Abstract
Following a previous study by de Courcy et al. ((2009) Interdiscip. Sci. Comput. Life Sci. 1, 55-60), we demonstrate in this contribution, using quantum chemistry, that metal cations exhibit a specific topological signature in the electron localization of their density interacting with ligands according to its “soft” or “hard” character. Introducing the concept of metal cation subvalence, we show that a metal cation can split its outer-shell density (the so-called subvalent domains or basins) according to it capability to form a partly covalent bond involving charge transfer. Such behaviour is investigated by means of several quantum chemical interpretative methods encompasing the topological analysis of the Electron Localization Function (ELF) and Bader's Quantum Theory of Atoms in Molecules (QTAIM) and two energy decomposition analyses (EDA), namely the Restricted Variational Space (RVS) and Constrained Space Orbital Variations (CSOV) approaches. Further rationalization is performed by computing ELF and QTAIM local properties such as electrostatic distributed moments and local chemical descriptors such as condensed Fukui Functions and dual descriptors. These reactivity indexes are computed within the ELF topological analysis in addition to QTAIM offering access to non atomic reactivity local index, for example on lone pairs. We apply this “subvalence” concept to study the cation selectivity in enzymes involved in blood coagulation (GLA domains of three coagulation factors). We show that the calcium ions are clearly able to form partially covalent charge transfer networks between the subdomain of the metal ion and the carboxylate oxygen lone pairs whereas magnesium does not have such ability. Our analysis also explains the different role of two groups (high affinity and low affinity cation binding sites) present in GLA domains. If the presence of Ca(II) is mandatory in the central “high affinity” region to conserve a proper folding and a charge transfer network, external sites are better stabilised by Mg(II), rather than Ca(II), in agreement with experiment. The central role of discrete water molecules is also discussed in order to understand the stabilities of the observed X-rays structures of the Gla domain. Indeed, the presence of explicit water molecules generating indirect cation-protein interactions through water networks is shown to be able to reverse the observed electronic selectivity occuring when cations directly interact with the Gla domain without the need of water.
Keywords: Coagulation, subvalence, QTAIM, Electron Localization Function (ELF), softness, hardness, charge transfer, electrostatic moments, Fukui functions, water networks, metals, biology
Introduction
Metal cations play a critical role in many biological systems. In most cases, they are specific in their ability to bind to proteins and thereby confer appropriate biological function or activity. For example, the presence of calcium cations is important in blood clotting, signal transduction, and cell division. Specifically, in the case of blood clotting, it has been experimentally observed that the presence of calcium is required for clot formation1. Indeed, calcium cations directly participate in the binding and folding of the γ-carboxyglutamic acid Gla rich domain that is common to the vitamin-K-dependent serine proteases present in the blood coagulation cascade1. Blood plasma does not coagulate in the sole presence of magnesium ions2,3, an effect attributed to the concomitant lack of binding of the GLA residue to negatively charged phospholipids when only magnesium ions are present. More precisely, recent X-ray crystal structures with a mixture of both Ca(II) and Mg(II) show that the N-terminus ω-loop segment that is thought to be the key determinant for the binding of GLA domain to membranes has a disordered structure when only magnesium ions are present4. In the presence of calcium ions, GLA domains have been crystallized and a strong GLA-calcium network has been observed with varying degree of calcium ion coordination. However, despite the mandatory presence of calcium ion to structure the GLA domain and allowing it to point the three hydrophobic (or anionic) residues forming “the keel” to the direction of negatively charged phospholipids found in cellular membranes, it has also been experimentally demonstrated that the presence of magnesium ions in addition with calcium enhance the affinity of the enzyme for both the membrane and cofactors5.
The ability of calcium ion to coordinate water molecules with flexible coordination is thought to be important for this function. At the theoretical level, recent first-principle Car-Parrinello6 and force-field simulation7 studies have reported in-depth description of the hydration shells and of the preferred coordination numbers for the calcium and magnesium cations. Although we know that the residence time for water on magnesium ion is substantially longer than that for calcium ion8, we as yet do not know the physical origins of the differences between binding of calcium and magnesium ion in biological systems as no clear electron structure-biological activity relation has been yet uncovered in realistic model systems. In this context, we proposed to apply modern quantum chemistry to study the nature of the binding of calcium and magnesium ions in model systems of Factors VII, IX and X, the structures of which are based on protein structures extracted from the Protein Data Bank.
Outline
In this work, we will first focus on the fundamental interactions occurring in such systems between the metal cations and their environment. Indeed, thanks to X-ray studies, we know that interactions occur between the calcium or magnesium cations and carboxylate moieties. We will then present an extensive ab initio study of the binding of several hard and soft metal cations to carboxylates in different position. Then, applying quantum topological approaches such as Atoms in Molecules (QTAIM)9 theory and the topological analysis of the Electron Localization Function (ELF)10, we shed light on the origin of the different behaviour of the electronic structure of metal cation-ligands so as to unravel the specific cation topological signature. The observed topological descriptions will be complemented by intermolecular interaction energy decomposition using the Restricted Variational Space approach (RVS)11a and the Constrained-Space Orbital variation (CSOV)11b which provide insights about the nature, covalent or electrostatic, of the bonding between the cations and carboxylate. To connect this work to conceptual Density Functional Theory (DFT), we will also provide a detailed analysis by means of local chemical descriptors such as the condensed Fukui functions. In a second part, we will present a study of models of the GLA domain from Factors IX, VII and X of the blood coagulation process using ELF computations complemented by multimolecular RVS energy decomposition analyses.
Method
a) Topological analysis of ELF
The topological analysis relies on a partition of the molecular space achieved in the framework of the theory of gradient dynamics applied to a a scalar potential function, say V(r), called “potential function” which contains the physical or chemical information. This partitioning gives rise to a set of non overlapping molecular volumes called basins localized around the maxima of the ELF (the attractors of the vector field). The—boundaries between these basins, the separatrices, are zero-flux surfaces satisfying the following condition that every point r is a unit vector normal to the surface. In the QTAIM theory of Bader9, the scalar function is the electron density distribution whose basins have their attractors located on the nuclei and which are therefore associated with the atoms that constitute the molecule. In order to recover a chemist's representation of a molecule consistent with Lewis's valence picture, one must use another “local” function that is able to describe the electron pair regions. For almost two decades, the topological analysis of the ELF has been extensively developed and used to analyze chemical bonding and to investigate chemical reactivity (for reviews, see refs12,13). The ELF can be interpreted as a signature of the electron pair distribution. The relationship of the kernel of ELF to pair functions has been established13 but, in contrast to these latter, the ELF values are confined in the [0,1] range by a Lorentzian transformation which facilitates the interpretation. The basins of the ELF are either core basins, labelled C(A) corresponding to the inner shells of atom A and encompassing its nucleus (if Z>2) or valence basins denoted by V(A, B, C...) where A, B, C are the element symbols. A valence basin can belong to a single atomic valence shell, in this case V(A) corresponds to a lone pair, or be shared by several atoms and associated to a bond V(A,B).
The ELF basins closely match the electronic domains of the VSEPR12 model and it has been shown that the interbasin repulsion provides a map onto the Gillespie-Nyholm rules which describe molecular geometry14,15. It has been recently shown that non-VSEPR structures which occur around neutral atoms belonging to the fourth and higher periods can be explained by considering the structure of the external core shell basins16 and hereafter referred to as subvalence basins. Details about the ELF analysis can also be found in a recent review paper dealing with the application of ELF to systems of biological interest17.
b) Integration of local properties within the ELF or QTAIM partition
i) Local electrostatic moments
From a quantitative point of view, a population analysis can be carried out by integrating the electron density distribution over the basin volumes. Recently, the distributed moments analysis based on the QTAIM partition9 and on the ELF basins (DEMEP)18a has been introduced which enables an extended discussion on the nature of bonding in molecules. In this paper, we use more specifically the First Moment (denoted as M1) which represents the local dipolar polarization of the density.
