An increased number of primer–template junctions generated by PCNA, Pol-δ, and Pol-ε at stalled replication forks activates Chk1.
Abstract
Stalled replication forks activate and are stabilized by the ATR (ataxia-telangiectasia mutated and Rad3 related)-mediated checkpoint, but ultimately, they must also recover from the arrest. Although primed single-stranded DNA (ssDNA) is sufficient for checkpoint activation, it is still unknown how this signal is generated at a stalled replication fork. Furthermore, it is not clear how recovery and fork restart occur in higher eukaryotes. Using Xenopus laevis egg extracts, we show that DNA replication continues at a stalled fork through the synthesis and elongation of new primers independent of the checkpoint. This synthesis is dependent on the activity of proliferating cell nuclear antigen, Pol-δ, and Pol-ε, and it contributes to the phosphorylation of Chk1. We also used defined DNA structures to show that for a fixed amount of ssDNA, increasing the number of primer–template junctions strongly enhances Chk1 phosphorylation. These results suggest that new primers are synthesized at stalled replication forks by the leading and lagging strand polymerases and that accumulation of these primers may contribute to checkpoint activation.
Introduction
DNA replication is a complex process that must be faithfully completed for genome integrity to be maintained. Replication forks can stall at lesions arising from exogenous DNA-damaging agents and endogenous metabolic byproducts or at replication fork barriers comprised of protein–DNA complexes or DNA secondary structures (Lambert and Carr, 2005; Tourrière and Pasero, 2007). Cells use many mechanisms to deal with the constant challenge of replication stress because persistent stalled forks are prone to collapse, leading to increased genome instability (Branzei and Foiani, 2005; Lambert and Carr, 2005; Paulsen and Cimprich, 2007; Friedel et al., 2009). The DNA damage and replication checkpoints act to stabilize and restart stalled replication forks as well as coordinate downstream cell cycle arrest with different repair processes (Branzei and Foiani, 2005; Paulsen and Cimprich, 2007; Friedel et al., 2009; Segurado and Tercero, 2009). In the absence of this checkpoint, cells fail to complete DNA replication, and genomic instability is increased. Cells also attempt to continue replication when forks stall using postreplication repair pathways to bypass lesions and, in some cases, restarting replication downstream of a stalled replisome (Friedberg, 2005; Heller and Marians, 2006; Chang and Cimprich, 2009).
The checkpoint kinase ATR (ataxia-telangiectasia mutated and Rad3 related) has a central role in the replication stress response, mediating checkpoint activation and other events (Shechter et al., 2004b; Cimprich and Cortez, 2008; Friedel et al., 2009). Several studies suggest that the ATR pathway is activated by primed single-stranded DNA (ssDNA) and that this structure is sufficient to stimulate ATR in Xenopus laevis egg extracts and in mammalian cell extracts (Michael et al., 2000; You et al., 2002; Zou and Elledge, 2003; MacDougall et al., 2007; Shiotani and Zou, 2009). The components of this structure, ssDNA and the ssDNA–double-stranded DNA (dsDNA) junction, act as a scaffold to recruit and colocalize ATR with other checkpoint proteins that facilitate its activation (Zou, 2007; Cimprich and Cortez, 2008). The ssDNA is bound by replication protein A (RPA), which can recruit ATR via the ATR-interacting protein (ATRIP; Zou and Elledge, 2003). The ssDNA–dsDNA junction is critical for the recruitment of the Rad9–Hus1–Rad1 (9-1-1) complex, a heterotrimeric clamp with structural homology to proliferating cell nuclear antigen (PCNA), which is the processivity factor for replicative polymerases (Bermudez et al., 2003; Ellison and Stillman, 2003; Zou et al., 2003; Majka et al., 2006). Loading of the 9-1-1 complex onto these junctions is mediated by the Rad17–RFC2-5 complex, which is stimulated by the presence of RPA. The presence of RPA may direct loading of the 9-1-1 complex to a 5′ ssDNA–dsDNA junction in vitro, although under some conditions, loading at the 3′ junction has also been observed (Ellison and Stillman, 2003; Zou et al., 2003; Majka et al., 2006). Interestingly, the 5′ ssDNA–dsDNA junction is necessary for checkpoint activation in Xenopus egg extracts when replication is blocked (MacDougall et al., 2007).
Another critical factor for checkpoint activation is TopBP1, a BRCA1 carboxy-terminal repeat–containing protein (Burrows and Elledge, 2008). TopBP1 interacts with both the 9-1-1 and ATR–ATRIP complexes, and it plays a role in loading and/or stabilizing the 9-1-1 complex on damaged chromatin (Parrilla-Castellar and Karnitz, 2003; Delacroix et al., 2007; Lee et al., 2007; Mordes et al., 2008; Yan and Michael, 2009a). In addition, TopBP1 directly stimulates the kinase activity of the ATR–ATRIP complex (Hashimoto et al., 2006; Kumagai et al., 2006; Yan et al., 2006; Mordes et al., 2008). This leads to phosphorylation of many proteins, including Chk1, one of the primary effectors of ATR and a necessary component of the arrest response (Kumagai et al., 2006; Segurado and Tercero, 2009). Thus, the ssDNA–dsDNA junction is critical for checkpoint activation because it facilitates recruitment of the TopBP1 activator to ATR. However, the amount of ssDNA adjacent to a primer–template junction is also important, as larger regions of ssDNA induce greater Chk1 phosphorylation (MacDougall et al., 2007).
Numerous activities are required for the successful completion of DNA replication (Waga and Stillman, 1998; Hubscher et al., 2002; Arias and Walter, 2007). DNA is unwound by the MCM2-7 (minichromosomal maintenance) helicase complex, and the unwound ssDNA is stabilized by RPA binding (Waga and Stillman, 1998; Bochman and Schwacha, 2008). On both strands, replication is initiated by the RNA primase and DNA polymerase activities of Pol-α, which synthesize an 8–12 ribonucleotide primer extended by the addition of ∼20 deoxyribonucleotides. PCNA loads onto the 3′ end of this primer, facilitating the recruitment of a new polymerase (Waga and Stillman, 1998; Hubscher et al., 2002). A study in yeast suggests that continuous and processive replication on the leading strand is mediated primarily by Pol-ε (Pursell et al., 2007). In contrast, lagging strand replication is discontinuous, with Pol-δ extending Pol-α primers to synthesize Okazaki fragments (Pursell et al., 2007; Kunkel and Burgers, 2008; Nick McElhinny et al., 2008). Consistent with these strand preferences, depletion of either Pol-δ or -ε from Xenopus egg extracts causes a similar reduction in DNA replication (Fukui et al., 2004).
Because ATR activation is critical for a proper response to stalled forks, it follows that the primed ssDNA structure that exists at normal replication forks would persist, increase, and/or change when forks stall to activate ATR. During normal replication, coupling of the helicase and polymerase activities prevents the accumulation of RPA-coated ssDNA and the primed ssDNA is transient. When forks stall at lesions that do not block helicase progression, these activities functionally uncouple, allowing continued unwinding, ssDNA accumulation, and checkpoint activation (Walter and Newport, 2000; Sogo et al., 2002; Pacek and Walter, 2004; Byun et al., 2005; Liu et al., 2007). This is accompanied by increased RPA recruitment to chromatin, which is thought to reflect the ssDNA that accumulates during unwinding (Michael et al., 2000; Mimura et al., 2000; Walter and Newport, 2000; Lupardus et al., 2002; Zou and Elledge, 2003). In response to etoposide or aphidicolin, Pol-α and the 9-1-1 complex are also recruited and/or stabilized onto chromatin in a TopBP1-dependent manner (Michael et al., 2000; Lupardus et al., 2002; You et al., 2002; Parrilla-Castellar and Karnitz, 2003; Yan and Michael, 2009a). The reason for this increase is not clear but could suggest that DNA synthesis continues on the accumulated ssDNA. It is important to understand whether this occurs, as continued DNA synthesis may contribute to checkpoint activation (Yan and Michael, 2009a). Indeed, on the leading strand, new DNA synthesis could be required to generate the 5′ ssDNA–dsDNA junction needed for checkpoint activation (Cimprich, 2007; Yan and Michael, 2009b).
Studies in mammalian cell extracts indicate that DNA synthesis may continue past a lesion because plasmids containing a single UV light–induced lesion can be fully replicated in the absence of repair (Svoboda and Vos, 1995; Carty et al., 1996). Although the mechanism of this process has not been well characterized, it is known in bacterial systems that the leading and lagging strand machinery can uncouple from each other such that replication can continue on the undamaged strand. Furthermore, DNA synthesis may continue on the damaged strand through ongoing synthesis of Okazaki fragments past a stalled lagging strand polymerase or through repriming downstream of a stalled leading strand polymerase, which is a process known as replication restart (Heller and Marians, 2006; Yao and O’Donnell, 2009). Analysis of replication forks isolated from UV-irradiated Saccharomyces cerevisiae suggests that replication can restart downstream of a lesion on both the leading and lagging strand (Lopes et al., 2006). However, this restart process has not been shown in higher eukaryotes, nor is it known how it is regulated.
The Xenopus egg extract system has been used to study many aspects of checkpoint activation in large part because these extracts faithfully recapitulate DNA replication in a manner similar to that seen in mammalian cells. Replication can be monitored using chromatin or plasmid templates in different types of extracts, and in all cases, this involves the formation of a prereplication complex, activation of the S-phase Cdks, and initiation of a single round of semiconservative DNA replication (Arias and Walter, 2004; Garner and Costanzo, 2009). Using this system, we studied the replication that occurs when replication forks stall, how this replication affects checkpoint activation, and how it relates to the generation of primed ssDNA. We also used synthetic DNA structures to probe the relationship between generation of the primed ssDNA structure and activation of the checkpoint. Collectively, our data suggest that new primer synthesis at a stalled fork occurs on both the leading and lagging strand and that this replication contributes to checkpoint activation.
