Defective mitochondrial quality control is shown to be a mechanism for neurodegeneration in some forms of Parkinson's disease.
Abstract
Parkinson's disease (PD) is a prevalent neurodegenerative disorder. Recent identification of genes linked to familial forms of PD such as Parkin and PINK1 (PTEN-induced putative kinase 1) has revealed that ubiquitylation and mitochondrial integrity are key factors in disease pathogenesis. However, the exact mechanism underlying the functional interplay between Parkin-catalyzed ubiquitylation and PINK1-regulated mitochondrial quality control remains an enigma. In this study, we show that PINK1 is rapidly and constitutively degraded under steady-state conditions in a mitochondrial membrane potential–dependent manner and that a loss in mitochondrial membrane potential stabilizes PINK1 mitochondrial accumulation. Furthermore, PINK1 recruits Parkin from the cytoplasm to mitochondria with low membrane potential to initiate the autophagic degradation of damaged mitochondria. Interestingly, the ubiquitin ligase activity of Parkin is repressed in the cytoplasm under steady-state conditions; however, PINK1-dependent mitochondrial localization liberates the latent enzymatic activity of Parkin. Some pathogenic mutations of PINK1 and Parkin interfere with the aforementioned events, suggesting an etiological importance. These results provide crucial insight into the pathogenic mechanisms of PD.
Introduction
Parkinson's disease (PD) is a very common movement disorder characterized by dopaminergic neuronal loss. The majority of PD cases are sporadic; however, the discovery of genes linked to rare familial forms of this disease has provided important insight into the molecular mechanisms of disease pathogenesis (Moore et al., 2005; Hardy et al., 2006). In 2000, we and others found that dysfunction of an E3 ubiquitin ligase (Imai et al., 2000; Shimura et al., 2000; Zhang et al., 2000) termed Parkin causes autosomal recessive juvenile Parkinsonism (Kitada et al., 1998). Since then, a multitude of papers have been published, but the mechanism by which dysfunction of Parkin causes autosomal recessive juvenile Parkinsonism has largely remained obscure, and claims of pathogenicity remain controversial (Lim, 2007; Matsuda and Tanaka, 2010). In addition, PINK1 (PTEN-induced putative kinase 1) was identified in 2004 as the gene responsible for another form of early-onset PD (Valente et al., 2004). PINK1 functions in mitochondrial maintenance, suggesting that mitochondrial integrity is another key factor in disease pathogenesis (Dodson and Guo, 2007; Schapira, 2008). Intriguingly, genetic studies using Drosophila melanogaster revealed that PINK1 and Parkin function in the same pathway, with PINK1 functioning upstream of Parkin (Clark et al., 2006; Park et al., 2006; Yang et al., 2006). Little is known about how PINK1 regulates Parkin, and our knowledge, especially in mammals, of their relationship is limited. In this study, we describe the mechanism underlying the functional interplay between ubiquitylation catalyzed by Parkin and mitochondrial quality control regulated by PINK1.
Results and discussion
Parkin localizes to and ubiquitylates mitochondria with low membrane potential
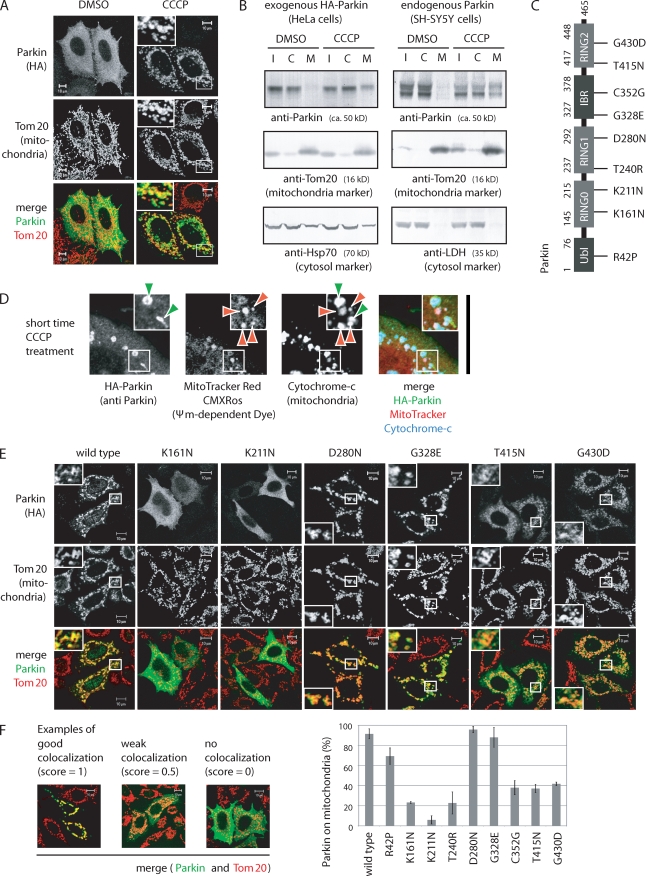
We initially sought to study the subcellular localization and E3 activity of Parkin using HeLa cells, which reportedly lack a functional Parkin gene (Denison et al., 2003). In support of that study, we found that endogenous Parkin was barely detectable in HeLa cells even when PRK8, the best-characterized specific anti-Parkin antibody (Pawlyk et al., 2003), was used (Fig. S1 A). Consequently, HA-Parkin was exogenously introduced into HeLa cells. Under steady-state conditions, HA-Parkin was diffusely localized throughout the cytosol and did not overlap with mitochondria, whereas Parkin was rapidly recruited to the mitochondria when HeLa cells were treated with the mitochondrial uncoupler CCCP (carbonyl cyanide m-chlorophenylhydrazone; Fig. 1 A), as reported by Narendra et al. (2008). Next we tried to confirm the redistribution of Parkin from the cytoplasm to the mitochondria using a biochemical approach. In fractionation experiments, detection of Parkin in the mitochondria-rich fraction was faint, probably because Parkin was weakly associated with the mitochondria and thus unstable during fractionation. Inclusion of the cross-linker DSP (dithiobis[succinimidyl propionate]) significantly strengthened the signal and further confirmed redistribution of exogenous (Fig. 1 B, left) and endogenous (Fig. 1 B, right) Parkin from the cytoplasm to a mitochondria-enriched fraction. (Note that endogenous Parkin in SH-SY5Y cells is detectable as a doublet, which is consistent with a previous study [Pawlyk et al., 2003].) To more convincingly demonstrate that Parkin is selectively recruited to depolarized mitochondria, we used MitoTracker red CMXRos, which accumulates in mitochondria with an intact membrane potential. Incomplete treatment with CCCP can generate cells in which healthy and damaged mitochondria coexist. Under these conditions, signals of Parkin and MitoTracker were mutually exclusive, and Parkin selectively localized on mitochondria with lower MitoTracker red staining (Fig. 1 D), indicating that Parkin was selectively targeted to mitochondria whose membrane potential had been lost.