That way, the Distributed Electrostatic Moments based on the ELF Partition (DEMEP) allows the calculation of local moments located at non-atomic centres such as lone pairs, σ bonds and π systems. Local dipole contributions have been shown to be useful to rationalize inductive polarization effects and typical hydrogen bond interactions. Moreover, bond quadrupole polarization moments being related to a π character enable to discuss bond multiplicities, and to sort families of molecules according to their bond order.
To summarize, the M0(Ω) monopole term corresponds to the negative of the population (denoted N):
| (1) |
The first moments or dipolar polarization components of the charge distribution are defined by three-dimensional integrals for a given basin Ω according to:
| (2) |
where Xc, Yc, and Zc are the Cartesian coordinates of the basin centres.
The five second-moment spherical tensor components can also be calculated and are defined as the quadrupolar polarization terms. They can be seen as the ELF basin equivalents to the atomic quadrupole moments introduced by Popelier9c in the case of an QTAIM analysis:
| (3) |
The first- or second-moment basin magnitude is then defined as the square root of the sum of squared components:
| (4) |
Thanks to the invariance of the magnitude of any multipole rank (|M1| or |M2|) with respect to the axis for a given bond or lone pair, the approach allows us to compare the dipolar or quadrupolar polarization of a given basin in different chemical environments.
ii) Fukui Functions as local chemical descriptors
Beyond the computations of local distributed electrostatic moments, it is also possible to access the topological partion of local chemical descriptors. Among the numerous chemical indicators, the Fukui functions18b,c, based on the relative properties of the Highest Occupied Molecular Orbital (HOMO) and the Lowest Unoccupied Molecular Orbital (LUMO, are interesting as they are particularly usefull for the interpretation of chemical reactivity, particularly toward nucleophiles or electrophiles18d. Indeed, following Parr and Yang, conceptual DFT provides such function defined in terms of the variation of chemical potential with respect to changes in the external potential v(r) or equivalently as the derivative of electron density with respect to changes in the number of electrons N.
| (5) |
Three Fukui function are usually evaluated: f+(r), f+(r) and f0(r)
| (6) |
They are sometime also associated to the computation of another value called dual descriptor (denoted Δf(r)18e,f) and calculated upon the f+(r) and the f-(r) functions:
| (7) |
The f+(r) function usually characterises the reactivity of a given species toward nucleophilic attack (in that case, Δf(r)>0) whereas the f-(r) function usually caracterises the reactivity of a given specie toward electrophilic (in that case, Δf(r)<0). When Δf(r) =0, the site's reactivity should be equilibrated. As an analytic expression of such function is not available it remains possible to compute them numerically using finite differences (see for example ref. 18g and references therein).
In that case, f+(r), f+(r) and f0(r) can be computed as follows:
| (8) |
qx(N) represents the atomic charges associated to atom x within the N-electron species. Recently, it has been shown18h-i that it is possible to use a QTAIM condensation scheme for Frontier Molecular Orbitals Fukui functions using finite differences within a topological partition. Such an approach has been shown to be particularly stable having some advantage on other atomic evaluations of the Fukui functions scheme:
| (9) |
where h denotes HOMO, and l, LUMO; and the subscript x under the integration sign indicates that the integration has to be performed only within the particular atomic domain of atom x.
As previouslsy demonstrated for the computations of electrostatic moments, any QTAIM local properties computations can be performed using a topological ELF analysis. In this contribution, following the studies by Fuenteabla et al.18j, we present a Fukui analysis performed at both QTAIM and ELF levels.
b) Computational procedures
The geometries of all formate-cation complexes were optimized using the hybrid functional B3LYP19a,b with the Jaguar 5.5 software20. The choice of the B3LYP functional was motivated by its observed good performance in the modelling of biomolecules containing Ca(II) and Mg(II) cations compared to MP219b,c. We report in Supplementary Information S1 some comparisons between different functionnals, MP2 and CCSD(T) on optimal Ca(II) (or Mg(II))-formate geometries confirming these findings. The LACV3P**21 basis set combining a pseudo-potential for the cation and the all-electron 6-311G** basis set for the other atoms was employed. All geometries obtained with this less accurate energy function were then optimized further using the hybrid functional B3LYP, but applying the all-electron 6-311++G** basis set22 to all atoms, as provided by the Gaussian 03 software23. For the coagulation factors studied in the second part of this paper, single point calculations were employed for the two optimzed malonate-cation complexes at the B3LYP/6-311++G** level of theory. All topological analysis were carried out using ELF grids of size 180 × 180 × 180 for moment analysis (300 × 300 × 300 for pictures) with the last version of the TopMoD9024 package coupled to the TopChem17 program providing DEMEP analysis. To compute the total molecular dipole, we have assumed as “global (or molecular) frame” the standard orientation provided by Gaussian 03, which computes molecular dipoles at the center of nuclear charges. B3LYP/CSOV computations were performed with the same basis set using an inhouse version of HONDO 95.325 whereas the GAMESS26 software provided the RVS results computed at the Hartree-Fock level.
Results
I. Theoretical description of hard and soft metal-cations interactions with carboxylate moities
For blood coagulation proteins, X-ray studies clearly show that the main interactions involved in the biological activity of such enzymes involves networks built on the interaction of calcium or magnesium cations with carboxylate groups27. More precisely, X-rays unravel direct malonate-Ca(II)/or Mg(II) interactions.
To start our quantum chemical description of such proteins, we present here results on the interactions of differents metal cations with formate which are simple malonate models. As both monodentate and bidendate formate-cations coordination are found in structures, we have investigated the two cases. In a second part, we will focus on the specific interaction of calcium and magnesium cations with more realistic models directly extracted from the PDB structures of the different available Factors.
