αE-catenin has cell–cell contact–dependent and –independent functions in regulating actin and membrane dynamics.
Abstract
αE-catenin binds the cell–cell adhesion complex of E-cadherin and β-catenin (β-cat) and regulates filamentous actin (F-actin) dynamics. In vitro, binding of αE-catenin to the E-cadherin–β-cat complex lowers αE-catenin affinity for F-actin, and αE-catenin alone can bind F-actin and inhibit Arp2/3 complex–mediated actin polymerization. In cells, to test whether αE-catenin regulates actin dynamics independently of the cadherin complex, the cytosolic αE-catenin pool was sequestered to mitochondria without affecting overall levels of αE-catenin or the cadherin–catenin complex. Sequestering cytosolic αE-catenin to mitochondria alters lamellipodia architecture and increases membrane dynamics and cell migration without affecting cell–cell adhesion. In contrast, sequestration of cytosolic αE-catenin to the plasma membrane reduces membrane dynamics. These results demonstrate that the cytosolic pool of αE-catenin regulates actin dynamics independently of cell–cell adhesion.
Introduction
The balance between cell–cell adhesion and cell migration is critical in development and is altered in disease states including metastatic cancers. During development, cells migrate to specific sites and then, upon contact with other cells, become stationary and differentiate into tissues (Baum et al., 2008). Subsequently, strong cell–cell adhesion is important in the maintenance of tissue integrity (Gumbiner, 2005; Halbleib and Nelson, 2006). However, changes in cell–cell adhesion complexes can lead to the reinitiation of cell migration during cell turnover or wound healing (Thiery and Sleeman, 2006) or enable metastatic cells to disperse to distant organs (for review see Benjamin and Nelson, 2008). Mechanisms linking cell–cell adhesion to the regulation of cell migration are poorly understood.
Cell–cell adhesion between migratory cells results in immediate and long-term changes in actin-based membrane dynamics (Adams et al., 1996; Krendel and Bonder, 1999; Vasioukhin et al., 2000; Yamada and Nelson, 2007). Membrane activity at the leading edge of cells, driven largely by Arp2/3 complex–mediated nucleation of branched filamentous actin (F-actin) networks, promotes cell movement (Pollard and Borisy, 2003) and is involved in the initiation of intercellular contacts (Ehrlich et al., 2002; Kovacs et al., 2002). After contact initiation in simple epithelial cells, the actin network associated with the plasma membrane is reorganized (Krendel and Bonder, 1999; Yamada and Nelson, 2007) and eventually forms bundled filaments oriented parallel to the lateral contact between cells (Hirokawa and Heuser, 1981). These changes in actin organization coincide with dampening of membrane dynamics and cell migration and the establishment of strong cell–cell adhesion (Ehrlich et al., 2002).
Members of the cadherin family of Ca2+-dependent cell–cell adhesion proteins have a major role in the initiation of early cell–cell contacts (Halbleib and Nelson, 2006) and regulation of actin cytoskeleton organization (Kobielak and Fuchs, 2004). In epithelial cells, E-cadherin binds to β-catenin (β-cat), which in turn binds to αE-catenin (Aberle et al., 1994). αE-catenin also binds and bundles actin filaments (Rimm et al., 1995), and therefore, it was assumed that αE-catenin statically links the cadherin–catenin complex to the actin cytoskeleton (Gates and Peifer, 2005). Additionally, other actin-associated proteins bind αE-catenin and could provide an indirect link to the actin cytoskeleton. These proteins include some that bind actin directly, such as vinculin (Watabe-Uchida et al., 1998), afadin (Tachibana et al., 2000; Pokutta et al., 2002), EPLIN (Abe and Takeichi, 2008), and ZO-1 (Itoh et al., 1997), and others that regulate actin polymerization, such as formins/mDia (Kobielak et al., 2004) and the Arp2/3 complex (Kovacs et al., 2002). However, in almost all cases, the molecular affinities and stoichiometries of these additional proteins in the cadherin–catenin complex have not been defined.
Studies of αE-catenin in vivo have also suggested functions independent of cadherin-mediated cell–cell adhesion. Depletion of αE-catenin from mouse epidermis (Vasioukhin et al., 2001) or neural progenitor cells of the developing cortex (Lien et al., 2006) reduces cadherin-based cell–cell contacts and induces formation of hyperplastic cell masses. Similarly, cultured αE-catenin–null keratinocytes have increased cell proliferation and migration (Vasioukhin et al., 2001). Furthermore, loss of both αE-catenin and E-cadherin in some human cancers correlates with worse patient prognosis than loss of either protein alone (Matsui et al., 1994; Mialhe et al., 1997; Kim et al., 2002; Scholten et al., 2006; Setoyama et al., 2007; for review see Benjamin and Nelson, 2008).
In vitro binding studies with purified proteins provided direct evidence that αE-catenin functions independent of the cadherin–catenin complex (Drees et al., 2005). Key findings were that binding to β-cat weakens the affinity of αE-catenin for F-actin and that the oligomeric state of αE-catenin regulates its affinity for F-actin; αE-catenin monomer binds β-cat, whereas αE-catenin homodimer preferentially binds to actin filaments and inhibits Arp2/3 complex–mediated F-actin polymerization (Drees et al., 2005; Yamada et al., 2005). Thus, the αE-catenin species with the strongest affinity for F-actin, the homodimer, is not bound to the cadherin complex. These results raised the possibility that the cadherin-free, cytosolic pool of αE-catenin could regulate actin-based membrane dynamics and potentially cell migration independently of the cadherin–catenin complex at the plasma membrane. In this study, we test this hypothesis by depleting the cytosolic pool of αE-catenin in simple epithelial (MDCK) cells and analyzing the effects on cell–cell adhesion, actin-based membrane dynamics, and cell migration. Our results identify cell–cell contact–dependent and –independent functions of αE-catenin in regulating actin-based membrane dynamics that control cell–cell adhesion and cell migration.
Results
αE-catenin exists as a monomer, homodimer, and a heterodimer with β-cat in MDCK cells
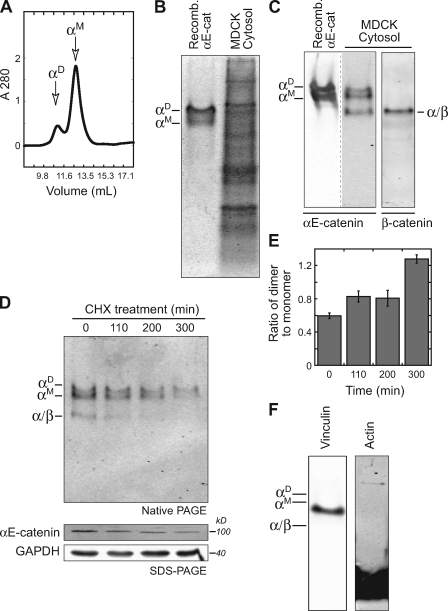
We first sought to characterize the distribution and molecular form (monomer or homodimer) of αE-catenin in the cytoplasm and membrane of MDCK cells. Recombinant αE-catenin expressed in bacteria eluted in two peaks from an S200 gel filtration column (Fig. 1 A) that were identified as monomer (αM) and homodimer (αD) by multiangle light scattering (Drees et al., 2005). Analysis of these peak fractions by native PAGE revealed two distinct protein bands: a faster migrating monomer and slower migrating homodimer (Fig. 1, B and C). To identify αE-catenin monomers and homodimers in normal epithelial cells, MDCK cell cytosol (supernatant) was separated from membranes and cytoskeleton (pellet) by high speed centrifugation and analyzed by native PAGE to maintain protein complexes (Fig. 1 B, lane 2) and immunoblotted for αE-catenin (Fig. 1 C, lane 2) and β-cat (Fig. 1 C, lane 3). Three prominent protein bands containing αE-catenin were detected: a pair of slower migrating bands that comigrated with recombinant αE-catenin monomer (αM) and homodimer (αD) and a faster migrating band that contained β-cat (α/β; referred to as heterodimer; Fig. 1 C). The turnover of these forms was analyzed after protein synthesis was halted with cyclohexamide; the monomer and heterodimer were degraded rapidly, and the homodimer was slightly more stable (Fig. 1, D and E). Several other proteins that bind αE-catenin, including the frequently cited vinculin (Watabe-Uchida et al., 1998) and actin (Rimm et al., 1995), did not comigrate with any of these three αE-catenin bands (Fig. 1 F), and binding to other high molecular weight proteins such as ZO-1 and afadin would be expected to cause a significant change in the electrophoretic mobility of αE-catenin monomer and homodimer. These results indicate that αE-catenin in MDCK cytosol comprises a monomer, homodimer, and heterodimer with β-cat.
Figure 1.
αE-catenin exists as a monomer and homodimer in MDCK cytosol. (A) Recombinant αE-catenin fractions collected from S200 gel filtration. The two peaks correspond to αE-catenin monomer (αM) and homodimer (αD). (B) Coomassie-stained native PAGE. Purified recombinant αE-catenin homodimer and monomer (αD/αM) served as markers for MDCK cytosol. (C) Recombinant αE-catenin homodimer and monomer and MDCK cell cytosol were run on a native PAGE gel and blotted for αE-catenin (left) and β-cat (right). αE-catenin monomer (αM), homodimer (αD), and heterodimer with β-cat (α/β) are marked. Note that recombinant αE-catenin and MDCK cytosol were run on the same gel but shown at different exposures, which are separated by a dotted line. (D, top) MDCK cytosol from cyclohexamide (CHX)-treated cells was separated by native PAGE and Western blotted for αE-catenin. (D, bottom) Total cell lysates were also separated by SDS-PAGE and blotted for αE-catenin and GAPDH. (E) Ratio of αE-catenin homodimer to monomer from experiment shown in D was determined by measuring band immunofluorescence intensity. (F) Native PAGE of MDCK cytosol blotted for vinculin and actin. Positions of αM, αD, and α/β complexes are marked for reference. Error bars represent SEM from two independent experiments.
We attempted to determine whether αE-catenin monomer and homodimer associated with the actin cytoskeleton by analyzing changes in the amount of αE-catenin forms in the cytosolic pool upon stabilizing (jasplakinolide) or depolymerizing (cytochalasin-B) the F-actin cytoskeleton. However, these conditions yielded only small and variable changes (<20%) in the amount of αE-catenin monomer and homodimer in the cytosol (unpublished data), indicating that the pool of αE-catenin bound directly to F-actin in MDCK cells is either relatively small or is unaffected by changes in the state of F-actin polymerization.
