Loss of β3 integrin enhances turnover of focal adhesions and cell migration speed due to increased β1 integrin–talin interactions.
Abstract
Integrins are fundamental to the control of protrusion and motility in adherent cells. However, the mechanisms by which specific members of this receptor family cooperate in signaling to cytoskeletal and adhesion dynamics are poorly understood. Here, we show that the loss of β3 integrin in fibroblasts results in enhanced focal adhesion turnover and migration speed but impaired directional motility on both 2D and 3D matrices. These motility defects are coupled with an increased rate of actin-based protrusion. Analysis of downstream signaling events reveals that loss of β3 integrin results in a loss of protein kinase A–dependent phosphorylation of the actin regulatory protein vasodilator-stimulated phosphoprotein (VASP). Dephosphorylated VASP in β3-null cells is preferentially associated with Rap1-GTP–interacting adaptor molecule (RIAM) both in vitro and in vivo, which leads to enhanced formation of a VASP–RIAM complex at focal adhesions and subsequent increased binding of talin to β1 integrin. These data demonstrate a novel mechanism by which αvβ3 integrin acts to locally suppress β1 integrin activation and regulate protrusion, adhesion dynamics, and persistent migration.
Introduction
Cell migration is a tightly regulated process involving the spatial and temporal coordination of protrusion, adhesion, and contraction (Ridley et al., 2003). Integrins are a family of heterodimeric transmembrane receptors that form the main cell–ECM contact point in many cell types. As such, integrins are one of the primary components in the control of cell adhesion and protrusion both from the ligand-binding side and the formation of signaling platforms at the cytoplasmic face of the plasma membrane (Hynes, 2002). In adherent cells, it is widely known that both β1 and β3 integrins contribute to both normal cell migration and cancer cell invasion (Ramsay et al., 2007; Caswell and Norman, 2008). β1 and β3 integrins are also proposed to act upon one another through transdominant inhibition, resulting in suppression of signaling, possibly through competition for key integrin activation proteins such as talin (Calderwood et al., 2004). As both receptors share extracellular ligands, as well as cytoplasmic binding partners such as the cytoskeletal proteins talin and filamin (Arnaout et al., 2007), the manner in which these subunits individually contribute to cell motility is unclear.
Previous studies have localized αvβ3 integrin within small dynamic focal complexes at the leading edge of motile fibroblasts, whereas β1 is found predominantly in larger focal adhesions that act to stabilize new actin-based membrane protrusions and the cell body (Zamir et al., 2000; Zaidel-Bar et al., 2003). Both adhesion types are required to disassemble in order to allow efficient cell migration to occur, with smaller focal complexes displaying more rapid turnover than focal adhesions (Broussard et al., 2008). In order for cells to undergo persistent motility, adhesion dynamics must also be coordinated with the assembly of new F-actin–containing protrusions, which are in turn under the control of several actin-nucleating and elongating molecules (Le Clainche and Carlier, 2008). One such family are the Enabled/protein vasodilator-stimulated phosphoprotein (Ena/VASP) proteins, which act to antagonize capping protein, induce actin filament elongation (Sechi and Wehland, 2004), and, additionally, localize to focal adhesions. Rap1-GTP–interacting adaptor molecule (RIAM) is a member of the Mig-10/RIAM/Lamellipodin (MRL) family of adaptor proteins (Lafuente et al., 2004). RIAM overexpression induces β1 and β2 integrin–mediated cell adhesion and cell spreading; RIAM knockdown reduces Rap1-dependent cell adhesion and integrin activation, and leads to a reduction in cellular F-actin content (Lafuente et al., 2004; Han et al., 2006). RIAM is a binding partner for both talin and VASP, and RIAM association with talin has recently been shown to modulate integrin activation (Lee et al., 2009). However, the role of the VASP–RIAM complex in regulating talin dynamics, integrin activation, and motility remains unknown.
Here, we provide evidence that αvβ3 integrin promotes efficient cell motility through two mechanisms. Fibroblasts lacking β3 integrin show increased F-actin content and enhanced rates of membrane protrusion. This is coincident with the presence of constitutively dephosphorylated VASP that associates with RIAM at peripheral focal adhesions and promotes enhanced formation of a RIAM–talin complex. Moreover, β3 expression and localization to the leading edge suppresses local activation of β1 integrin. Loss of β3 results in an increase in VASP/RIAM-dependent peripheral active β1 integrin–containing adhesions that show increased rates of assembly and disassembly compared with those in wild-type (WT) cells. These findings suggest that αvβ3 integrin contributes toward regulation of actin dynamics at the leading edge as well as negatively regulating β1 activation, adhesion dynamics, and stable protrusion.
Results
Fibroblasts lacking β3 integrin have altered adhesion localization and dynamics
To determine whether β3 integrin is required for correct spatial adhesion organization, fibroblasts isolated from WT and β3-deficient (β3−/−) animals (Hodivala-Dilke et al., 1999) were plated on fibronectin (FN) and stained with antibodies to β1 and β3 and the focal adhesion markers vinculin (Fig. 1 A), zyxin, paxillin, or talin (not depicted). Confocal images and quantitative analysis revealed that cells lacking β3 integrin formed small, more peripheral focal adhesions and fewer large adhesions beneath the cell body as compared with WT cells (Fig. 1, A and B), but did not show a difference in total spread area on FN (not depicted). WT phenotypes were restored in all cases in β3−/− rescued with stable expression of a human β3-GFP construct. Western blotting showed an increase in levels of talin in cells lacking β3, which were also restored to WT levels in β3-GFP cells (Fig. S1 B). No other changes in levels of other focal adhesion proteins or other integrin subunits were observed (Fig. S1 A).
Figure 1.
Loss of β3 leads to formation of peripheral adhesions and enhanced migration. (A) Confocal images of WT, β3−/−, or β3-GFP cells plated on FN and costained for (top panels) β3 (FITC or GFP, green) and β1 (Alexa Fluor 568, red) or vinculin (bottom; shown in black and white). Bar, 10 µm. (B) Quantification of focal adhesion length, percentage of peripheral adhesions, and area/cell from confocal images. Adhesions were quantified from at least 100 cells over three independent experiments. Data are shown as mean ± SEM. (C, left) Confocal images of WT and β3−/− plated on CDMs and costained for active β1 (9EG7-FITC, green), paxillin (Alexa Fluor 568, red), and FN (Alexa Fluor 633, blue). Insets show enlarged views of the boxed regions. Bar, 10 µm. (right) The bar graph shows quantification of the percentage of peripheral adhesions from confocal images. Adhesions were quantified from at least 30 cells over three independent experiments. Data are shown as mean ± SEM. (D and E) Rate constants of focal adhesion dynamics in WT, β3−/−, or β3-GFP cells plated on 2D FN (D) or CDM (E). Data were quantified from time-lapse videos of cells expressing GFP- or mCherry-talin. Data are expressed as min−1 ± SEM. Data were quantified from a minimum of 200 adhesions per cell type over four experiments. (F) WT, β3−/−, and β3-GFP cells were fixed and stained for cell surface levels of total or active β1 integrin (MB1.2 or 9EG7, respectively) and analyzed by FACS. Data are presented as the mean percentage of active cell surface integrin as compared with WT cells ± SEM. (G and H) Quantification of migration speed (G) and directional persistence (H) of WT, β3−/−, or β3-GFP cells plated on 2D monolayers of FN or CDM. Data were quantified from >120 cells per matrix type over three independent experiments. Data are shown as mean ± SEM (indicated by error bars). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Control of cell adhesion formation in vivo is regulated by the architecture of the surrounding ECM. To determine whether the differences in adhesion localization in the β3−/− cells were also evident in a more complex 3D environment, we plated both WT and β3−/− cells on cell-derived matrices (CDMs; Cukierman et al., 2001). Confocal images of fixed cells in CDM stained for active β1 integrin, paxillin, and FN also showed increased numbers of peripheral matrix–associated adhesions in β3−/− (Fig. 1 C), which suggests that spatial control of adhesion formation is altered in these cells irrespective of matrix cues. To investigate whether the altered adhesion localization also translated to a functional change in adhesion turnover, WT and β3−/− cells expressing low levels of GFP-talin (Fig. 1 D) or GFP-vinculin (not depicted) were plated on 2D FN and imaged by time-lapse fluorescence microscopy to monitor adhesion dynamics. Analysis of adhesion intensity over time and calculation of rate constants revealed that loss of β3 resulted in a higher rate of adhesion assembly and disassembly compared with WT or β3-GFP rescued β3-null cells (Fig. 1 D). Additional quantification of the number of dynamic adhesions seen during the imaging period demonstrated that significantly more adhesions disassembled in β3−/− compared with WT cells (Fig. S2 A). To determine whether the ECM environment could alter this phenomenon, we also performed analysis of adhesion dynamics in cells in CDM. Interestingly, data revealed two populations of adhesions in both cell types in CDM (Fig. 1 E): one “slow” population showing rate constants of assembly and disassembly in the order of 0.1–0.3 min−1, and a “fast” population of 0.4–0.9 min−1. Of note, the fast population showed significantly faster adhesion dynamics than all adhesion in cells on 2D FN. Importantly, β3−/− cells showed higher rates of both assembly and disassembly in CDM, and this difference was more striking within the slow adhesion population. This data suggests that β3 can act to stabilize focal adhesions in fibroblasts.
