. This effect is pronounced when the overall abundance of miRNAs varies, e.g., in experiments affecting the miRNA processing machinery, or in comparisons involving multiple tissues (such as demonstrated by Sato et al. 2009) or combinations of tissues and cell lines. In such cases, it is advisable to perform qPCR measurements of numerous miRNAs, including those identified as stably expressed, to obtain a measure of the linear correlation intercept prior to assignment of validated DE values. Alternatively, microarrays and NGS can be used for mutual validation, circumventing the need for external references.
To our knowledge this is the first systematic study scrutinizing the relative performance of miRNA microarrays, NGS, and qPCR across several well-studied biological samples. While our analysis is not intended to serve as a recommendation for any particular platform, we present practical criteria and metrics to evaluate the reproducibility, specificity, and reliability of methods measuring miRNA expression.
MATERIALS AND METHODS
Preparation of total RNA and small-RNA enriched samples
A pool of commercial normal breast tissue (hereafter termed Normal) total RNAs was created from 78 μg comprising a five-donor pool (BioChain Institute, lot no. A512460), 130 μg Hm breast total RNA (Ambion AM6952, lot no. 02060262), and 75 μg MVP human adult breast total RNA (Stratagene 540045-41, lot no. 0870161). The breast cancer cell lines PMC42 (a gift from Michael O'Hare, University College London) (Whitehead et al. 1983, 1984) and MCF7 (from ATCC) (Soule et al. 1973) were cultured in RPMI or DMEM media (Invitrogen), respectively, supplemented with 10% bovine calf serum (Invitrogen). RNA was extracted from subconfluent cultures (estimated 85% density) that were refed with fresh medium 24 h prior to harvesting. In brief, cultures were washed once with cold phosphate-buffered saline (PBS). Upon complete removal of the PBS, cells were lysed directly in 8.4 mL of QIAzol (Qiagen), and total RNA was extracted using 10 miRNeasy columns (Qiagen) according to manufacturer's recommendations.
Several 100 μg aliquots from each total RNA were further separated into large- and small-RNA enriched fractions (cutoff ∼200 nt) using the miRNeasy columns and reagents. The yield and quality of the total RNA were monitored by spectrophotometry at 260, 280, and 230 nm, by Agarose gel electrophoresis, and on a Bioanalyzer Eukaryote Total RNA Nano Series II chip (Agilent). RNA integrity number (RIN) values were 9.4 (MCF7), 10.0 (PMC42), and 7.6 (Normal). The yield and quality of the small-RNA enriched fraction (sRef) were monitored by spectrophotometry (as above), urea/polyacrylamide gel electrophoresis (Git et al. 2008), and on a Bioanalyzer Small RNA Series II chip (Agilent). sRef were extracted with a near 100% efficiency, contained predominantly tRNA and small rRNA, and comprised a different but reproducible proportion of the total RNA in each sample: 14 ± 1% in MCF7, 12.5 ± 0.5% in PMC42, and 6 ± 0.2% in normal breast tissue. The miRNA contained within these fractions was <0.5 ng per 10 μg total RNA (Git et al. 2008; data not shown).
Microarray study design
For single-channel platforms (Agilent, Illumina), each sample was hybridized in quadruplicate (samples are termed M, MCF7; P, PMC42; and N, Normal throughout the text). For dual-channel platforms, a balanced-dye design was employed in which quadruplicate hybridizations were set up in the following combinations: Cy3-MCF7 with Cy5-PMC42 (sample MP), Cy3-PMC42 with Cy5-Normal (sample PN), and Cy3-Normal with Cy5-MCF7 (sample NM).
The hybridizations for each quadruplicate were carried out on two different days. Where possible replicates that were labeled side-by-side were hybridized on different days, and those labeled on different days were hybridized side by side. For microarray platforms requiring near immediate application of the labeled samples (Combimatrix, Exiqon), independent labeling reactions were carried out on the day of the hybridization. For Ambion assays, two independent sets of duplicate dried polyadenylation reactions were frozen for <48 h, and labeling of individual replicates was completed immediately prior to hybridization.
RNA labeling and microarray hybridization
RNA input and labeling kits were chosen and used according to the recommendations of each microarray manufacturer (Kreatech labeling for Combimatrix arrays and the manufacturers' labeling kits for others). Arrays were hybridized for 16–20 h in an Agilent G2545A hybridization oven and washed according to the manufacturer's instructions. To minimize bias due to seasonal changes in ultraviolet light and ambient ozone, we completed all in-house experimental work at one location over a span of 6 wk.