A. Topological study of hard and soft metals cations: the subvalence concept
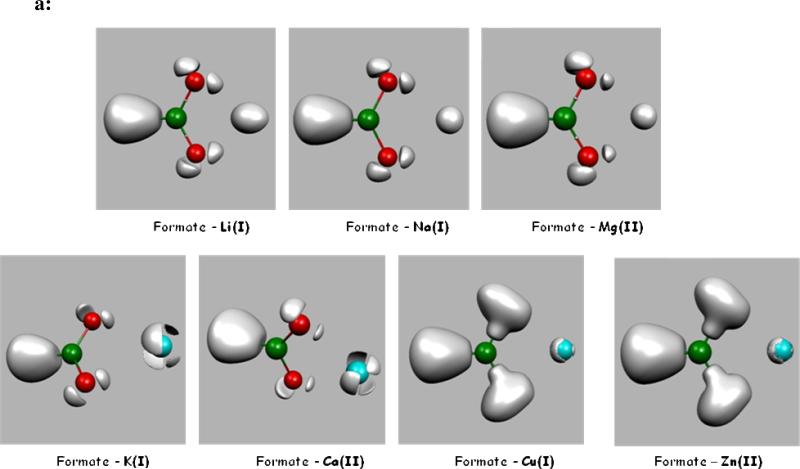
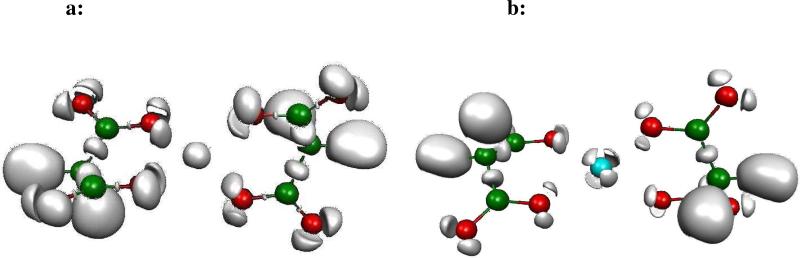
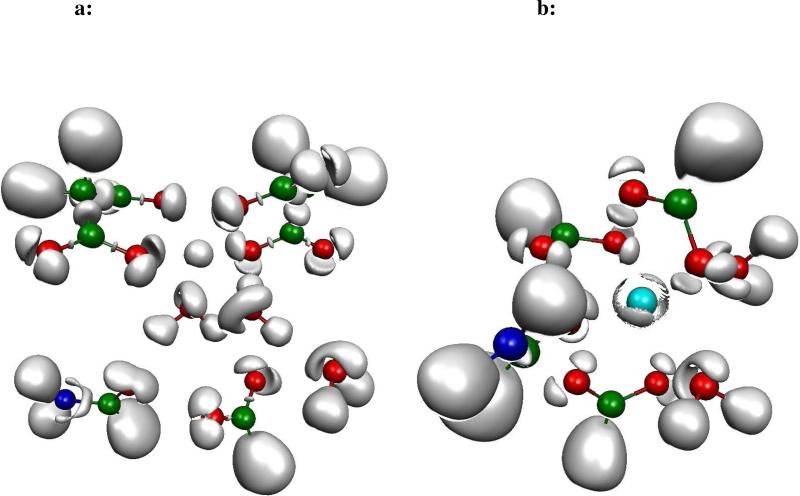
In order to study the differences between metal cations, we have performed ELF analysis upon DFT computations on several metal-cation-formate complexes encompassing monovalent cations such as Li(I), Na(I), K(I), Cu(I) and divalent cations, namely Mg(II), Ca(II), Zn(II). They are displayed in Figure 1a and 1b. Indeed, in a recent study28, we showed that the density of Zn(II) exhibits a striking plasticity. On the opposite, Zn(II) binding ligands were shown to be able to adapt/redistribute their density according to their nature: sulphur atoms were shown to be the softest, being able to spatially delocalize their lone pairs as oxygen and nitrogen mainly contract their lone pairs volumes. The present study intends to generalize such observations. Indeed, striking differences can be observed by visually analyzing the obtained ELF topological pictures that can be associated to the well known Parr and Pearson hardness concept linked to the resistance of an atom to change or deform. One can see (Figure 1a and 1b) that the expected “hard cations” (high value of the ηA hardness parameter, see Table III of ref. 29) such as Li(I), Na(I) or Mg(II) have a spatial localization of their electron density condensed around the nucleus position. “Soft” cations, usually associated to lower ηA values, exhibit specific splits within their outer-shell densities.
Figure 1a. ELF representation of formate interacting in a bidentate mode with metal cations.
Topological analysis of interactions between a formate and seven metal cations revealed at the isosurface coefficient of 0.87 (except for Cu(I) and Zn(II) complexes where it is 0.77). As seen in these pictures, electron densities remain condensed around the nucleus position for hard cations such as Li(I), Na(I) and Mg(II), whereas electron densities are split in four distinct subunits (basins), avoiding oxygen lone pairs, for softer cations such as K(I) and Ca(II), and split in two distinct subunits (basins), one of which inserted between oxygen lone pairs and the cation, for soft cations such as Cu(I) and Zn(II). In soft cation complexes, a blue sphere describes the core electrons of the cations. In hard cation complexes, the spherical subvalence obscures the core electrons so that the blue sphere is not visible.
Figure 1b. ELF representation of formate interacting in a monodentate mode with metal cations.
Isosurface coefficient used to make theses pictures are the same as the ones used for Figure 1a. A very similar pattern is observed as for the bidentate formate-cation complexes. The Mg(II) electron density stays spherical. In the Ca(II) complex, three basins have merged into an annular one still avoiding oxygen lone pairs. The same split as for the bidentate complex is shown for Zn(II).
It is then possible to use the concept of a “subvalence” associated to outer-shell core basins which can be seen as the topological signature for a given hard or soft behavior of the metal. A quick look at the observed topological structures shows that the observed subvalent ELF basins are more numerous for soft cations reflecting a more covalent character of their bonding to formate anions (see Figure 1).
B. Subvalence: understanding physics at play
To have a deeper understanding of the physics taking place in such interactions, it is possible to extend the ELF analysis to the computation of local electrostatic moments (see Table I) and Fukui functions (see Tables III) and to correlate them to RVS energy decompositions (see Table II).
Table I. QTAIM and ELF M1 and dipole moments of formate-cation complexes.
Values of cation's M1 and dipole moment (μ espressed in Debye) for formate-cation complexes in both modentate (mono) and bidentate (bi) binding modes. M1 is the polarization component of the total dipole moment (See text and SI). Concerning a cation it is computed as the gap to the sphericity: the more a cation exhibits a spherical subvalence, the less its polarization and M1 value are, and reciprocally. Two sets of M1 are reported: QTAIM values where all the electrons are gathered around the nucleus Ω(M), and ELF values where electrons are spread over the core basin C(M) and the subvalence basin V(M). QTAIM and ELF values for the total dipole moment are also reported; abinitio dipole moments computed with Gaussian G03 software are given in parenthesis for comparison.
| M1 | μ(D) | |||||
|---|---|---|---|---|---|---|
| QTAIM | ELF | QTAIM | ELF | |||
| Ω(M) | C(M) | V(M) | ||||
| Formate | 0.92 | 0.90 | ||||
| Li | ||||||
| mono | 0.01 | 0 | - | 9.41 | 9.40 (9.39) | |
| bi | 0 | 0 | - | 3.95 | 3.95 (3.96) | |
| Na | ||||||
| mono | 0 | 0.07 | - | 11.60 | 11.67 (11.69) | |
| bi | 0 | 0.08 | - | 6.03 | 6.06 (6.07) | |
| K | ||||||
| mono | 0.21 | 0.31 | - | 12.50 | 12.60 (12.70) | |
| bi | 0.20 | 0.31 | - | 7.20 | 7.30 (7.30) | |
| Mg | ||||||
| mono | 0.15 | 0.06 | 0.19 | 13.30 | 13.30 (13.30) | |
| bi | 0.02 | 0.06 | - | 7.30 | 7.30 (7.30) | |
| Ca | ||||||
| mono | 0.19 | 0.37 | - | 14.80 | 14.90 (14.90) | |
| bi | 0.16 | 0.35 | - | 8.06 | 8.10 (8.11) | |
| Cu | ||||||
| mono | 0.18 | 0.47 | - | 8.08 | 8.10 (8.13) | |
| bi | 0.23 | 0.48 | - | 2.97 | 3.01 (3.04) | |
| Zn | ||||||
| mono | 0.23 | 0.36 | 0.45 | 6.41 | 6.44 (6.45) | |
| bi | 0.04 | 0.39 | - | 3.06 | 3.09 (3.10) | |
Table II. RVS energy components for selected formate-metal cations complexes.