Talin is known to bind directly to the cytoplasmic domain of β1 integrin, leading to inside-out activation (Tadokoro et al., 2003), and changes in dynamic behavior of talin in the absence of β3 may contribute toward the increased activation of β1. To determine whether loss of β3 led to changes β1 activation in these cells, we performed cell surface FACS analysis using antibodies to detect either total or active (9EG7) β1. Interestingly, despite similar levels of total protein and membrane levels of β1, β3−/− cells showed significantly higher active β1 at the cell surface compared with WT cells (Fig. 1 F). These data suggest that β3 integrin may act to suppress the activation of β1 in fibroblasts.
β3 integrin regulates fibroblast migration speed and persistence
Previous studies have shown that localization and recycling of β3 integrin is important in dictating polarized cell movement (Ballestrem et al., 2001; Danen et al., 2005; White et al., 2007). To determine whether genetic loss of β3 in fibroblasts affects migration, we imaged WT and β3−/− cells plated on 2D FN or in CDM by phase-contrast time-lapse microscopy, and quantified the speed and persistence of migration. Fig. 1 (G and H) and Fig. S2 B show quantification from videos of WT and β3−/− cells on FN (Videos 1 and 2), which demonstrates that β3−/− cells display an increase in migration speed but a decreased persistence of movement. This phenotype was also evident in cells migrating within CDM, where β3−/− cells show multiple protrusions and WT cells display an elongated morphology and migrate persistently along matrix fibers (seen as black lines in the phase-contrast images; Videos 3 and 4). These data are in agreement with previous studies in proposing a role for β3 in negative regulation of migration speed (Danen et al., 2005; Reynolds et al., 2005; White et al., 2007) and suggest that β3 additionally functions to promote directional persistence of motility.
Loss of β3 integrin results in increased cellular F-actin
Adhesion dynamics can contribute to the stability of the F-actin cytoskeleton and in turn regulate mechanical coupling of the cell to the ECM during cell motility (Vicente-Manzanares et al., 2009). To address whether the changes in motility in β3−/− cells were coupled with altered actin assembly, we measured total F-actin levels in WT and β3−/− cells by both FACS and Western blotting. Surprisingly, both types of analyses revealed a significant increase in insoluble or F-actin in β3−/− cells compared with WT (Fig. 2, A and B), which suggests that loss of β3 leads to stabilization of actin filaments. To study membrane protrusion in more detail, WT, β3-GFP, and β3−/− cells were imaged by phase-contrast time-lapse microscopy over short time periods and subjected to kymograph analysis to quantify membrane protrusion rates. Live imaging revealed that β3−/− cells exhibit multiple unstable protrusions, and quantification demonstrated a significant increase in both the protrusion rate and the amount of protrusion per cell area in β3−/− cells compared with WT or β3-GFP rescue cells (Fig. 2 C). This altered protrusive behavior was also evident in β3−/− cells transfected with GFP-actin and monitored by fluorescence time-lapse microscopy (Fig. 2 D and Videos 5 and 6). Moreover, analysis of Alexa Fluor 568–labeled G-actin incorporation in live WT or β3−/− cells also revealed an increase in free barbed end formation in cells lacking β3 integrin (Fig. 2 E). Together, these data suggest that the presence of β3 can act to suppress and stabilize the formation of F-actin protrusions. These data combined with the observed increases in β1 integrin activation in the absence of β3 provide one possible explanation for the enhanced migration speed and loss of persistent movement seen in these cells. Furthermore, this data agrees with a recent study showing that newly polymerizing actin can mobilize active β1 integrin at the leading edge of motile cells (Galbraith et al., 2007).
Figure 2.
Loss of β3 results in formation of unstable actin protrusions. (A) WT and β3−/− cells analyzed for total F-actin content by FACS analysis of phalloidin staining intensity. Data are presented as the percentage intensity of WT control cells ± SEM pooled from three independent experiments (indicated by error bars). (B) WT, β3−/−, or β3-GFP cell lysates from TX-100 insoluble fractions analyzed by Western blotting for β-actin or GAPDH as a loading control. Graphs are representative of four experiments. (C) Kymography analysis of protrusion rate and the percentage of cell area protruding/frame from time-lapse videos. Still images and example of kymographs obtained from the example white lines are shown on the left. Six lines were quantified from 12 cells over three independent experiments. Data are mean ± SEM (indicated by error bars). (D) WT and β3−/− cells transfected with GFP-actin and imaged by fluorescence time-lapse microscopy. Stills are shown from an example of each cell type, enlarged images of protruding regions of membrane over time (boxes) are shown on the right. (E) Quantification and example images of free barbed end formation in WT and β3−/− cells. Live cells were pulsed with Alexa Fluor 568–G-actin (red) for 1 min, fixed, and stained with phalloidin-488 (green). The intensity of G-actin incorporation was quantified from at least 100 cells from three independent experiments. Data are shown as the mean percentage intensity ± SEM compared with WT control (indicated by error bars). **, P < 0.01. Bars, 10 µm.
β3 integrin promotes VASP phosphorylation
For cells to undergo persistent motility, adhesion dynamics must also be coordinated with assembly of new F-actin–containing protrusions, which are in turn under the control of several actin-nucleating and elongating molecules (Le Clainche and Carlier, 2008). One such family are the Ena/VASP proteins, which act to antagonize capping protein to induce actin filament elongation (Sechi and Wehland, 2004) and localize to focal adhesions. To identify the potential signaling proteins responsible for regulating actin dynamics downstream of β3, we adopted a candidate approach to test for levels and localization of several adhesion and actin regulatory proteins in WT and β3−/− cells. One of the most striking differences was in the phosphorylation of VASP. Western blot analysis for VASP demonstrated an almost total loss of S153 phosphorylation in cells lacking β3 compared with that in WT cells or β3−/− cells rescued with human β3-GFP (Fig. 3 A; quantified in Fig. 3 B). VASP is known to be serine-phosphorylated by protein kinase A (PKA) during cell spreading (Howe et al., 2002). Phosphorylation of VASP at both S153 in mice (or S157 in humans) and S235 has also been shown to promote actin filament assembly in vitro (Barzik et al., 2005). More recent studies have also shown that VASP phosphorylated at S157 accumulates at the cell periphery in platelets and in spreading cells (Wentworth et al., 2006; Benz et al., 2009). However, the role of VASP phosphorylation in regulating focal adhesion protein behavior is unclear. Western blot analysis additionally showed that VASP S153 phosphorylation levels were reduced after PKA inhibition by H89, and an activator of PKA signaling (Forskolin) was able to in part rescue S153 phosphorylation of VASP in β3−/− cells (Fig. 3, A and B). Conversely, phosphorylation of VASP S235 was unchanged between cell types (Fig. S3 B). Total levels of PKA RII regulatory subunit and PKA-dependent phospho-substrates were unchanged between cells types (Fig. S3 A), which suggests that β3 regulates local rather than global PKA activation. Interestingly, β1 integrin–null cells did not show changes in VASP phosphorylation. Moreover, phosphorylation of the related family member Mena was unchanged between WT and β1−/− or β3−/− cells (as also detected by phospho-specific antibodies), which suggests that VASP may be specifically acting downstream of β3 integrin (Fig. 3 A and not depicted). Knockdown of endogenous β3 by siRNA in human dermal fibroblasts also resulted in decreased levels of phosphorylated VASP, which suggested that the differences observed in the β3−/− cells were not caused by compensatory mechanisms (Fig. S3 C).
Figure 3.