Figure 1.
Mitochondrial localization of Parkin is etiologically important. (A) HeLa cells expressing HA-Parkin were treated with CCCP or DMSO (control) and then immunostained with the indicated antibodies. (B) HeLa cells stably expressing HA-Parkin or intact SH-SY5Y cells were treated with CCCP or DMSO and subjected to fractionation experiments. I, C, and M indicate input, cytosol-rich supernatant, and mitochondria-rich membrane pellet, respectively. (C) Schematic diagram of disease-relevant mutants of Parkin used in this study. IBR, in between RING; Ubl, ubiquitin like. (D) Polarized mitochondria stained with MitoTracker red (red arrowheads) were not labeled by Parkin. In contrast, damaged mitochondria marked by Parkin (green arrowheads) were not stained with MitoTracker red. (E) HeLa cells expressing HA-Parkin with various pathogenic mutations were treated with CCCP, followed by immunocytochemistry. (A, D, and E) Higher magnification views of the boxed areas are shown in the insets. (F) Parkin colocalization with mitochondria was analyzed in >100 cells per mutation. Example figures indicative of robust colocalization (counted as 1), weak colocalization (counted as 0.5), and no colocalization (counted as 0) are shown. Error bars represent the mean ± SD values of at least three experiments. Bars: (A, E, and F) 10 µm; (D) 30 µm.
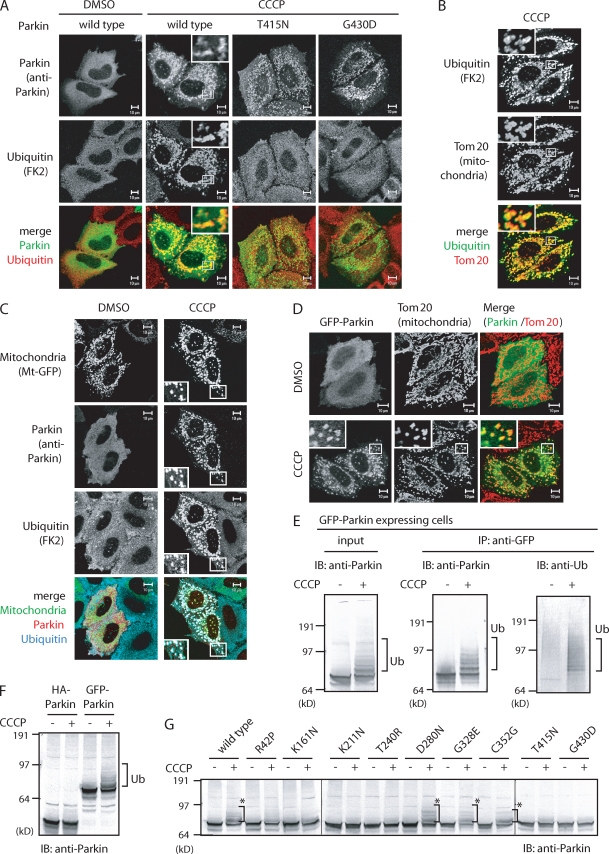
Subsequently, we performed immunofluorescence staining using an antiubiquitin antibody. Under normal conditions, the ubiquitin signal was spread throughout the cell. In contrast, when cells were treated with CCCP, the ubiquitin signal was concentrated in the mitochondria (Fig. 2, A and B). Mitochondrial ubiquitylation was only observed in Parkin-expressing cells (Fig. 2 A and Fig. S1 B) and disappeared when Parkin mutants deficient in E3 activity (T415N and G430D) were introduced (Fig. 2 A). Triple staining using mitochondria-targeting GFP (Mt-GFP), anti-Parkin, and antiubiquitin antibodies further confirmed the colocalization of Parkin, ubiquitin, and mitochondria after CCCP treatment (Fig. 2 C). Staining with single antibodies or Mt-GFP alone indicated that the aforementioned merged data were not derived from channel cross talk (Fig. S1, C and D). These results demonstrate that Parkin ubiquitylates mitochondria in response to a reduction in mitochondrial membrane potential.