Agilent
One hundred nanograms of total RNA samples was dephosphorylated, 3′ end-labeled with Cy3-pCp, purified on Micro Bio-Spin columns, dried, and hybridized using miRNA Microarray System labeling kit and arrays (Agilent) (Wang et al. 2007).
Ambion
sRef samples equivalent to 10 μg total RNA were polyadenylated, purified, dried to completion, coupled to Cy3 or Cy5 amine-reactive dyes (GE Healthcare), purified, dried, and hybridized using mirVana miRNA Labeling and Bioarrays Version 2 (Ambion) (Shingara et al. 2005).
Combimatrix
sRef samples equivalent to 10 μg total RNA were coupled to Cy3- or Cy5-ULS reagent using ULS Small RNA Labeling kit (Kreatech) and hybridized to MicroRNA 4X2K Microarrays (Combimatrix).
Exiqon
One microgram total RNA samples was dephosphorylated, Hy3- or Hy5- end-labeled, and hybridized using miRCURY LNA microRNA Array Power Labeling kit and microarray kit (Exiqon).
Illumina
Two hundred nanograms of total RNA samples were processed by Illumina using a Sentrix Array Matrix 96-well MicroRNA Expression Profiling Assay v1 (Chen et al. 2008). In brief, samples are polyadenylated and reverse-transcribed, and the cDNA is hybridized to a specific primer pool and extended to incorporate address tags and universal sequences. PCR-amplified samples are then hybridized to address-coded beads on a solid support.
Invitrogen
One microgram of total RNA samples was polyadenylated, 3′ splint-ligated to Cy3- or Cy5-labeled oligonucleotides, and hybridized using NCode Rapid miRNA Labeling System and NCode Multi-Species miRNA Microarray v2 (Invitrogen).
Microarray scanning and feature extraction
Illumina bead-based arrays were processed at the manufacturer's facility in San Diego, California. In brief, arrays were scanned on a BeadScan instrument, and fluorescence intensities were extracted and summarized using the BeadStudio software (Illumina), resulting in a set of summarized fluorescence measurements (Supplemental file “MPN_miRNA_Illumina”). Agilent, Ambion, Exiqon, and Invitrogen arrays were scanned on a G2505B Microarray Scanner (Agilent Technologies), and Combimatrix arrays were scanned on the InnoScan700 (Innopsys). Feature recognition and alignment of all in-house scanned images were carried out using GenePix Pro 6.1 and, where necessary, adjusted manually by the same operator. To minimize variation in alignment correction, arrays from each platform were processed in a single session. Data from Agilent, Ambion, Combimatrix, Exiqon, and Invitrogen arrays have been deposited in ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/; accession E-MTAB-96).
Microarray normalization and processing
Data analyses were carried out within the R statistical computing framework version 2.8.0 (http://www.R-project.org) (R Development Core Team 2008). Following quality control assessment, two out of the 60 arrays hybridized in-house were excluded (see Fig. 1B) due to either low intensity (Exiqon, sample M) or array-specific artifacts (Combimatrix, sample PN). The overall Cy5 intensities in Exiqon arrays were too low for reliable analysis, and the data from the Cy3 channel was treated as a single-channel assay.
The limma package (Smyth 2005) was used for microarray processing. Different methods for background correction were tested for all platforms except Illumina (none, subtract, half, minimum, movingmin, normexp) and normalization (none, vsn, quantile), depending on whether the platforms were used for a single- or dual-channel assay. Ultimately, normexp was chosen due to its superior performance in correcting spatial artifacts, maximizing the uniformity of foreground and background signal, and minimizing the variability within and between arrays. All platforms were background-corrected using normexp, except for Combimatrix, where minimum was used due to constraints specific to the array layout. Dual-channel platforms (Ambion, Combimatrix, Invitrogen) were normalized using loess spatial correction within arrays, and single-channel platforms (Agilent, Exiqon) were quantile normalized between arrays. Spike-in controls were not used for normalization purposes as they were only available for some of the platforms, and where present were too few to be reliably utilized.
SNR was calculated using the SSDR method (He and Zhou 2008), μi/(σiF + σiB) (where μ equals spot intensity; σ, pixel standard deviation; i, spot; F, foreground; and B, background).