β3 regulates phosphorylation of VASP. (A) WT, β3−/−, or β3-GFP cells either untreated or treated with 1 µM H89 (PKA signaling inhibitor) or 1 µM forskolin (PKA signaling activator) were analyzed by Western blotting for total VASP, phospho-S153 VASP, or total Mena. WT and β1−/− cells were lysed and analyzed by Western blotting for total VASP. (B) Quantification of blots from WT, β3−/−, or β3-GFP cells shown in A. Data are mean ± SEM from four independent experiments (indicated by error bars). ***, P < 0.001 compared with WT control levels. (C) Confocal images of WT and β3−/− cells plated on 2D FN (left; the dotted lines indicate the edge of the cells) or CDM (right) and stained for VASP (FITC, green), vinculin (Alexa Fluor 568, red), and FN (CDM only, Alexa Fluor 633, blue). Merged, enlarged images are shown. Bars: (left) 5 µm; (right) 10 µm. (D) WT and β3−/− cells transfected with WT-VASP-GFP or S153A-VASP-GFP and subjected to FRAP analysis. τ1/2 values (s) were obtained from normalized recovery data for each construct in each cell type. Data are shown as mean values ± SEM (indicated by error bars) from at least 10 cells from four independent experiments. **, P < 0.01; *, P < 0.05 compared with WT VASP recovery in WT cells. (E) WT and β3−/− cells transfected with talin-GFP or vinculin-GFP and subjected to FRAP analysis. τ1/2 values (s) were obtained from normalized recovery data for each construct in each cell type. Data are shown as mean values ± SEM from at least 11 cells from three independent experiments. **, P < 0.01 compared with recovery in WT cells. (F) FRAP analysis of talin-mCherry recovery at GFP-positive adhesions in β1-GFP or β3-GFP cells. τ1/2 values (s) were obtained from normalized recovery data for each construct in each cell type. Data are shown as mean values ± SEM from at least 14 cells from four independent experiments (indicated by error bars). **, P < 0.01 compared with recovery in β1-GFP cells.
Confocal analysis of the localization of endogenous VASP in WT and β3−/− cells showed that VASP was recruited to small, peripheral adhesions in the WT cells, whereas VASP was absent from these adhesions in cells lacking β3 (Fig. 3 C). Confocal analysis of endogenous phospho-VASP also demonstrated predominant localization to the periphery of WT and β3-GFP cells, but very low signal in β3−/− cells (Fig. S3 D). To analyze the dynamics of VASP, WT, and β3−/−, cells were transfected with GFP-WT VASP, and the protein was subjected to FRAP analysis. Surprisingly, analysis of recovery in peripheral adhesions in each cell type revealed significantly slower recovery (τ1/2) of VASP in β3−/− cells (Fig. 3 D). To determine the contribution of S153 phosphorylation to the dynamics of VASP, FRAP analysis was also performed on both WT and β3−/− cells expressing GFP-S153A VASP (to mimic a de-phospho form of VASP). A significant increase in the τ1/2 recovery time of S153A VASP was seen compared with WT VASP in WT cells (Fig. 3 D), which suggests that dephosphorylation of VASP at this site results in increased residence time of the protein within peripheral adhesions. To determine whether loss of β3 also affected dynamics of other focal adhesion proteins, we performed FRAP analysis on both WT and β3−/− cells expressing GFP-talin or GFP-vinculin. Interestingly, data demonstrated a significant increase in the τ1/2 recovery time of talin in the β3−/− cells compared with WT (Fig. 3 E). Conversely, vinculin demonstrated similar recovery kinetics in both cell types (Fig. 3 E). This suggests that β3 integrin is specifically involved in regulating VASP and talin dynamics at adhesions. To further examine the relationship between specific integrin populations and talin recovery, we performed FRAP analysis on talin-mCherry expressed in β1-GFP or β3-GFP (expressed in respective null integrin backgrounds)–positive adhesions. Data revealed that talin recovery was significantly slower at β1- versus β3-rich adhesion sites (Fig. 3 F), which suggests that integrin-specific signals can act to differentially regulate talin dynamics.
β3 integrin restricts VASP binding to RIAM
To better understand how phosphorylation of VASP at S153 may be affecting subcellular localization of VASP, we expressed GFP-tagged WT, S153D, or S153A VASP in WT cells and analyzed localization with respect to active β1 integrin in fixed cells. Confocal images demonstrate partial colocalization between both WT and S153D VASP and active integrin at focal adhesions but not at the leading edge of new protrusions (Fig. 4 A). S153A VASP, however, showed markedly higher colocalization, with the majority of active integrin seen within focal adhesions, and this was confirmed by quantification using Pearson's correlation coefficient analysis (Fig. 4 A). VASP is a known binding partner for several focal adhesion proteins, including vinculin, αII spectrin, zyxin, and the MRL family protein RIAM (Brindle et al., 1996; Lafuente et al., 2004; Benz et al., 2008). As our data demonstrates that loss of β3 promoted retention of VASP within β1-containing focal adhesions, we reasoned that this could be caused by increased binding of VASP to a focal adhesion protein. To test this hypothesis, we immunoprecipitated endogenous VASP from WT, β3−/−, or β3-GFP rescued cells and probed the bound fractions for associated focal adhesion proteins. Vinculin, αII spectrin, and zyxin coimmunoprecipitated with VASP at equal levels in all cell types (Fig. S4 A). Conversely, levels of RIAM were strikingly higher in complex with VASP in lysates from β3−/− cells as compared with WT or β3-GFP–rescued cells, whereas total RIAM levels were unchanged between cell lysates (Fig. 4 B). To test whether the increased binding between VASP and RIAM seen in β3−/− cells was caused by loss of phosphorylation on VASP S153, ectopically expressed GFP-tagged WT, S153A, or S153D VASP was immunoprecipitated from cells, and complexes were probed for the presence of endogenous RIAM. Data showed that RIAM was associated at higher levels with S153A versus WT VASP, which supports our hypothesis that phosphorylation of this residue can indeed regulate binding to RIAM (Fig. S3 E).
Figure 4.
VASP-RIAM binding is enhanced in the absence of β3. (A, right) Confocal images of β3-GFP cells transfected with WT, S153A, or S153D VASP-mCherry fixed and stained for active β1 (9EG7–Alexa Fluor 568, blue). Merged images are shown. Bars, 10 µm. (A, left) The histogram shows Pearson's correlation coefficient analysis of colocalization between VASP and 9EG7. Error bars indicate ± SEM, n = 12 for each. **, P < 0.01. (B) WT, β3−/−, or β3-GFP cells were lysed and subjected to immunoprecipitation with anti-VASP or control antibodies. Bound and input lysates were Western blotted for RIAM or VASP. Blots are representative of similar results from three independent experiments. (C, left) Example images of WT and β3−/− cells transfected with GFP-VASP alone (left, control) or in combination with mCherry-RIAM (right) subjected to FRET analysis by FLIM. Images show the GFP multiphoton intensity image and (where appropriate) corresponding wide-field image of the acceptor. Lifetime images mapping spatial FRET in cells are depicted using a pseudocolor scale (blue, normal lifetime; red, FRET). (C, right) Histogram of quantification of FRET efficiency from FLIM data. The graph represents mean FRET efficiency of >14 cells of each type over four independent experiments. Error bars indicate ± SEM. **, P < 0.01. (D) Example FRET images for WT and β3−/− cells transfected with GFP-RIAM and mCherry-talin. Quantification of FRET efficiency from FLIM data as in C. The graph represents mean FRET efficiency of >12 cells of each type over three independent experiments. Error bars indicate ± SEM. *, P < 0.05. Bars, 10 µm.
To analyze this enhanced interaction in vivo, we coexpressed GFP-VASP and RIAM-mCherry in WT and β3−/− cells and analyzed direct interactions between these proteins by fluorescence resonance energy transfer (FRET) using fluorescence lifetime imaging microscopy (FLIM). This technique allows analysis of protein–protein interactions between a donor (GFP) and acceptor (mCherry) by measuring the fluorescence lifetime decay of the donor by time-correlated single photon counting. When donor and acceptor are in close proximity (<10 nm), energy transfer results in a shortening of the GFP fluorescence lifetime (Parsons et al., 2008; Worth and Parsons, 2008). Analysis of FRET lifetime images and cumulative FRET efficiency data (Fig. 4 C) confirmed that direct binding between VASP-RIAM was enhanced in β3−/− cells when compared with WT. Moreover, the interacting (FRET) population in β3−/− cells was predominantly at the cell periphery, compared with within discrete regions beneath the cell body in WT cells. Confocal analysis of endogenous active β1 and RIAM localization in β3-GFP rescue cells also revealed colocalization between β1 and RIAM, whereas both proteins were absent from β3-containing peripheral adhesions (Fig. S4 B). Furthermore, analysis of FRET between RIAM-GFP and talin-mCherry in WT and β3−/− cells also demonstrated a significant increase in the interaction between these proteins in cells lacking β3 (Fig. 4 D). These data together suggest that loss of β3 promotes enhanced association of a complex containing VASP, RIAM, and talin.