Values are given for the two monodentate (mono) and bidentate (bi) cation binding mode. Elec. is the Coulomb electrostatic energy, Exch. is the exchange repulsion; the sum of the two constitutes the first order term E1. Epol and ECT are the polarization and charge transfer components of the second order term E2, Etot being the sum of E1 and E2. A RVS decomposition energy allows us to separate the second order terms over the constitutive fragments of a system. The individual polarization of each cation is reported. The more spherical the subvalence of a cation is, the less Epol. is. The same pattern is observed for ECT of each complex. It can be seen that ECT for the monodentate formate-Mg(II) complex is double that of the bidentate formate-Mg(II) complex. This is to be put in context with the appearance of the additional subvalence basin in the monodentate formate-Mg(II) complex (Figure 3).
| Kcal/mol | Elec. | Exch. | E1 | Epol | Epol(cation) | ECT | E2 | Etot | |
|---|---|---|---|---|---|---|---|---|---|
| Formate Li(I) | |||||||||
| mono | -158.3 | 27.8 | -130.4 | -17.6 | -0.2 | -4.5 | -22.1 | -152.6 | |
| bi | -179.2 | 28.1 | -151.1 | -14.5 | -0.1 | -6.7 | -21.2 | -172.3 | |
| Formate Na(I) | |||||||||
| mono | -138.0 | 20.9 | -117.1 | -10.1 | -0.3 | -0.2 | -10.3 | -127.4 | |
| bi | -159.2 | 22.7 | -136.5 | -8.3 | -0.2 | -1.0 | -9.3 | -145.8 | |
| Formate K(I) | |||||||||
| mono | -125.9 | 26.4 | -99.5 | -8.7 | -1.8 | -1.3 | -9.9 | -109.5 | |
| bi | -147.0 | 30.2 | -116.8 | -7.1 | -1.7 | -1.6 | -8.6 | -125.4 | |
| Formate Mg(II) | |||||||||
| mono | -300.0 | 43.7 | -256.3 | -54.4 | -0.4 | -7.8 | -62.1 | -318.4 | |
| bi | -354.7 | 52.7 | -302.0 | -48.4 | -0.3 | -15.8 | -64.1 | -366.1 | |
| Formate Ca(II) | |||||||||
| mono | -287.7 | 76.3 | -211.5 | -39.8 | -2.3 | -17.8 | -57.7 | -269.1 | |
| bi | -335.9 | 82.4 | -253.5 | -32.8 | -2.0 | -18.4 | -51.2 | -304.7 | |
| Formate Cu(I) | |||||||||
| mono | -177.6 | 68.1 | -109.5 | -29.3 | -15.3 | 8.4 | -20.9 | -130.4 | |
| bi | -186.9 | 50.7 | -136.2 | -19.1 | -8.1 | 3.3 | -15.8 | -152.1 | |
| Formate Zn(I) | |||||||||
| mono | -319.5 | 66.7 | -252.8 | -64.9 | -4.5 | -6.7 | -71.6 | -324.4 | |
| bi | -371.0 | 72.1 | -298.9 | -56.0 | -3.4 | -18.8 | -74.8 | -373.7 | |
From these Tables, we see that the more covalent the bonding character of the formate-cation intermolecular bond is, the greater the RVS cation polarization energy and local ELF cation dipole moment are. For example, a hard cation such as Na(I), which is poorly polarizable (weak polarizability), is involved in interactions dominated by electrostatics (Table II) and does not show any split within its subvalence whereas cations exhibiting stronger polarization and charge transfer interactions possess a higher number of basins. Moreover, for a covalently bonded very soft cation such as Zn(II), a subvalent Zn-O basin is observed between the formate oxygen lone pair and the metal, a hint of electron sharing. Ca(II), which is less soft and less covalently bonded, still exhibits a split but subvalent basins remain distributed around the nucleus (Figure 1) and do not form any bond. Correlated CSOV energy decomposition computations have also been performed and are fully in line with HF RVS results. Details can be found in the Supplementary Information S2.
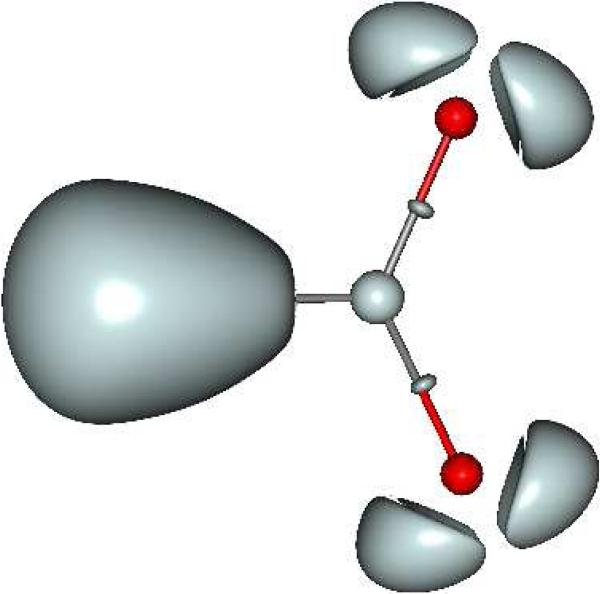
Observations of both monodentate and bidentate coordination modes show that “soft cation” subvalent basins clearly have the ability to orient themselves towards the formate oxygen lone pairs. That way, depending on its electron structure, each cation shows a specific topological signature which enables one to predict specific abilities of the cation to interact with its immediate environment thanks to the plasticity of its valence electron spatial organization. An indirect measurement of the soft/hard nature of the cations can be appraised by studying the volumes and density values of the formate oxygen lone pairs when interacting with cations. Hard cations such as Li(I) or Mg(II) clearly act on the lone pair densities which appear lower when compared to softer cations. Figure 2 exhibits the four oxygen lone pairs as they are when no metal cation interacts with the formate. Volume and density values reveal a dissymmetry between the internal and the external lone pairs, internal ones being less populated and more contracted than the external ones. This is due to the fact that the internal lone pairs interact with each other because of the shorter distance between them. From Figures 1 and 2, it is possible to appraise the electronic redistribution within the oxygen lone pairs according to the presence or not of a binding metal cation and its hardness or softness. Overall, it is important to point out that trends are conserved between ELF observations and Parr's hardness concept29. However, ELF pictures the final state of the cation electronic structure within the complex after cation-ligand orbital mixing and metal density relaxation (therefore a feature also linked to its polarizability).
Figure 2. ELF representation of a formate without a binding cation.
This picture presents a formate in an uncomplexed state, where densities are not rearranged through interactions with a cation. Volumes of the oxygen lone pairs can be compared to the ones of formatecations complexes shown in Figures 1a and 1b.