We could not determine the molecular forms of αE-catenin associated with the cadherin complex in the plasma membrane fraction pelleted by high speed centrifugation. Detergent extraction, required to release the cadherin complex from the insoluble pellet for analysis, alters protein migration in native PAGE; we also do not have a purified ternary complex of full-length E-cadherin bound to the αE-catenin/β-cat heterodimer to run as a standard. However, we assume that αE-catenin is bound to the cadherin complex as a monomer because the binding interfaces for αE-catenin homodimerization and heterodimerization with β-cat overlap almost completely (Pokutta and Weis, 2000). Thus, at a structural level, these two forms of αE-catenin are mutually exclusive.
Changing the distribution of different pools of endogenous αE-catenin in MDCK cells
The aforementioned results show that αE-catenin monomer and homodimer are not an artifact of purified recombinant αE-catenin but exist in the cytosol of MDCK cells along with a membrane-associated pool bound to E-cadherin (likely monomer) and possibly a small actin-associated pool. To test the hypothesis that the cytosolic pool of αE-catenin regulates actin dynamics independently of cadherin, we sought to deplete the cytosolic pool of αE-catenin without affecting the cadherin-bound pool.
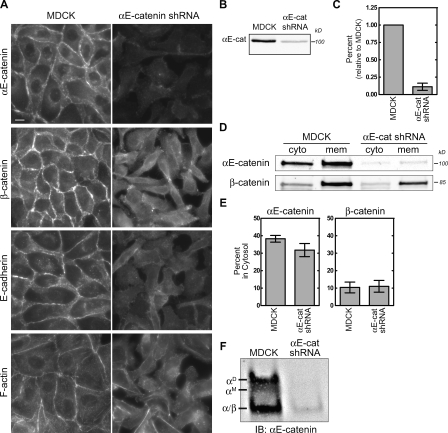
Stable expression of a short hairpin RNA (shRNA) against αE-catenin in MDCK cells (referred to as αE-cat shRNA) successfully depleted αE-catenin levels, which disrupted cell–cell adhesion and altered cell morphology (Fig. 2 A). Western blot analysis revealed that αE-catenin levels were reduced to <20% of control (Fig. 2, B and C). However, both the cytosolic and membrane pools of αE-catenin were reduced (Fig. 2, D and E), and both αE-catenin monomer and homodimer were reduced by ∼80% compared with control (Fig. 2 F). Therefore, we devised methods to reduce only the cytosolic pool of αE-catenin by targeting αE-catenin to mitochondria or to membranes without affecting either the overall level or cadherin-bound pool of αE-catenin.
Figure 2.
shRNA-mediated knockdown of αE-catenin depletes membrane and cytosolic pools. (A) Control MDCK and stable αE-catenin knockdown MDCK cells (αE-cat shRNA) were stained for αE-catenin, β-cat, E-cadherin, and F-actin. Bar, 10 µm. (B) Whole cell lysates from MDCK and αE-cat shRNA cells separated by SDS-PAGE and blotted for αE-catenin. (C) Band intensities shown in B were measured, normalized to MDCK control levels, and plotted. (D) Cytosol (cyto) and membrane (mem) fractions from MDCK and αE-cat shRNA cells were separated by SDS-PAGE and blotted for αE-catenin and β-cat. (E) Individual band intensities shown in D were measured, and percent distribution was plotted. (F) Native PAGE of MDCK and αE-catenin knockdown cytosol blotted for αE-catenin. αE-catenin monomer (αM), homodimer (αD), and heterodimer with β-cat (α/β) are marked. IB, immunoblot. Error bars represent SEM from at least three independent experiments.
Targeting αE-catenin to mitochondria
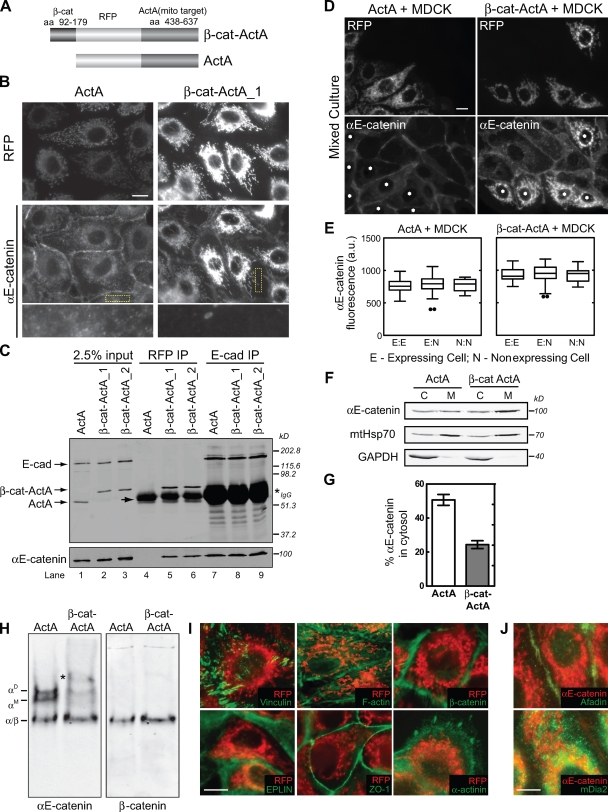
Endogenous αE-catenin was targeted to mitochondria by expressing a chimeric protein comprising a minimal αE-catenin–binding region of β-cat (aa 92–179) fused to monomeric RFP (mRFP) and the C-terminal region (aa 438–637) of the mitochondrial binding domain of the Listeria monocytogenes protein ActA (referred to as β-cat–ActA; Fig. 3 A; Niebuhr et al., 1997); this domain of ActA lacks the actin-, Ena/VASP-, and Arp2/3-binding motifs (Niebuhr et al., 1997). In vitro binding studies show that binding of α-catenin to β-cat is enhanced >10-fold when β-cat is bound to cadherin (unpublished data); thus, this β-cat fragment should bind cytosolic αE-catenin but not compete with αE-catenin in the cadherin–β-cat complex on the plasma membrane. A similar construct lacking the αE-catenin–binding domain of β-cat was used as a control (referred to as ActA; Fig. 3 A).
Figure 3.
Targeting endogenous αE-catenin to mitochondria depletes the cytosolic αE-catenin pool. (A) Schematic of αE-catenin mitochondrial-targeting constructs. β-Cat–ActA, minimal αE-catenin–binding domain of β-cat (aa 92–179) fused to mRFP and the mitochondrial targeting region (aa 436–637) of ActA. ActA, control construct containing mRFP fused to aa 436–637 of ActA. (B) RFP and αE-catenin staining of β-cat–ActA and ActA MDCK cell lines. Yellow boxes highlight magnified regions shown in the bottom row. (C) Coimmunoprecipitation of αE-catenin from β-cat–ActA and ActA lysates using either RFP (lanes 4–6) or E-cadherin (lanes 7–9) antibodies. Blots probed for E-cadherin and RFP (top) and αE-catenin (bottom) are shown. Note that ActA migrates slightly faster than the IgG heavy chain (arrow). Asterisk marks cross reacting IgG heavy chain. β-Cat–ActA_1 and β-cat–ActA_2 were two independent, stable cell lines. (D) ActA- or β-cat–ActA-expressing MDCK cells (asterisks) mixed with wild-type MDCK cells and stained for αE-catenin. (E) The mean level of αE-catenin at cell–cell contacts was quantified between two expressing cells (E/E), an expressing and nonexpressing MDCK cell (E/N), and two nonexpressing MDCK cells (N/N). Results are presented in a box and whisker format. The ends of the box mark the upper and lower quartiles, the horizontal line in the box indicates the median, and the whiskers outside the box extend to the highest and lowest value within 1.5 times the interquartile range. Outliers are represented as dots. About 30 cell–cell contacts for each condition were measured. (F) Cytosol (C) and membrane (M) fractions from β-cat–ActA and ActA stable cells were separated by SDS-PAGE and blotted for αE-catenin, mtHsp70, and GAPDH. (G) αE-catenin band intensities shown in F were measured, and the percentage of αE-catenin in the cytosol fraction is graphed. (H) Native PAGE of ActA and β-cat–ActA cytosol blotted for αE-catenin (left) and β-cat (right). An additional slow-migrating band (asterisk) present in β-cat–ActA cytosol cross reacted with RFP (not depicted) and is presumed to be an αE-catenin/β-cat–ActA heterodimer synthesized in the cytoplasm that binds posttranslationally to mitochondria. (I) Immunofluorescence of EPLIN, ZO-1, α-actinin, β-cat, F-actin, vinculin (all shown in green), and RFP (red) in β-cat–ActA cells. (J) Immunofluorescence of αE-catenin (red) and afadin and mDia2 (green) in β-cat–ActA cells. Bars, 10 µm. Error bars represent SEM from three independent experiments.
Stable MDCK cell lines expressing these constructs were generated, and two β-cat–ActA clones and one ActA clone were chosen randomly. These clones were used in parallel, but data from only one are presented as being representative. The levels of E-cadherin, αE-catenin, β-cat, actin, tubulin, and mtHsp70 (mitochondrial heat shock protein 70) were similar in control cells and cell clones expressing β-cat–ActA (Fig. S1 A). The growth rates of both cell types were also similar (Fig. S1 B).
Immunofluorescence microscopy of MDCK cell clones showed that β-cat–ActA colocalized with mtHsp70 (unpublished data) and recruited αE-catenin to mitochondria (Fig. 3 B). The ActA control construct also localized to mitochondria but did not recruit αE-catenin (Fig. 3 B). To further verify αE-catenin binding to β-cat–ActA and not ActA, mRFP was immunoprecipitated from Triton X-100–extracted membranes. αE-catenin coimmunoprecipitated with β-cat–ActA but not ActA (Fig. 3 C, lanes 4–6). In contrast, similar amounts of αE-catenin coimmunoprecipitated with E-cadherin from both β-cat–ActA and ActA cells (Fig. 3 C, lanes 7–9). Thus, sequestering cytosolic αE-catenin to mitochondria did not affect the plasma membrane (E-cadherin bound) pool of αE-catenin. Consistent with this, quantification of αE-catenin immunofluorescence in mixed populations of ActA or β-cat–ActA (expressing) and wild-type (nonexpressing) MDCK cells revealed no statistically significant differences in αE-catenin intensity at cell–cell contacts between any combination of these cells (Fig. 3, D and E).