Figure 2.
Parkin exerts E3 activity only when the mitochondrial membrane potential is decreased. (A) HeLa cells expressing wild-type Parkin or E3-inactivating mutations were treated with CCCP and then immunostained with the indicated antibodies. When E3-inactivating mutations were introduced into Parkin, the mitochondrial ubiquitylation signal disappeared. (B and C) HeLa cells expressing HA-Parkin (B) or expressing both Mt-GFP and HA-Parkin (C) were treated with CCCP or DMSO (control) and then immunostained with the indicated antibodies. (D) Localization of GFP-Parkin to the mitochondria after CCCP treatment. (A–D) Higher magnification views of the boxed areas are shown in the insets. (E) HeLa cell lysates expressing GFP-Parkin were immunoprecipitated by anti-GFP antibody, followed by immunoblotting with the indicated antibodies. (F) Straight immunoblotting of HA- and GFP-Parkin in the absence or presence of CCCP. Note the slower migrating ladders derived from ubiquitylation (Ub) in only the GFP-Parkin with CCCP lane. (G) GFP-Parkin–expressing HeLa cells with various pathogenic mutations (Fig. 1 C) were treated with CCCP and subjected to immunoblotting. Asterisks show ubiquitylation of GFP-Parkin. Vertical black lines indicate that intervening lanes have been spliced out. IB, immunoblot; IP, immunoprecipitation. Bars, 10 µm.
Disease-relevant mutations of Parkin impair mitochondrial localization
To confirm that translocation of Parkin to depolarized mitochondria is etiologically important, we selected nine pathogenic mutations and examined their subcellular localization (Fig. 1 C). In vitro experiments have previously shown that two of the mutations (T415N and G430D) in the RING2 domain abolish E3 activity of Parkin, whereas E3 activity is unaffected by the other mutations (Hampe et al., 2006; Matsuda et al., 2006). These mutants were serially introduced into HeLa cells, followed by CCCP treatment, and their subcellular localization was examined. Parkin with the D280N or G328E mutation in RING1 or the IBR (in between RING) domain, respectively, was recruited to the mitochondria in a manner similar to wild-type Parkin (Fig. 1, E and F). In contrast, the other pathogenic mutations altered to some degree the mitochondrial localization of Parkin; in particular, the K161N, K211N, and T240R mutations, which lie in or near the RING0 domain in the linker region (Hristova et al., 2009), severely compromised the mitochondrial localization of Parkin (Fig. 1, E and F). The aforementioned results suggest that mitochondrial localization of Parkin is pathologically significant and that the RING0 domain is important for the translocation of Parkin to the damaged mitochondria.
Parkin exerts E3 activity only when the mitochondrial membrane potential decreases
As shown in Fig. 2 C and Fig. S1 B, mitochondrial ubiquitylation was dependent on Parkin translocation to the mitochondria. Thus, we tried to determine whether the subcellular localization of Parkin modulates its E3 activity. To address this issue, we monitored the E3 activity of Parkin using an artificial substrate fused to Parkin. In vitro experiments have shown that Parkin can ubiquitylate an N-terminally fused lysine-rich protein as a pseudosubstrate (Matsuda et al., 2006). Similar ubiquitylation of pseudosubstrates even in the cytoplasm under normal conditions in vivo would be evidence that the E3 activity is constitutive; otherwise, E3 activity of Parkin is dependent on mitochondrial retrieval. A GFP tag is lysine rich and thus a good candidate for an in vivo pseudosubstrate, whereas an HA tag possesses no lysine residues and thus cannot function as a pseudosubstrate. GFP- and HA-Parkin expressed in HeLa cells treated with CCCP were both recruited to damaged mitochondria (Figs. 1 A and 2 D), but interestingly, a higher molecular mass population of only GFP-Parkin was observed (Fig. 2 F). Immunoprecipitation experiments demonstrated that GFP-Parkin was indeed ubiquitylated (Fig. 2 E). This was not based on autoubiquitylation of Parkin itself because mitochondria-associated HA-Parkin did not undergo ubiquitylation after CCCP treatment (Fig. 2 F and not depicted). Moreover, ubiquitylation of GFP-Parkin was absent in the T415N and G430D mutants, which lack E3 activities, suggesting that ubiquitylation of GFP-Parkin is not derived from other E3s (Fig. 2 G). The K161N and K211N mutants that impaired mitochondrial localization also inhibited ubiquitylation of GFP-Parkin (Fig. 2 G). Collectively, the aforementioned results indicate that Parkin ubiquitylates fused GFP only when it is retrieved to the mitochondria, suggesting that the latent E3 activity of Parkin is dependent on decreased mitochondrial membrane potential.
PINK1 localization is stabilized by damaged mitochondria
Recessive mutations in the human PINK1 gene are also the cause of autosomal recessive early-onset PD (Valente et al., 2004). We next examined whether the subcellular localization of PINK1 was affected by mitochondrial membrane potential. As reported previously (Valente et al., 2004; Beilina et al., 2005; Takatori et al., 2008), N-terminal Myc– or N-terminal Flag–tagged PINK1 clearly localized to the mitochondria, whereas C-terminal Flag– or C-terminal V5–tagged PINK1 mainly localized to the cytoplasm (Fig. 3 A and not depicted). Exogenous nontagged PINK1 also localized to the cytoplasm under steady-state conditions (Fig. 3 B), suggesting that mitochondrial localization of PINK1 is an artifact of the N-terminal epitope. More importantly, similar to Parkin, untagged PINK1 and C-terminal Flag– or C-terminal V5–tagged PINK1 localized to the mitochondria after CCCP treatment (Fig. 3, A and B; and not depicted). These results suggest that the subcellular localization of PINK1 is also regulated by the mitochondrial membrane potential.