Microarray probe reannotation
All probe sequences were mapped to mature human and mouse miRNA sequences from miRBase version 12 (Griffiths-Jones et al. 2008) using WU-BLAST (Lopez et al. 2003). Ungapped alignment was performed, using word length shorter than the default when necessary. For “long probe” platforms (Ambion, Invitrogen, Combimatrix, Illumina), all perfect match hits with length greater than 15 were retained and filtered as described below. For “short probe” platforms (Agilent, Exiqon), probes with length greater than 15 were treated similar to the “long probes” platforms, whereas for short probes only perfect match hits with alignment length equal to probe length were considered. Where alignments ≥20 bases were found, shorter alignments were discarded. For alignments <20 bases, the longest was assigned to the given probe, or multiple miRNAs in the case of matches of equal length. For alignments ≥20, there were occasionally several possible miRNAs targets, and these were all assigned to the probe to account for potential cross-hybridization. A complete list of reannotated probes can be found in Supplemental file “Reannotation.”
In cases where a probe sequence aligned to both a human and mouse miRNA, targets were assigned to the probe under the following priorities: human perfect match > human with one mismatch > mouse miRNAs. Probes were finally grouped into the following categories: PosControl, NegControl, and Novel: positive or negative controls and putative novel miRNAs, respectively, as defined by array manufacturer; Empty indicates spots with no printed probes; Human and MM_human, probes targeting human miRNAs with perfect complementarity or with a single internal mismatch, respectively; Mouse, probes targeting mouse, but not human, miRNAs; Obsolete, probes that were designed to target miRNAs but do not map to targets in the current version of miRBase; and Miscellaneous, probes outside the aforementioned categories, such as probes targeting miRNAs from other species, spike-in controls, and all unidentifiable probes.
We examined the signal intensities across probes of different lengths and GC content. Some variation was observed (data not shown), but since the binding kinetics for individual platforms are affected by numerous factors, we did not attempt to correct for these in the analysis.
Putative polycistronic miRNAs were defined as sets of miRNAs sharing a genomic locus with no more than 500 bases between any two adjacent miRNAs, and were obtained via the Clusters interface of miRGen (Megraw et al. 2007).
Assignment of microarray present and absent spots
Spots were called as “absent,” “marginally present,” or “present” using a modified version of the R package “panp” (Warren et al. 2007). A probability distribution of signal intensities from empty and negative control spots was calculated, and the cumulative distribution function (CDF) generated. Each spot was called as present or absent based on expression value cutoffs defined from the survivor distribution (1-CDF) for each individual array, using P-values of 0.05 (present) and 0.1 (marginally present). For dual-channel arrays (Ambion, Combimatrix, and Invitrogen), each channel was treated separately, and the percentage of present calls for each miRNA was taken across both the Cy3- and Cy5-labeled fluorescence data.
Identification of differentially expressed miRNAs
For single-channel platforms, M-values were calculated based on the individual Cy3 data from each sample (hereafter also referred to as M-values). Ratio compression was taken as the slope of the linear least-squares regression of microarray versus qPCR across all three biological comparisons. All M- and P-values are available in Supplemental file “M_pValue_204probes.” The empirical Bayes moderated f-statistics implemented in the R package limma was used. Differentially expressed genes were identified using the limma nestedF procedure, applying a significance threshold of 0.05 in combination with Benjamini–Hochberg false-discovery rate control and unless otherwise specified, a minimal cutoff of 2. Where multiple probes targeting the same miRNA did not agree, one of two approaches was chosen for clarity of presentation: For Figure 4D, we have assigned the corresponding miRNA with an “NA” value, while for Figure 4E, the miRNA was assigned with the value of the probe that showed differential expression, as long as the two calls were not contradictory (up- and down-regulated) in which case the miRNA was assigned “NA.”