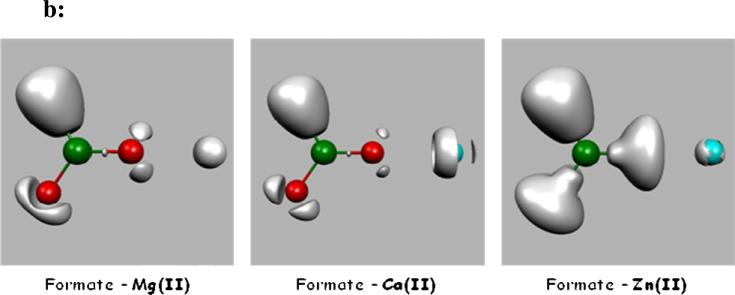
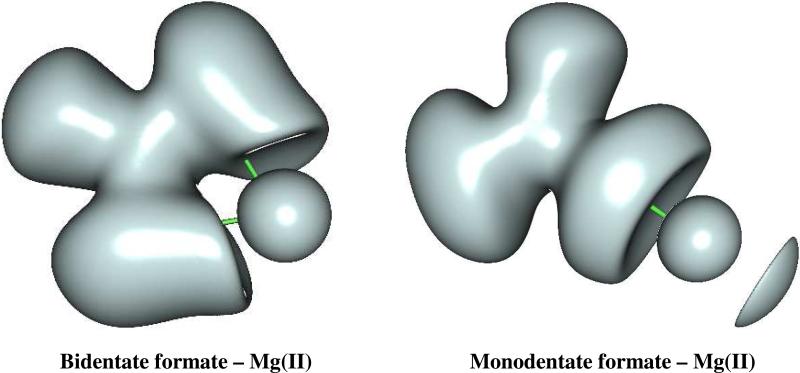
Concerning the specific Ca(II)/Mg(II) differences, our results demonstrate that overall less flexibility occurs in Mg(II) density compared to Ca(II) which tends to adjust to its immediate ligands. However, for Mg(II) in the monodentate binding mode, a slight increase of cation polarization associated to a topological split of its outer shell density is noted and this leads to the fact that Mg(II) could act slightly differently from usual hard cations. As we will see, this will have some consequences. Figure 3 shows a ELF representation of both monodentate and bidentate formate-Mg(II) complexes. A well-separated additional basin is found in the monodentate complex, where a partial charge is transferred. It worth noticing that the Sr(II) cation which is sometimes found to substitute calcium in certain conditions (ref. 30 and references therein) exhibits the same topological pattern as Ca(II).
Figure 3. ELF representation of formate-Mg(II) complexes at the isosurface coefficient of 0,22.
The electron density of the two formate-Mg(II) complexes are compared in this picture. Contrary to the bidentate formate-Mg(II), where the subvalence stays spherical, in the monodentate formate-Mg(II) an additional basin appears at the opposite side of the coordination to the oxygen. This is consistent with the augmentation of both M1 shown in Table 1, and charge transfer energy shown in table 2 for this complex.
To conclude, as the concepts of softness and hardness are involved, it is of importance to also consider the possibility of computing other popular local reactivity indicators, usually utilized to rationalize such phenomena. That way, we propose here an evaluation of the different “local” Fukui functions at both QTAIM and ELF levels. First, we computed such functions on an isolated formate molecule (see Table IIIa and IIIb). Again, the ELF Fukui analysis clearly shows the non-equivalence of the formate oxygen lone pairs, the external basins having different indicators from internal lone pairs. If QTAIM tends to show a uniform Fukui descriptor, (the dual descriptor Δf is always positive), ELF does not, as it provides a Δf negative value on the C-H bond providing insights about a possible different reactivity. Concerning the metal cations, Table IIIc brings interesting informations. Let's consider first the isolated metal cations. If all cations exhibit f- values of 1, they have very different f+ values. Again, following chemical intuition, strong differences occur between hard and soft cations. Hard cations such as Li(I), exhibits low values of f+, whereas soft cation such as Zn(II) have f+ values tending toward 1. The dual descriptor appears then a good global indicator reflecting the cations ranking as Δf tends to 0 with increasing cation's softness. Table IIIc depicts the results for selected bidentate formate cations. Global trends are preserved despite strong different bonding modes. Again, the softer cations exhibit smaller Δf values (here more negative) than harder ones. If they agree well, one difference between ELF and QTAIM values is observed: ELF tend to show more difference between Ca(II) and Mg(II) than QTAIM. This is reflected by a difference of 0.2 (QTAIM) vs. 0.3 (ELF) in the Δf values between the two cations.
Table IIIa.
a) ELF Integrated Fukui functions and dual descriptor values for an isolated formate molecule
| Formate | Basin | f-(ELF) | f+(ELF) | f0(ELF) | Af(ELF) |
|---|---|---|---|---|---|
| C(C) | 0,00 | 0,00 | 0,00 | 0,00 | |
| C(O1) | 0,02 | 0,00 | 0,01 | 0,02 | |
| C(O2) | 0,02 | 0,00 | 0,01 | 0,02 | |
| V(C,H) | 0,22 | 0,30 | 0,26 | -0,08 | |
| V(C,O2) | 0,04 | 0,01 | 0,02 | 0,03 | |
| V(C,O1) | 0,04 | 0,01 | 0,02 | 0,03 | |
| V(O1) | 0,20 | 0,01 | 0,10 | 0,19 | |
| V(O1) | 0,13 | 0,03 | 0,08 | 0,10 | |
| V(O2) | 0,14 | 0,03 | 0,09 | 0,11 | |
| V(O2) | 0,20 | 0,01 | 0,10 | 0,19 |
Table IIIb.
b) QTAIM Integrated Fukui functions and dual descriptor values for an isolated formate molecule
| Formate | Atom | f−(QTAIM) | f+(QTAIM) | f0(QTAIM) | Δf(QTAIM) |
|---|---|---|---|---|---|
| H | 0,17 | 0,15 | 0,16 | 0,02 | |
| C | 0,05 | 0,04 | 0,05 | 0,02 | |
| O1 | 0,38 | 0,03 | 0,21 | 0,35 | |
| O2 | 0,38 | 0,03 | 0,21 | 0,35 |
Table IIIc.
c) ELF and QTAIM Fukui functions and dual descriptor values for selected metal cations. Values are given for the isolated cations and for a cation within a bidentate formate-Metal complex
| Cations | f−(QTAIM) | f+(QTAIM) | f0(QTAIM) | Δf(QTAIM) isolated | Δf(ELF) Isolated | Δf(QTAIM) complexed | Δf(ELF) complexed |
|---|---|---|---|---|---|---|---|
| Li | 1,00 | 0,23 | 0,61 | 0,77 | 0,70 | -0,14 | -0,11 |
| Na | 1,00 | 0,44 | 0,72 | 0,56 | 0,45 | -0,34 | -0,37 |
| K | 1,00 | 0,46 | 0,73 | 0,54 | 0,43 | -0,30 | -0,36 |
| Mg | 1,00 | 0,59 | 0,79 | 0,41 | 0,32 | -0,67 | -0,24 |
| Ca | 1,00 | 0,98 | 0,99 | 0,02 | 0,00 | -0,55 | -0,59 |
| Zn | 1,00 | 0,91 | 0,96 | 0,09 | 0,04 | -0,69 | -0,50 |
Of course, the numerical values strongly depend on the (never unique) topological partion scheme and therefore on the density attribution to atoms/centers18h but the ELF and AIM approaches are not subject to strong basis set/diffuse functions dependence18a,h and are quite stable. Overall, beyond the numbers, an interesting qualitative agreement is observed and supports the previously depicted ELF subvalence basins and the numerical values extracted from others interpretatives techniques described above.