Figure 3.
PINK1 is constitutively degraded in a mitochondrial membrane potential–dependent manner and localizes to depolarized mitochondria. (A) The number of HeLa cells with N-terminal– or C-terminal–tagged PINK1 localized to the mitochondria was counted in >100 cells. (B and C) Exogenous nontagged PINK1 (B) or endogenous PINK1 (C) in HeLa cells was immunostained with the indicated antibodies. (D) The number of HeLa cells with endogenous PINK1 localized to the mitochondria was counted as in A. (A and D) Error bars represent the mean ± SD values of least three experiments. (E) Endogenous PINK1 gradually accumulated after CCCP treatment. The first through the fifth lanes show endogenous PINK1, and the sixth lane shows overexpressed untagged PINK1. Note that the asterisk indicates a cross-reacting band because it was not affected by overproduction of untagged PINK1. (F) Subcellular fractionation of endogenous PINK1. Intact SH-SY5Y cells were treated with CCCP or DMSO and subjected to fractionation experiments (same sample as Fig. 1 B). I, C, and M indicate input, cytosol-rich supernatant, and the mitochondria-rich membrane pellet, respectively. (G) HeLa cells were treated with CCCP and cycloheximide as depicted, followed by immunoblotting with the indicated antibodies. LDH, lactate dehydrogenase. (F and G) Asterisks indicate a cross-reacting band. (H) N-terminal 34 aa of PINK1 recruited GFP to the mitochondria both in the absence and presence of CCCP. The top panel shows control HeLa cells expressing only GFP. (B, C, and H) Higher magnification views of the boxed areas are shown in the insets. Bars, 10 µm.
We next sought to determine the subcellular localization of endogenous PINK1. Immunocytochemical experiments showed, as reported previously (Zhou et al., 2008), that the endogenous PINK1 signal was barely detectable in HeLa cells under steady-state conditions. However, a decrease in mitochondrial membrane potential resulted in a mitochondria-associated PINK1 signal (Fig. 3, C and D). We found that CCCP treatment promoted the gradual accumulation of endogenous PINK1 in immunoblots as well (Fig. 3 E) and the presence of endogenous PINK1 in a mitochondria-enriched fraction (Fig. 3 F). More importantly, when CCCP was washed out, the accumulated endogenous PINK1 rapidly disappeared (within 30 min) both in the presence and absence of cycloheximide (Fig. 3 G and not depicted). Moreover, the N-terminal 34 aa of PINK1 sufficiently recruited GFP to the mitochondria even in the absence of CCCP (Fig. 3 H). These results support the hypothesis in which PINK1 is constantly transported to the mitochondria but is rapidly degraded in a membrane potential–dependent manner. We speculate that PINK1 is stabilized by a decrease in mitochondrial membrane potential and, as a result, accumulates in depolarized mitochondria.
PINK1 normally exists as either a long (∼60 kD) or a short (∼50 kD) protein. Because the canonical mitochondria-targeting signal (matrix-targeting signal) is cleaved after import into the mitochondria, the long form has been designated as the precursor and the short form as the mature PINK1 (Beilina et al., 2005; Silvestri et al., 2005). The short (processed) form of PINK1 was clearly detected when untagged PINK1 was overexpressed (Fig. 3 E, sixth lane); however, this form of endogenous PINK1 was rarely detectable after CCCP treatment (Fig. 3 E, the first through the fifth lanes). Our subcellular localization study of endogenous PINK1 after CCCP treatment showed that the long form was recovered in the mitochondrial fraction (Fig. 3 F), suggesting that it is not the preimport precursor form. Moreover, by monitoring the degradation process of PINK1 after recovery of membrane potential, we realized that the short form of PINK1 transiently appeared soon after CCCP was washed out and then later disappeared (Fig. 3 G), suggesting that the processed form of PINK1 is an intermediate in membrane potential–dependent degradation. In conclusion, these results imply that PINK1 cleavage dose not reflect a canonical maturation process accompanying mitochondrial import as initially thought.