Because both the overall level of αE-catenin and the amount of membrane-bound αE-catenin were unaltered in β-cat–ActA cells, it was likely that the cytosolic pool of αE-catenin had been sequestered to mitochondria. Indeed, immunofluorescence revealed very low cytoplasmic staining of αE-catenin in β-cat–ActA cells compared with ActA cells (Fig. 3 B, bottom). To quantify this difference, postnuclear supernatants from ActA and β-cat–ActA cells were separated into membrane (pellet) and cytosol fractions: ∼50% of αE-catenin was present in the cytosol of ActA cells compared with ∼25% in β-cat–ActA cells (Fig. 3, F and G). Importantly, native PAGE of cytosolic proteins showed that the combined amount of αE-catenin monomer and homodimer was reduced by ∼60–70% (n = 6) from the cytosol of β-cat–ActA cells (Fig. 3 H, left; and Fig. S1 C). Because monomer and homodimer separate poorly from each other in native PAGE, it was not possible to measure their relative levels accurately. The amount of heterodimer appeared slightly increased in β-cat–ActA cytosol (Fig. 3 H, right; and Fig. S1 C).
We examined whether other reported αE-catenin–binding partners were cosequestered with αE-catenin to mitochondria in β-cat–ActA cells. Immunofluorescence of vinculin, ZO-1, α-actinin, F-actin, β-cat, EPLIN, afadin, and mDia2 (and mDia1, not depicted) revealed that none of these proteins colocalized with αE-catenin on mitochondria (Fig. 3, I and J; and Fig. S2); note that the space-filling, punctate-staining pattern of mDia2 (and mDia1, not depicted) throughout the cytoplasm and in the perinuclear region was similar in ActA and β-cat–ActA cells (Fig. S2 G). Furthermore, we did not detect actin assembly or actin filaments associated with mitochondria in live cells expressing actin-GFP (unpublished data). Thus, the observed phenotypes in β-cat–ActA cells are caused by the sequestration of cytosolic αE-catenin to the mitochondria and not to the mislocalization of these actin-binding proteins.
Overall, these data show that in β-cat–ActA cells, αE-catenin is bound to the plasma membrane by the native E-cadherin–β-cat complex and sequestered to mitochondria by β-cat–ActA. Although the amount of αE-catenin associated with E-cadherin at the plasma membrane was unchanged compared with control cells, the cytosolic pool of αE-catenin monomer and homodimer was depleted.
Effects of sequestering the cytosolic pool of αE-catenin on cell–cell adhesion and cell migration
The effects of sequestering the cytosolic pool of αE-catenin to mitochondria on cell–cell adhesion and cell migration were tested in β-cat–ActA cells. For comparison, we also examined αE-catenin knockdown cells in which both the membrane and cytosolic pools of αE-catenin were depleted.
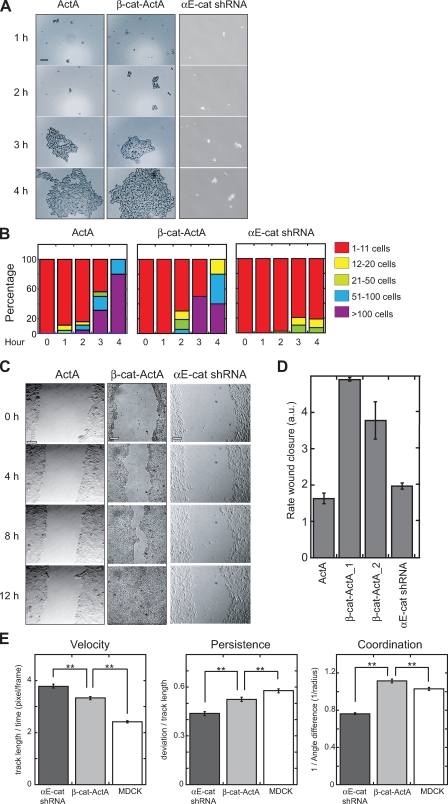
The rate and strength of cell–cell adhesion was measured with a hanging drop assay (Fig. 4 A). β-cat–ActA and ActA cells formed trituration-resistant cell aggregates at similar rates (Fig. 4 B). In contrast, αE-catenin knockdown cells failed to form even small cell clusters (Fig. 4, A and B), indicating a significant loss of cell–cell adhesion.
Figure 4.
Sequestration of cytosolic αE-catenin to mitochondria increases cell migration but does not disrupt cell–cell adhesion. (A) ActA, β-cat–ActA, and αE-cat shRNA cell suspensions were triturated, fixed, and imaged at the indicated times. (B) Cell clusters were binned into the following classes: 1–11, 12–20, 21–50, 51–100, or >100 cells, and the percentage of cell clusters in a bin class at a given time point is shown. Data shown are a representative example from two independent experiments. (C) Confluent monolayers were scratched (0 h) and imaged over time to track wound closure. (D) Rate of wound closure was measured and plotted as the mean width of the wound over time and defined in arbitrary units (au). Data shown are a representative example from two independent experiments. (E) 250–400 individual cells from each cell type were tracked for 2 h during wound closure. Velocity is defined as length of track/time. Coordination is defined as 1/radius (the inverse of the mean difference of angle between a target cell and neighbors; the lowest possible value is 2/π [∼0.64], and the highest is infinity). Persistence is defined as deviation/track length (the lowest possible value is 0, and the highest is 1). Error bars indicate SEM. **, P < 0.002 (Mann-Whitney test). Bars, 100 µm.
Next, we used a wound-healing assay to examine cell sheet migration (Fig. 4 C). The rate of wound closure was similar for ActA and control MDCK cells (Fig. 4 D). In contrast, β-cat–ActA cells closed the wound about two to three times faster. Because no defects in growth rate (Fig. S1 B) or cell–cell adhesion (Fig. 4, A and B) were observed for β-cat–ActA cells, the increased rate of wound closure was not caused by filling in by increased cell division or loss of cell–cell adhesion. We ruled out effects of defective cell polarization on altered wound healing as the centrosome, a marker of polarized cell migration, polarized normally relative to the wound edge (unpublished data).
Surprisingly, αE-catenin knockdown cells closed the wound at a rate similar to control cells (Fig. 4 D). This result was not caused by decreased cell division, as αE-catenin knockdown did not significantly affect cell proliferation in MDCK cells (Fig. S3). Close inspection of cell movements indicated that cells were moving around rapidly but not in a directed manner. Indeed, automated single-cell tracking of scratched monolayers revealed that αE-catenin knockdown cells migrated even faster than β-cat–ActA cells (Fig. 4 E). However, migration of αE-catenin knockdown cells was statistically less persistent and uncoordinated compared with both β-cat–ActA and MDCK control cells (see Materials and methods; Fig. 4 E), thereby counteracting the increased migration rate of these cells.
Changes in plasma membrane dynamics by sequestering the cytosolic pool of αE-catenin to mitochondria
Next, we examined whether increased cell migration of β-cat–ActA and αE-catenin knockdown cells reflected increased plasma membrane dynamics as a result of changes in actin dynamics. Kymograph analysis of lamellipodial dynamics in β-cat–ActA and ActA cells showed that ActA control cells produced thin, undynamic lamellipodia, whereas β-cat–ActA cells had highly dynamic lamellipodia with many rapid protrusions and retractions (Fig. 5 A). Both the velocity and frequency of membrane protrusions were significantly higher in β-cat–ActA cells than in ActA control cells (Fig. 5 B). αE-catenin knockdown caused a similar increase in protrusion velocity (Fig. 5, C and D) but a less-marked increase in protrusion number relative to MDCK controls.
Figure 5.
Cytosolic αE-catenin regulates actin-dependent membrane dynamics. (A and B) Representative kymographs of EGFP-actin in membrane protrusions from ActA and β-cat–ActA cells. (A) 2-pixel-wide kymographs were compiled parallel to protrusion direction over 10 min. (B) Number of protrusions per 10-min window (left) and the mean speed of protrusions (right) were measured in ActA and β-cat–ActA cells and shown using a box and whisker plot. 11 cells with 30 protrusions in ActA and 11 cells with 50 protrusions in β-cat–ActA cells were quantified. (C and D) Representative kymographs of membrane protrusions from MDCK and αE-cat shRNA cells (C) and quantification (D). 75 protrusions from 12 MDCK cells and 116 protrusions from 15 αE-cat shRNA cells were measured. Results are presented in a box and whisker format. The ends of the box mark the upper and lower quartiles, the horizontal line in the box indicates the median, and the whiskers outside the box extend to the highest and lowest value within 1.5 times the interquartile range. Outliers are represented as dots. *, P < 0.02; ***, P < 0.0002 (Mann-Whitney test). Bars, 5 µm.
These results show that altering the distribution of αE-catenin in MDCK cells has broad effects on plasma membrane dynamics. Selective depletion of cytosolic αE-catenin in β-cat–ActA cells increased plasma membrane dynamics and the rate of cell migration without affecting cell–cell adhesion. Depleting both cytosolic and membrane pools of αE-catenin by shRNA knockdown also increased plasma membrane dynamics and the rate of cell migration but reduced coordinated cell movements because of a lack of cadherin-mediated cell–cell adhesion.