II. Metal cation's electron structure / biological activity relationship in coagulation proteins
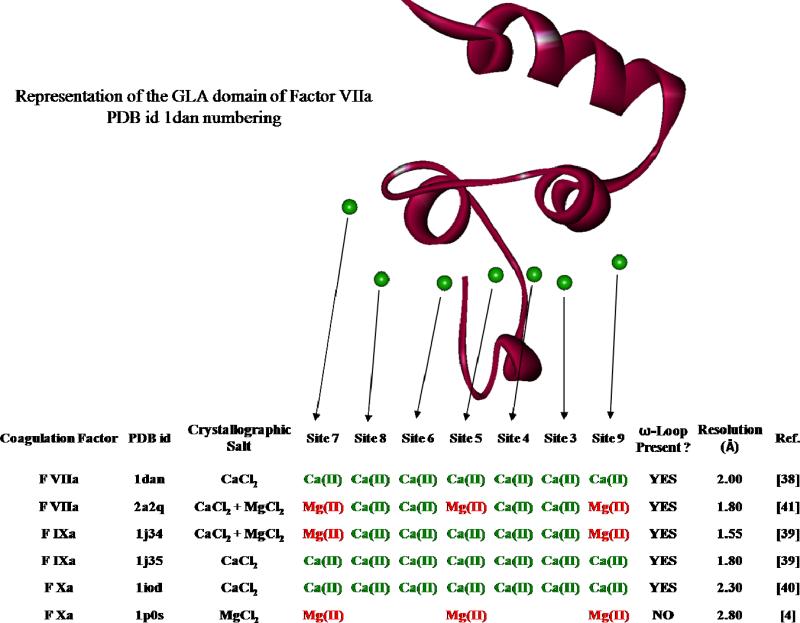
We use here the commonly accepted terminology to first differentiate the γ-carboxyglutamic acid itself identified as (Gla) from the γ-carboxyglutamic acid-rich-domain identified as (GLA), secondly to name the first 11 residues at the N-terminal extremity of the ω-loop, and third to specifically identify residues 4, 5 and 8 (5, 6 and 9 for FIX) from the previous sequence as the keel. In addition, since we have to compare several GLA domain crystal structures each having their Ca(II) or Mg(II) positions differently numbered, we will use for clarity purpose the same numbering for all, that is, the one found in the crystal structure of the human Factor VII (1DAN, see ref. 38)
Metal cation's electron structure / biological activity relationship in coagulation proteins
When a blood vessel is injured, a cascade of protein-protein interactions1 rapidly occurs leading to the formation of a cross-linked fibrin clot, eventually restoring the integrity of the circulatory system. A number of proteins involved at the early stage of the process are vitamin K-dependent zymogens of serine proteases31. Among this family of coagulation factors, especially factor VII, once activated and bound to tissue factor (TF), activates zymogen factors IX and X into functional factors IXa and Xa. These in turn transform prothrombin in active thrombin which is ultimately responsible for the conversion of fibrinogen to fibrin32. A cell-based model of coagulation, which incorporates the important roles of endothelial cells and platelets, has recently been introduced and this more physiological view appears to be gaining acceptance33. Many other factors and cofactors are necessary for the completion of the mechanism, but we will focus only on two steps1.
Structural analysis and biological activity
The above mentioned three factors are comprised of four domains. At the N extremity, we find a GLA domain, followed by two Epidermal Growth Factor (EGF) like domains, namely EGF1 and EGF2, terminated by a Serine Protease (SP) domain. The primary structures of GLA domains (residues 1 to 48) are highly conserved34: only a few residues differ along the sequences of factor VII, IX and X. Remarkably, the first nine Gla residues, as well as two cysteines are always found precisely at the same place. Accordingly, for all factors, two hydrophobic residues are adjacent to the first group of two Gla residues: Phe4 and Leu5 for FVIIa and FXa factor, Leu6 and Phe9 for FIXa. We can also notice the presence of an Asn residue in the second position of the sequence. Moreover, analysis of the pdb files for these factors show that the secondary structures are also very similar and essentially superimposable. From the N-extremity to the end of the domain we find successively the ω-loop with its two (mentioned above) hydrophobic amino-acids along with either another hydrophobic residue (FVIIa - Leu8 and FXa - Met8) or an cationic one (FIXa – Lys6) pointing their side chains toward the exterior of the protein. Three α-helices are also found. The first two are parallel and maintained so by a disulfide bridge established by the two cysteines cited before. All Gla residues are distributed along these three sub-units: Gla6 and 7 are at the top of the central loop of the ω-loop; Gla14, 16, 19 and 20 belong to the first α helix whereas Gla25, 26 and 29 belong to the second. The third α-helix links the GLA domain to the remaining part (3rd helix) of the protein. A Gla residue can also be found in this part of the domain. In all examined X-ray structures, eight cations are present38. One cation is located at the hinge between the third α helix and the first chain of EGF1 domain. The other seven cations are found aligned at the interstice between the base of the two parallel α helices, and the top of the ω-loop. In this zone, two groups of cation binding sites can be defined. The first group is called “high affinity” Ca(II) binding sites39 and is constituted by the 5 inner Ca(II) sites (numbered 3, 4, 5, 6, 8, respectively)38. Within these sites, Ca(II) was found coordinated 6, 7, 7, 7 and 3 times respectively at distances included in the 2.4 – 2.8 Å range in the very first GLA domain ever structurally determined (1992), namely the GLA domain of Ca-Prothrombin Fragment I (see Figure 4 and Table III of ref. 35). Subsequently several authors36,37,41 reported very similar coordination numbers and distances for the factors studied here. The second group is called “low affinity” Ca(II) binding sites define the two external sites. At these positions, cations can be either Ca(II) or Mg(II). Figure 4 lists the cation binding sites present in different PDB structures of the studied factors GLA domains. Occupation by, either Ca(II) or Mg(II), in all binding sites seems to be due to details in the preparation of the protein crystal during the X ray crystallization process. In fact, F VIIa (1DAN)38, F IXa (1J35)39 and F Xa (1IOD)40 were prepared using only CaCl2 as crystallographic salt. It is noticeable that all the binding sites are occupied by calcium, solely present in the environment. When MgCl2 is added to CaCl2, at physiological concentration, for the preparation of F IXa (1J34)39, all the inner binding sites are occupied by calcium, while the external binding sites are occupied by magnesium. But, prepared in the same conditions, a recent X-ray structure deposited in the Protein Data Bank by Bajaj et al. shows a Mg(II) residing in the central site number 5, replacing a Ca(II) in the GLA domain of F VIIa (2A2Q)41. However, when only MgCl2 is used for the preparation of F Xa (1P0S)4, the two external sites contain a Mg(II) cation, but the sole central site number 5 shows a third Mg(II) cation. In addition, contrary to the five other structures that contain it, no ω-loop is present in the structure of this domain, its constitutive peptide seeming to “float” in the environment. From these observations, it may be deducted that Ca(II) cations are necessary in the central zone to structure the ω-loop, and that the external sites are usually occupied by magnesium. Close examination of the binding of the ω-loop to the rest of the GLA domain reveals the network of interactions between the amino acids borne by the ω-loop, the cations and the aminoacids present in the two anti-parallel α helices. In all factors where the ω-loop is present, except for F VIIa with Mg(II) in the central site, the NH3+ extremity of Ala1 (or Tyr1 for F IXa) establishes three H-bonds with the surrounding residues, namely carbonyl O of Gln21 (F VIIa), Ala21 (F Xa) or Lys22 (F IXa), Oε4 of Gla20 (F VIIa and F Xa) or Gla21 (F IXa) and Oε4 of Gla26 (F VIIa and F Xa) or Gla27 (F IXa). In F VIIa (2A2Q), however, a single H-bond remains with Oε2 of Gla26. Comparison between coordination numbers and coordination distances for each cation of the two structures of F VIIa (1DAN vs. 2A2Q), as given in Table 2 of ref. 41 by Bajaj et al., reveals that when only Ca(II) is present in the five inner sites, direct coordinations of cations by the carbonyl O of Ala1 (or Tyr1 for F IXa), Oδ1 of Asn2, Oε1 and Oε4 of Gla6 and, Oε1, Oε2 and Oε4 of Gla7 are observed. By contrast, in F VIIa (2A2Q), where the central Ca(II) is substituted by Mg(II) with two water molecules (S209 and S363) completing the coordination number of this cation, the same interactions are observed, but this time through a network of eight water molecules (S209, S262, S363, S411, S508, S602, S694 and S722), present in between the cations and the ω-loop. As a consequence, the ω-loop has moved down approximately 0.5 Å in order to leave enough space for water to insert (See Figure 2A of ref. 41).