PINK1 retrieves Parkin from the cytoplasm to the mitochondria
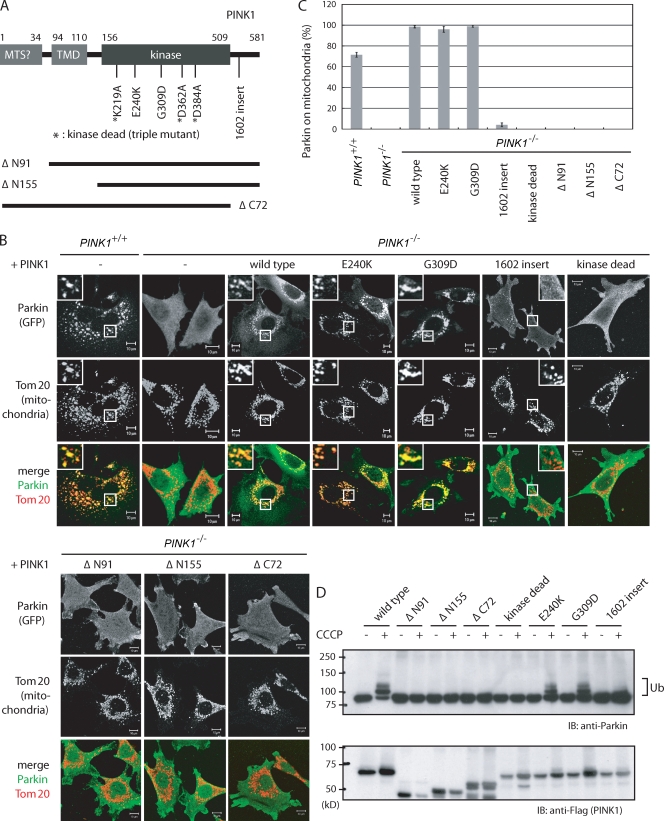
Because previous studies revealed that PINK1 functions upstream of Parkin (Clark et al., 2006; Park et al., 2006; Yang et al., 2006; Exner et al., 2007), we next examined the potential role of PINK1 in the mitochondrial recruitment of Parkin. To obtain clear-cut conclusions, we set up our experimental system using mouse embryonic fibroblasts (MEFs) derived from control (PINK1+/+) or PINK1 knockout (PINK1−/−) mouse (Gautier et al., 2008). Endogenous Parkin is undetectable in MEFs (Fig. S1 A); consequently, HA- or GFP-Parkin was introduced into these cells by retroviral transfection (Kitamura et al., 2003). In control MEFs (PINK1+/+), Parkin was selectively recruited to the mitochondria after CCCP treatment (Fig. 4 A) and subsequently resulted in the disappearance of the mitochondria (Fig. 4, D and E). This mitochondrial clearance was considerably impeded by Atg7 (essential gene for autophagy) knockout (Fig. S2; Komatsu et al., 2005), suggesting that Parkin degrades mitochondria by selective autophagy as reported previously (Narendra et al., 2008). In sharp contrast, Parkin was not translocated to the mitochondria in PINK1 knockout (PINK1−/−) MEFs after CCCP treatment (Fig. 4, A and B). Subsequent activation of Parkin and mitochondrial degradation were also completely impeded (Fig. 4, C–E). To exclude the possible role of retroviral integration of Parkin in the aforementioned phenotype, we checked whether reintroduction of PINK1 rescued this phenotype. Untagged or C-terminal Flag–tagged PINK1 complemented the mislocalization of Parkin in PINK1−/− MEFs (Fig. 5, B and C; and not depicted), confirming that the aforementioned defects were caused by the loss of PINK1.
Figure 4.
PINK1 recruits cytoplasmic Parkin to damaged mitochondria. (A) PINK1 knockout (PINK1−/−) or control (PINK1+/+) MEFs were transfected with HA-Parkin, treated with CCCP, and subjected to immunocytochemistry with the indicated antibodies. Higher magnification views of the boxed areas are shown in the insets. (B) The number of MEFs with Parkin localized to the mitochondria was counted as in Fig. 3 A. (C and D) Neither activation of Parkin nor mitochondrial degradation was observed in PINK1−/− MEFs. MEFs stably expressing GFP-Parkin were treated with CCCP for 4 h and subjected to immunoblotting (C) or for 24 h, followed by immunocytochemistry (D). IB, immunoblot; Ub, ubiquitylation. (E) The number of MEFs without anti-Tom20 antibody–detectable mitochondria was counted as in Fig. 3 A. In the example figure (left), blue arrowheads indicate cells without anti-Tom20 antibody–detectable mitochondria, and red arrowheads indicate cells harboring anti-Tom20 antibody–detectable mitochondria. (B and E) Error bars represent the mean ± SD values of least three experiments. Bars: (A) 10 µm; (D) 50 µm; (E) 150 µm.
Figure 5.
Kinase activity and mitochondrial targeting of PINK1 is imperative for mitochondrial localization of Parkin. (A) Schematic depiction of pathogenic and deletion mutants of PINK1 used in this study. MTS, mitochondria-targeting sequence; TMD, transmembrane domain. (B) Subcellular localization of Parkin in PINK1−/− cells complemented by various pathogenic and deletion mutants of PINK1-Flag. Cells were treated with CCCP. Higher magnification views of the boxed areas are shown in the insets. (C) The number of cells with Parkin-positive mitochondria was counted as in Fig. 3 A. Error bars represent the mean ± SD values of least three experiments. (D) PINK1−/− MEFs complemented by various PINK1 mutants were treated with CCCP and subjected to immunoblotting using anti-Parkin or anti-Flag (tag of PINK1) antibodies. IB, immunoblot; Ub, ubiquitylation. Bars, 10 µm.
To examine whether pathogenic mutations of PINK1 affect its mitochondrial localization, we expressed PINK1 mutants harboring the missense mutations E240K and G309D, or a CAA nucleotide insertion behind C1602 (referred to hereafter as 1602-insert) in PINK1−/− MEFs. Similar to wild-type PINK1, these PINK1 mutants colocalized with mitochondria after CCCP treatment (Fig. S3). Next, GFP-Parkin was introduced into these cells to examine whether pathogenic mutations of PINK1 affect the mitochondrial localization and activation of Parkin. The E240K and G309D mutants restored the mitochondrial localization and activation of Parkin as well as wild-type PINK1, whereas recruitment of Parkin to the mitochondria by the 1602-insert mutant was abolished (Fig. 5, B–D), suggesting that the pathology of this PINK1 mutation is mislocalization and consequent inactivation of Parkin.