Results
Small DNA products accumulate in response to aphidicolin
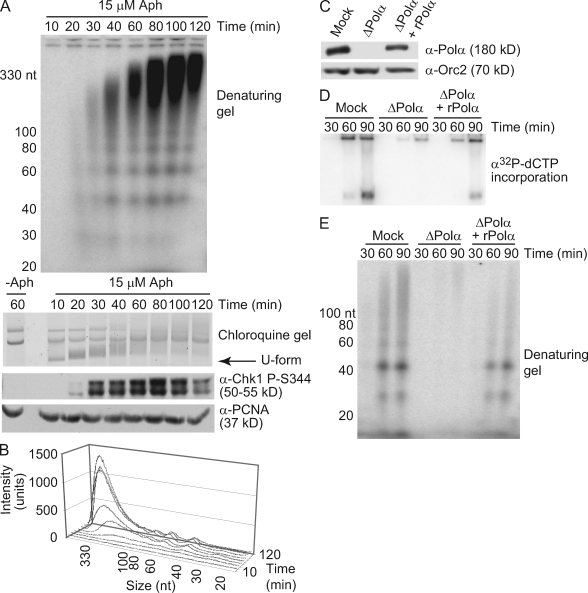
To study the effects of fork stalling on replication, we took advantage of the effect of aphidicolin on DNA replication in Xenopus egg extracts. Aphidicolin blocks the activity of the replicative polymerases (α, δ, and ε) by binding in the polymerase active site. In doing so, it stalls all replication forks, allowing for detection of continued DNA synthesis after fork stalling without the background of undamaged replication products. First, we examined the replication of plasmid DNA, which occurs upon sequential incubation of a plasmid with high speed extract (HSE) and nucleoplasmic extract (NPE; Walter et al., 1998; Walter and Newport, 2000). HSE allows prereplication complex assembly on plasmid or chromatin templates, and NPE provides the Cdk activity needed for initiation. We asked whether new DNA synthesis could be detected during widespread fork stalling by monitoring nucleotide incorporation into low molecular weight intermediates in the presence of a concentration of aphidicolin known to reduce polymerase activity and slow replication forks. Intermediates as short as ∼25–30 nt were observed on a denaturing polyacrylamide gel in response to aphidicolin treatment (Fig. 1, A and B). Interestingly, the products were of relatively defined sizes, occurring primarily at 20-nt intervals on both plasmid and chromatin templates (Fig. S1 A). Although the reason for this regular distribution is not clear, similar patterns have been observed during early SV40 replication, suggesting that these intermediates may represent transient replication products that accumulate with aphidicolin (Nethanel et al., 1988).
Figure 1.
Small DNA products accumulate in response to aphidicolin. (A) A 9-kb plasmid was incubated in HSE and NPE containing α-[32P]dCTP with or without aphidicolin (15 µM Aph). Three samples were taken at the indicated times after NPE addition. DNA was isolated from the first sample, run on a denaturing polyacrylamide gel, and autoradiographed (top). The second was run on a chloroquine agarose gel and stained with SybrGold (middle; U-form refers to unwound DNA). The third was run on an SDS-PAGE gel and immunoblotted for Chk1 phosphorylation with PCNA as a loading control. (B) Lengths and intensities of DNA intermediates in A were calculated using ImageQuant software (n = 3). (C–E) Xenopus sperm chromatin (2,500 nuclei/µl) and α-[32P]dCTP were added to mock- or Pol-α–depleted LSE. Recombinant Pol-α complex was added where indicated. (C) Depletion was verified by immunoblotting with Orc2 as a loading control. (D) Aliquots taken at the indicated times were analyzed for replication. (E) Mock and depleted extracts were treated with 15 µM aphidicolin, and samples were analyzed for continued synthesis as in A.
To determine how the appearance of these products is related to other events that occur upon replication fork stalling, such as checkpoint activation and helicase–polymerase uncoupling, plasmid unwinding and Chk1 phosphorylation were analyzed in parallel (Walter and Newport, 2000; Byun et al., 2005). As shown previously, aphidicolin treatment led to rapid unwinding of the plasmid, which was seen as a highly supercoiled band on a chloroquine gel (Fig. 1 A, U-form arrow; Walter and Newport, 2000). Chk1 phosphorylation was detected ∼20 min after U-form appearance but decreased at later times, likely because of adaptation in the extract (Fig. 1 A; Yoo et al., 2004). Interestingly, the radiolabeled products that we observed appeared after the formation of U-form and were concomitant with Chk1 phosphorylation and U-form disappearance. This sequence is consistent with the idea that some DNA synthesis on the unwound template is required for checkpoint activation and suggests that the small products we observed may represent this synthesis.
To confirm that these intermediates are related to fork progression, we asked whether their synthesis is dependent on replisome activity. Geminin prevents minichromosomal maintenance loading and origin firing (McGarry and Kirschner, 1998). The addition of recombinant geminin to the extract prevented replication and accumulation of these small DNA intermediates after aphidicolin treatment. These intermediates were also not detected in the absence of aphidicolin, suggesting that origin firing is needed for their synthesis and that they only accumulate during fork stalling (Fig. S1, B and C). We then asked whether Pol-α is required for synthesis of these intermediates by immunodepleting Pol-α from extract (Fig. 1 C). Replication and the aphidicolin-dependent accumulation of small DNA intermediates were lost after Pol-α depletion, and addition of the recombinant Pol-α complex rescued these effects (Fig. 1, D and E). These observations suggest that Pol-α is required for the synthesis of these small DNA intermediates. We next asked whether these intermediates contain a 5′ RNA component, normally synthesized by Pol-α. RNaseA/T1 treatment of purified intermediates led to loss of 3–10 nt in a fraction of the products, although the mobility of others was unchanged (Fig. S1 D). The variable presence of a 5′ RNA component has previously been observed in other systems during replication and is attributed to RNA degradation during primer maturation (DePamphilis and Wassarman, 1980; Méchali and Harland, 1982). We have also observed rapid degradation of an RNA primer annealed to ssDNA in NPE (MacDougall et al., 2007). Collectively, these observations suggest that the DNA intermediates observed after aphidicolin treatment are nascent, replication-dependent RNA-DNA primers synthesized by Pol-α.
Primer synthesis and elongation continues at a stalled fork
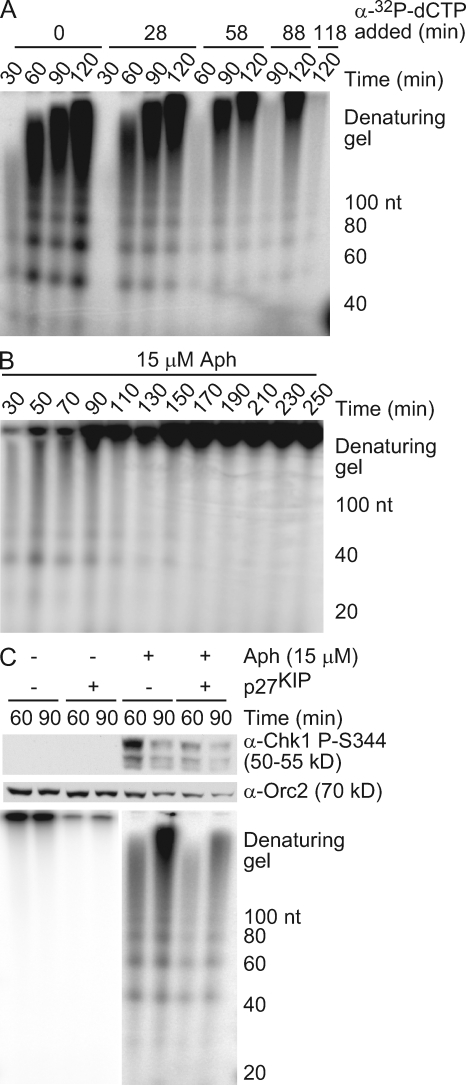
We noticed that although longer DNA intermediates accumulate at later time points, shorter nascent DNAs also persisted (quantified in Fig. 1 B). These kinetics suggest that small nascent DNAs are elongated over time, whereas new primers continue to be synthesized and elongated. To test the idea that new primer synthesis continues, α-[32P]deoxy-CTP (dCTP) was added into the extract at different times after aphidicolin treatment, and the radiolabeled products were analyzed as in Fig. 1 A. Radioactivity was incorporated into both short and long products for up to 2 h after aphidicolin addition, indicating that new RNA-DNA primers continue to be synthesized and elongated after unwinding (Fig. 2 A). However, when total radionucleotide incorporation was analyzed over an extended time frame, small nascent DNAs were not observed at later time points (Fig. 2 B). Collectively, these data suggest that small nascent DNA synthesis occurs through the generation of new RNA-DNA primers and their elongation on the rapidly unwound DNA but that primer synthesis stops when the replisome reaches the end of the unwound region.
Figure 2.
Primer synthesis and elongation continues at a stalled fork. (A and B) A 9-kb plasmid was replicated in HSE and NPE with 15 µM aphidicolin (Aph). Radionucleotides were added at the indicated times after NPE addition (A) or at the start of the reaction (B), and samples were analyzed as in Fig. 1 A. (C) Sperm chromatin (4,000 nuclei/µl) was replicated in HSE and NPE with 15 µM aphidicolin. 30 min after NPE addition, p27KIP was added, and 5 min later, α-[32P]dCTP was added. Samples were taken at the indicated times and analyzed for Chk1 phosphorylation and small nascent DNAs as in Fig. 1 A.