Next-generation sequencing
sRef samples equivalent to 2 μg total RNA were ligated to a preadenylylated 3′ adapter v1.5 (5′-rApp-[desoxy]ATCTCGTATGCCGTCTTCTGCTTG-[didesoxy]ddC-3′; Illumina or Dharmacon) in 1× T4 RNL2 truncated reaction buffer (NEB), 10 mM MgCl2 (Ambion), 20 units of RNaseOUT (Invitrogen), and 300 units truncated T4 RNA ligase 2 for 1 h at 22°C. The reactions were then supplemented with 12.5 nmol 5′ adapter (all RNA; GUUCAGAGUUCUACAGUCCGACGAUC; Dharmacon), 1 mM ATP (Ambion), and 20 units of T4 RNA ligase (NEB) and the second ligation allowed to proceed for 6 h at 20°C. The double-ligation products were reverse-transcribed by SuperScriptII reverse transcriptase (Invitrogen) in the presence of primer GX1 (all desoxy; CAAGCAGAAGACGGCATACGA; Sigma) following manufacturer's instructions. The cDNA was PCR-amplified by Phusion DNA Polymerase with Primers GX1 and GX2 (all DNA; AATGATACGGCGACCACCGACAGGTTCAGAGTTCTACAGTCCGA; Sigma) for 19 cycles of [10 sec at 98°C, 30 sec at 60°C, and 15 sec at 72°C]. The amplification products were separated on a Novex 6% TBE gel (Invitrogen), and the 90–100 base-pair bands were excised, eluted into 0.3 M NaCl, and ethanol precipitated. Following quality control on a Bioanalyzer 1000 DNA chip (Agilent), the purified DNA fragments were used directly for two independent repeats of sequencing via 36 alternating cycles of enzymatic synthesis and optical interrogation using the Illumina Cluster Station and GAII Genome Analyzer following manufacturer's protocols. Sequencing reads were extracted from the image files generated by Genome Analyzer II using the GAPipeline software, version 1.4 (Illumina).
NGS data analysis
3′ adapters were trimmed from sequencing reads using an in-house script (available upon request). Reads of length <15 nt after adapter trimming and comprising more than 50% polyA stretches were excluded from further analyses. The remaining reads were mapped to known mature miRNAs (miRBases version 12) using the “ssaha2” program (Ning et al. 2001), where 100% identity between reads and known miRNAs sequences was required. miRNAs with an aggregate count of less than 10 in all samples were eliminated (see Supplemental file “GA_Read_Counts”); then the total read count for each lane was scaled relative to the library size (total number of reads that mapped to known miRNAs). Read counts of technical replicates were then merged, and log2 (fold-change) values were calculated for each miRNA. P-values were subsequently calculated using a binomial approximation to Fisher's exact test for each miRNA.
Real-time RT-PCR (qPCR)
For SYBR green-based assays, sRef samples equivalent to 10 μg total RNA were polyadenylated, reverse-transcribed using a tagged and anchored oligo-dT primer, and then amplified using a gene-specific forward primer and universal reverse primer (see Supplemental file “qPCR Primers”) in the presence of SYBR green as described by Git et al. (2008). For TaqMan assays (ABI), total RNA samples were reverse-transcribed using a pool of gene-specific primers and amplified using individual gene-specific assays. All RT reactions were performed with three different RNA inputs, and all PCR reactions were carried out in triplicate. RNU48 and 5S rRNA were used as non-miRNA reference genes for TaqMan and SYBR green qPCR, respectively. The measured Ct values were M:11.24, P:11.63, N:11.70 (RNU48) and M:16.20, P:16.50, N:16.27 (5S rRNA), and the magnitude of the variance did not warrant ΔΔCt normalization. Where miRNAs were tested by both methods, the average correlation was 0.94.
iMLE algorithm
The input for the algorithm is a table of discrete DE calls (up-regulated, +1; not DE, 0; down-regulated, −1) for each miRNA/comparison combination (rows) made by each experimental platform at a particular threshold value with P-value <0.05 (columns). An initial “truth” value was assigned for each row according to the majority of calls. A matrix (i,j) was then generated for each platform, representing the proportion of cases where the assay called various j for each Truth i, e.g., P(−1,−1) + P(−1,0) + P(−1,+1) = 1. Subsequently the algorithm reiterated two steps until Truth values converged: (1) selected for each row the Truth (−1/0/+1) with the highest maximal likelihood estimate [MLE, defined as the product of all platform probabilities to have given this Truth call under the existing (i,j) parameters], followed by (2) a recalculation of the platform matrices.
To determine the optimal cutoffs for each platform, the iMLE was performed in an iterative fashion, where cutoffs were fixed for all but one tested platform at a time, for which a series of discrete cutoffs was tested, and the cutoff that yielded the highest number of correct calls was fixed as a temporary optimum. This was repeated across all platforms until each platform cutoff converged to a stable value. The following measures were then extracted from the platform matrices: TP [the average of (1,1) and (−1,−1)]; TN (0,0); FP [average of (0,1) and (0, −1)]; FN [average of (1,0) and (−1,0)]; reverse [average of (1, −1) and (−1,1)].