Phosphorylation of VASP regulates β1-talin binding
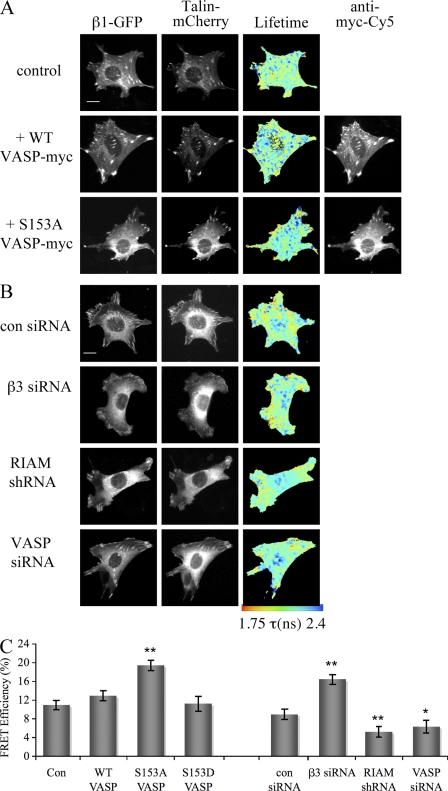
To further investigate whether VASP regulation of RIAM–talin binding had any downstream effect upon talin–integrin association, we used our previously established model to measure FRET between β1-GFP (expressed in β1−/− fibroblasts) and talin-mCherry (Parsons et al., 2008). As shown in lifetime images and FRET efficiency histograms (Fig. 5, A and C), coexpression of a myc-tagged form of S153A VASP in these cells led to a significant increase in the interaction between β1-GFP and talin compared with cells expressing endogenous or overexpressed WT VASP-myc. To reconfirm our findings that a β3 integrin–dependent VASP-RIAM signaling event acts to negatively regulate β1-talin binding, we knocked down endogenous β3, VASP, or RIAM in β1-GFP cells using siRNA/small hairpin RNA (Fig. S3 F) and measured binding between β1 and talin by FRET. Lifetime images and cumulative FRET efficiency analysis both demonstrate that a reduction in levels of β3 integrin promoted binding between β1 and talin; conversely, reduction of either RIAM or VASP led to a reduction in β1–talin binding (Fig. 5, B and C). Western blotting of β1-GFP cells expressing β3 siRNA also demonstrated a decrease in the higher molecular weight migrating species of VASP, confirming our findings that loss of β3 leads to loss of phosphorylated VASP at S153 (Fig. 3, A and B; and Fig. S3 F).
Figure 5.
VASP regulates the interaction between β1 and talin. (A) Example FLIM images of β1-GFP–expressing fibroblasts cotransfected with mCherry-talin and either WT-VASP-myc or S153A VASP-myc. Cells were fixed and stained with an anti–myc-Cy5 antibody to detect VASP expression. Cells were then subjected to FRET analysis by FLIM. Images show the GFP multiphoton intensity image and (where appropriate) corresponding wide-field image of the acceptor and myc-tagged VASP. Lifetime images mapping spatial FRET across the cells are depicted using a pseudocolor scale (blue, normal lifetime; red, FRET). Bars, 10 µm. (B) Example FLIM images of β1-GFP–expressing fibroblasts cotransfected with mCherry-talin and either control siRNA (con) or siRNA directed against β3 integrin, RIAM, or VASP. Cells were fixed and subjected to FRET analysis by FLIM. Images show the GFP multiphoton intensity image and corresponding wide-field image of the talin acceptor. Lifetime images mapping spatial FRET across the cells are depicted using a pseudocolor scale (blue, normal lifetime; red, FRET). (C) Quantification of FRET efficiency from FLIM data in A and B. The graphs represent mean FRET efficiency of >12 cells of each type over three independent experiments. Error bars indicate ± SEM. *, P < 0.05; **, P < 0.01.
To determine whether the effects of VASP on integrin–talin binding translated into functional regulation of β1 integrin dynamics, we performed FRAP analysis on β1-GFP alone or in cells coexpressing WT, S153A, or S153D VASP-mCherry (Fig. 6, A and B). Interestingly, data demonstrated a significant decrease in the τ1/2 recovery time of β1 in cells coexpressing S153A VASP compared with either control, WT, or S153D VASP-transfected cells (Fig. 6 B), which suggests that VASP phosphorylation status can alter both talin binding to β1 and β1 dynamics at adhesions. As loss of β3 integrin and/or loss of VASP S153 phosphorylation leads to increased activation of β1, this would suggest that active β1 demonstrates reduced τ1/2 recovery time in FRAP experiments. This is in contrast to the increased τ1/2 recovery time seen for talin in cells lacking β3 (Fig. 3 E). Interestingly, additional analysis of the immobile fraction of β1-GFP in these experiments revealed an increase in the immobile fraction of β1 in cells coexpressing S153A VASP compared with WT VASP (62.6% vs. 43.7%, respectively; P = 0.005). These novel observations also suggest a model whereby increased residence time of proteins within focal adhesions may be required to induce integrin activation, leading to increased dynamic recovery of the integrin itself within the same adhesion site.
Figure 6.
VASP phosphorylation regulates β1 integrin dynamics. (A) Example images of FRAP analysis of β1-GFP before and after bleaching. The arrow denotes the region of interest subjected to photobleaching. Bar, 10 µm. (B) Quantification of FRAP analysis of β1-GFP recovery in mock transfected (control) cells or those coexpressing WT, S153A, or S153D VASP-mCherry. τ1/2 values (s) were obtained from normalized recovery data for β1-GFP in each condition. Data are shown as mean values ± SEM from 12 cells from three independent experiments (indicated by error bars). **, P < 0.01 compared with recovery in control cells.
VASP phosphorylation regulates adhesion dynamics and cell migration speed
Our data demonstrates that loss of β3 results in increased adhesion turnover coupled with increased cell migration speed but with lower directional persistence. These phenotypes in β3−/− cells are coincident with decreased levels of S153-phosphorylated VASP. To determine whether β3-mediated signaling to VASP plays a role in fibroblast adhesion dynamics, we measured rate constants of adhesion assembly and disassembly of GFP-talin expressed in WT, β3−/−, or β3−/− cells expressing S153D VASP. Analysis revealed that the adhesion-enhanced assembly and disassembly seen in β3−/− cells was restored to WT levels in β3−/− cells expressing S153D VASP (Fig. 7 A). This suggests that forcing a balance of phosphorylated and nonphosphorylated VASP is sufficient to restore normal adhesion dynamics in cells lacking β3 integrin. To determine whether this rescue of adhesion kinetics was coupled with a rescue in migration, we performed phase-contrast time-lapse microscopy on WT or β3−/− cells expressing S153A or S153D VASP mCherry. Data shown in Fig. 7 B and Fig. S3 G demonstrate that expression of S153D VASP did indeed partially decrease enhanced migration speeds seen in β3−/− cells. However, the decreased directional persistence in β3−/− cells was unchanged by expression of either VASP mutant. Interestingly, expression of S153A but not S153D resulted in increased speed and decreased migration persistence in WT cells. To determine whether this S153D VASP-induced rescue in migration speed was coupled with changes in activation of β1, we analyzed 9EG7 intensity in β3, VASP, or RIAM-depleted cells and β3−/− cells expressing VASP mutants. Data demonstrated that β3 depletion in WT cells resulted in increased active β1, whereas expression of S153D VASP in β3−/− cells lead to reduced active β1 compared with nonexpressing β3−/− cells in the same samples (Fig. 7 C). This suggests that β3-dependent regulation of adhesion dynamics, β1 activation, and migration speed are controlled at least in part through regulation of VASP phosphorylation. However, directional migration appears to be regulated by an additional β3-dependent pathway.
Figure 7.
Regulation of cell migration speed by VASP phosphorylation. (A) Rate constants of focal adhesion dynamics in WT and β3−/− cells or β3−/− cells cotransfected with S153D VASP-mCherry. Data are quantified from time-lapse videos of cells expressing GFP-talin. Data are expressed as min−1 ± SEM. Data were quantified from a minimum of 200 adhesions per condition over three experiments. (B) WT or β3−/− cells were mock transfected or transfected with S153A or S153D VASP-mCherry, plated onto FN-coated plates, and imaged using phase-contrast time-lapse microscopy over 16 h. Frames were taken every 10 min. Cells were tracked and migration speed and directional persistence were quantified as in Fig. 1. Data were quantified from >75 cells per condition over four independent experiments. Data are shown as mean ± SEM (indicated by error bars). (C) Analysis of intensity levels of 9EG7 (active β1) in cells expressing specified constructs. Example images of 9EG7 staining are shown underneath the graph for each sample type analyzed. Bars, 10 µm. 10 Error bars indicate ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (D) Proposed model of β3 regulation of β1 activation through control of VASP, RIAM, and talin. In normal fibroblasts (left), the β3 is located predominantly at the periphery of the cell, where it acts to maintain a local phosphorylated pool of VASP. In the absence of β3 (right), VASP is predominantly dephosphorylated at S153 (or S157), leading to enhanced VASP binding to RIAM. This results in increased RIAM association with talin and enhanced talin binding to β1 integrin, which leads to increased activation of β1 at peripheral adhesion sites, increased cell protrusion, and faster, less persistent cell migration.