Although global origin firing is inhibited upon damage-induced checkpoint activation, local, dormant origins near a stalled fork are activated to complete replication (Blow and Ge, 2008). Thus, the small nascent DNAs we observed may be caused by the activation of these dormant origins. To distinguish continued synthesis of new primers at a previously stalled fork from initiation of a new fork at a dormant origin, we used p27KIP to inhibit Cdk activity and subsequent origin firing. Extracts were treated with or without aphidicolin to stall replication forks and then incubated with p27KIP to inhibit further origin firing. α-[32P]dCTP was added 5 min after p27KIP addition, ensuring that any detectable products arose from previously fired origins. When p27KIP was added to extracts lacking aphidicolin, replication decreased, indicating that Cdk activity and further origin firing were blocked (Fig. 2 C). New DNA synthesis and Chk1 phosphorylation also decreased after aphidicolin and p27KIP treatment, suggesting that dormant origin firing partially contributes to the synthesis we observe. Nevertheless, small nascent DNAs were still detected in the presence of p27KIP, indicating that primer synthesis and elongation continue to occur on the unwound DNA at the existing stalled replication fork.
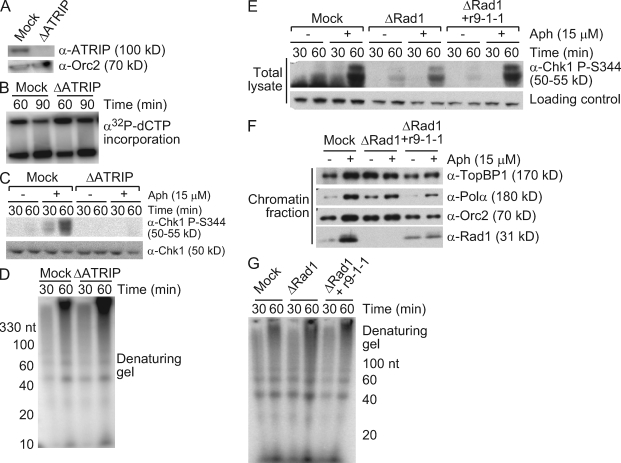
The ATR–9-1-1 pathway is not required for primer synthesis
The ATR pathway is involved in the stabilization and recovery of stalled forks, and it has also been tied to postreplication repair, suggesting that checkpoint activation might be needed for new primer synthesis at stalled forks (Chang and Cimprich, 2009). To test whether ATR activity is involved in this process, ATRIP was depleted from the extract (Fig. 3 A). There was little effect of ATRIP depletion on small nascent DNA synthesis after aphidicolin treatment, and in fact, a slight increase was observed (Fig. 3, B–D). This increase may reflect the release of late firing origins in the absence of ATR activity, an effect more evident at higher concentrations of chromatin (Yanow et al., 2003; Luciani et al., 2004; Shechter et al., 2004a). We also asked whether Rad1, which may have functions independent of ATR activity, was required for the synthesis of small nascent DNA. Depletion of Rad1 strongly reduced Chk1 phosphorylation, as previously reported (Lupardus and Cimprich, 2006), but had no effect on small nascent DNA accumulation (Fig. 3, E–G). These observations indicate that although nascent DNA synthesis coincides with checkpoint activation, the checkpoint does not regulate continued primer synthesis.
Figure 3.
The ATR–9-1-1 pathway is not required for primer synthesis. (A–D) Sperm chromatin (2,000 nuclei/µl) was replicated in mock- or ATRIP-depleted LSE with or without 15 µM aphidicolin (Aph). (A) Samples were immunoblotted to verify depletion. (B–D) Aliquots were taken at the indicated times and analyzed for replication (B), Chk1 phosphorylation (C), and primer synthesis (D) as in Fig 1. (E–G) Sperm chromatin (2,500 nuclei/µl) was replicated in mock- or Rad1-depleted LSE containing recombinant 9-1-1 complex and 15 µM aphidicolin as indicated. (E and G) Aliquots were analyzed for Chk1 phosphorylation (E) and primer synthesis (G) as in Fig. 1 A. (F) In parallel, chromatin was isolated from a third aliquot, and bound proteins were immunoblotted with the indicated antibodies.
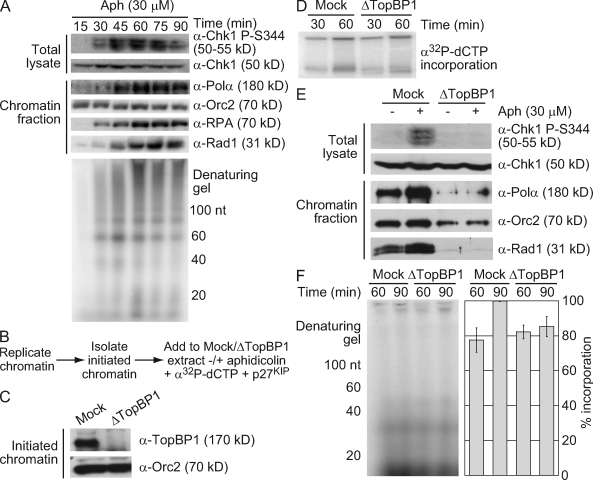
Relationship between nascent DNA synthesis, Pol-α hyperloading, and TopBP1 function
The need for Pol-α in the synthesis of small nascent DNAs could indicate that Pol-α hyperloading plays a role in this synthesis. Alternatively, continued synthesis could arise from the recycling of a low level of chromatin-bound Pol-α, which is similar to a proposed model for Okazaki fragment synthesis during replication in Escherichia coli (Hamdan and Richardson, 2009). To establish whether there is a correlation between the synthesis of small nascent DNAs and Pol-α hyperloading, we first compared the kinetics of these two events (Fig. 4 A). Levels of chromatin-bound Pol-α rapidly increased in response to aphidicolin, peaking at 60 min and then leveling off (Fig. 4 A). Although the combined synthesis of small nascent DNAs (<100 nt) reached a maximum at 45 min, different maximums were seen for different primer sizes.
Figure 4.
Relationship between nascent DNA synthesis, Pol-α hyperloading, and TopBP1 function. (A) Sperm chromatin (2,500 nuclei/µl) was replicated in LSE containing 30 µM aphidicolin (Aph). At the indicated times, parallel samples were analyzed for Chk1 phosphorylation and chromatin binding as in Fig. 3 (E and F). 5 min before each time point, an aliquot was incubated with α-[32P]dCTP and analyzed for nascent DNA synthesis as in Fig. 1 A. (B) Experimental schematic for C–F. Sperm chromatin (4,000 nuclei/µl) was replicated in LSE and isolated after 45 min to yield initiated chromatin. Initiated chromatin was replicated in mock- or TopBP1-depleted LSE containing α-[32P]dCTP and p27KIP with or without 30 µM aphidicolin. (C) Initiated chromatin was blotted for chromatin-bound proteins. (D) Replication was analyzed in mock- and TopBP1-depleted extracts lacking aphidicolin as in Fig. 1 D. (E) Parallel samples were taken 60 min after the addition of initiated chromatin and analyzed for Chk1 phosphorylation or chromatin binding as in Fig. 3 (E and F). (F) Primer synthesis was analyzed in aphidicolin-treated extracts as in Fig. 1 A, and intensities were calculated using ImageQuant software and normalized relative to the 90-min sample from mock-depleted extract. Error bars represent standard error (n = 8).
TopBP1 is needed for Pol-α hyperloading after etoposide and aphidicolin treatment (Parrilla-Castellar and Karnitz, 2003; Yan and Michael, 2009a). To directly test the requirement for Pol-α hyperloading, we analyzed the effect of TopBP1 depletion on small nascent DNA synthesis. Because TopBP1 has a separate function in replication initiation (Hashimoto et al., 2006; Yan and Michael, 2009a), we used an established chromatin transfer protocol to remove TopBP1 from initiated chromatin without affecting elongation (Fig. 4 B; Yan and Michael, 2009a). Chromatin replication was initiated in undepleted extract, and then chromatin was isolated in a high salt buffer to remove chromatin-bound TopBP1 (Fig. 4 C). The initiated chromatin was then added to mock- or TopBP1-depleted extracts in the presence or absence of aphidicolin, and small nascent DNA synthesis was analyzed as in Fig. 1 A. To block further initiation, p27KIP was added to the mock- or TopBP1-depleted extracts. Consistent with published results, replication of initiated chromatin was unaffected by loss of TopBP1, and when initiated chromatin was added into TopBP1-depleted extracts with aphidicolin, Pol-α and Rad1 hyperloading as well as Chk1 phosphorylation were lost (Fig. 4, D and E; Yan and Michael, 2009a). Under these conditions, we observed a slight decrease in the accumulation of small nascent DNAs (Fig. 4 F). This indicates that although TopBP1-mediated Pol-α hyperloading may be required for the synthesis of a subset of these small nascent DNAs, the bulk of ongoing primer synthesis at a stalled fork does not require this hyperloading.