Targeting αE-catenin to the plasma membrane independently of E-cadherin
Normally, αE-catenin is bound to the E-cadherin–β-cat complex on the plasma membrane. Therefore, we questioned whether increasing the membrane-associated pool of αE-catenin would affect plasma membrane dynamics and cell migration. Endogenous αE-catenin was targeted to membranes, including the plasma membrane, independently of E-cadherin by expressing a chimeric protein comprising the same minimal αE-catenin–binding region of β-cat (aa 92–179) used in the aforementioned mitochondrial targeting construct but fused instead to mCherry and a 10-aa palmitoylation/myristoylation sequence from Lyn tyrosine kinase (Kovárová et al., 2001) that targets the chimeric protein to membranes (referred to as Lyn–β-cat; Fig. 6 A). As an additional control, two tyrosine residues (Y120 and Y142) in the β-cat sequence required for β-cat binding to αE-catenin (Aberle et al., 1996) were mutated to alanine (referred to as Lyn–β-cat mutant; Fig. 6 A). Two independent cell lines of each construct were tested and gave equivalent results. In addition, two chimeric proteins were constructed in which mCherry was replaced with the Fc domain from human IgG1 (termed Lyn-Fc–β-cat and Lyn-Fc–β-cat mutant) for coimmunoprecipitation experiments. No changes in the total cellular levels of αE-catenin, E-cadherin, actin, or tubulin were detected in Lyn–β-cat cells compared with control cells (Fig. S4 A)
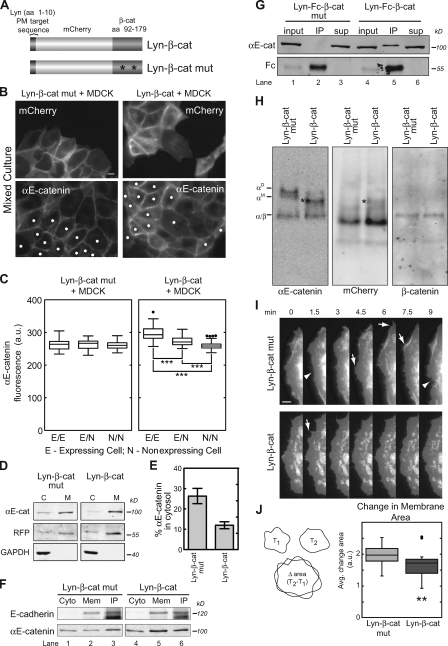
Figure 6.
Depletion of αE-catenin cytosolic pool by cadherin-independent recruitment to membranes. (A) Schematic of αE-catenin membrane-targeting constructs. Lyn–β-cat, minimal αE-catenin–binding domain of β-cat (aa 92–179) fused to mCherry and a 10-aa palmitoylation and myristoylation (PM) sequence from Lyn. Lyn–β-cat mutant, similar to Lyn–β-cat but containing two mutated residues in the β-cat fragment (asterisks), shown to eliminate αE-catenin binding. (B) Cells expressing Lyn–β-cat or Lyn–β-cat mutant (asterisks) were mixed with wild-type MDCK cells and stained for αE-catenin. (C) The mean level of endogenous αE-catenin at cell–cell contacts was quantified as described in Fig. 3 E and graphed. 30–50 cell–cell contacts for each of the three conditions were measured. ***, P < 0.0002 (Mann-Whitney test). Results are presented in a box and whisker format. The ends of the box mark the upper and lower quartiles, the horizontal line in the box indicates the median, and the whiskers outside the box extend to the highest and lowest value within 1.5 times the interquartile range. Outliers are represented as dots. (D) Cytosol (C) and membrane (M) fractions from Lyn–β-cat and Lyn–β-cat mutant cells separated by SDS-PAGE and blotted for αE-catenin, mCherry, and GAPDH. (E) Percentage of αE-catenin in cytosol fraction from experiment shown in D was measured and plotted. (F) E-cadherin immunoprecipitates from Lyn–β-cat and Lyn–β-cat mutant cell lysates were blotted for E-cadherin and αE-catenin. Cyto, cytosol; mem, membrane. (G) Fc immunoprecipitates (IP) from Lyn-Fc–β-cat and Lyn-Fc–β-cat mutant cell lysates were blotted for αE-catenin and Fc. Sup, supernatant. (H) Native PAGE of Lyn–β-cat mutant and Lyn–β-cat cytosol blotted for αE-catenin (left), mCherry (middle), and β-cat (right). An additional band (asterisks) in Lyn–β-cat cytosol cross reacted with mCherry and αE-catenin and is likely an αE-catenin/Lyn–β-cat heterodimer. (I and J) Representative time-lapse montage of mCherry-labeled membrane protrusions in Lyn–β-cat mutant and Lyn–β-cat cells. (I) Arrows, membrane extensions; arrowheads, membrane retractions. (J) Quantification of membrane dynamics in 29 Lyn–β-cat mutant and 24 Lyn–β-cat cells. Change in membrane area was calculated as the mean difference in area between two 10-s frames normalized for cell area (left, schematic; right, quantification). **, P < 0.002 (Mann-Whitney test). Error bars indicate SEM from three independent experiments. Bars: (B) 10 µm; (I) 5 µm.
Lyn–β-cat but not the Lyn–β-cat mutant recruited additional αE-catenin to the plasma membrane (Fig. 6 B). The level of αE-catenin at the plasma membrane in Lyn–β-cat cells was quantified using the aforementioned mixed culture assay in which Lyn–β-cat or Lyn–β-cat mutant cell lines were mixed with wild-type MDCK cells (Fig. 6, B and C). Two contacting Lyn–β-cat cells had significantly more αE-catenin at the plasma membrane compared with a Lyn–β-cat cell contacting a wild-type MDCK cell, which in turn had significantly more αE-catenin at the plasma membrane compared with two contacting wild-type MDCK cells (Fig. 6 C). No change in αE-catenin levels at the plasma membrane was noted in Lyn–β-cat mutant cells, as expected (Fig. 6 C). Recruitment of αE-catenin to the membrane from the cytosol in Lyn–β-cat cells was also quantified by separation of membrane and cytosol, as described for β-cat–ActA cells (Fig. 3, F and G). The amount of cytosolic αE-catenin decreased from ∼40% of total αE-catenin in control cells to <10% in Lyn–β-cat cells (n = 3; Fig. 6, D and E). Note that the amount of αE-catenin bound to the E-cadherin complex was similar in Lyn–β-cat and Lyn–β-cat mutant cells (Fig. 6 F, compare lane 3 with lane 6) even though the overall amount of αE-catenin was increased at the plasma membrane (Fig. 6 F, compare lane 2 with lane 5). Thus, the additional number of β-cat–binding sites provided by Lyn–β-cat increased αE-catenin recruitment to the plasma membrane independently of binding sites on the E-cadherin–β-cat complex. Finally, binding of αE-catenin to the membrane-anchored fragment of β-cat was verified by immunoprecipitation. The Fc chimeric proteins (Lyn-Fc–β-cat and Lyn-Fc–β-cat mutant) were transiently expressed, and protein complexes immunoprecipitated with anti–human IgG1 antibodies from Triton X-100–extracted membranes. As expected, αE-catenin coimmunoprecipitated with Lyn-Fc–β-cat but not with the Lyn-Fc–β-cat mutant (Fig. 6 G, lanes 2 and 5).
Native PAGE analysis revealed that, similar to β-cat–ActA cells, αE-catenin monomer and homodimer were reduced by ∼70% in the cytosol of Lyn–β-cat cells compared with control cytosol (n = 3; Fig. 6 H, left), whereas the amount of heterodimer was unchanged (Fig. 6 H, right). An mCherry signal was detected in cytosol of Lyn–β-cat and Lyn–β-cat mutant cells (Fig. 6 H) and likely represents newly translated Lyn constructs before lipid modification and membrane localization.
We tested whether increased amounts of αE-catenin at the plasma membrane affected the distribution of EPLIN, an actin-binding protein recently reported to bind αE-catenin (Abe and Takeichi, 2008). EPLIN was localized to sites of cell–cell contact and actin stress fibers (Abe and Takeichi, 2008), but enrichment of EPLIN at the plasma membrane was not observed in Lyn–β-cat cells (Fig. S3, B and C). Also, we did not observe additional recruitment of putative αE-catenin–binding partners vinculin, ZO-1, α-actinin, F-actin, or β-cat to the plasma membrane (unpublished data). Therefore, similar to the β-cat mitochondrial strategy, functional differences between Lyn–β-cat and control cells can be attributed to changes in αE-catenin localization and not to the redistribution of these actin-binding proteins.
We tested the effects of increasing αE-catenin binding to membranes on cell–cell adhesion and cell migration. In the hanging drop assay, Lyn–β-cat and Lyn–β-cat mutant cells formed trituration-resistant cell aggregates at similar rates (Fig. S4, D and E), indicating that cell–cell adhesion was unaffected. Likewise, the rate of wound closure was similar for Lyn–β-cat, Lyn–β-cat mutant cells, and control MDCK cells (Fig. S4, F and G) in the wound-healing assay, demonstrating that cell migration was unperturbed. However, time-lapse images revealed that the lamellipodial dynamics of Lyn–β-cat cells were significantly reduced compared with Lyn–β-cat mutant control cells (Fig. 6, I and J).
Effects of changes in endogenous αE-catenin localization on Arp2/3 and F-actin distributions at the plasma membrane
We next examined whether changes in plasma membrane dynamics and cell migration in β-cat–ActA and Lyn–β-cat cells corresponded to differences in Arp2/3 complex localization, F-actin assembly, and F-actin ultrastructure, as predicted from the inhibitory effect of αE-catenin on Arp2/3-mediated actin polymerization in vitro (Drees et al., 2005).
Arp2/3 complex localization in Lyn–β-cat, ActA, and β-cat–ActA cells was visualized by staining for the p34 subunit of the complex (Fig. 7 A). Relative to the sharp and discrete staining along the leading edge of ActA controls, p34 localization was noticeably broader and more continuous in β-cat–ActA cells (Fig. 7 A and Fig. S5). Increased F-actin staining was also observed along the leading edge of p34-rich lamellipodia in β-cat–ActA cells (Fig. 7 A). To quantify p34 and F-actin fluorescence intensity, we measured pixel intensities along line scans originating at the cell edge and extending into the cell cortex (Fig. 7 B). Both p34 and F-actin signals were distributed more broadly in β-cat–ActA protrusions relative to ActA controls, confirming visual observations (Fig. 7 B). This broader enrichment of Arp2/3 complex at the leading edge of ruffling β-cat–ActA cells was also observed in live cell imaging of cells expressing Arp3-GFP (unpublished data). In contrast, little or no enrichment of p34 along the leading edge was observed in Lyn–β-cat cells; instead, p34 levels in membrane protrusions were contiguous with levels in the cortex (Fig. 7, A and B). Note that changes in Arp2/3 complex fluorescent profiles similar to those observed in Lyn–β-cat cells were found upon injection of skeletal tropomyosin, another inhibitor of the Arp2/3 complex, into cells (Gupton et al., 2005), further supporting a role for αE-catenin in regulating the Arp2/3 complex and thereby actin organization.