Discussion
Our studies of β3-null fibroblasts have revealed that this integrin plays a role in locally suppressing activation of β1 integrin to promote efficient persistent cell protrusion and migration. We show that β3−/− cells have increased migration speed and rates of adhesion assembly/disassembly, coupled with increased cell surface levels of active β1 integrin and decreased levels of phosphorylated VASP. Expression of a S153A mutant of VASP promotes formation of complexes between VASP, RIAM, and talin; increased residence times of talin within adhesions; and increased talin-dependent activation of β1 integrin. S153D VASP expression partially rescued levels of active β1 and migration speed defects in β3−/− cells; however, no effect was seen on the loss of persistent migration. Based on this evidence, we propose a model (Fig. 7 D) whereby in normal migrating fibroblasts, β3 integrin localizes predominantly to peripheral adhesion sites, where it retains a pool of phospho-VASP through a PKA-dependent mechanism. Under these conditions, very little VASP is associated with RIAM, and RIAM instead associates with talin (and other binding partners) at β1-rich focal adhesions. However, loss of β3 results in the loss of S153-phosphorylated VASP, thus promoting formation of a VASP–RIAM–talin complex at peripheral β1-containing adhesions, which results in enhanced activation of β1, more rapid adhesion turnover, and increased speed of migration.
Transdominant inhibition between integrin β subunits has previously been suggested to occur as a result of competition for binding to talin (Calderwood et al., 2004). Our data would suggest that β3 is able to act transdominantly upon β1 integrin in fibroblasts, in part through a surprisingly direct pathway under the control of VASP that results in restricted spatial association between β1 and talin. This data favors an active signaling-dependent rather than passive stoichiometry model that governs the local association between integrins and talin. Whether such a model also applies to the other recently characterized family of integrin-activation proteins, the kindlins, remains to be tested. Recent studies have suggested that β1 and β3 integrins require association with both talin and kindlins to assume a fully activated conformation (Ma et al., 2008; Harburger et al., 2009). Furthermore, filamin is an additional β integrin–binding protein that potentially competes with talin for binding to the integrin tails (Kiema et al., 2006) and can also bind to migfilin (Lad et al., 2008). The cooperation between these alternative integrin-binding partners in complex with one another remains poorly understood, and it is likely that the balance between such complexes plays a key role in regulating kinetics of integrin activation. It will be important to address in future studies whether β3 also plays a role in regulating spatial interactions between these proteins.
Our data demonstrates a role for VASP phosphorylation in regulating both binding of VASP to RIAM and control of integrin activation. Although VASP phosphorylation has been previously implicated in the regulation of actin polymerization in vitro, little is known about functional regulation of VASP binding partners after phosphorylation by PKA (Barzik et al., 2005). Interestingly, a recent study has shown that dephosphorylation of VASP at S153 may be required for the growth of adhesion strength during membrane retraction in microglia, which supports a role for VASP in regulation of integrin function (Lee and Chung, 2009). Previous biochemical studies have shown that RIAM is a binding partner for talin, and that formation of this complex can have a direct effect upon integrin activation (Lafuente et al., 2004; Han et al., 2006; Lee et al., 2009). More recently, an N-terminal helical motif within RIAM was identified as the putative binding sequence for talin, and coupling of this minimal sequence to the membrane-targeting domain of Rap1 was sufficient to induce activation of overexpressed αIIbβ3 integrin (Lee et al., 2009). The same study reported that the VASP-binding site in RIAM (previously mapped to the C terminus of RIAM; Lafuente et al., 2004) was not required for RIAM to bind talin in vitro. However, as this study did not compare the ability of full-length versus VASP-binding mutants of RIAM to activate integrins, the observed positive contribution of VASP reported here remains a possible mechanism of regulation in other model systems. Furthermore, we cannot rule out the possibility that β3-dependent changes in Rap1 signaling may also be occurring in our model and possibly contributing to the enhanced β1–talin binding we observe in β3-null cells. Further detailed analysis of spatial activation of Rap1 will be required to determine the relative contribution of these pathways to β3-dependent regulation of RIAM and talin.
Our data also suggests that the integrin-dependent cycling of phospho-VASP may act to partition VASP into different compartments within the cell. Spatial control of VASP phosphorylation may in part be mediated through localized PKA activation. PKA can be activated in an integrin-dependent manner at the leading edge of migrating cells, and in the case of α4β1 integrin, this effect is mediated directly through the ability of the integrin to act as an A-kinase anchoring protein (AKAP; Lim et al., 2007; Lim et al., 2008). In addition, β1 is known to activate and recruit phosphatases (Mutz et al., 2006; Gonzalez et al., 2008) and may therefore promote dephosphorylation of VASP. Our data suggests that this would lead to increased VASP residence time at adhesions, resulting in a RIAM-dependent positive feedback loop to increase β1 activation and adhesion turnover through cooperation with talin (Lafuente et al., 2004; Han et al., 2006; Lee et al., 2009). This model further strengthens our hypothesis that crosstalk between integrins is tightly regulated within discrete subcellular structures and that relatively minor changes to the dynamics of these key adhesion proteins can result in dramatic changes to cell protrusion and migration.
Data presented here also shows that β3−/− cells have increased formation of free barbed ends and enhanced assembly of F-actin–containing protrusions. The mechanism by which β3 regulates actin assembly is currently unclear. Ena/VASP proteins have previously been shown to have potent effects on free barbed end formation (Furman et al., 2007; Philippar et al., 2008). The combined activity of VASP coupled to increased association of VASP with RIAM in the absence of β3 provides one possible explanation for the increased barbed end formation in these cells. Rho GTPases are also primary targets for candidate downstream regulators, and in particular, regulation of the actin-severing protein cofilin by Rho represents one potential pathway by which integrins can regulate actin assembly. Conflicting studies in the literature suggest that engagement of β3 integrin can both activate and inactivate cofilin under different conditions (Danen et al., 2005; Falet et al., 2005; White et al., 2007). Further experiments to study spatial regulation of cofilin and other actin regulatory proteins in integrin-null cells may provide more clues as to how these dynamic processes are regulated to separate adhesion assembly by integrin signaling during motility. Interestingly, our data would suggest that β3 can control focal adhesion assembly and migration speed in part through VASP, but that β3-dependent regulation of directed motility operates through an alternative and yet-undefined pathway. Uncoupling of these two distinct parameters has highlighted an additional layer of complexity in the cooperation between β1 and β3 integrins. Very few specific signaling events have been attributed to these seemingly functionally overlapping integrin β subunits within the same cellular context. Of note, one candidate that has received recent attention is the nonreceptor tyrosine kinase Src, which has previously been shown to bind directly to the cytoplasmic tail of β3 (Arias-Salgado et al., 2003). This interaction is proposed to regulate β3-dependent cell spreading and contractility as well as oncogenic transformation and anchorage-independent tumor cell growth (Flevaris et al., 2007; Huveneers et al., 2007; Desgrosellier et al., 2009). Src can phosphorylate several adhesion and cytoskeletal-associated substrates, and as such represents an attractive potential mediator downstream of β3 in the regulation of both F-actin assembly and migration persistence. Furthermore, proteomic-based screens to identify integrin-specific signaling complexes such as the one recently reported for α4β1 and α5β1 (Humphries et al., 2009) will provide additional novel targets for the dissection of functional differences between members of this receptor family within the same cell.
In summary, we have uncovered a novel VASP-dependent signaling mechanism downstream of β3 integrin that contributes toward local inhibition of β1 activation and regulates adhesion assembly and fibroblast migration speed. Our findings highlight the importance of dissecting such intricate, dynamic, and highly spatially regulated signaling networks to better understand the regulation of integrin-dependent adhesion and motility.
Materials and methods
Antibodies and reagents
Antibodies were obtained from the following suppliers: mouse anti–β-actin, anti-vinculin, anti-zyxin, and anti-talin (all from Sigma-Aldrich); mouse anti–total β1 and anti–rat active β1 (9EG7; both from Millipore); rabbit anti-β3 (AbD Serotec); mouse anti-paxillin (BD); FAK, FAK397(P), FAK 407(P), and FAK576(P) (all from Invitrogen); αII spectrin (a gift from P. Bennett, King's College London, London, England, UK); rabbit anti-FN (2413; Abcam); control IgG (Dako); glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Sigma-Aldrich); rabbit anti-S157(P) VASP (3111; Cell Signaling Technology); rabbit anti-VASP (2010) and mouse anti-RIAM (5541) as described in Lafuente et al. (2004); rabbit anti-PKA substrate antibody (Cell Signaling Technology); mouse anti-PKA RII antibody (Abcam); secondary HRP antibodies for Western blotting (all from Dako); and secondary fluorescently labeled antibodies (all from Invitrogen). G-actin-568 was obtained from Sigma-Aldrich, phalloidin-488 was obtained from Invitrogen, H89 and Forskolin were obtained from EMD; and human plasma FN was obtained from BD.