An outline of the algorithm is included in Supplemental file “iMLE Algorithm,” and the code for the implementation of the algorithm is available from the authors upon request.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Yoav Git for advice in algorithm design; and Sarah Moffatt, Nick Matthews, and Rory Stark for help with sequencing the small RNA libraries. C.C., J.H. and A.G. conceived and coordinated the study; A.G. extracted and labeled the RNA, extracted feature intensities after scanning and participated in the analysis; M.O. carried out the hybridizations and scans; C.K. prepared the small RNA libraries; H.D. and M.S.-D. analyzed the data with advice from P.B.; and A.G., H.D., and M.S.-D. drafted the manuscript; which was approved by all authors. This work was supported by the University of Cambridge, Cancer Research UK, Hutchison Whampoa Limited, the European Molecular Biology Laboratory, and the Swiss National Science Foundation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1947110.
REFERENCES
- Ach RA, Wang H, Curry B 2008. Measuring microRNAs: Comparisons of microarray and quantitative PCR measurements, and of different total RNA prep methods. BMC Biotechnol 8: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbato C, Giorgi C, Catalanotto C, Cogoni C 2008. Thinking about RNA? MicroRNAs in the brain. Mamm Genome 19: 541–551 [DOI] [PubMed] [Google Scholar]
- Baumbusch LO, Aaroe J, Johansen FE, Hicks J, Sun H, Bruhn L, Gunderson K, Naume B, Kristensen VN, Liestol K, et al. 2008. Comparison of the Agilent, ROMA/NimbleGen, and Illumina platforms for classification of copy number alterations in human breast tumors. BMC Genomics 9: 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissels U, Wild S, Tomiuk S, Holste A, Hafner M, Tuschl T, Bosio A 2009. Absolute quantification of microRNAs by using a universal reference. RNA 15: 2375–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno MJ, de Castro IP, Malumbres M 2008. Control of cell proliferation pathways by microRNAs. Cell Cycle 7: 3143–3148 [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. 2005. Real-time quantification of microRNAs by stem–loop RT-PCR. Nucleic Acids Res 33: e179. doi: 10.1093/nar/gni178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lozach J, Garcia EW, Barnes B, Luo S, Mikoulitch I, Zhou L, Schroth G, Fan JB 2008. Highly sensitive and specific microRNA expression profiling using BeadArray technology. Nucleic Acids Res 36: e87. doi: 10.1093/nar/gkn387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Git A, Spiteri I, Blenkiron C, Dunning MJ, Pole JC, Chin SF, Wang Y, Smith J, Livesey FJ, Caldas C 2008. PMC42, a breast progenitor cancer cell line, has normal-like mRNA and microRNA transcriptomes. Breast Cancer Res 10: R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: Tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T 2008. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 44: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Zhou J 2008. Empirical evaluation of a new method for calculating signal-to-noise ratio for microarray data analysis. Appl Environ Microbiol 74: 2957–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Eberhart JK, Postlethwait JH 2009. MicroRNAs and micromanaging the skeleton in disease, development, and evolution. J Cell Mol Med 13: 606–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy E, O'Driscoll L 2008. Molecular medicine of microRNAs: Structure, function, and implications for diabetes. Expert Rev Mol Med 10: e24. doi: 10.1017/S1462399408000781 [DOI] [PubMed] [Google Scholar]
- Hua YJ, Tu K, Tang ZY, Li YX, Xiao HS 2008. Comparison of normalization methods with microRNA microarray. Genomics 92: 122–128 [DOI] [PubMed] [Google Scholar]
- Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y 2009. miR2Disease: A manually curated database for microRNA deregulation in human disease. Nucleic Acids Res 37: D98–D104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Li W, Gordon DB, Bhattacharjee A, Curry B, Ghosh J, Brizuela L, Carroll JS, Brown M, Flicek P, et al. 2008. Systematic evaluation of variability in ChIP-chip experiments using predefined DNA targets. Genome Res 18: 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic M, Hengartner MO 2006. miRNAs and apoptosis: RNAs to die for. Oncogene 25: 6176–6187 [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J, Stoffel M 2006. MicroRNAs: A new class of regulatory genes affecting metabolism. Cell Metab 4: 9–12 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858 [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD 2008. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA 14: 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ruan K 2009. MicroRNA detection by microarray. Anal Bioanal Chem 394: 1117–1124 [DOI] [PubMed] [Google Scholar]