Figure 4. Description of metal cations coordination sites of six GLA domains.
The picture is a representation of the GLA domain of F VIIa (1DAN), exhibiting its constitutive parts including the seven cation binding sites. For each site, a comparison of six X-ray geometries shows its occupation by either a Mg(II) or a Ca(II), according to the crystallographic salt used for the preparation.
The existence of the ω-loop is of much importance in that the very first step of the coagulation process is for coagulation factors to co-localise on cell surfaces42. In that step, the biological function of the GLA domain is directly first responsible for the linkage of the factors to the membranes, and ultimately to the fibrin clot. This can be done thanks to the three hydrophobic residues of the ω-loop43 (or two hydrophobic residues and a cationic one in the F IXa), oriented in such a way that they are able to dive deeply inside the membrane and interact through hydrophobic bonds with neighbouring lipids43,44, in addition probably through a buried salt bridge involving Lys5 of F IXa43. The deep insertion of the GLA domain inside the membrane also allows the creation of an interaction between a phosphatidylserine (PS) of the membrane and the Ca(II) cation present in the binding site number 8, strengthening the anchorage of the coagulation factor inside the cellular membrane45. Inhibition of the GLA domain by direct ligand bonding to the ω-loop (such as snake venom protein, see ref. 39) is responsible for the loss of membrane linking with a subsequent loss of the coagulation process.
Theoretical study of interactions of Ca(II) vs Mg(II) with malonate groups
In this section we systematically supplement ELF analysis with detailed RVS energy decomposition results.
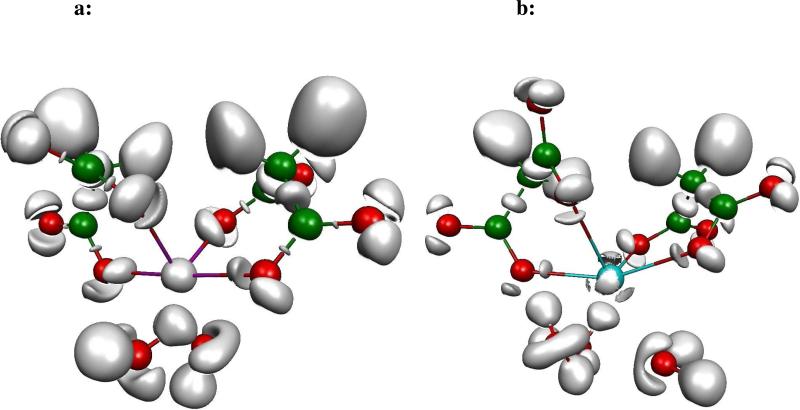
In GLA domains, the transformation of glutamic acids in Gla is realised by the action of vitamin K and several specific enzymes with the addition of a carboxylate group to the γ-carbon of the glutamate46; two carboxylate groups borne by the same γ carbon constitutes a malonate group. Two malonates coordinating a metal cation is one of the unit structures observed in GLA domains, displaying as many as four monodentate formate-cation interactions. Two different computations at the same level of theory (B3LYP/6-311G++G**) have been performed on these systems: a single point calculation using directly extracted geometries from PDB structures on which H atoms were added, and geometry optimisations using the previous systems as starting points. At the end of the optimisation process, the obtained complexes show the two approximate planes of malonates perpendicular to each other around the metal cation, itself four times tetrahedrally coordinated with coordination distances of approximately 2.1 Å for the magnesium, and 2.4 Å for the calcium. A calcium complex was extracted from the PDB structure of F Xa (1IOD), whereas the magnesium complex was extracted from the PDB structure of F IXa (1J34). From these two systems, external binding sites (number 7 or 9) are selected, in which cations are surrounded by solvation shell water molecules: two for Mg(II) and three for Ca(II). Figures 5a and 5b shows the ELF topological analysis on the two optimized geometries. As expected, Mg(II) (Figure 5a) does not exhibit any split of its subvalence, whereas Ca(II) (Figure 5b) has its subvalence split into four well separated basins, oriented in such a manner that no oxygen lone pair faces a cation basin. The same pattern is observed in Figure 6a for Mg(II) and Figure 6b for Ca(II) as the one described above, within the extracted geometries. Thus, upon increasing the coordination number from two in the bidentate formate-cation complexes, to four in the optimized geometries shown in Figures 5a and 5b and ultimately to their maximum in the extracted complexes, each cation consistently exhibits the same behaviour. Table IV reports the results of the RVS analysis disclosing the individual contributions of the interaction energies for the two malonates-cation complexes shown in Figures 5a and 5b, 6a and 6b. These results show first a greater electrostatic term for Mg(II) less compensated by the repulsion term than for Ca(II). As a result, the excess of electrostatic energy reflected by the negative first order term of Mg(II) over Ca(II) is of -90 kcal/mol for the optimized complexes and less than -77 kcal/mol for the extracted geometries, reflecting the hardness of the Mg(II) cation. Since a RVS analysis can separate the second order energies for each constitutive monomer of a many-body system, the individual polarization energies of the cations are also reported. It can thus be observed that, as Ca(II) moves away from its optimal geometry, its polarization increases, and that Mg(II) is essentially not polarized. The charge transfer contribution in the Ca(II) complexes decreases upon moving away from the optimal geometry but remains significant whereas it is null for the Mg(II) optimized complex, and slightly increases in the complex extracted from X-ray crystallography. It can be deduced that, contrary to Mg(II), Ca(II) is polarized as it is in the bidentate formate-cation type of complex, and that the charge transfer contribution becomes selective of one cation to another in such complexes. Thus Mg(II) enters in a more electrostatic type of interaction, whereas a polarized Ca(II), generating a stronger charge transfer, enters in a more covalent type of interaction. It is important to point out that we also perform such analysis in the presence of an additional PCM implicit solvent. We did not observe any changes in the topology of the system. Such results can be found as Supplementary Information S3.
Figure 5.
a Optimized geometry of 2 malonates-Mg(II) complex.
b Optimized geometry of 2 malonates-Ca(II) complex.
All the following fictures were revealed at the 0.87 isosurface coefficient. These pictures exhibit the perfect tetrahedral binding mode of the cations. Mg(II) subvalence is spherical, Ca(II) one is split in four well separated basins recalling the bidentate formate-cations complexes.
Figure 6.
a Extracted 2 malonates-Mg(II) “deformed tetrahedral” complex.
b Extracted 2 malonates-Ca(II) “deformed tetrahedral” complex.
The Mg(II) complex is the binding site n° 7 of F IXa (1J34), the Ca(II) one is the binding site n° 9 of F Xa (1IOD). Water molecules complete the coordination sphere of each cation: 6 for Mg(II) and 7 for Ca(II). Cation coordinations are also disclosed in order to evidence the “deformed tetrahedral” binding mode of the cations. Each cation behavior dos not change as the number of coordination increases.
Table IV. Theoretical RVS analysis of tetrahedral cation binding sites.