Constructs and siRNA
WT and mutant VASP constructs described previously (Barzik et al., 2005) were subcloned in GFP-C1 (Takara Bio Inc.) or mCherry-C1 (a gift from R. Tsien, University of California, San Diego, La Jolla, CA) for imaging. GFP- and mCherry-talin were gifts from K. Yamada (National Institutes of Health, Bethesda, MD), and mRFP-vinculin was as described in Hashimoto et al. (2007). GFP-actin was as in Zicha et al. (2003). β3-GFP retrovirus was generated using human β3-N1-GFP (a gift from N. Kieffer, Centre National de la Recherche Scientifique/GDRE-ITI, University of Luxembourg, Luxembourg). The full β3-GFP sequence was amplified by PCR from the backbone using the following primers: (forward) 5′-CCCAAGCTTATGCGAGCGCGGCCG-3′ and (reverse) 5′-CCCATCGATTTACTTGTACAGCTCG-3′. The PCR product was digested and ligated into the HindIII site (5′) and ClaI site (3′) in pLCPX retroviral vector (Takara Bio Inc.). The construct was verified by sequencing before use. Control and On-target siRNA oligos against mouse VASP and mouse or human β3 integrin were obtained from Thermo Fisher Scientific. The small hairpin RNA construct to RIAM was as described in Lafuente et al. (2004).
Cell culture and CDM preparation
Fibroblasts were isolated from WT and β3-null mice and immortalized in culture as described previously (Hodivala-Dilke et al., 1999). Cells were cultured in DME containing 10% FCS plus penicillin, streptomycin, glutamine, and 20 U/ml IFN-γ (all from Sigma-Aldrich) and maintained at 33°C and 5% CO2. Cells isolated from at least five different animals for both WT and β3−/− were used throughout the studies. WT, β1-GFP, and β1−/− cells were derived and cultured in DME containing 10% FCS plus penicillin, streptomycin, glutamine, and 20 U/ml IFN-γ, as described previously (Parsons et al., 2008). Immortalized human dermal fibroblasts were cultured in α-MEM (Sigma-Aldrich) containing 10% FCS plus penicillin and streptomycin. CDMs were prepared as described previously (Cukierman et al., 2001; Caswell et al., 2007). In brief, gelatin-coated coverslips were cross-linked with glutaraldehyde, quenched, and equilibrated in DME containing 10% FCS. Primary human dermal fibroblasts (TCS Cellworks) were seeded at confluence and grown for 10–14 d in DME containing 10% FCS and 50 µg/ml ascorbic acid. Matrices were denuded of living cells by incubation with PBS containing 20 mM NH4OH and 0.5% Triton X-100, and DNA residue was removed by incubation with DNaseI. Matrices were blocked with 0.1% heat-denatured BSA before the seeding of cells.
Transfections and β3-GFP stable cell line generation
All transfections were performed using FuGene 6 reagent (Roche) or Dharmafect1 (for siRNA; Thermo Fisher Scientific) according to the manufacturer's instructions. Cells were plated at 50% confluency in 6-well plates and transfected using 0.5–2 µg/well of DNA (depending on the construct used) or 5 nmol of siRNA suspended in Optimem (Invitrogen). β3-GFP stable cells were generated by infecting β3−/− cells with β3-GFP retrovirus using AM12 packaging cells as described previously (Parsons et al., 2008). GFP-positive cells were selected by FACS and maintained under identical conditions to parent cells.
Immunoprecipitation and Western blotting
Cells were lysed in RIPA buffer (10 mM Tris, pH7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 10 µM sodium fluoride, and 1 µM okadaic acid with protease inhibitor complex [EMD]). Cells were centrifuged at 13,000 rpm for 5 min, and lysate protein concentration was determined with the BCA assay (Thermo Fisher Scientific). For Western analysis, protein was loaded onto polyacrylamide gels and transferred to polyvinylidene fluoride membranes. Membranes were blocked in TBS-T and 5% nonfat milk, and incubated with the specified primary antibodies in TBS-T overnight at 4°C. The immunoblots were washed in TBS-T and incubated for 2 h with HRP-conjugated secondary antibodies. Membranes were then washed with TBS-T and visualized with ECL substrate reagent (Thermo Fisher Scientific). For immunoprecipitation, lysates were precleared with protein A/G beads (Santa Cruz Biotechnology, Inc.) and incubated with the indicated antibodies followed by incubation with protein A/G beads for 2 h at 4°C. Beads were washed three times in lysis buffer before being analyzed by Western blotting.
FACS
Cells were scraped into PBS on ice and centrifuged, and then the supernatant was removed. The cells were resuspended and incubated with phalloidin (1:100; Invitrogen) or 9EG7 (Millipore) on ice for 60 min. Cells were washed and resuspended where necessary in secondary antibody (Dako) and incubated on ice for 30 min. The supernatant was removed and the cells were resuspended in 250 µl of FACS Flow ready to be read by FACS Calibur (BD) using the CellQuest Pro version 4.0.2 software (BD).
Free barbed end assay and quantification of immunostaining
Analysis of actin free barbed-end formation was performed as described previously (Furman et al., 2007). In brief, cells were permeabilized with 0.125 mg/ml saponin in the presence of 0.5 mM Alexa Fluor 568–conjugated G-actin (Invitrogen). After a 2-min labeling period, samples were fixed in 0.5% glutaraldehyde, permeabilized with 0.5% Triton X-100, and blocked in the presence of Alexa Fluor 488–phalloidin (Invitrogen). Images were acquired using a microscope (A1R; Nikon) equipped with a CFI Plan-Fluor 40× oil objective lens. For quantification of 9EG7 intensity, coverslips were imaged using identical laser settings as for the free barbed-end assay. In both experimental setups, look-up tables were used to ensure that pixel saturation was not reached, and the intensity per pixel was quantified using ImageJ software according to common background subtraction.
Live cell imaging and kymography
Phase-contrast imaging of cells was performed on a microscope (Axio100; Carl Zeiss, Inc.) equipped with a charge-coupled device camera (Sensicam; PCO), motorized stage (Ludl), excitation/emission filters (Chroma Technology Corp.) and filter wheels (Ludl). Images were acquired using a 40× oil objective (for single cell analysis) or 10× phase objective (for random motility). Kymography analysis of protrusion was performed using the ImageJ plugin (http://rsb.info.nih.gov/ij/). Six lines were quantified from 12 cells over three independent experiments.
Confocal microscopy, adhesion dynamics, and FRAP analysis
For all confocal experiments, cells were plated on glass coverslips coated with either 10 µg/ml FN or CDM as specified in the figure legends. Confocal microscopy and FRAP experiments were performed on a microscope (A1R; Nikon) equipped with a CFI Plan-Fluor 40× oil objective lens. Images were captured, analyzed, and exported using AR software (NIS Elements). Analysis of FRAP data was performed as in Humphries et al. (2007). In brief, raw intensity data were corrected for background fading during imaging and plotted as the percentage recovery over time, and τ1/2 values were calculated from the resultant curves. Data were pooled from at least 12 cells over five independent experiments. Focal adhesion dynamics were imaged and analyzed in transfected cells as described previously (Hashimoto et al., 2007). Rate constants of assembly and disassembly were determined from videos of cells expressing GFP-talin plated on either 2D FN or within CDM. Images were acquired on a confocal microscope (A1R). In the case of cells within CDM, rapid z stacks were acquired throughout the entire cell, and 3D projections were assembled before analysis. Rates of assembly and disassembly of adhesions were calculated by measuring the incorporation or loss of fluorescent signal of the protein over time. An increase in signal is a result of adhesion assembly and growth, whereas disassembly results in a loss of fluorescent signal. These intensity values were plotted over time on semilogarithmic graphs to provide a profile of intensity ratios over time. These ratios were calculated using the formula In(I/I0) for assembly and In(I0/I) for disassembly (where I0 is the initial fluorescence intensity value and I is the intensity value for the relevant time point). Rates were then calculated from the gradient of the line of best fit.