- Linsen SE, de Wit E, Janssens G, Heater S, Chapman L, Parkin RK, Fritz B, Wyman SK, de Bruijn E, Voest EE, et al. 2009. Limitations and possibilities of small RNA digital gene expression profiling. Nat Methods 6: 474–476 [DOI] [PubMed] [Google Scholar]
- Lopez R, Silventoinen V, Robinson S, Kibria A, Gish W 2003. WU-Blast2 server at the European Bioinformatics Institute. Nucleic Acids Res 31: 3795–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina PP, Slack FJ 2008. microRNAs and cancer: An overview. Cell Cycle 7: 2485–2492 [DOI] [PubMed] [Google Scholar]
- Megraw M, Sethupathy P, Corda B, Hatzigeorgiou AG 2007. miRGen: A database for the study of animal microRNA genomic organization and function. Nucleic Acids Res 35: D149–D155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Z, Cox AJ, Mullikin JC 2001. SSAHA: A fast search method for large DNA databases. Genome Res 11: 1725–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, et al. 2004. Identification of virus-encoded microRNAs. Science 304: 734–736 [DOI] [PubMed] [Google Scholar]
- Pradervand S, Weber J, Thomas J, Bueno M, Wirapati P, Lefort K, Dotto GP, Harshman K 2009. Impact of normalization on miRNA microarray expression profiling. RNA 15: 493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2008. R: A language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Roush S, Slack FJ 2008. The let-7 family of microRNAs. Trends Cell Biol 18: 505–516 [DOI] [PubMed] [Google Scholar]
- Sato F, Tsuchiya S, Terasawa K, Tsujimoto G 2009. Intra-platform repeatability and inter-platform comparability of microRNA microarray technology. PLoS One 4: e5540. doi: 10.1371/journal.pone.0005540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y, Engstrom PG, Lenhard B 2007. Mammalian microRNA prediction through a support vector machine model of sequence and structure. PLoS One 2: e946. doi: 10.1371/journal.pone.0000946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R, Chiang VL 2005. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques 39: 519–525 [DOI] [PubMed] [Google Scholar]
- Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, et al. 2006. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol 24: 1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingara J, Keiger K, Shelton J, Laosinchai-Wolf W, Powers P, Conrad R, Brown D, Labourier E 2005. An optimized isolation and labeling platform for accurate microRNA expression profiling. RNA 11: 1461–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK 2005. Limma: Linear models for microarray data. In Bioinformatics and computational biology solutions using R and bioconductor (ed. Gentleman R, et al. ), pp. 397–420 Springer, New York [Google Scholar]
- Soule HD, Vazguez J, Long A, Albert S, Brennan M 1973. A human cell line from a pleural effusion derived from a breast carcinoma. J Natl Cancer Inst 51: 1409–1416 [DOI] [PubMed] [Google Scholar]
- Stefani G, Slack FJ 2008. Small noncoding RNAs in animal development. Nat Rev Mol Cell Biol 9: 219–230 [DOI] [PubMed] [Google Scholar]
- van den Berg A, Mols J, Han J 2008. RISC-target interaction: Cleavage and translational suppression. Biochim Biophys Acta 1779: 668–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ach RA, Curry B 2007. Direct and sensitive miRNA profiling from low-input total RNA. RNA 13: 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren P, Taylor D, Martini PGV, Jackson J, Bienkowska J 2007. PANP−a new method of gene detection on oligonucleotide expression arrays. In 7th IEEE International Conference on Bioinformatics and Bioengineering, 2007, 108–115, Boston, MA [Google Scholar]
- Web-report 2009. ABRF study explores microRNA array platforms. GenomeWeb daily news http://www.genomeweb.com/print/911534?page=show.
- Whitehead RH, Bertoncello I, Webber LM, Pedersen JS 1983. A new human breast carcinoma cell line (PMC42) with stem cell characteristics. I. Morphologic characterization. J Natl Cancer Inst 70: 649–661 [PubMed] [Google Scholar]
- Whitehead RH, Quirk SJ, Vitali AA, Funder JW, Sutherland RL, Murphy LC 1984. A new human breast carcinoma cell line (PMC42) with stem cell characteristics. III. Hormone receptor status and responsiveness. J Natl Cancer Inst 73: 643–648 [PubMed] [Google Scholar]
- Willenbrock H, Salomon J, Sokilde R, Barken KB, Hansen TN, Nielsen FC, Moller S, Litman T 2009. Quantitative miRNA expression analysis: Comparing microarrays with next-generation sequencing. RNA 15: 2028–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin JQ, Zhao RC, Morris KV 2008. Profiling microRNA expression with microarrays. Trends Biotechnol 26: 70–76 [DOI] [PubMed] [Google Scholar]