Mitochondrial localization and kinase activity of PINK1 are essential for translocation of Parkin to damaged mitochondria
Finally, we investigated the role of various PINK1 domains (Fig. 5 A) in the mitochondrial recruitment of Parkin. PINK1 is composed of an atypical N-terminal mitochondrial localization signal and transmembrane domain, a kinase domain in the middle, and a conserved C-terminal domain (Zhou et al., 2008). Deletion of the N-terminal 91 aa abolished the mitochondrial localization of PINK1 (Zhou et al., 2008). We also confirmed that the ΔN91 and ΔN155 mutants did not target to the mitochondria even after CCCP treatment (Fig. S3). We also generated a mutant containing the triple K219A, D362A, and D384A mutations that abolish kinase activity (Beilina et al., 2005) and a C-terminal domain deletion mutant associated with PINK1 dysfunction (Sim et al., 2006; Yang et al., 2006). The kinase-dead and ΔC72 mutants of PINK1 colocalized with damaged mitochondria similar to wild type (Fig. S3). When introduced into PINK1−/− cells harboring GFP-Parkin, the mutants were unable to complement the mislocalization and inactivation of Parkin (Fig. 5, B–D), even though the mutant PINK1 proteins were expressed (Fig. 5 D and Fig. S3). These results indicate that the kinase activity and mitochondrial targeting of PINK1 are essential for the mitochondrial recruitment of Parkin.
Conclusion
In summary, we have shown that (a) PINK1 is a Parkin-recruitment factor that recruits Parkin from the cytoplasm to damaged mitochondria in a membrane potential–dependent manner for mitochondrial degradation, (b) endogenous PINK1 is constitutively degraded at the mitochondria, but its localization is specifically linked to a decrease in membrane potential, (c) under steady-state conditions, the E3 activity of Parkin is repressed in the cytoplasm but is liberated by PINK1-dependent mitochondrial localization, and (d) the aforementioned phenomena are presumably etiologically important in part because they were impeded for the most part by disease-linked mutations of PINK1 or Parkin. We believe that these results provide solid insight into the molecular mechanisms of PD pathogenesis not only for familial forms caused by Parkin and PINK1 mutations but also major sporadic forms of PD.
Materials and methods
Cell culture and transfection
MEFs derived from embryonic day 12.5 embryos of PINK1 knockout mice (provided by J. Shen, Harvard Medical School, Boston, MA) were mechanically dispersed by repeated passage through a P1000 pipette tip and plated with MEF media containing DME, 10% FCS, β-mercaptoethanol (Sigma-Aldrich), 1× nonessential amino acids, and 1 mM l-glutamine. Various stable transformants of MEFs were established by infecting MEFs with recombinant retroviruses. HA-Parkin, GFP-Parkin, PINK1 (provided by Y. Nakamura, T. Iwatsubo, and S. Takatori, University of Tokyo, Bunkyo-ku, Tokyo, Japan), or various PINK1 mutants were cloned into a pMXs-puro vector. Retrovirus packaging cells, PLAT-E (provided by T. Kitamura, University of Tokyo; Kitamura et al., 2003), were transfected with the aforementioned vectors and were cultured at 37°C for 24 h. After changing the medium, cells were further incubated at 37°C for 24 h, and the viral supernatant was collected and used for infection. MEFs were plated on 35-mm dishes at 24 h before infection, and the medium was replaced with the aforementioned undiluted viral supernatant with 8 µg/ml polybrene (Sigma-Aldrich). 2 d later, transformants were selected by the medium containing 10 µg/ml puromycin.
Cell fractionation and immunoprecipitation
To depolarize the mitochondria, HeLa and SH-SY5Y cells were treated with 10 µM CCCP, and MEFs were treated with 30 µM CCCP. For fractionation experiments, HeLa and SH-SY5Y cells were treated with CCCP for 1–5 h and subsequently treated with 1 mM DSP (Thermo Fisher Scientific) in PBS for 1 h on ice, inactivated by 10 mM glycine in PBS three times, and suspended in chappell-perry buffer (0.15 M KCl, 20 mM Hepes-NaOH, pH 8.1, 5 mM MgCl2, and protease and phosphatase inhibitor [Roche]). Cells were disrupted by five passages through a 25-gauge needle (with 1-ml syringe), debris was removed by centrifugation at 1,000 g for 7 min, and the supernatant was subjected to 10,000 g for 10 min to separate the mitochondria-rich fraction from the cytosol-rich fraction. Immunoblotting and immunoprecipitation were performed by conventional methods. To detect the ubiquitylation of GFP-Parkin, the cell lysate of HeLa cells (10 µM CCCP for 1 h) or MEFs (30 µM CCCP for 3 h) was collected in the presence of 10 mM N-ethylmaleimide to protect ubiquitylated Parkin from deubiquitylation enzymes. To monitor the degradation of endogenous PINK1, HeLa cells were treated with 10 µM CCCP and 50 µg/ml cycloheximide as depicted in Fig. 3 G and were subjected to immunoblotting.