Figure 7.
Redistribution of cytosolic and membrane-associated αE-catenin pools affects Arp2/3 complex enrichment in lamellipodia. (A) Representative images from two ActA, β-cat–ActA, and Lyn–β-cat cells fixed and stained with anti-p34 antibody (Arp2/3 complex) and Alexa Fluor–labeled phalloidin (F-actin). Bar, 10 µm. (B) Fluorescence intensity of p34 and F-actin signals in lamellipodia was measured by line scan analysis. Mean fluorescence <3 µm extending from the cell edge (0) in the cell cortex was plotted. 40–50 protrusions from each cell type were measured at three separate points and averaged.
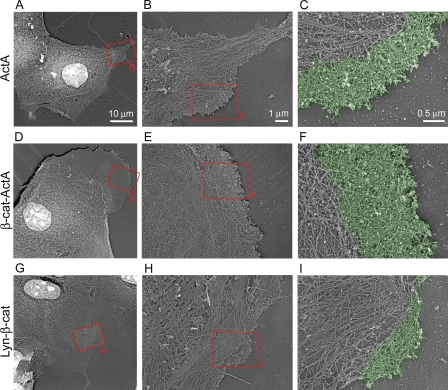
The ultrastructural organization of F-actin at the plasma membrane was examined by electron microscopy after critical point drying and platinum/carbon coating of detergent-extracted and chemically fixed cells (Fig. 8). The major difference between cell types was in formation of lamellipodia, which could be recognized by the presence of a dense, branched network of relatively short actin filaments at the cell leading edges. β-Cat–ActA cells tended to have larger and more continuous lamellipodial regions at the leading edge (Fig. 8, D–F) compared with control cells (Fig. 8, A–C). This was in contrast to Lyn–β-cat cells, which typically had decreased lamellipodial area and actin network density (Fig. 8, G–I) compared with control cells. In addition, the lamella, a sparser region behind lamellipodia containing long actin filaments, actin bundles, microtubules, and intermediate filaments, was much broader in β-cat–ActA cells compared with Lyn–β-cat and ActA cells (Fig. 8, A, D, and G). These data, in conjunction with those obtained from Arp2/3 complex staining and the barbed-end actin nucleation assay, indicate that differences in cell migration and membrane dynamics between β-cat–ActA, Lyn–β-cat, and control cells correlate with differences in both actin polymerization and organization.
Figure 8.
Perturbation of cytosolic and membrane-associated αE-catenin pools alters actin ultrastructure. (A–I) Platinum replica electron microscopy of membrane protrusions from control ActA (A–C), β-cat–ActA (D–F), and Lyn–β-cat (G–I) cells. Red boxes denote magnified regions shown in the indicated panel. Pseudo coloring in C, F, and I highlight lamellipodia.
Discussion
In vitro studies with purified proteins showed that αE-catenin is an allosteric protein in which the monomer binds the cadherin–catenin complex and homodimer preferentially binds actin filaments and inhibits Arp2/3 complex–mediated actin polymerization (Drees et al., 2005; Yamada et al., 2005). These results intimated roles for αE-catenin in the regulation of actin-based membrane dynamics in addition to and perhaps independent of αE-catenin roles in cell–cell adhesion. In this study, we tested key predictions of these in vitro results in epithelial cells.
We identified multiple forms of αE-catenin in MDCK cells. A membrane-bound pool of αE-catenin was identified in the E-cadherin–catenin complex; formally, we do not know whether this pool is a monomer, homodimer, or both, although it is likely to be a monomer based upon structural evidence (Pokutta and Weis, 2000). In the cytosol, we identified αE-catenin monomer and homodimer, which had a relatively fast turnover rate. We also observed αE-catenin in complex with β-cat (heterodimer) in the cytosol. The source and function of this heterodimer pool is unclear. One possibility is that the heterodimer becomes dissociated from the cadherin–catenin complex during cytosol preparation. Alternatively, β-cat in excess of E-cadherin–binding sites might be sequestered by cytosolic αE-catenin and degraded rapidly (Fig. 1 D). Finally, we attempted to determine the form of αE-catenin associated with the actin cytoskeleton, but we could not biochemically distinguish proteins that pelleted with F-actin from those bound to membranes.
Based upon in vitro experiments, we suggested that αE-catenin, rather than solely serving as a static link between cadherin and actin, has an essential and active role in regulating F-actin organization during the formation and maintenance of cell–cell contacts (Drees et al., 2005). We proposed that during cell–cell contact formation, recruitment of αE-catenin to the E-cadherin–β-cat complex present on the lamellipodial membrane would produce a local high concentration of cytosolic αE-catenin that would inhibit Arp2/3 complex–mediated branched actin polymerization, thereby dampening membrane dynamics and allowing the development of strong cell–cell adhesion. We tested this hypothesis by reducing the cytosolic pool of αE-catenin.
shRNA-mediated knockdown of αE-catenin in MDCK cells depleted both plasma membrane and cytosolic pools equally and greatly reduced cell–cell adhesion while increasing cell migration. These effects are similar to genetic deletion of αE-catenin in mice, which causes increased cell migration and tumor formation (Vasioukhin et al., 2001; Lien et al., 2006), and in humans, results in tumors with very poor prognosis (for review see Benjamin and Nelson, 2008), even though levels of E-cadherin can remain high in both cases.
The effects of shRNA-induced knockdown of αE-catenin indicate that both the plasma membrane and cytosolic pools of αE-catenin function together to control cell–cell adhesion and cell migration. We sought to separate functions of the cytosolic and membrane-bound pools of αE-catenin by sequestering the cytosolic pool to mitochondria without affecting the membrane-bound pool or increasing the membrane-bound pool at the expense of the cytosolic pool.
Selective depletion of only the cytosolic pool of αE-catenin to mitochondria in β-cat–ActA cells increased plasma membrane dynamics, generated broader and more extensive Arp2/3 complex staining at the leading edge of lamellipodia, and increased the areas of lamellipodia and lamellae compared with controls. It is likely that changes in the distribution of αE-catenin directly affected F-actin polymerization rather than inducing off-target effects, as we did not detect changes in the levels of E-cadherin or β-cat (and therefore presumably other E-cadherin–bound proteins such as p120), the amount of αE-catenin associated with the cadherin complex (Fig. 3 C), or sequestration of vinculin, EPLIN, α-actinin, actin, afadin, mDia, or ZO-1 to mitochondria (Fig. 3, I and J; and Fig. S2). We suggest that cytosolic levels of αE-catenin in β-cat–ActA cells were too low to inhibit Arp2/3 complex–mediated actin polymerization, and consequentially, increased Arp2/3 complex activity increased the rate of cell migration. Thus, consistent with our previous in vitro study (Drees et al., 2005), the cytosolic pool of αE-catenin regulates actin-based membrane dynamics at a distance from cell–cell contacts and thereby normally contributes to the suppression of cell migration.
When we increased the membrane-bound pool of αE-catenin in Lyn–β-cat cells, there was a concomitant decrease in actin-based plasma membrane dynamics, Arp2/3 complex staining, and actin-rich lamellipodia compared with control cells. This was expected from our earlier hypothesis that increasing the level of αE-catenin at the plasma membrane might result in a rapid flux between αE-catenin between membrane-bound and cytosolic pools, thereby locally inhibiting Arp2/3 complex activity (Drees et al., 2005). Alternatively, a higher concentration of membrane-associated αE-catenin in Lyn–β-cat cells could further dampen actin and membrane dynamics by favoring direct associations between the cadherin–catenin complex and the actin cytoskeleton, possibly through other actin regulatory proteins as suggested previously (Gates and Peifer, 2005). Although we did not detect an increase in EPLIN or vinculin associated with the plasma membrane in Lyn–β-cat cells, other actin-associated proteins localized to adherens junctions (Hildebrand, 2005; Pilot et al., 2006; Sawyer et al., 2009) could mediate these events independently of the cadherin–catenin complex at the plasma membrane. Further studies are required to examine these interactions biochemically and in cell-based assays.
Cavey et al. (2008) also suggested roles for different pools of α-catenin in the Drosophila melanogaster embryonic epidermis. They found immobile clusters of E-cadherin at cell–cell contacts that appeared to be linked to actin, probably in an α-catenin–independent manner; however, the localization of these clusters depended on an actin network that required α-catenin (Cavey et al., 2008). They suggested that α-catenin dynamically associates with the cadherin–β-cat complex and with actin to restrict the localization of cadherins to cell–cell contact sites (Cavey et al., 2008). Placing these previous studies in the context of the cell-based assays reported in this study supports roles for αE-catenin that are cadherin dependent at the plasma membrane and cadherin independent in the cytosol. The latter roles involve regulating actin-based membrane dynamics that control the balance between cell–cell adhesion and cell migration, which is critical in development and is altered in diseases including metastatic cancers (Gumbiner, 2005; Halbleib and Nelson, 2006; Thiery and Sleeman, 2006; Baum et al., 2008; for review see Benjamin and Nelson, 2008). It has long been appreciated that αE-catenin functions as the primary effector of cadherin engagement on the actin cytoskeleton; this newly defined role as a regulator of actin dynamics independently of cell–cell adhesion offers new perspective on αE-catenin function in development and disease.
Materials and methods
Generation of constructs
Generation of Lyn-Fc–β-cat construct.
Lyn-Fc–β-cat was constructed using a multistep cloning process starting with a myristoylated and palmitoylated GFP in pcDNA 3.1 vector (Lyn-GFP; provided by T. Meyer, Stanford University, Stanford, CA). (a) GFP in Lyn-GFP was replaced with the cytosolic domain of E-cadherin using KpnI and XbaI sites (New England Biolabs, Inc.). Primers used to amplify and clone the cytosolic domain of E-cadherin were 5′-GCCGGGGTACCTCAGAACGGTGGTCAAAG-3′ and 5′-CGGCCTCTAGACTAGTCGTCCTCACCACCGC-3′. The resulting plasmid was termed Lyn–E-cad. (b) Monomeric Fc from human IgG (Chen and Nelson, 1996) was inserted upstream of E-cadherin in the Lyn–E-cad plasmid using a single KpnI site (New England Biolabs, Inc.). Primers used to amplify and clone Fc were 5′- GCCGGGGTACCTGCTCGAGCTCGACAAAAC-3′ and 5′-GCCGGTACCGGATCCCCCCCGGAGACAGGGAG-3′ (note that the 3′ primer contained sequences to introduce a novel BamHI site directly upstream of KpnI). The resulting plasmid was termed Lyn-Fc–E-cad. (c) The cytosolic domain of E-cadherin in Lyn-Fc–E-cad was replaced with five glycine repeats directly followed by aa 92–179 of β-cat using BamHI and XbaI sites (New England Biolabs, Inc.). Primers used to amplify and clone the β-cat fragment were 5′-GGCCGGATCCGCGGAGGAGGAGGAGGAGCTCAGAGGGTCCG-3′ and 5′-GGCCTCTAGACTAGGAAAGCTGATGGAC-3′. The resulting plasmid was termed Lyn-Fc–β-cat.