FRET/FLIM
Fluorescence lifetime imaging was performed and data were analyzed as described previously (Parsons et al., 2008). We used FLIM to measure FRET between protein pairs, which allows the determination of spatial protein interactions. Time-domain FLIM was performed with a multiphoton microscope system as described previously (Parsons et al., 2008). Fluorescence lifetime imaging capability was provided by time-correlated single photon counting electronics (SPC 700; Becker & Hickl). A 40× objective was used throughout (CFI60 Plan-Fluor, NA 1.3; Nikon) and data were collected at 500 ± 20 nm through a bandpass filter (35–5040; Coherent, Inc.). Laser power was adjusted to give mean photon counting rates of the order 104–105 photons s − 1 (0.0001–0.001 photon counts per excitation event) to avoid pulse pileup. Acquisition times up to 300 s at low excitation power were used to achieve sufficient photon statistics for fitting, while avoiding either pulse pileup or significant photobleaching. Excitation was at 890 nm. Wide-field acceptor (mRFP) or Cy5 images were acquired using a charge-coupled device camera (Hamamatsu Photonics) at <100 ms exposure times.
Data were analyzed as described previously (Parsons et al., 2008) using TRI2 software (developed by P. Barber, University of Oxford, Oxford, England, UK). Histogram data presented here are plotted as mean FRET efficiency from >10 cells per sample. Mean FRET efficiency is shown ± SEM. Analysis of variance (ANOVA) was used to test statistical significance between different populations of data. Lifetime images of example cells are presented using a pseudocolor scale whereby blue depicts normal GFP lifetime (no FRET) and red depicts lower GFP lifetime (areas of FRET).
Online supplemental material
Fig. S1 shows levels of focal adhesion proteins in WT and β3−/− cells. Fig. S2 shows the percentage of dynamic adhesions in WT and β3−/− cells. Fig. S3 shows regulation of VASP phosphorylation in WT and β3−/− or β3-depleted cells. Fig. S4 shows regulation of ASP binding partners by β3 integrin. Video 1 shows random migration of WT cells on 2D FN. Video 2 shows random migration of β3−/− cells on 2D FN. Video 3 shows migration of WT cells on CDM. Video 4 shows migration of β3−/− on CDM. Video 5 shows GFP-actin dynamics in WT cells on 2D FN. Video 6 shows GFP-actin dynamics in β3−/− cells on 2D FN. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200912014/DC1.
Acknowledgments
We would like to thank Nelly Kieffer and Ken Yamada for the kind gifts of reagents and Marjolein Snippe for technical assistance.
This work was supported by funding from the Royal Society (University Research Fellowship to M. Parsons), the Biotechnology and Biological Sciences Research Council (BBSRC; to M. Parsons and D.C. Worth) and National Institutes of Health grant GM50081 (to F.B. Gertler).
Footnotes
Abbreviations used in this paper:
- CDM
- cell-derived matrix
- Ena/VASP
- Enabled/protein vasodilator-stimulated phosphoprotein
- FLIM
- fluorescence lifetime imaging microscopy
- FN
- fibronectin
- FRET
- fluorescence resonance energy transfer
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- RIAM
- Rap1-GTP–interacting adaptor molecule
- WT
- wild type
References
- Arias-Salgado E.G., Lizano S., Sarkar S., Brugge J.S., Ginsberg M.H., Shattil S.J. 2003. Src kinase activation by direct interaction with the integrin beta cytoplasmic domain. Proc. Natl. Acad. Sci. USA. 100:13298–13302 10.1073/pnas.2336149100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaout M.A., Goodman S.L., Xiong J.P. 2007. Structure and mechanics of integrin-based cell adhesion. Curr. Opin. Cell Biol. 19:495–507 10.1016/j.ceb.2007.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballestrem C., Hinz B., Imhof B.A., Wehrle-Haller B. 2001. Marching at the front and dragging behind: differential alphaVbeta3-integrin turnover regulates focal adhesion behavior. J. Cell Biol. 155:1319–1332 10.1083/jcb.200107107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzik M., Kotova T.I., Higgs H.N., Hazelwood L., Hanein D., Gertler F.B., Schafer D.A. 2005. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J. Biol. Chem. 280:28653–28662 10.1074/jbc.M503957200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz P.M., Blume C., Moebius J., Oschatz C., Schuh K., Sickmann A., Walter U., Feller S.M., Renné T. 2008. Cytoskeleton assembly at endothelial cell-cell contacts is regulated by αII-spectrin-VASP complexes. J. Cell Biol. 180:205–219 10.1083/jcb.200709181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz P.M., Blume C., Seifert S., Wilhelm S., Waschke J., Schuh K., Gertler F., Münzel T., Renné T. 2009. Differential VASP phosphorylation controls remodeling of the actin cytoskeleton. J. Cell Sci. 122:3954–3965 10.1242/jcs.044537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindle N.P., Holt M.R., Davies J.E., Price C.J., Critchley D.R. 1996. The focal-adhesion vasodilator-stimulated phosphoprotein (VASP) binds to the proline-rich domain in vinculin. Biochem. J. 318:753–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broussard J.A., Webb D.J., Kaverina I. 2008. Asymmetric focal adhesion disassembly in motile cells. Curr. Opin. Cell Biol. 20:85–90 10.1016/j.ceb.2007.10.009 [DOI] [PubMed] [Google Scholar]
- Calderwood D.A., Tai V., Di Paolo G., De Camilli P., Ginsberg M.H. 2004. Competition for talin results in trans-dominant inhibition of integrin activation. J. Biol. Chem. 279:28889–28895 10.1074/jbc.M402161200 [DOI] [PubMed] [Google Scholar]
- Caswell P., Norman J. 2008. Endocytic transport of integrins during cell migration and invasion. Trends Cell Biol. 18:257–263 10.1016/j.tcb.2008.03.004 [DOI] [PubMed] [Google Scholar]
- Caswell P.T., Spence H.J., Parsons M., White D.P., Clark K., Cheng K.W., Mills G.B., Humphries M.J., Messent A.J., Anderson K.I., et al. 2007. Rab25 associates with alpha5beta1 integrin to promote invasive migration in 3D microenvironments. Dev. Cell. 13:496–510 10.1016/j.devcel.2007.08.012 [DOI] [PubMed] [Google Scholar]
- Cukierman E., Pankov R., Stevens D.R., Yamada K.M. 2001. Taking cell-matrix adhesions to the third dimension. Science. 294:1708–1712 10.1126/science.1064829 [DOI] [PubMed] [Google Scholar]
- Danen E.H., van Rheenen J., Franken W., Huveneers S., Sonneveld P., Jalink K., Sonnenberg A. 2005. Integrins control motile strategy through a Rho-cofilin pathway. J. Cell Biol. 169:515–526 10.1083/jcb.200412081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desgrosellier J.S., Barnes L.A., Shields D.J., Huang M., Lau S.K., Prévost N., Tarin D., Shattil S.J., Cheresh D.A. 2009. An integrin alpha(v)beta(3)-c-Src oncogenic unit promotes anchorage-independence and tumor progression. Nat. Med. 15:1163–1169 10.1038/nm.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falet H., Chang G., Brohard-Bohn B., Rendu F., Hartwig J.H. 2005. Integrin alpha(IIb)beta3 signals lead cofilin to accelerate platelet actin dynamics. Am. J. Physiol. Cell Physiol. 289:C819–C825 10.1152/ajpcell.00587.2004 [DOI] [PubMed] [Google Scholar]
- Flevaris P., Stojanovic A., Gong H., Chishti A., Welch E., Du X. 2007. A molecular switch that controls cell spreading and retraction. J. Cell Biol. 179:553–565 10.1083/jcb.200703185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman C., Sieminski A.L., Kwiatkowski A.V., Rubinson D.A., Vasile E., Bronson R.T., Fässler R., Gertler F.B. 2007. Ena/VASP is required for endothelial barrier function in vivo. J. Cell Biol. 179:761–775 10.1083/jcb.200705002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith C.G., Yamada K.M., Galbraith J.A. 2007. Polymerizing actin fibers position integrins primed to probe for adhesion sites. Science. 315:992–995 10.1126/science.1137904 [DOI] [PubMed] [Google Scholar]
- Gonzalez A.M., Claiborne J., Jones J.C. 2008. Integrin cross-talk in endothelial cells is regulated by protein kinase A and protein phosphatase 1. J. Biol. Chem. 283:31849–31860 10.1074/jbc.M801345200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Lim C.J., Watanabe N., Soriani A., Ratnikov B., Calderwood D.A., Puzon-McLaughlin W., Lafuente E.M., Boussiotis V.A., Shattil S.J., Ginsberg M.H. 2006. Reconstructing and deconstructing agonist-induced activation of integrin alphaIIbbeta3. Curr. Biol. 16:1796–1806 10.1016/j.cub.2006.08.035 [DOI] [PubMed] [Google Scholar]