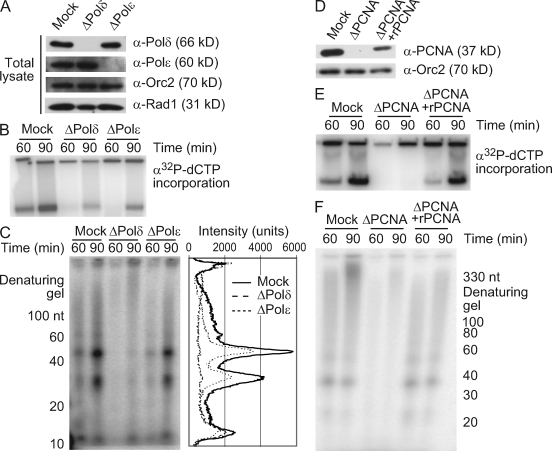
Pol-δ, Pol-ε, and PCNA are required for the accumulation of small nascent DNAs
The small intermediates that accumulate in the presence of aphidicolin may represent nascent, stalled lagging strand products, replication restart on the leading strand, or a combination of both. Because these intermediates are longer than the size normally synthesized by Pol-α, a polymerase switch to Pol-δ or -ε may be needed for their formation. As Pol-δ and -ε are thought to be the major lagging and leading strand polymerases, respectively, involvement of either polymerase could indicate the strand of the unwound fork on which new DNA synthesis occurs (Kunkel and Burgers, 2008).
To determine the role of these polymerases in the synthesis of small nascent DNAs, we individually depleted each one from the extract (Fig. 5 A). As shown previously, Pol-δ or -ε depletion reduced chromatin replication (Fig. 5 B; Fukui et al., 2004; Waga et al., 2001). More importantly, the synthesis of small nascent DNA dramatically decreased after Pol-δ depletion, whereas Pol-ε depletion had a more modest effect (Fig. 5 C). The relative contributions of Pol-δ and -ε to new DNA synthesis suggest that the majority of the small nascent DNAs that form at stalled forks accumulate on the lagging strand in a Pol-δ–dependent manner, with a smaller, Pol-ε–dependent fraction on the leading strand. This also suggests that continued progression of the replisome after fork stalling and downstream synthesis of small nascent DNA may require the switch to Pol-δ or -ε. Consistent with this idea, we did not observe the synthesis of small nascent DNA when PCNA was depleted, and this effect could be rescued by the addition of recombinant PCNA (Fig. 5, D–F).
Figure 5.
Pol-δ, Pol-ε, and PCNA are required for the accumulation of small nascent DNAs. (A–F) Sperm chromatin (2,500 nuclei/µl) was added to mock-, Pol-δ–, Pol-ε–, or PCNA-depleted LSE with or without 15 µM aphidicolin (n = 3). (A and D) Depletion was verified by immunoblotting. (B and E) Replication was monitored in the absence of aphidicolin as in Fig. 1 D. (C) Synthesis of small nascent DNAs was monitored in aphidicolin-treated extracts and quantitated as in Fig. 1 B. (F) Small nascent DNAs were monitored as in C.
We also noticed that although short primers (∼10–30 nt) were still detected after Pol-δ and -ε depletion, they did not further accumulate as might be expected if Pol-α continued to synthesize primers that cannot be elongated. Small primers also did not accumulate in the absence of PCNA. One possible explanation for these findings is that primers are unstable in the absence of elongation. To test this idea, we allowed small nascent DNAs to accumulate and then added a concentration of aphidicolin that fully blocks polymerase activity. We reasoned that when further synthesis and elongation are inhibited in this manner, the stability of the labeled intermediates could be assessed. Under these conditions, the population of small nascent DNAs decreased over time, suggesting that these structures are not completely stable in the absence of elongation (Fig. S2).
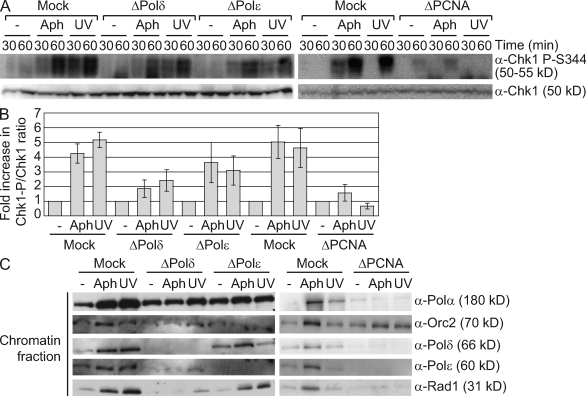
Because the primer–template junction is a critical component of the checkpoint-activating signal, we examined the effect of Pol-δ or -ε depletion on the checkpoint pathway. Pol-δ or -ε depletion resulted in similar decreases in Chk1 phosphorylation (Fig. 6, A and B). We also observed a decrease in Rad1 hyperloading, most dramatically after Pol-δ depletion (Fig. 6 C). Although there is partial reduction in total Rad1 when Pol-δ is depleted (Fig. 5 A), this modest effect is unlikely to account for the defect in Rad1 loading observed. It seems more likely that the reduction in Rad1 loading arises from the loss of Pol-δ–dependent primer synthesis. Depletion of Pol-δ and -ε also reduced UV irradiation–induced Chk1 phosphorylation and Rad1 loading, suggesting that these effects are not specific to aphidicolin treatment (Fig. 6, A–C). Similarly, PCNA depletion caused a complete loss of Chk1 phosphorylation and Rad1 hyperloading in response to UV irradiation or aphidicolin treatment (Fig. 6, A–C). Altogether, these observations indicate that PCNA-mediated recruitment of Pol-δ and -ε is needed for optimal phosphorylation of Chk1.
Figure 6.
Pol-δ, Pol-ε, and PCNA are required for optimal Chk1 phosphorylation. (A–C) Untreated or UV-irradiated (1,000 J/m2) sperm chromatin (2,500 nuclei/µl) was added to mock-, Pol-δ–, Pol-ε–, or PCNA-depleted LSE in the presence or absence of 15 µM aphidicolin (Aph). (A) Chk1 phosphorylation was analyzed as in Fig. 1 A. (B) Chk1 phosphorylation at 60 min was normalized to total Chk1 levels and quantitated using FluorChem analysis software. Error bars indicate standard error (n = 3). (C) Samples were taken at 30 min and analyzed for chromatin binding as in Fig. 3 F.
Primer synthesis contributes strongly to Chk1 phosphorylation
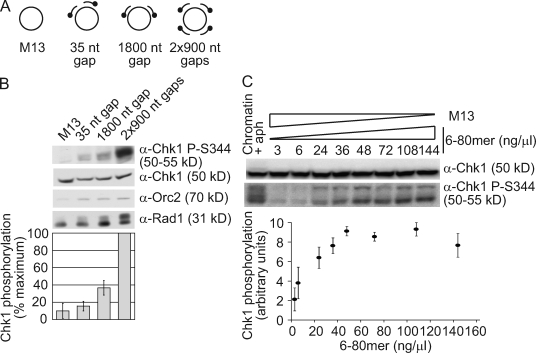
To directly determine whether additional primers affect checkpoint activation on a defined length of ssDNA, we took advantage of a system we previously developed to monitor checkpoint activation with defined DNA structures (MacDougall et al., 2007). This system allows us to clearly identify and compare the relative contributions of different components of the checkpoint-activating structure with Chk1 phosphorylation. In addition, the structures have been shown to activate a physiologically relevant checkpoint response. We designed a set of DNA structures in which the amount of ssDNA and number of primers varied (Fig. 7 A). Biotinylated primers were annealed to circular ssM13 to form a 35-nt gap, an 1800-nt gap, or two 900-nt gaps (2 × 900 nt), and the contribution of ssDNA outside of the gap was eliminated by blocking the outer primer ends with biotin–streptavidin complexes as described previously (MacDougall et al., 2007). Thus, the structures with a 35- and 1800-nt gap contain an equal number of primer–template junctions but vary in the amount of ssDNA adjacent to the primer ends. In contrast, the structures with an 1800-nt gap or two 900-nt gaps have an equal amount of total ssDNA adjacent to primer ends, but the structure with two 900-nt gaps contains twice as many ends. We reasoned that if the amount of ssDNA was the primary determinant for checkpoint activation, there would be little difference in the Chk1 phosphorylation induced by these two structures. However, if the primer end contributed more, the structure with two 900-nt gaps would induce stronger Chk1 phosphorylation.
Figure 7.
Primer synthesis contributes strongly to Chk1 phosphorylation. (A) Schematic of purified ssM13 structures. Biotinylated ends are represented by a dot. (B) Purified ssM13 structures were coupled to streptavidin and added at 6 ng/µl to NPE containing 300 µM aphidicolin. Samples were taken after 20 min and blotted for the proteins indicated. Chk1 phosphorylation was quantitated using Photoshop. (C) Purified ssM13 and 6–80-mer structures were added to NPE containing 300 µM aphidicolin (aph). The indicated concentration of the 6–80 mer is shown, and ssM13 was added so the total concentration of DNA was 144 ng/µl. Sperm chromatin (2,500 nuclei/µl) was also replicated in HSE and NPE containing 15 µM aphidicolin. Samples were analyzed at 20 (structures) or 60 min (chromatin) for Chk1 phosphorylation and quantitated as in Fig. 6 B. (B and C) Error bars indicate standard error (n = 3).
Consistent with our previous findings, ssM13 alone did not activate the checkpoint, whereas Chk1 phosphorylation induced by the 1800-nt gap structure was greater than that from the 35-nt gap structure (Fig. 7 B; MacDougall et al., 2007). Importantly, the structure with two 900-nt gaps induced stronger Chk1 phosphorylation than the 1800-nt gap structure, and this increase was much larger than the relative effect caused by increasing the gap size. Rad1 phosphorylation also increased when two gaps were present, although, as previously noted, this was unaffected by gap size (MacDougall et al., 2007). These results clearly indicate that the additional primer directly contributes to checkpoint activation and has a more significant effect on Chk1 phosphorylation than ssDNA accumulation.