Generation of Lyn-Fc–β-cat mutant construct.
Lyn-Fc–β-cat mutant was derived through a two-step site-directed mutagenesis. To generate the Y142A mutation, the entire Lyn-Fc–β-cat plasmid was amplified (PfuTurbo; Agilent Technologies) using forward primer 5′-GTCAATTTGATTAACGCTCAGGATGACGCGGAACTTGC-3′ and reverse primer 5′-GCAAGTTCCGCGTCATCCTGAGCGTTAATCAAATTGAC-3′ (underlined regions correspond to mismatched residues). The resulting PCR was digested for 1 h at 37°C with DpnI (New England Biolabs, Inc.) to destroy original template DNA and was transfected into competent cells (Agilent Technologies). Colonies were grown, and DNA was isolated (QIAGEN) and sequenced (Sequetech). The resulting plasmid was introduced to a second round of mutagenesis to generate the Y120A mutation as described using forward primer 5′-GACGCTGCTCATCCCGCTAATGTCCAGCGC-3′ and reverse primer 5′-GCGCTGGACATTAGCGGGATGAGCAGCGTC-3′.
Generation of Lyn-mCherry–β-cat and mutant constructs.
Lyn-mCherry–β-cat and Lyn-mCherry–β-cat mutant constructs were cloned from Lyn-Fc–β-cat and a Lyn-Fc–β-cat mutant, respectively. Fc was replaced with mCherry using XhoI and BamHI sites. Primers used to amplify and clone mCherry were 5′-GGGCTCGAGATGGTGAGCAAGGGC-3′ and 5′-CCCGGATCCCCTTGTACAGCTCGTC-3′.
Generation of β-cat–mRFP-ActA construct.
Amino acids 92–179 of β-cat directly followed by five glycine repeats were cloned into mRFP-N1-ActA (Takara Bio Inc.; provided by J. Theriot, Stanford University, Stanford, CA) by using EcoRI and BamHI sites (New England Biolabs, Inc.). Primers used to amplify and clone the β-cat fragment were 5′-GGGCAGAATTCCGCCACCATGGCTCAGAGGGTCCG-3′ and 5′-GGCAGGATCCCCTCCTCCTCCTCCGGAAAGCTGATGGAC-3′. The resulting plasmid was termed Lyn-Fc–β-cat.
Generation of αE-catenin knockdown.
Transient canine αE-catenin depletion was obtained by shRNA interference with annealed primers expressed in a pSuper vector (provided by T. Brummelkamp, Massachusetts Institute of Technology, Cambridge, MA; Brummelkamp et al., 2002). Primers for stable shRNA knockdown of αE-catenin in MDCK cells were replicated from Capaldo and Macara (2007) and are 5′ sense strand, 5′-GATCCCCGGCTAACAGAGACCTGATATTCAAGAGATATCAGGTCTCTGTTAGCCTTTTTGGAAA-3′, and 3′ antisense strand, 5′-AGCTTTTCCAAAAAGGCTAACAGAGACCTGATATCTCTTGAATATCAGGTCTCTGTTAGCCGGG-3′. Annealed primers were cloned using BglII and HindIII sites into a pSuper/pEGFP-C1 hybrid vector (provided by B. Grill, University of Minnesota, Minneapolis, MN) that permits both EGFP and shRNA expression and confers neomycin resistance. To generate a stable knockdown line, this construct was transfected into MDCK cells and cells cultured under selection in 400 µg/ml G418. After 1 wk of selection, individual clones were isolated by serial dilution, expanded, and screened for EGFP expression and reduced levels (<25%) of αE-catenin by immunostaining and Western blotting.
Cell lines
MDCK G type II cells were maintained in DME with 1 g/L sodium bicarbonate, 10% fetal bovine serum (Atlas Biologicals), penicillin, streptomycin, and kanamycin. To generate stable cell lines, Lyn–β-cat, Lyn–β-cat mutant, β-cat–ActA, or ActA constructs were transfected (Lipofectamine 2000; Invitrogen) into MDCK cells and grown under selection in 500 µg/ml G418 (Invitrogen). After 1 wk of selection, individual clones were isolated by serial dilution, expanded, and screened for RFP or mCherry expression. Lyn–β-cat and Lyn–β-cat mutant cell lines were FACS sorted before isolating single clones to enrich for mCherry expression.
Growth curve
100,000 cells were plated on multiple 35-mm dishes (day 0). Cells were trypsinized and counted from three separate dishes on each day (days 1– 6), and the mean number of cells/dish was plotted.
Hanging drop assay
The assay was performed as described previously (Kim et al., 2000; Ehrlich et al., 2002). In brief, MDCK cells were grown at low density, and cells were trypsinized, centrifuged, and resuspended as single-cell suspensions at 2.5 × 105 cells/ml. 20-µl drops of cell suspension were pipetted onto the inside surface of 35-mm culture dish lids, and dishes were filled with 2 ml media to prevent evaporation. At each time point, the lid was inverted, and drops were triturated 10 times through a 20-µl pipet. 4 µl 16% PFA was added, and each drop was spread onto a glass slide. The entire coverslip was scanned and photographed with an inverted microscope with a 10× objective (Axiovert 200M; Carl Zeiss, Inc.), and numbers and sizes of clusters were counted.
Wound-healing assay
Cells were trypsinized, resuspended in media containing 5 µM Ca2+, and 3 × 106 cells were plated at confluency on collagen-coated 35-mm glass-bottom dishes (MaTek). Cells were allowed to adhere to the substratum for 75 min, after which the media were replaced with DME containing normal 1.8 mM Ca2+. After a 3-h incubation, a scratch was made along the length of the dish using a 1,000-µl pipette tip, and the media were replaced with live cell imaging buffer (DME without phenol red supplemented with 25 mM Hepes; Invitrogen). Cells were imaged at 10× magnification every 15 min for 12 h at 37°C on an inverted microscope (Axiovert 200M) controlled with Slidebook software (Intelligent Imaging Innovations); three to five sites were imaged along the wound edge. Wound healing was quantified as the change in mean wound width (area wound/wound height) over time and measured using ImageJ software (National Institutes of Health).
Tracking analysis
Cells were prepared as described for the wound-healing analysis except cells were treated with 5 µg/ml Hoechst for 15 min before scratch formation and imaged every 5 min for 100 min. Individual nuclei were tracked to determine the coordination, velocity, and persistence of cell movements using custom software (Vitorino and Meyer, 2008). Persistence was measured as the fraction of time cells spent migrating toward the monolayer edge (± 45°) and plotting the mean value as a function of cell distance from the open edge; coordination was measured by calculating the mean angular difference between the directions of a pair of cell trajectories and plotting the inverted value as a function of the distance between cell pairs (Vitorino and Meyer, 2008).
Kymograph and membrane analysis
Kymograph analysis.
Cells were transfected with EGFP-actin (β-cat–ActA and ActA) or mCherry-actin (MDCK and αE-cat–shRNA). Cells were trypsinized, resuspended in live cell imaging buffer (DME without phenol red supplemented with 25 mM Hepes; Invitrogen), and plated at single-cell density (<5 × 104 cells) on collagen-coated 35-mm glass-bottom dishes (MaTek). Cells were imaged at 100× magnification with a microscope (Axiovert) every 10 s for 10 min. Kymographs were generated by generating a time-lapse montage of a single 1–2-pixel-wide frame rectangle (perpendicular to cell edge) for each frame of the video (ImageJ). The resulting image represented membrane activity (y axis) over time (x axis). Protrusion activity was defined as the number of peaks extending >0.5 µm and persisting for >30 s formed in 10 min (membrane extension and retraction). Protrusion velocity was defined as the rate of membrane extension (mean slope of peaks). For visual presentation, kymographs were orientated, inverted, smoothed, despeckled, and shadowed from the north in ImageJ.
Whole cell membrane analysis.
Lyn–β-cat mutant and Lyn–β-cat–expressing stable cell lines were trypsinized, plated, and imaged as described for kymograph analysis. Change in membrane area is defined as a measurement of membrane activity. The cell area difference between sequential 10-s frames for each cell was averaged and normalized (to cell area). Custom-made macros in ImageJ were used to quantify changes in cell area.
Immunofluorescence
Cells were processed for immunofluorescence as follows (unless otherwise described): cells were fixed for 15 min in 3% PFA (Electron Microscopy Sciences) and rinsed with PBS (Sigma-Aldrich). Cells were blocked and stained with primary and secondary antibodies in a PBS solution containing 1 mM MgCl2, 2 mM EGTA, 2% BSA, 1% goat serum, and 0.05% saponin and mounted in Vectashield + DAPI (Vector Laboratories). Primary antibodies used were vinculin (1:100; Sigma-Aldrich), αE-catenin (1:100; Enzo Life Sciences, Inc.), EPLIN (1:100; BD), ZO-1 (1:100; Invitrogen), β-cat (1:500; Näthke et al., 1994), α-actinin (1:100; Sigma-Aldrich), afadin (1:500; Sigma-Aldrich), and mDia1 and 2 (1:400; provided by A. Alberts, Van Andel Institute, Grand Rapids, MI). Secondary antibodies used were goat/donkey anti–mouse or –rabbit IgG labeled with FITC or rhodamine red-X/Cy5 (1:200; Jackson ImmunoResearch Laboratories, Inc.). Fluorescently labeled phalloidin was used to visualize actin (1:500; Invitrogen). Coverslips were imaged using an inverted microscope (Axiovert 200) and AxioVision software (Carl Zeiss, Inc.).
Arp2/3 staining.