- Harburger D.S., Bouaouina M., Calderwood D.A. 2009. Kindlin-1 and -2 directly bind the C-terminal region of beta integrin cytoplasmic tails and exert integrin-specific activation effects. J. Biol. Chem. 284:11485–11497 10.1074/jbc.M809233200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y., Parsons M., Adams J.C. 2007. Dual actin-bundling and protein kinase C-binding activities of fascin regulate carcinoma cell migration downstream of Rac and contribute to metastasis. Mol. Biol. Cell. 18:4591–4602 10.1091/mbc.E07-02-0157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodivala-Dilke K.M., McHugh K.P., Tsakiris D.A., Rayburn H., Crowley D., Ullman-Culleré M., Ross F.P., Coller B.S., Teitelbaum S., Hynes R.O. 1999. Beta3-integrin-deficient mice are a model for Glanzmann thrombasthenia showing placental defects and reduced survival. J. Clin. Invest. 103:229–238 10.1172/JCI5487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe A.K., Hogan B.P., Juliano R.L. 2002. Regulation of vasodilator-stimulated phosphoprotein phosphorylation and interaction with Abl by protein kinase A and cell adhesion. J. Biol. Chem. 277:38121–38126 10.1074/jbc.M205379200 [DOI] [PubMed] [Google Scholar]
- Humphries J.D., Wang P., Streuli C., Geiger B., Humphries M.J., Ballestrem C. 2007. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J. Cell Biol. 179:1043–1057 10.1083/jcb.200703036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries J.D., Byron A., Bass M.D., Craig S.E., Pinney J.W., Knight D., Humphries M.J. 2009. Proteomic analysis of integrin-associated complexes identifies RCC2 as a dual regulator of Rac1 and Arf6. Sci. Signal. 2:ra51 10.1126/scisignal.2000396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huveneers S., van den Bout I., Sonneveld P., Sancho A., Sonnenberg A., Danen E.H. 2007. Integrin alpha v beta 3 controls activity and oncogenic potential of primed c-Src. Cancer Res. 67:2693–2700 10.1158/0008-5472.CAN-06-3654 [DOI] [PubMed] [Google Scholar]
- Hynes R.O. 2002. Integrins: bidirectional, allosteric signaling machines. Cell. 110:673–687 10.1016/S0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- Kiema T., Lad Y., Jiang P., Oxley C.L., Baldassarre M., Wegener K.L., Campbell I.D., Ylänne J., Calderwood D.A. 2006. The molecular basis of filamin binding to integrins and competition with talin. Mol. Cell. 21:337–347 10.1016/j.molcel.2006.01.011 [DOI] [PubMed] [Google Scholar]
- Lad Y., Jiang P., Ruskamo S., Harburger D.S., Ylänne J., Campbell I.D., Calderwood D.A. 2008. Structural basis of the migfilin-filamin interaction and competition with integrin beta tails. J. Biol. Chem. 283:35154–35163 10.1074/jbc.M802592200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafuente E.M., van Puijenbroek A.A., Krause M., Carman C.V., Freeman G.J., Berezovskaya A., Constantine E., Springer T.A., Gertler F.B., Boussiotis V.A. 2004. RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev. Cell. 7:585–595 10.1016/j.devcel.2004.07.021 [DOI] [PubMed] [Google Scholar]
- Le Clainche C., Carlier M.F. 2008. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol. Rev. 88:489–513 10.1152/physrev.00021.2007 [DOI] [PubMed] [Google Scholar]
- Lee S., Chung C.Y. 2009. Role of VASP phosphorylation for the regulation of microglia chemotaxis via the regulation of focal adhesion formation/maturation. Mol. Cell. Neurosci. 42:382–390 10.1016/j.mcn.2009.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.S., Lim C.J., Puzon-McLaughlin W., Shattil S.J., Ginsberg M.H. 2009. RIAM activates integrins by linking talin to ras GTPase membrane-targeting sequences. J. Biol. Chem. 284:5119–5127 10.1074/jbc.M807117200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C.J., Han J., Yousefi N., Ma Y., Amieux P.S., McKnight G.S., Taylor S.S., Ginsberg M.H. 2007. Alpha4 integrins are type I cAMP-dependent protein kinase-anchoring proteins. Nat. Cell Biol. 9:415–421 10.1038/ncb1561 [DOI] [PubMed] [Google Scholar]
- Lim C.J., Kain K.H., Tkachenko E., Goldfinger L.E., Gutierrez E., Allen M.D., Groisman A., Zhang J., Ginsberg M.H. 2008. Integrin-mediated protein kinase A activation at the leading edge of migrating cells. Mol. Biol. Cell. 19:4930–4941 10.1091/mbc.E08-06-0564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.Q., Qin J., Wu C., Plow E.F. 2008. Kindlin-2 (Mig-2): a co-activator of β3 integrins. J. Cell Biol. 181:439–446 10.1083/jcb.200710196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutz D., Weise C., Mechai N., Hofmann W., Horstkorte R., Brüning G., Danker K. 2006. Integrin alpha3beta1 interacts with I1PP2A/lanp and phosphatase PP1. J. Neurosci. Res. 84:1759–1770 10.1002/jnr.21078 [DOI] [PubMed] [Google Scholar]
- Parsons M., Messent A.J., Humphries J.D., Deakin N.O., Humphries M.J. 2008. Quantification of integrin receptor agonism by fluorescence lifetime imaging. J. Cell Sci. 121:265–271 10.1242/jcs.018440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippar U., Roussos E.T., Oser M., Yamaguchi H., Kim H.D., Giampieri S., Wang Y., Goswami S., Wyckoff J.B., Lauffenburger D.A., et al. 2008. A Mena invasion isoform potentiates EGF-induced carcinoma cell invasion and metastasis. Dev. Cell. 15:813–828 10.1016/j.devcel.2008.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay A.G., Marshall J.F., Hart I.R. 2007. Integrin trafficking and its role in cancer metastasis. Cancer Metastasis Rev. 26:567–578 10.1007/s10555-007-9078-7 [DOI] [PubMed] [Google Scholar]
- Reynolds L.E., Conti F.J., Lucas M., Grose R., Robinson S., Stone M., Saunders G., Dickson C., Hynes R.O., Lacy-Hulbert A., Hodivala-Dilke K. 2005. Accelerated re-epithelialization in beta3-integrin-deficient- mice is associated with enhanced TGF-beta1 signaling. Nat. Med. 11:167–174 10.1038/nm1165 [DOI] [PubMed] [Google Scholar]
- Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., Horwitz A.R. 2003. Cell migration: integrating signals from front to back. Science. 302:1704–1709 10.1126/science.1092053 [DOI] [PubMed] [Google Scholar]
- Sechi A.S., Wehland J. 2004. ENA/VASP proteins: multifunctional regulators of actin cytoskeleton dynamics. Front. Biosci. 9:1294–1310 10.2741/1324 [DOI] [PubMed] [Google Scholar]
- Tadokoro S., Shattil S.J., Eto K., Tai V., Liddington R.C., de Pereda J.M., Ginsberg M.H., Calderwood D.A. 2003. Talin binding to integrin beta tails: a final common step in integrin activation. Science. 302:103–106 10.1126/science.1086652 [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Choi C.K., Horwitz A.R. 2009. Integrins in cell migration—the actin connection. J. Cell Sci. 122:199–206 10.1242/jcs.018564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentworth J.K., Pula G., Poole A.W. 2006. Vasodilator-stimulated phosphoprotein (VASP) is phosphorylated on Ser157 by protein kinase C-dependent and -independent mechanisms in thrombin-stimulated human platelets. Biochem. J. 393:555–564 10.1042/BJ20050796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White D.P., Caswell P.T., Norman J.C. 2007. α v β3 and α5β1 integrin recycling pathways dictate downstream Rho kinase signaling to regulate persistent cell migration. J. Cell Biol. 177:515–525 10.1083/jcb.200609004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worth D.C., Parsons M. 2008. Adhesion dynamics: mechanisms and measurements. Int. J. Biochem. Cell Biol. 40:2397–2409 10.1016/j.biocel.2008.04.008 [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar R., Ballestrem C., Kam Z., Geiger B. 2003. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J. Cell Sci. 116:4605–4613 10.1242/jcs.00792 [DOI] [PubMed] [Google Scholar]
- Zamir E., Katz M., Posen Y., Erez N., Yamada K.M., Katz B.Z., Lin S., Lin D.C., Bershadsky A., Kam Z., Geiger B. 2000. Dynamics and segregation of cell-matrix adhesions in cultured fibroblasts. Nat. Cell Biol. 2:191–196 10.1038/35008607 [DOI] [PubMed] [Google Scholar]
- Zicha D., Dobbie I.M., Holt M.R., Monypenny J., Soong D.Y., Gray C., Dunn G.A. 2003. Rapid actin transport during cell protrusion. Science. 300:142–145 10.1126/science.1082026 [DOI] [PubMed] [Google Scholar]