In the context of these structures, the effect of an additional primer on Chk1 phosphorylation is surprisingly strong, given that loss of Pol-δ or -ε causes a noticeable reduction in the synthesis of small nascent DNAs but a relatively small decrease in Chk1 phosphorylation (Figs. 5 C and 6 A). One possible reason for this apparent discrepancy is that the effect of primer accumulation on Chk1 phosphorylation is saturable. Primer accumulation may have less of an effect on Chk1 phosphorylation in the chromatin experiments because there are more primers present, whereas in the structure-based experiments, the total concentration of primers may be lower so the effect of adding more primers is greater. Consistent with this idea, the effect of Pol-δ depletion on Chk1 phosphorylation was greater at lower chromatin concentrations, as would be predicted if there were fewer primer ends (Fig. S3). To further test this model, we annealed six unmodified primers to ssM13 (6–80 mer) and assessed the effect of increasing amounts of this structure on Chk1 phosphorylation. Unmodified ssM13 was also added to maintain the total amount of DNA at a constant level. At low concentrations of the 6–80 mer, similar to that used with the aforementioned gapped structures, we observed a strong concentration-dependent increase in Chk1 phosphorylation (Fig. 7 C). However, as the concentration of the 6–80-mer structure increased, Chk1 phosphorylation plateaued. These observations suggest that the effect of additional primer–template junctions on Chk1 phosphorylation is dependent on the total concentration of primer–template junctions. Collectively, our results also indicate that continued DNA synthesis at a stalled fork may be an effective mechanism for increasing the strength of Chk1 phosphorylation.
Discussion
In this study, we took advantage of the Xenopus egg extract system and the effect of aphidicolin on polymerase activity to assess the interplay between fork stalling, DNA replication, and checkpoint activation. By characterizing the intermediates formed upon inhibition of fork progression, we show that new RNA-DNA primers continue to be synthesized and elongated on the unwound ssDNA downstream of a stalled replisome. Synthesis of these small nascent DNAs is dependent on PCNA, as well as both the leading and lagging strand polymerases, Pol-ε and -δ. Furthermore, loss of PCNA, Pol-ε, or Pol-δ leads to a decrease in Chk1 phosphorylation, suggesting that primer synthesis may affect checkpoint activation. Using defined DNA structures, we unambiguously define the contribution of primers to checkpoint activation, showing that a single additional primer can dramatically influence the phosphorylation of Chk1. Collectively, these data suggest that cells use the replication machinery to synthesize additional primers and that the initial accumulation and elongation of these primers at a stalled fork has a significant effect on checkpoint activation.
Continued primer synthesis at stalled forks
Our data are consistent with findings in bacteria and yeast that show that DNA replication uncouples on both the leading and lagging strand after a replication block (Heller and Marians, 2006; Langston and O’Donnell, 2006). In support of this model, we find that some small nascent DNAs are synthesized after depletion of Pol-δ or -ε, which are thought to function as the lagging and leading strand polymerases, respectively (Kunkel and Burgers, 2008). We also find that there is a greater decrease in small nascent DNA synthesis after Pol-δ depletion than Pol-ε depletion, suggesting that the bulk of new primers are made on the lagging strand. Indeed, we and others have observed increased chromatin binding of Pol-δ but not Pol-ε in response to aphidicolin and UV irradiation, as might be expected if Pol-δ plays a greater role than Pol-ε in this synthesis (Fig. 6 C; Sasakawa et al., 2006). Although it is possible that Pol-δ and -ε can act redundantly, this difference in synthesis suggests that there may be larger stretches of ssDNA on the leading strand between the stalled polymerase and the newly synthesized primer than on the lagging strand. Indeed, published electron microscopy images of yeast DNA containing irreparable UV light–induced lesions revealed gaps of 3 kb on the leading strand versus 400 nt on the lagging strand (Lopes et al., 2006).
The short nascent DNAs we observed most likely represent new primers synthesized in the presence of aphidicolin. Although small primer-like structures could result from the repair of a double-strand break or the resection of replication products, this does not seem likely because the primers we detect are elongated over time, are observed upon pulse labeling with α-[32P]dCTP, and are geminin sensitive. Our observations also suggest that continued synthesis of these primers involves recycling of chromatin-bound Pol-α. First, we find that although unwinding is rapid, new DNA synthesis is a slow and ongoing process. Second, depletion of TopBP1 suppresses Pol-α hyperloading (Parrilla-Castellar and Karnitz, 2003; Yan and Michael, 2009a) but has only a modest effect on the synthesis of small nascent DNAs, suggesting that synthesis does not require this hyperloading. This may indicate that the chromatin-bound Pol-α recycles, synthesizing new primers as it moves away from the stalled polymerase. Such recycling has been observed during lagging strand replication in E. coli, T4, and T7 bacteriophage replication systems (Hamdan and Richardson, 2009). Interestingly, we find that small RNA-DNA primers do not accumulate after Pol-δ, Pol-ε, or PCNA depletion, as might be expected if Pol-α continues to recycle and prime but elongation is blocked. This could indicate that the recruitment of PCNA, Pol-δ, and Pol-ε is needed to release Pol-α and allow its recycling. It is also possible that accumulation of the small RNA-DNA primers is not observed because degradation occurs in the absence of elongation. Indeed, our data suggest that small nascent DNAs are slowly degraded when elongation is blocked by aphidicolin. This degradation may be further accelerated after PCNA, Pol-δ, or Pol-ε depletion because the 3′ primer ends are unprotected.
Our data may also suggest that Pol-α recycling occurs specifically on the lagging strand, whereas a different mechanism is involved in reestablishing synthesis on the leading strand. Because TopBP1 mediates Pol-α hyperloading in response to replication stress, an interesting hypothesis is that TopBP1 specifically directs or stabilizes Pol-α on the leading strand to mediate replication restart (Parrilla-Castellar and Karnitz, 2003; Yan and Michael, 2009a,b). We observed similar decreases in small nascent DNA synthesis after TopBP1 or Pol-ε depletion, as would be predicted if both proteins were needed for priming on the leading but not the lagging strand. Given the gap sizes reported in yeast in response to UV irradiation (Lopes et al., 2006), one would expect the ratio of primers synthesized on the lagging and leading strands to be ∼7.5:1. Therefore, a lagging strand defect would result in a more significant loss in accumulation of small nascent DNAs, whereas a loss of leading strand restart would have less of an effect. The effects we observed after TopBP1, Pol-δ, and Pol-ε depletion are consistent with these ideas, although additional work will be needed to conclusively show whether the synthesis we observed represents replication restart on the leading strand.
Primer synthesis and checkpoint activation
We have previously shown that primed ssDNA is a sufficient structure for checkpoint activation (MacDougall et al., 2007). In this study, we used defined DNA structures to extend these experiments and show that an additional primer on a fixed length of ssDNA can strongly increase Chk1 phosphorylation. This may be particularly relevant in physiological situations when a small number of forks stall. However, when multiple forks stall, as would occur with aphidicolin treatment, the contribution of additional primers to Chk1 phosphorylation may be diminished because the effect is saturable. It is unclear whether this saturation occurs as the result of the total number of primers present or a signal-limiting checkpoint protein. Interestingly, although the effect of primer ends on Chk1 phosphorylation reaches a maximum, these structures alone do not result in maximal Chk1 phosphorylation because aphidicolin-treated chromatin causes a higher level of Chk1 phosphorylation (Fig. 7 C). This suggests that there are other features of the stalled replication fork or chromatin in addition to primer ends that can further contribute to Chk1 phosphorylation.
New primer synthesis may also have a context-dependent effect on checkpoint activation, depending on whether it occurs on the leading or lagging strand. Studies with purified proteins and in Xenopus extracts suggest that accumulated ssDNA must be adjacent to a 5′ primer end to activate the checkpoint when replication is blocked, and there appears to be a preference for loading of the 9-1-1 complex on this end (Ellison and Stillman, 2003; Majka et al., 2006; MacDougall et al., 2007). On the lagging strand, discontinuous replication would result in the formation of a gap between a stalled polymerase and the 5′ end of the previous Okazaki fragment, creating the primed ssDNA structure needed for checkpoint activation. Therefore, additional primer synthesis would not be required for checkpoint activation from this strand, but it could still contribute, as our data suggest. However, on the leading strand, the accumulated ssDNA is adjacent to the 3′ end of the stalled replicating strand. Thus, replication restart on the leading strand may be needed to generate a 5′ end for checkpoint activation. It is also possible that the 3′ end can signal checkpoint activation in some circumstances. In at least one case, the 9-1-1 complex has been reported to load on the 3′ end (Zou et al., 2003), and we have observed checkpoint activation with a structure containing a replicating 3′ end (and blocked 5′ end; MacDougall et al., 2007).
We have found that both Pol-δ and -ε depletions cause decreases in Chk1 phosphorylation. The effect of Pol-ε is consistent with published data in yeast demonstrating a role for this polymerase in S-phase checkpoint activation and survival in response to UV irradiation (Navas et al., 1996). Our structure experiments strongly suggest that at least a portion of the observed effect on checkpoint activation is caused by the loss of small nascent DNAs synthesized by these polymerases. Furthermore, it seems likely that the reduction in Rad1 loading is also caused by this loss. However, the effect of polymerase depletion on Chk1 phosphorylation is not proportional to the reduction in synthesis of small nascent DNAs; fewer DNA intermediates are lost after Pol-ε depletion, but the decrease in Chk1 phosphorylation is comparable with that seen with Pol-δ depletion. One possibility is that leading strand and lagging strand primers have different contributions to Chk1 phosphorylation because of different gap sizes on these strands. We have previously shown that the amount of ssDNA adjacent to a primer end contributes to the strength of Chk1 phosphorylation (MacDougall et al., 2007), and published data in UV-irradiated yeast indicate that larger gaps exist on the leading strand than on the lagging strand (Lopes et al., 2006).