Cells were fixed in PFA/PHEM buffer (Strasser et al., 2004), washed with PBS, blocked for 1 h at RT in PBS + 10% BSA, washed three times in PBS, incubated with primary in PBS + 1% BSA for 1 h at RT, washed three times in PBS, incubated with secondary in PBS + 1% BSA for 1 h at RT, washed three times in PBS, and mounted in Fluoromount G (SouthernBiotech). Arp2/3 was stained using anti–p34-Arc/ARPC2 (1:100; Millipore), and F-actin was stained using Alexa Fluor phalloidin (Invitrogen). Cells were imaged on a widefield deconvolution system (Deltavision; Applied Precision).To measure fluorescence, line scans (3-pixels wide; mean fluorescence) were drawn perpendicular to the leading edge, extending 3 µm into the cell center, and pixel intensities were quantified using Metamorph software (MDS Analytical Technologies).
Quantification of αE-catenin and EPLIN at sites of cell–cell contact.
Cells were prepared and stained as described in Immunofluorescence. Using an inverted microscope (Axiovert 200M) controlled with Slidebook software, z-stack sections of contacting cells were imaged, and a mean z-stack projection image was generated (Slidebook). Quantification of mean intensity (fluorescence) at cell contacts was analyzed by tracing cell–cell contacts (ImageJ). A cell–cell contact between two expressing cells was defined as an E/E contact. Contacts between an expressing and nonexpressing MDCK cell were defined as E/N contacts, and contacts between two MDCK cells were defined as N/N contacts.
Platinum replica EM
After brief washing with DPBS, cells were extracted for 3 min at room temperature with 1% Triton X-100 in PEM buffer (100 mM Pipes-KOH, pH 6.9, 1 mM MgCl2, and 1 mM EGTA) containing 2% polyethyleneglycol (35,000 molecular weight) and 10 µM phalloidin. After extraction, cells were washed with PEM buffer containing 10 µM phalloidin three times then fixed with 2% glutaraldehyde for 20 min. Samples for platinum replica EM were processed as described previously (Svitkina and Borisy, 1998) and analyzed using a transmission electron microscope (JEM 1011; JEOL) operated at 100 kV. Images were captured by a charge-coupled device camera (ORIUS 835.10W; Gatan) and presented in inverted contrast.
Cell extraction
Cells were grown to ∼70% confluence by plating 1.5 × 106 cells on a 10-cm dish 2 d before harvesting. Total protein concentration was quantified using either a BCA protein determination kit (Thermo Fisher Scientific) or a Bradford assay (Bio-Rad Laboratories).
Whole cell lysate preparation.
One to two 10-cm dishes at 70% confluency were rinsed with PBS and scraped with 0.5 ml of SDS buffer (1% SDS, 10 mM Tris-HCL, pH 7.5, and 2 mM EDTA). Cell lysates was transferred to a 1.5-ml tube and boiled for 10 min at 100°C. ∼30 µg total protein was loaded for each sample.
Cell fractionation.
6–12 10-cm dishes at ∼70% confluence were rinsed with PBS and harvested in CSK buffer (50 mM NaCl, 300 mM sucrose, 10 mM Pipes, pH 6.8, 3 mM MgCl2, and 1 mM pefabloc [Roche]). Cells were homogenized in the cold using a custom-made ball-bearing homogenizer (Varian Physics Machine Shop, Stanford University, Stanford, CA) and spun at ∼1,000 g for 10 min to pellet nuclei (Eppendorf). Postnuclear supernatant was subjected to ultracentrifugation at ∼128,000 g for 45 min at 4°C (TLA 100.3; Beckman Coulter). Membranes (pellet) were resuspended in a volume of CSK buffer + 0.5% Triton X-100 (Sigma-Aldrich) equal to that of the cytosol using a pestle and microtube (VWR). Cytosol and membrane fractions were either separated directly by native PAGE (cytosol only), by SDS-PAGE gels, or by SDS-PAGE after immunoprecipitation.
Immunoprecipitation.
Antibodies (E-cadherin; 1:100; Marrs et al., 1993), RFP (1:100; Rockland), and Fc (1:100; Sigma-Aldrich) were added to Triton X-100 resuspended membrane fractions, and tubes were rotated at 4°C for 1 h. 25 µl protein A–Sepharose 4B beads (50 µl slurry; GE Healthcare) was added, and tubes were rotated for 1 h at 4°C. Beads were washed in CSK buffer + Triton X-100 twice and once with CSK buffer and resuspended in SDS loading buffer.
Western blotting
10–12% polyacrylamide (Protogel) SDS-PAGE gels and 5% polyacrylamide native PAGE gels were prepared identically (electrophoresis systems; Bio-Rad Laboratories) except for the absence of SDS in native PAGE gels. Samples loaded onto native PAGE gels were supplemented with 100 µM DTT and bromophenol blue (to track the dye front). Native PAGE was performed at 4°C at 50 V constant until ∼30 min after the dye front ran off the gels. Gels were transferred onto nitrocellulose (GE Healthcare) overnight (∼1,000 mAmp h) and blocked in 5% milk (1% goat serum and 1% BSA, optional) in Tris-buffered saline. Primary and secondary antibodies were prepared in blocking buffer + 0.1% Tween-20. Primary antibodies used were vinculin (1:1,500; Sigma-Aldrich), αE-catenin (1:1,000; Enzo Life Sciences, Inc.), EPLIN (1:1,000; BD), β-cat (1:1,000; Näthke et al., 1994), E-cadherin (1:1,000; Marrs et al., 1993), mtHsp70 (1:500; Thermo Fisher Scientific), tubulin (1:1,000; Santa Cruz Biotechnology, Inc.), and GAPDH (1:4,000; Abcam). Secondary antibodies used were goat anti–mouse 800CW (1:15,000; LI-COR Biosciences) and goat anti–rabbit Alexa Fluor (1:15,000; Invitrogen). Membranes were scanned using an Odyssey imager and software (LI-COR Biosciences), and bands were quantified using ImageJ.
Online supplemental material
Fig. S1 shows that sequestration of cytosolic αE-catenin to mitochondria has no effect on cellular protein levels or cell proliferation while effectively reducing the amount of cytosolic αE-catenin monomer and homodimer. Fig. S2 shows that targeting αE-catenin to mitochondria does not recruit additional αE-catenin ligands to mitochondria. Fig. S3 shows that knockdown of αE-catenin does not affect cell proliferation in MDCK cells. Fig. S4 shows that cadherin-independent recruitment of αE-catenin to the plasma membrane does not affect protein levels, cell–cell adhesion, cell migration, or EPLIN recruitment. Fig. S5 shows that Arp2/3 complex staining in MDCK cells is altered upon redistribution of cytosolic and membrane-associated αE-catenin. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200910041/DC1.
Acknowledgments
We would like to thank Feng-Chiao Tsai, who performed the single-cell tracking analysis in Fig. 4, and Dr. Art Alberts for the mDia1 and mDia2 antibodies.
J.M. Benjamin was supported by a Stanford Graduate Fellowship and Cancer Biology Training grant (5 T32 CA009151-34). A.V. Kwiatkowski was supported by a Ruth L. Kirschstein National Research service award (5T32 CA09302). This work was also supported by the National Institutes of Health (grants GM35527 to W.J. Nelson, GM56169 to W.I. Weis, and GM70898 to T. Svitkina).