Immunocytochemistry
To depolarize the mitochondria, HeLa cells (provided by A. Tanaka and R. Youle, National Institutes of Health, Bethesda, MD) were treated with 10 µM CCCP for 1 h (exogenous Parkin and PINK1) or 5 h (endogenous PINK1), and MEFs were treated with 30 µM CCCP for 3–4 h (Figs. 4 A and 5 B; and Fig. S3 B) or 24 h (Fig. 4 D and Fig. S2 A). For immunofluorescence experiments, cells were fixed with 4% paraformaldehyde, permeabilized with 50 µg/ml digitonin, and stained with primary antibodies described in the next section and with the following secondary antibodies: mouse and/or rabbit Alexa Fluor 488, 568, and 647 (Invitrogen). N-terminal 34 aa of PINK1 were fused to GFP to stain mitochondria in the triple staining experiments. To monitor the mitochondrial membrane potential, MEFs were treated with 50 nM MitoTracker red CMXRos (Invitrogen) for 15 min, washed three times, and incubated for an additional 10 min before fixation, as reported previously (Narendra et al., 2008). Cells were imaged using a laser-scanning microscope (LSM510 META; Carl Zeiss, Inc.) with a Plan-Apochromat 63× NA 1.4 oil differential interference contrast objective lens. Image contrast and brightness were adjusted in Photoshop (Adobe).
Antibodies
Antibodies used in this study are as follows: antiactin (AC-40; Sigma-Aldrich), anti–cytochrome c (6H2.B4; BD), anti-Flag (M2; Sigma-Aldrich), anti-GFP (3E6 [Wako Chemicals USA, Inc.]; or A6455 [Invitrogen]), anti-HA (12CA5; Roche), anti-Hsp70 (MBL), anti–lactate dehydrogenase (Abcam), anti-Parkin (#2132 [Cell Signaling Technology] for immunocytochemistry; or PRK8 [Sigma-Aldrich] for immunoblotting), anti-PINK1 (Novus), anti-Tom20 (FL-145 and F-10; Santa Cruz Biotechnology, Inc.), antiubiquitin (P4D1 [Santa Cruz Biotechnology, Inc.]; or FK2 [MBL]), and anti-V5 (Invitrogen).
Online supplemental material
Fig. S1 shows various control experiments for immunoblotting and immunocytochemistry. Fig. S2 shows that Parkin promoted degradation of depolarized mitochondria via autophagy. Fig. S3 shows the subcellular localization of pathogenic and deletion mutants of PINK1 after CCCP treatment. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200910140/DC1.
Acknowledgments
We thank Dr. T. Kitamura for providing the PLAT-E cells, Drs. A. Tanaka and R. Youle for NIH HeLa cells, Dr. J. Shen for PINK1 knockout MEFs, and Drs. Y. Nakamura, T. Iwatsubo, and S. Takatori for plasmids.
This work was supported by Grants-in-Aid for Young Scientist (B) (to N. Matsuda, K. Shiba, and S. Saiki) and by Specially Promoted Research (to K. Tanaka) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and Heath and Labor Science Research grants (to N. Hattori and K. Tanaka).
Note added in review. While our manuscript was under review, Geisler et al. (2010), Narendra et al. (2010), and Vives-Bauza et al. (2010) independently published results that are consistent with those described herein.
Footnotes
Abbreviations used in this paper:
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- MEF
- mouse embryonic fibroblast
- Mt-GFP
- mitochondria-targeting GFP
- PD
- Parkinson's disease
References
- Beilina A., Van Der Brug M., Ahmad R., Kesavapany S., Miller D.W., Petsko G.A., Cookson M.R. 2005. Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability. Proc. Natl. Acad. Sci. USA. 102:5703–5708 10.1073/pnas.0500617102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark I.E., Dodson M.W., Jiang C., Cao J.H., Huh J.R., Seol J.H., Yoo S.J., Hay B.A., Guo M. 2006. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 441:1162–1166 10.1038/nature04779 [DOI] [PubMed] [Google Scholar]
- Denison S.R., Wang F., Becker N.A., Schüle B., Kock N., Phillips L.A., Klein C., Smith D.I. 2003. Alterations in the common fragile site gene Parkin in ovarian and other cancers. Oncogene. 22:8370–8378 10.1038/sj.onc.1207072 [DOI] [PubMed] [Google Scholar]
- Dodson M.W., Guo M. 2007. Pink1, Parkin, DJ-1 and mitochondrial dysfunction in Parkinson's disease. Curr. Opin. Neurobiol. 17:331–337 10.1016/j.conb.2007.04.010 [DOI] [PubMed] [Google Scholar]
- Exner N., Treske B., Paquet D., Holmström K., Schiesling C., Gispert S., Carballo-Carbajal I., Berg D., Hoepken H.H., Gasser T., et al. 2007. Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by parkin. J. Neurosci. 27:12413–12418 10.1523/JNEUROSCI.0719-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier C.A., Kitada T., Shen J. 2008. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc. Natl. Acad. Sci. USA. 105:11364–11369 10.1073/pnas.0802076105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S., Holmström K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., Springer W. 2010. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12:119–131 10.1038/ncb2012 [DOI] [PubMed] [Google Scholar]
- Hampe C., Ardila-Osorio H., Fournier M., Brice A., Corti O. 2006. Biochemical analysis of Parkinson's disease-causing variants of Parkin, an E3 ubiquitin-protein ligase with monoubiquitylation capacity. Hum. Mol. Genet. 15:2059–2075 10.1093/hmg/ddl131 [DOI] [PubMed] [Google Scholar]
- Hardy J., Cai H., Cookson M.R., Gwinn-Hardy K., Singleton A. 2006. Genetics of Parkinson's disease and parkinsonism. Ann. Neurol. 60:389–398 10.1002/ana.21022 [DOI] [PubMed] [Google Scholar]