Overall, our results suggest a model in which leading and lagging strand replication uncouple at an unwound fork. On the lagging strand, primers accumulate through recycling of Pol-α and are elongated by Pol-δ. On the leading strand, TopBP1 recruits or stabilizes Pol-α, which synthesizes a primer that is extended by Pol-ε. On both strands, continued synthesis allows recruitment of the 9-1-1 complex and TopBP1 to a 5′ end, which stimulates activation of ATR on the adjacent ssDNA. Leading strand primers may be required to generate the 5′ end needed for Chk1 and may also have a greater effect on the Chk1 signal because of the larger sized gap. Additional studies will be needed to elucidate the precise contributions of leading and lagging strand primers to checkpoint activation and whether this continued synthesis contributes to genome stabilization by preventing the accumulation of ssDNA and ensuing fork collapse. Nevertheless, our findings further expand upon the close ties between replication and checkpoint activation and provide valuable insight to the relationship between these processes at stalled forks.
Materials and methods
Xenopus egg extracts and replication assays
Xenopus sperm chromatin was obtained through removal and fractionation of male Xenopus frogs testes, and the resulting chromatin was demembranated after lysolecithin treatment (Murray, 1991). Low speed extract (LSE) was prepared by centrifugation of eggs from Xenopus females and isolation of the cytoplasmic layer (LSE). To generate HSE, LSE was further clarified through high speed centrifugation to remove membranes. NPE was prepared by isolation of nuclei from LSE containing replicating chromatin through centrifugation and then further fractionation and isolation of the NPE as previously described (Tutter and Walter, 2006; Lupardus et al., 2007). Indicated concentrations of sperm chromatin or a 9-kb plasmid (14 ng/µl) were replicated in HSE for 30 min, followed by the addition of an equal volume of NPE. For replication assays, radiolabeled samples prepared by the addition of α-[32P]dCTP to the extract were incubated with replication stop buffer (0.5% SDS, 20 mM EDTA, pH 8.0, and 500 µg/ml proteinase K) and analyzed by agarose gel electrophoresis and autoradiography (Walter and Newport, 2000; Lupardus et al., 2007). Samples for U-form analysis were isolated in a similar manner, with an additional phenol/chloroform extraction and ethanol precipitation before electrophoresis. U-form samples were analyzed on a 0.7% Tris/borate/EDTA agarose gel containing 4 µM chloroquine (Sigma-Aldrich; Walter and Newport, 2000), and the gel was stained with SybrGold (Invitrogen). Aphidicolin (Sigma-Aldrich) was dissolved in DMSO to 30 mM and diluted to the indicated concentrations. RNase Out (Invitrogen) was added to HSE at 4 U/µl where indicated. All experiments have been performed at least three times, with the following exceptions in which results are representative of two independent experiments: Fig. 1 (C–E), Fig. 2 (A and C), Fig. 3 (E–G), and Fig. S1 D.
Antibodies and recombinant proteins
Chk1 (G-4) antibody was obtained from Santa Cruz Biotechnology, Inc. The Chk1 P-S344 antibody was used as previously described (Chang et al., 2006) or purchased from Cell Signaling Technology. Pol-δ (p66 and p125) and Pol-ε (p60) antibodies were used as previously described (Waga et al., 2001; Fukui et al., 2004). Orc2 antibody was provided by J.C. Walter (Harvard Medical School, Boston, MA; Walter and Newport, 1997). Rad1, ATRIP, and PCNA antibodies have been described previously (Byun et al., 2005; Lupardus and Cimprich, 2006; Kochaniak et al., 2009). Rabbit polyclonal antibodies against the carboxy-terminal 333 aa of TopBP1 and the 180-kD subunit of Pol-α were raised at Josman, LLC. Purification of recombinant x9-1-1 complex has been previously described (Lupardus and Cimprich, 2006). His6-xPCNA was cloned and expressed as previously described (Chang et al., 2006). Recombinant Pol-α was prepared as previously reported using baculovirus from T. Wang (Stanford University, Stanford, CA; Stadlbauer et al., 1994). Geminin and p27 were produced as previously described and added to extracts at 2 µM (Lupardus et al., 2002).
Immunodepletions, immunoprecipitations, chromatin binding, and transfer
To deplete Pol-α, serum was incubated with nProtein A–Sepharose Fast Flow beads (GE Healthcare) at a 1:1 ratio, and the resulting beads were mixed with extract at a 2:1 extract/bead ratio for three 1-h incubations at 4°C. 10 mg/ml rabbit IgG (Sigma-Aldrich) was incubated with nProtein A–beads for mock depletions at a 1:10 ratio. Immunodepletions of Pol-δ (1:2), Pol-ε (1:5), ATRIP (1:1), TopBP1 (4:1), Rad1 (3:1), and PCNA (3:1) were performed essentially as the aforementioned Pol-α depletion but with the indicated serum/bead coupling ratios (Fukui et al., 2004; Byun et al., 2005; Lupardus and Cimprich, 2006; MacDougall et al., 2007), although for these depletions, the antibody and control IgG were cross-linked to the beads using dimethyl pimelimidate (Sigma-Aldrich). To isolate chromatin-bound proteins, nuclei were isolated by centrifugation of extract through a sucrose cushion, resuspended in chromatin extraction buffer (egg-laying buffer with 0.5% NP-40), and recentrifuged through a second sucrose cushion to obtain the purified chromatin (Lupardus et al., 2002; Zhu et al., 2007). Isolation of TopBP1-free initiated chromatin was performed by diluting replicating extract with a high salt nuclear isolation buffer and then isolating chromatin through a sucrose cushion as with the chromatin-bound protein protocol (Yan and Michael, 2009a). Where indicated, Western blots were developed and quantitated using a FluorChem HD2 MultiImage II (Alpha Innotech) or Photoshop image analysis software (Adobe).
Primer detection and RNase treatment
Samples were added to replication stop buffer (0.5% SDS and 20 mM EDTA) and treated with 0.3 mg/ml proteinase K (Sigma-Aldrich) at 37°C for 30 min. DNA was extracted in 25:24:1 phenol/chloroform/isoamyl alcohol, ethanol precipitated, and resuspended in denaturing dye (95% formamide, 18 mM EDTA, 0.025% SDS, 0.025% xylene cyanol, and 0.025% bromophenol blue). Samples were run at 20 W on a prerun 15% polyacrylamide gel containing 7 M urea and autoradiographed on a Typhoon 9410 (GE Healthcare). Primer lengths were calculated against the DNA ladder and plotted against pixel intensity using ImageQuant software (GE Healthcare). To purify primers for RNase treatment, the radiolabeled bands were excised from the gel and incubated overnight in 150 µl Tris/EDTA on a rotator at 4°C. Primers were subsequently ethanol precipitated, resuspended in Tris/EDTA, and treated with or without an RNaseA/T1 cocktail (0.025 U RNaseA/µl and 1 U RNaseT1/µl; Applied Biosystems) in 10 mM Hepes, pH 7.2, 20 mM NaCl, 0.1% Triton X-100, and 1 mM EDTA for 4 h at 37°C and reanalyzed on a denaturing gel as described above in this section.
DNA structure preparation and structure assays
20 µg/ml of DNA oligonucleotides was annealed to 1 mg/ml M13 mp18 ssDNA and purified as previously described (MacDougall et al., 2007). Oligonucleotides for the 35-nt gap structure have been previously described (MacDougall et al., 2007). The 1800-nt gap structure was made using the oligonucleotides 5′-/5Bio/GCTGATAAATTAATGCCGGAGAGGGTAGCTATTTTTGAGAGATCTACAAAGGCTATCAGGTCATTGCCTGAGAGTCTGGA-3′ and 5′-AGATTCACCAGTCACACGACCAGTAATAAAAGGGACATTCTGGCCAACAGAGATAGAACCCTTCTGACCTGAAAGCGTAA/3Bio/-3′. The four oligonucleotides used to make the structure with two 900-nt gaps were 5′-AGATTCACCAGTCACACGACCAGTAATAAAAGGGACATTCTGGCCAACAGAGATAGAACCCTTCTGACCTGAAAGCGTAA/3Bio/-3′, 5′-/5Bio/GCTAACTCACATTAATTGCGTTGCGCTCACTGCCCGCTTTCCAGTCGGGAAACCTGTCGTGCCAGCTGCATTAATGAATC-3′, 5′-/5Bio/CCAGCAAAATCACCAGTAGCACCATTACCATTAGCAAGGCCGGAAACGTCACCAATGAAACCATCGATAGCAGCACCGTA-3′, and 5′-TAAACAACTTTCAACAGTTTCAGCGGAGTGAGAATAGAAAGGAACAACTAAAGGAATTGCGAATAATAATTTTTTCACGT/3Bio/-3′. The six oligonucleotides used for the 6–80-mer structure were 5′-GCTGATAAATTAATGCCGGAGAGGGTAGCTATTTTTGAGAGATCTACAAAGGCTATCAGGTCATTGCCTGAGAGTCTGGA-3′, 5′-ATTGGGCTTGAGATGGTTTAATTTCAACTTTAATCATTGTGAATTACCTTATGCGATTTTAAGAACTGGCTCATTATACC-3′, 5′-TTAGTACCGCCACCCTCAGAACCGCCACCCTCAGAACCGCCACCCTCAGAGCCACCACCCTCATTTTCAGGGATAGCAAG-3′, 5′-AGACTCCTTATTACGCAGTATGTTAGCAAACGTAGAAAATACATACATAAAGGTGGCAACATATAAAAGAAACGCAAAGA-3′, 5′-GGAAACAGTACATAAATCAATATATGTGAGTGAATAACCTTGCTTCTGTAAATCGTCGCTATTAATTAATTTTCCCTTAG-3′, and 5′-AAAGGAAGGGAAGAAAGCGAAAGGAGCGGGCGCTAGGGCGCTGGCAAGTGTAGCGGTCACGCTGCGCGTAACCACCACAC-3′. DNA oligonucleotides were PAGE purified and either 3′ biotinylated (/3Bio/) or 5′ biotinylated (/5Bio/) where indicated by Integrated DNA Technologies. Structures were preincubated with streptavidin (Sigma-Aldrich) before addition into NPE containing 300 µM aphidicolin as previously described (MacDougall et al., 2007).