Footnotes
Abbreviations used in this paper:
- β-cat
- β-catenin
- F-actin
- filamentous actin
- mRFP
- monomeric RFP
- shRNA
- short hairpin RNA
References
- Abe K., Takeichi M. 2008. EPLIN mediates linkage of the cadherin catenin complex to F-actin and stabilizes the circumferential actin belt. Proc. Natl. Acad. Sci. USA. 105:13–19 10.1073/pnas.0710504105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aberle H., Butz S., Stappert J., Weissig H., Kemler R., Hoschuetzky H. 1994. Assembly of the cadherin-catenin complex in vitro with recombinant proteins. J. Cell Sci. 107:3655–3663 [DOI] [PubMed] [Google Scholar]
- Aberle H., Schwartz H., Hoschuetzky H., Kemler R. 1996. Single amino acid substitutions in proteins of the armadillo gene family abolish their binding to alpha-catenin. J. Biol. Chem. 271:1520–1526 10.1074/jbc.271.3.1520 [DOI] [PubMed] [Google Scholar]
- Adams C.L., Nelson W.J., Smith S.J. 1996. Quantitative analysis of cadherin-catenin-actin reorganization during development of cell-cell adhesion. J. Cell Biol. 135:1899–1911 10.1083/jcb.135.6.1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum B., Settleman J., Quinlan M.P. 2008. Transitions between epithelial and mesenchymal states in development and disease. Semin. Cell Dev. Biol. 19:294–308 10.1016/j.semcdb.2008.02.001 [DOI] [PubMed] [Google Scholar]
- Benjamin J.M., Nelson W.J. 2008. Bench to bedside and back again: molecular mechanisms of alpha-catenin function and roles in tumorigenesis. Semin. Cancer Biol. 18:53–64 10.1016/j.semcancer.2007.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards R., Agami R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science. 296:550–553 10.1126/science.1068999 [DOI] [PubMed] [Google Scholar]
- Capaldo C.T., Macara I.G. 2007. Depletion of E-cadherin disrupts establishment but not maintenance of cell junctions in Madin-Darby canine kidney epithelial cells. Mol. Biol. Cell. 18:189–200 10.1091/mbc.E06-05-0471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavey M., Rauzi M., Lenne P.F., Lecuit T. 2008. A two-tiered mechanism for stabilization and immobilization of E-cadherin. Nature. 453:751–756 10.1038/nature06953 [DOI] [PubMed] [Google Scholar]
- Chen Y.T., Nelson W.J. 1996. Continuous production of soluble extracellular domain of a type-I transmembrane protein in mammalian cells using an Epstein-Barr virus Ori-P-based expression vector. Anal. Biochem. 242:276–278 10.1006/abio.1996.0466 [DOI] [PubMed] [Google Scholar]
- Drees F., Pokutta S., Yamada S., Nelson W.J., Weis W.I. 2005. Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly. Cell. 123:903–915 10.1016/j.cell.2005.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich J.S., Hansen M.D., Nelson W.J. 2002. Spatio-temporal regulation of Rac1 localization and lamellipodia dynamics during epithelial cell-cell adhesion. Dev. Cell. 3:259–270 10.1016/S1534-5807(02)00216-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates J., Peifer M. 2005. Can 1000 reviews be wrong? Actin, alpha-Catenin, and adherens junctions. Cell. 123:769–772 10.1016/j.cell.2005.11.009 [DOI] [PubMed] [Google Scholar]
- Gumbiner B.M. 2005. Regulation of cadherin-mediated adhesion in morphogenesis. Nat. Rev. Mol. Cell Biol. 6:622–634 10.1038/nrm1699 [DOI] [PubMed] [Google Scholar]
- Gupton S.L., Anderson K.L., Kole T.P., Fischer R.S., Ponti A., Hitchcock-DeGregori S.E., Danuser G., Fowler V.M., Wirtz D., Hanein D., Waterman-Storer C.M. 2005. Cell migration without a lamellipodium: translation of actin dynamics into cell movement mediated by tropomyosin. J. Cell Biol. 168:619–631 10.1083/jcb.200406063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbleib J.M., Nelson W.J. 2006. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 20:3199–3214 10.1101/gad.1486806 [DOI] [PubMed] [Google Scholar]
- Hildebrand J.D. 2005. Shroom regulates epithelial cell shape via the apical positioning of an actomyosin network. J. Cell Sci. 118:5191–5203 10.1242/jcs.02626 [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Heuser J.E. 1981. Quick-freeze, deep-etch visualization of the cytoskeleton beneath surface differentiations of intestinal epithelial cells. J. Cell Biol. 91:399–409 10.1083/jcb.91.2.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M., Nagafuchi A., Moroi S., Tsukita S. 1997. Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to α catenin and actin filaments. J. Cell Biol. 138:181–192 10.1083/jcb.138.1.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.B., Islam S., Kim Y.J., Prudoff R.S., Sass K.M., Wheelock M.J., Johnson K.R. 2000. N-Cadherin extracellular repeat 4 mediates epithelial to mesenchymal transition and increased motility. J. Cell Biol. 151:1193–1206 10.1083/jcb.151.6.1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.T., Choi E.K., Kim J.W., Kim D.K., Kim S.H., Yang W.I. 2002. Expression of E-cadherin and alpha-, beta-, gamma-catenin proteins in endometrial carcinoma. Yonsei Med. J. 43:701–711 [DOI] [PubMed] [Google Scholar]
- Kobielak A., Fuchs E. 2004. Alpha-catenin: at the junction of intercellular adhesion and actin dynamics. Nat. Rev. Mol. Cell Biol. 5:614–625 10.1038/nrm1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobielak A., Pasolli H.A., Fuchs E. 2004. Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nat. Cell Biol. 6:21–30 10.1038/ncb1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs E.M., Goodwin M., Ali R.G., Paterson A.D., Yap A.S. 2002. Cadherin-directed actin assembly: E-cadherin physically associates with the Arp2/3 complex to direct actin assembly in nascent adhesive contacts. Curr. Biol. 12:379–382 10.1016/S0960-9822(02)00661-9 [DOI] [PubMed] [Google Scholar]
- Kovárová M., Tolar P., Arudchandran R., Dráberová L., Rivera J., Dráber P. 2001. Structure-function analysis of Lyn kinase association with lipid rafts and initiation of early signaling events after Fcepsilon receptor I aggregation. Mol. Cell. Biol. 21:8318–8328 10.1128/MCB.21.24.8318-8328.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krendel M.F., Bonder E.M. 1999. Analysis of actin filament bundle dynamics during contact formation in live epithelial cells. Cell Motil. Cytoskeleton. 43:296–309 [DOI] [PubMed] [Google Scholar]
- Lien W.H., Klezovitch O., Fernandez T.E., Delrow J., Vasioukhin V. 2006. alphaE-catenin controls cerebral cortical size by regulating the hedgehog signaling pathway. Science. 311:1609–1612 10.1126/science.1121449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs J.A., Napolitano E.W., Murphy-Erdosh C., Mays R.W., Reichardt L.F., Nelson W.J. 1993. Distinguishing roles of the membrane-cytoskeleton and cadherin mediated cell-cell adhesion in generating different Na+,K(+)-ATPase distributions in polarized epithelia. J. Cell Biol. 123:149–164 10.1083/jcb.123.1.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui S., Shiozaki H., Inoue M., Tamura S., Doki Y., Kadowaki T., Iwazawa T., Shimaya K., Nagafuchi A., Tsukita S., et al. 1994. Immunohistochemical evaluation of alpha-catenin expression in human gastric cancer. Virchows Arch. 424:375–381 10.1007/BF00190559 [DOI] [PubMed] [Google Scholar]
- Mialhe A., Louis J., Montlevier S., Peoch M., Pasquier D., Bosson J.L., Rambeaud J.J., Seigneurin D. 1997. Expression of E-cadherin and alpha-,beta- and gamma-catenins in human bladder carcinomas: are they good prognostic factors? Invasion Metastasis. 17:124–137 [PubMed] [Google Scholar]
- Näthke I.S., Hinck L., Swedlow J.R., Papkoff J., Nelson W.J. 1994. Defining interactions and distributions of cadherin and catenin complexes in polarized epithelial cells. J. Cell Biol. 125:1341–1352 10.1083/jcb.125.6.1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebuhr K., Ebel F., Frank R., Reinhard M., Domann E., Carl U.D., Walter U., Gertler F.B., Wehland J., Chakraborty T. 1997. A novel proline-rich motif present in ActA of Listeria monocytogenes and cytoskeletal proteins is the ligand for the EVH1 domain, a protein module present in the Ena/VASP family. EMBO J. 16:5433–5444 10.1093/emboj/16.17.5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilot F., Philippe J.M., Lemmers C., Lecuit T. 2006. Spatial control of actin organization at adherens junctions by a synaptotagmin-like protein Btsz. Nature. 442:580–584 10.1038/nature04935 [DOI] [PubMed] [Google Scholar]
- Pokutta S., Weis W.I. 2000. Structure of the dimerization and beta-catenin-binding region of alpha-catenin. Mol. Cell. 5:533–543 10.1016/S1097-2765(00)80447-5 [DOI] [PubMed] [Google Scholar]
- Pokutta S., Drees F., Takai Y., Nelson W.J., Weis W.I. 2002. Biochemical and structural definition of the l-afadin- and actin-binding sites of alpha-catenin. J. Biol. Chem. 277:18868–18874 10.1074/jbc.M201463200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., Borisy G.G. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465 10.1016/S0092-8674(03)00120-X [DOI] [PubMed] [Google Scholar]
- Rimm D.L., Koslov E.R., Kebriaei P., Cianci C.D., Morrow J.S. 1995. Alpha 1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc. Natl. Acad. Sci. USA. 92:8813–8817 10.1073/pnas.92.19.8813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer J.K., Harris N.J., Slep K.C., Gaul U., Peifer M. 2009. The Drosophila afadin homologue Canoe regulates linkage of the actin cytoskeleton to adherens junctions during apical constriction. J. Cell Biol. 186:57–73 10.1083/jcb.200904001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholten A.N., Aliredjo R., Creutzberg C.L., Smit V.T. 2006. Combined E-cadherin, alpha-catenin, and beta-catenin expression is a favorable prognostic factor in endometrial carcinoma. Int. J. Gynecol. Cancer. 16:1379–1385 10.1111/j.1525-1438.2006.00406.x [DOI] [PubMed] [Google Scholar]
- Setoyama T., Natsugoe S., Okumura H., Matsumoto M., Uchikado Y., Yokomakura N., Ishigami S., Aikou T. 2007. alpha-catenin is a significant prognostic factor than E-cadherin in esophageal squamous cell carcinoma. J. Surg. Oncol. 95:148–155 10.1002/jso.20610 [DOI] [PubMed] [Google Scholar]
- Strasser G.A., Rahim N.A., VanderWaal K.E., Gertler F.B., Lanier L.M. 2004. Arp2/3 is a negative regulator of growth cone translocation. Neuron. 43:81–94 10.1016/j.neuron.2004.05.015 [DOI] [PubMed] [Google Scholar]
- Svitkina T.M., Borisy G.G. 1998. Correlative light and electron microscopy of the cytoskeleton of cultured cells. Methods Enzymol. 298:570–592 10.1016/S0076-6879(98)98045-4 [DOI] [PubMed] [Google Scholar]
- Tachibana K., Nakanishi H., Mandai K., Ozaki K., Ikeda W., Yamamoto Y., Nagafuchi A., Tsukita S., Takai Y. 2000. Two cell adhesion molecules, nectin and cadherin, interact through their cytoplasmic domain-associated proteins. J. Cell Biol. 150:1161–1176 10.1083/jcb.150.5.1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery J.P., Sleeman J.P. 2006. Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 7:131–142 10.1038/nrm1835 [DOI] [PubMed] [Google Scholar]
- Vasioukhin V., Bauer C., Yin M., Fuchs E. 2000. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 100:209–219 10.1016/S0092-8674(00)81559-7 [DOI] [PubMed] [Google Scholar]
- Vasioukhin V., Bauer C., Degenstein L., Wise B., Fuchs E. 2001. Hyperproliferation and defects in epithelial polarity upon conditional ablation of alpha-catenin in skin. Cell. 104:605–617 10.1016/S0092-8674(01)00246-X [DOI] [PubMed] [Google Scholar]
- Vitorino P., Meyer T. 2008. Modular control of endothelial sheet migration. Genes Dev. 22:3268–3281 10.1101/gad.1725808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe-Uchida M., Uchida N., Imamura Y., Nagafuchi A., Fujimoto K., Uemura T., Vermeulen S., van Roy F., Adamson E.D., Takeichi M. 1998. α-Catenin-vinculin interaction functions to organize the apical junctional complex in epithelial cells. J. Cell Biol. 142:847–857 10.1083/jcb.142.3.847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S., Nelson W.J. 2007. Localized zones of Rho and Rac activities drive initiation and expansion of epithelial cell-cell adhesion. J. Cell Biol. 178:517–527 10.1083/jcb.200701058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S., Pokutta S., Drees F., Weis W.I., Nelson W.J. 2005. Deconstructing the cadherin-catenin-actin complex. Cell. 123:889–901 10.1016/j.cell.2005.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]