- Hristova V.A., Beasley S.A., Rylett R.J., Shaw G.S. 2009. Identification of a novel Zn2+-binding domain in the autosomal recessive juvenile Parkinson-related E3 ligase parkin. J. Biol. Chem. 284:14978–14986 10.1074/jbc.M808700200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Soda M., Takahashi R. 2000. Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J. Biol. Chem. 275:35661–35664 10.1074/jbc.C000447200 [DOI] [PubMed] [Google Scholar]
- Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. 1998. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 392:605–608 10.1038/33416 [DOI] [PubMed] [Google Scholar]
- Kitamura T., Koshino Y., Shibata F., Oki T., Nakajima H., Nosaka T., Kumagai H. 2003. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol. 31:1007–1014 [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., et al. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169:425–434 10.1083/jcb.200412022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.L. 2007. Ubiquitin-proteasome system dysfunction in Parkinson's disease: current evidence and controversies. Expert Rev. Proteomics. 4:769–781 10.1586/14789450.4.6.769 [DOI] [PubMed] [Google Scholar]
- Matsuda N., Tanaka K. 2010. Does impairment of the ubiquitin-proteasome system or the autophagy-lysosome pathway predispose individuals to neurodegenerative disorders such as Parkinson's disease? J. Alzheimers Dis. 19:1–9 [DOI] [PubMed] [Google Scholar]
- Matsuda N., Kitami T., Suzuki T., Mizuno Y., Hattori N., Tanaka K. 2006. Diverse effects of pathogenic mutations of Parkin that catalyze multiple monoubiquitylation in vitro. J. Biol. Chem. 281:3204–3209 10.1074/jbc.M510393200 [DOI] [PubMed] [Google Scholar]
- Moore D.J., West A.B., Dawson V.L., Dawson T.M. 2005. Molecular pathophysiology of Parkinson's disease. Annu. Rev. Neurosci. 28:57–87 10.1146/annurev.neuro.28.061604.135718 [DOI] [PubMed] [Google Scholar]
- Narendra D., Tanaka A., Suen D.F., Youle R.J. 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183:795–803 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., Youle R.J. 2010. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8:e1000298 10.1371/journal.pbio.1000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Lee S.B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J.M., Chung J. 2006. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 441:1157–1161 10.1038/nature04788 [DOI] [PubMed] [Google Scholar]
- Pawlyk A.C., Giasson B.I., Sampathu D.M., Perez F.A., Lim K.L., Dawson V.L., Dawson T.M., Palmiter R.D., Trojanowski J.Q., Lee V.M. 2003. Novel monoclonal antibodies demonstrate biochemical variation of brain parkin with age. J. Biol. Chem. 278:48120–48128 10.1074/jbc.M306889200 [DOI] [PubMed] [Google Scholar]
- Schapira A.H. 2008. Mitochondria in the aetiology and pathogenesis of Parkinson's disease. Lancet Neurol. 7:97–109 10.1016/S1474-4422(07)70327-7 [DOI] [PubMed] [Google Scholar]
- Shimura H., Hattori N., Kubo S., Mizuno Y., Asakawa S., Minoshima S., Shimizu N., Iwai K., Chiba T., Tanaka K., Suzuki T. 2000. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 25:302–305 10.1038/77060 [DOI] [PubMed] [Google Scholar]
- Silvestri L., Caputo V., Bellacchio E., Atorino L., Dallapiccola B., Valente E.M., Casari G. 2005. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum. Mol. Genet. 14:3477–3492 10.1093/hmg/ddi377 [DOI] [PubMed] [Google Scholar]
- Sim C.H., Lio D.S., Mok S.S., Masters C.L., Hill A.F., Culvenor J.G., Cheng H.C. 2006. C-terminal truncation and Parkinson's disease-associated mutations down-regulate the protein serine/threonine kinase activity of PTEN-induced kinase-1. Hum. Mol. Genet. 15:3251–3262 10.1093/hmg/ddl398 [DOI] [PubMed] [Google Scholar]
- Takatori S., Ito G., Iwatsubo T. 2008. Cytoplasmic localization and proteasomal degradation of N-terminally cleaved form of PINK1. Neurosci. Lett. 430:13–17 10.1016/j.neulet.2007.10.019 [DOI] [PubMed] [Google Scholar]
- Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A.R., Healy D.G., et al. 2004. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 304:1158–1160 10.1126/science.1096284 [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L., Kim J., May J., Tocilescu M.A., Liu W., Ko H.S., et al. 2010. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. USA. 107:378–383 10.1073/pnas.0911187107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Gehrke S., Imai Y., Huang Z., Ouyang Y., Wang J.W., Yang L., Beal M.F., Vogel H., Lu B. 2006. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl. Acad. Sci. USA. 103:10793–10798 10.1073/pnas.0602493103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Gao J., Chung K.K., Huang H., Dawson V.L., Dawson T.M. 2000. Parkin functions as an E2-dependent ubiquitin- protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc. Natl. Acad. Sci. USA. 97:13354–13359 10.1073/pnas.240347797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Huang Y., Shao Y., May J., Prou D., Perier C., Dauer W., Schon E.A., Przedborski S. 2008. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc. Natl. Acad. Sci. USA. 105:12022–12027 10.1073/pnas.0802814105 [DOI] [PMC free article] [PubMed] [Google Scholar]