Online supplemental material
Fig. S1 shows that replication-dependent primers accumulate on aphidicolin-treated chromatin and that these primers contain a variable 5′ RNA component. Fig. S2 shows that small nascent DNAs are unstable when polymerase activity is inhibited by high concentrations of aphidicolin. Fig. S3 shows that the effect of Pol-δ depletion on Chk1 phosphorylation is dependent on chromatin concentration. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200909105/DC1.
Acknowledgments
This work was supported by an American Cancer Society grant (RSG-05-028-01) and a National Institutes of Health grant (ES016486) to K.A. Cimprich and a Department of Defense Breast Cancer research grant (04-1-0311) and Stanford Graduate Fellowship to C. Van.
Footnotes
Abbreviations used in this paper:
- ATR
- ataxia-telangiectasia mutated and Rad3 related
- ATRIP
- ATR-interacting protein
- dCTP
- deoxy-CTP
- dsDNA
- double-stranded DNA
- HSE
- high speed extract
- LSE
- low speed extract
- NPE
- nucleoplasmic extract
- PCNA
- proliferating cell nuclear antigen
- RPA
- replication protein A
- ssDNA
- single-stranded DNA
References
- Arias E.E., Walter J.C. 2004. Initiation of DNA replication in Xenopus egg extracts. Front. Biosci. 9:3029–3045 10.2741/1457 [DOI] [PubMed] [Google Scholar]
- Arias E.E., Walter J.C. 2007. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 21:497–518 10.1101/gad.1508907 [DOI] [PubMed] [Google Scholar]
- Bermudez V.P., Lindsey-Boltz L.A., Cesare A.J., Maniwa Y., Griffith J.D., Hurwitz J., Sancar A. 2003. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc. Natl. Acad. Sci. USA. 100:1633–1638 10.1073/pnas.0437927100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow J.J., Ge X.Q. 2008. Replication forks, chromatin loops and dormant replication origins. Genome Biol. 9:244 10.1186/gb-2008-9-12-244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman M.L., Schwacha A. 2008. The Mcm2-7 complex has in vitro helicase activity. Mol. Cell. 31:287–293 10.1016/j.molcel.2008.05.020 [DOI] [PubMed] [Google Scholar]
- Branzei D., Foiani M. 2005. The DNA damage response during DNA replication. Curr. Opin. Cell Biol. 17:568–575 10.1016/j.ceb.2005.09.003 [DOI] [PubMed] [Google Scholar]
- Burrows A.E., Elledge S.J. 2008. How ATR turns on: TopBP1 goes on ATRIP with ATR. Genes Dev. 22:1416–1421 10.1101/gad.1685108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun T.S., Pacek M., Yee M.C., Walter J.C., Cimprich K.A. 2005. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19:1040–1052 10.1101/gad.1301205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carty M.P., Lawrence C.W., Dixon K. 1996. Complete replication of plasmid DNA containing a single UV-induced lesion in human cell extracts. J. Biol. Chem. 271:9637–9647 10.1074/jbc.271.16.9637 [DOI] [PubMed] [Google Scholar]
- Chang D.J., Cimprich K.A. 2009. DNA damage tolerance: when it’s OK to make mistakes. Nat. Chem. Biol. 5:82–90 10.1038/nchembio.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D.J., Lupardus P.J., Cimprich K.A. 2006. Monoubiquitination of proliferating cell nuclear antigen induced by stalled replication requires uncoupling of DNA polymerase and mini-chromosome maintenance helicase activities. J. Biol. Chem. 281:32081–32088 10.1074/jbc.M606799200 [DOI] [PubMed] [Google Scholar]
- Cimprich K.A. 2007. Probing ATR activation with model DNA templates. Cell Cycle. 6:2348–2354 [DOI] [PubMed] [Google Scholar]
- Cimprich K.A., Cortez D. 2008. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9:616–627 10.1038/nrm2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacroix S., Wagner J.M., Kobayashi M., Yamamoto K., Karnitz L.M. 2007. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 21:1472–1477 10.1101/gad.1547007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis M.L., Wassarman P.M. 1980. Replication of eukaryotic chromosomes: a close-up of the replication fork. Annu. Rev. Biochem. 49:627–666 10.1146/annurev.bi.49.070180.003211 [DOI] [PubMed] [Google Scholar]
- Ellison V., Stillman B. 2003. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 1:E33 10.1371/journal.pbio.0000033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E.C. 2005. Suffering in silence: the tolerance of DNA damage. Nat. Rev. Mol. Cell Biol. 6:943–953 10.1038/nrm1781 [DOI] [PubMed] [Google Scholar]
- Friedel A.M., Pike B.L., Gasser S.M. 2009. ATR/Mec1: coordinating fork stability and repair. Curr. Opin. Cell Biol. 21:237–244 10.1016/j.ceb.2009.01.017 [DOI] [PubMed] [Google Scholar]
- Fukui T., Yamauchi K., Muroya T., Akiyama M., Maki H., Sugino A., Waga S. 2004. Distinct roles of DNA polymerases delta and epsilon at the replication fork in Xenopus egg extracts. Genes Cells. 9:179–191 10.1111/j.1356-9597.2004.00716.x [DOI] [PubMed] [Google Scholar]
- Garner E., Costanzo V. 2009. Studying the DNA damage response using in vitro model systems. DNA Repair (Amst.). 8:1025–1037 10.1016/j.dnarep.2009.04.015 [DOI] [PubMed] [Google Scholar]
- Hamdan S.M., Richardson C.C. 2009. Motors, switches, and contacts in the replisome. Annu. Rev. Biochem. 78:205–243 10.1146/annurev.biochem.78.072407.103248 [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Tsujimura T., Sugino A., Takisawa H. 2006. The phosphorylated C-terminal domain of Xenopus Cut5 directly mediates ATR-dependent activation of Chk1. Genes Cells. 11:993–1007 10.1111/j.1365-2443.2006.00998.x [DOI] [PubMed] [Google Scholar]
- Heller R.C., Marians K.J. 2006. Replisome assembly and the direct restart of stalled replication forks. Nat. Rev. Mol. Cell Biol. 7:932–943 10.1038/nrm2058 [DOI] [PubMed] [Google Scholar]
- Hubscher U., Maga G., Spadari S. 2002. Eukaryotic DNA polymerases. Annu. Rev. Biochem. 71:133–163 10.1146/annurev.biochem.71.090501.150041 [DOI] [PubMed] [Google Scholar]
- Kochaniak A.B., Habuchi S., Loparo J.J., Chang D.J., Cimprich K.A., Walter J.C., van Oijen A.M. 2009. Proliferating cell nuclear antigen uses two distinct modes to move along DNA. J. Biol. Chem. 284:17700–17710 10.1074/jbc.M109.008706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A., Lee J., Yoo H.Y., Dunphy W.G. 2006. TopBP1 activates the ATR-ATRIP complex. Cell. 124:943–955 10.1016/j.cell.2005.12.041 [DOI] [PubMed] [Google Scholar]
- Kunkel T.A., Burgers P.M. 2008. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 18:521–527 10.1016/j.tcb.2008.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S., Carr A.M. 2005. Checkpoint responses to replication fork barriers. Biochimie. 87:591–602 10.1016/j.biochi.2004.10.020 [DOI] [PubMed] [Google Scholar]
- Langston L.D., O’Donnell M. 2006. DNA replication: keep moving and don’t mind the gap. Mol. Cell. 23:155–160 10.1016/j.molcel.2006.05.034 [DOI] [PubMed] [Google Scholar]
- Lee J., Kumagai A., Dunphy W.G. 2007. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J. Biol. Chem. 282:28036–28044 10.1074/jbc.M704635200 [DOI] [PubMed] [Google Scholar]
- Liu E., Lee A.Y., Chiba T., Olson E., Sun P., Wu X. 2007. The ATR-mediated S phase checkpoint prevents rereplication in mammalian cells when licensing control is disrupted. J. Cell Biol. 179:643–657 10.1083/jcb.200704138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M., Foiani M., Sogo J.M. 2006. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 21:15–27 10.1016/j.molcel.2005.11.015 [DOI] [PubMed] [Google Scholar]
- Luciani M.G., Oehlmann M., Blow J.J. 2004. Characterization of a novel ATR-dependent, Chk1-independent, intra-S-phase checkpoint that suppresses initiation of replication in Xenopus. J. Cell Sci. 117:6019–6030 10.1242/jcs.01400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupardus P.J., Cimprich K.A. 2006. Phosphorylation of Xenopus Rad1 and Hus1 defines a readout for ATR activation that is independent of Claspin and the Rad9 carboxy terminus. Mol. Biol. Cell. 17:1559–1569 10.1091/mbc.E05-09-0865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupardus P.J., Byun T., Yee M.C., Hekmat-Nejad M., Cimprich K.A. 2002. A requirement for replication in activation of the ATR-dependent DNA damage checkpoint. Genes Dev. 16:2327–2332 10.1101/gad.1013502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupardus P.J., Van C., Cimprich K.A. 2007. Analyzing the ATR-mediated checkpoint using Xenopus egg extracts. Methods. 41:222–231 10.1016/j.ymeth.2006.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall C.A., Byun T.S., Van C., Yee M.C., Cimprich K.A. 2007. The structural determinants of checkpoint activation. Genes Dev. 21:898–903 10.1101/gad.1522607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka J., Binz S.K., Wold M.S., Burgers P.M. 2006. Replication protein A directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J. Biol. Chem. 281:27855–27861 10.1074/jbc.M605176200 [DOI] [PubMed] [Google Scholar]
- McGarry T.J., Kirschner M.W. 1998. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 93:1043–1053 10.1016/S0092-8674(00)81209-X [DOI] [PubMed] [Google Scholar]
- Méchali M., Harland R.M. 1982. DNA synthesis in a cell-free system from Xenopus eggs: priming and elongation on single-stranded DNA in vitro. Cell. 30:93–101 10.1016/0092-8674(82)90015-0 [DOI] [PubMed] [Google Scholar]
- Michael W.M., Ott R., Fanning E., Newport J. 2000. Activation of the DNA replication checkpoint through RNA synthesis by primase. Science. 289:2133–2137 10.1126/science.289.5487.2133 [DOI] [PubMed] [Google Scholar]
- Mimura S., Masuda T., Matsui T., Takisawa H. 2000. Central role for cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells. 5:439–452 10.1046/j.1365-2443.2000.00340.x [DOI] [PubMed] [Google Scholar]
- Mordes D.A., Glick G.G., Zhao R., Cortez D. 2008. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 22:1478–1489 10.1101/gad.1666208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A.W. 1991. Cell cycle extracts. Methods Cell Biol. 36:581–605 10.1016/S0091-679X(08)60298-8 [DOI] [PubMed] [Google Scholar]
- Navas T.A., Sanchez Y., Elledge S.J. 1996. RAD9 and DNA polymerase epsilon form parallel sensory branches for transducing the DNA damage checkpoint signal in Saccharomyces cerevisiae. Genes Dev. 10:2632–2643 10.1101/gad.10.20.2632 [DOI] [PubMed] [Google Scholar]
- Nethanel T., Reisfeld S., Dinter-Gottlieb G., Kaufmann G. 1988. An Okazaki piece of simian virus 40 may be synthesized by ligation of shorter precursor chains. J. Virol. 62:2867–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny S.A., Gordenin D.A., Stith C.M., Burgers P.M., Kunkel T.A. 2008. Division of labor at the eukaryotic replication fork. Mol. Cell. 30:137–144 10.1016/j.molcel.2008.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M., Walter J.C. 2004. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 23:3667–3676 10.1038/sj.emboj.7600369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrilla-Castellar E.R., Karnitz L.M. 2003. Cut5 is required for the binding of Atr and DNA polymerase alpha to genotoxin-damaged chromatin. J. Biol. Chem. 278:45507–45511 10.1074/jbc.C300418200 [DOI] [PubMed] [Google Scholar]
- Paulsen R.D., Cimprich K.A. 2007. The ATR pathway: fine-tuning the fork. DNA Repair (Amst.). 6:953–966 10.1016/j.dnarep.2007.02.015 [DOI] [PubMed] [Google Scholar]
- Pursell Z.F., Isoz I., Lundström E.B., Johansson E., Kunkel T.A. 2007. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 317:127–130 10.1126/science.1144067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakawa N., Fukui T., Waga S. 2006. Accumulation of FFA-1, the Xenopus homolog of Werner helicase, and DNA polymerase delta on chromatin in response to replication fork arrest. J. Biochem. 140:95–103 10.1093/jb/mvj130 [DOI] [PubMed] [Google Scholar]
- Segurado M., Tercero J.A. 2009. The S-phase checkpoint: targeting the replication fork. Biol. Cell. 101:617–627 10.1042/BC20090053 [DOI] [PubMed] [Google Scholar]
- Shechter D., Costanzo V., Gautier J. 2004a. ATR and ATM regulate the timing of DNA replication origin firing. Nat. Cell Biol. 6:648–655 10.1038/ncb1145 [DOI] [PubMed] [Google Scholar]
- Shechter D., Costanzo V., Gautier J. 2004b. Regulation of DNA replication by ATR: signaling in response to DNA intermediates. DNA Repair (Amst.). 3:901–908 10.1016/j.dnarep.2004.03.020 [DOI] [PubMed] [Google Scholar]
- Shiotani B., Zou L. 2009. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol. Cell. 33:547–558 10.1016/j.molcel.2009.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo J.M., Lopes M., Foiani M. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 297:599–602 10.1126/science.1074023 [DOI] [PubMed] [Google Scholar]
- Stadlbauer F., Brueckner A., Rehfuess C., Eckerskorn C., Lottspeich F., Förster V., Tseng B.Y., Nasheuer H.P. 1994. DNA replication in vitro by recombinant DNA-polymerase-alpha-primase. Eur. J. Biochem. 222:781–793 10.1111/j.1432-1033.1994.tb18925.x [DOI] [PubMed] [Google Scholar]
- Svoboda D.L., Vos J.M. 1995. Differential replication of a single, UV-induced lesion in the leading or lagging strand by a human cell extract: fork uncoupling or gap formation. Proc. Natl. Acad. Sci. USA. 92:11975–11979 10.1073/pnas.92.26.11975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H., Pasero P. 2007. Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair (Amst.). 6:900–913 10.1016/j.dnarep.2007.02.004 [DOI] [PubMed] [Google Scholar]
- Tutter A.V., Walter J.C. 2006. Chromosomal DNA replication in a soluble cell-free system derived from Xenopus eggs. Methods Mol. Biol. 322:121–137 10.1007/978-1-59745-000-3_9 [DOI] [PubMed] [Google Scholar]
- Waga S., Stillman B. 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67:721–751 10.1146/annurev.biochem.67.1.721 [DOI] [PubMed] [Google Scholar]
- Waga S., Masuda T., Takisawa H., Sugino A. 2001. DNA polymerase epsilon is required for coordinated and efficient chromosomal DNA replication in Xenopus egg extracts. Proc. Natl. Acad. Sci. USA. 98:4978–4983 10.1073/pnas.081088798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J., Newport J.W. 1997. Regulation of replicon size in Xenopus egg extracts. Science. 275:993–995 10.1126/science.275.5302.993 [DOI] [PubMed] [Google Scholar]
- Walter J., Newport J. 2000. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol. Cell. 5:617–627 10.1016/S1097-2765(00)80241-5 [DOI] [PubMed] [Google Scholar]
- Walter J., Sun L., Newport J. 1998. Regulated chromosomal DNA replication in the absence of a nucleus. Mol. Cell. 1:519–529 10.1016/S1097-2765(00)80052-0 [DOI] [PubMed] [Google Scholar]
- Yan S., Michael W.M. 2009a. TopBP1 and DNA polymerase-α directly recruit the 9-1-1 complex to stalled DNA replication forks. J. Cell Biol. 184:793–804 10.1083/jcb.200810185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S., Michael W.M. 2009b. TopBP1 and DNA polymerase alpha-mediated recruitment of the 9-1-1 complex to stalled replication forks: implications for a replication restart-based mechanism for ATR checkpoint activation. Cell Cycle. 8:2877–2884 [DOI] [PubMed] [Google Scholar]
- Yan S., Lindsay H.D., Michael W.M. 2006. Direct requirement for Xmus101 in ATR-mediated phosphorylation of Claspin bound Chk1 during checkpoint signaling. J. Cell Biol. 173:181–186 10.1083/jcb.200601076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanow S.K., Gold D.A., Yoo H.Y., Dunphy W.G. 2003. Xenopus Drf1, a regulator of Cdc7, displays checkpoint-dependent accumulation on chromatin during an S-phase arrest. J. Biol. Chem. 278:41083–41092 10.1074/jbc.M307144200 [DOI] [PubMed] [Google Scholar]
- Yao N.Y., O’Donnell M. 2009. Replisome structure and conformational dynamics underlie fork progression past obstacles. Curr. Opin. Cell Biol. 21:336–343 10.1016/j.ceb.2009.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H.Y., Kumagai A., Shevchenko A., Shevchenko A., Dunphy W.G. 2004. Adaptation of a DNA replication checkpoint response depends upon inactivation of Claspin by the Polo-like kinase. Cell. 117:575–588 10.1016/S0092-8674(04)00417-9 [DOI] [PubMed] [Google Scholar]
- You Z., Kong L., Newport J. 2002. The role of single-stranded DNA and polymerase alpha in establishing the ATR, Hus1 DNA replication checkpoint. J. Biol. Chem. 277:27088–27093 10.1074/jbc.M204120200 [DOI] [PubMed] [Google Scholar]
- Zhu W., Ukomadu C., Jha S., Senga T., Dhar S.K., Wohlschlegel J.A., Nutt L.K., Kornbluth S., Dutta A. 2007. Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev. 21:2288–2299 10.1101/gad.1528707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L. 2007. Single- and double-stranded DNA: building a trigger of ATR-mediated DNA damage response. Genes Dev. 21:879–885 10.1101/gad.1550307 [DOI] [PubMed] [Google Scholar]
- Zou L., Elledge S.J. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 300:1542–1548 10.1126/science.1083430 [DOI] [PubMed] [Google Scholar]
- Zou L., Liu D., Elledge S.J. 2003. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. USA. 100:13827–13832 10.1073/pnas.2336100100 [DOI] [PMC free article] [PubMed] [Google Scholar]