RVS energies decomposition has been performed upon theoretical models of tetrahedral binding sites. They are build on the same number of atoms (no water molecules are considered) in order to directly compare the energy contributions. Optimized geometries are the extracted ones on which optimisation process was realised. Mg(II) complexes first order terms are greater than the ones of Ca(II) complexes, whereas Ca(II) cations are more polarized and generate a greater charge transfer than for Mg(II). This is consistent with Mg(II) cation with a spherical subvalence entering in more electrostatic type of interactions, and Ca(II) cation splitting its subvalence and create more covalent type of interactions.
| kcal/mol | 2 malonates + Mg(II) | 2 malonates + Mg(II) | 2 malonates + Ca(II) | 2 malonates + Ca(II) |
|---|---|---|---|---|
| Geometry | Optimised | F IXa 1j34 | Optimised | F Xa 1iod |
| Figure number | 5a | 6a | 5b | 6b |
| Electrostatic | -777.7 | -703.3 | -709.0 | -636.8 |
| Repulsion | 90.8 | 68.2 | 111.5 | 78.2 |
| First order energies (e1) | -686.9 | -635.1 | -597.5 | -558.6 |
| Polarization of cations | -0.1 | -0.3 | -0.6 | -2.2 |
| Polarization | -94.2 | -90.5 | -63.0 | -61.5 |
| Charge transfer | 0.0 | -1.4 | -11.2 | -9.7 |
| Second order energies (e2) | -94.2 | -91.9 | -74.2 | -71.2 |
| Total e1 + e2 | -781.1 | -720.9 | -671.7 | -629.8 |
Theoretical study of the selectivity Ca(II)/ Mg(II)
a) Is Mg(II) cation strictly hard in the gas phase?
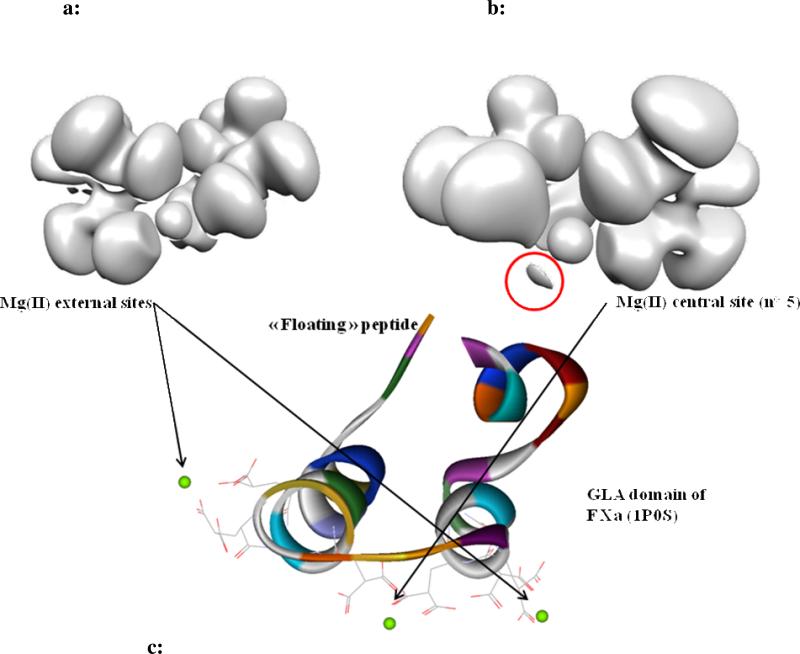
In the GLA domain of F Xa (1P0S), in which only magnesium ions are present in the environment, a Mg(II) is observed to be three times coordinated to oxygen atoms of Gla 16 and 26, in the 2.3 -2.5 range of distances. Figure 7a and 7b compare the topology of Mg(II) in two positions: the first one (Figure 7a) coming from an external cation site (site n°7 of F IXa 1J34), the second one (Figure 7b) is the central site (n°5). Due to the low resolution (2.8 Å) of the crystal structure of F Xa (1P0S), no water molecule is resolved near the cation in the central site n°5. Despite the fact that water must be present within the coordination sphere of Mg(II) in a real enzyme, no water molecule is considered in the present theoretical gas phase study, so that the coordination number is four for the external site, and three for the central site. As can be seen in Figure 7a, the cation does not exhibit any split of its subvalence, wheras in Figure 7b, the presence of the additional basin (red circle), recalling the monodentate formate-Mg(II) binding mode, clearly suggests that Mg(II) transfers a part of its density in this additional basin. This is confirmed by the magnitude of the RVS charge transfer contribution (see FXa 1P0S in Table V), namely -7.6 kcal/mol, instead of a null charge transfer in the optimized geometry. In this particular case, it is important to point out that Mg(II) does not have a hard cation behaviour since it exhibits some subvalence capabilities. However, if such a finding is important to understand the electronic distribution in Mg(II) complexes, could it have an impact when considering more realistic condensed phase GLA domains containing explicit solvent molecules?
Figure 7.
a “Hard” Mg(II) binding mode.
b “Soft” Mg(II) binding mode.
c Localization of the two binding modes within F Xa (1P0S).
Electron densities are revealed at the isosurface coefficient of 0.22. The additional basin shown in the red circle is confirmed by the increase of the charge transfer energy illustrated in Table 4 (-7.6 kcal/mol). As it is the case when only Mg(II) is present in a Vitamin K-dependent coagulation factor environment, the cations occupy only the three tetrahedral binding sites, being unable to structure the ω-loop, leaving its constitutive peptide floating.
Table V. RVS analysis of selected cation binding sites, extracted from X-ray structures.
RVS energies decomposition has been performed upon various extracted geometries in order to confirm the theoretical results on realistic systems. Thus all the components, including water molecules, were taken into consideration. The number of water molecules is detailed for each complex, as well as the X-ray structure of which the system was extracted, and the corresponding figure reference. The same trend is observed, as for theoretical tetrahedral systems, concerning electrostatic energies favouring Mg(II) complexes, and the Ca(II) cation being more polarized producing a greater charge transfer.
| kcal/mol | 2 malonates + Mg(II) | 2 malonates + Mg(II) | Mg(II) hexa coordinated | 2 malonates + Ca(II) | Ca(II) octo coordinated |
|---|---|---|---|---|---|
| Water molecules | 2 | 0 | 3 | 3 | 2 |
| PDB id | F IXa 1j34 | F Xa 1p0s | F VIIa 2a2q | F Xa 1iod | F IXa 1j34 |
| Figure number | 6a | 7b | 8a | 6b | 8b |
| Electrostatic | -696.7 | -577.9 | -746.5 | -690.6 | -598.5 |
| Repulsion | 130.0 | 23.7 | 184.5 | 188.3 | 111.1 |
| First order energies (e1) | -566.7 | -554.3 | -562.0 | -502.3 | -487.4 |
| Polarization of cations | -0.4 | -0.4 | -0.1 | -1.8 | -0.5 |
| Polarization | -122.8 | -66.5 | -92.6 | -95.0 | -33.0 |
| Charge transfer | -9.8 | -7.6 | -2.7 | -20.7 | -4.9 |
| Second order energies (e2) | -132.5 | -74.1 | -95.3 | -115.7 | -37.9 |
| Total e1 + e2 | -699.2 | -628.4 | -657.3 | -618.0 | -525.3 |
b) Selectivity of Ca(II) vs. Mg(II) cations within the six Gla domains
From Figure 4, we have found three possible binding sites for Mg(II), namely sites n° 7 and 9 (external) and 5 (central), the latter exhibiting either a Mg(II) or a Ca(II). The other sites are exclusively occupied by Ca(II) cations, a necessity to structure the ω-loop in its functional geometry. These observations have been confirmed by several experiments, in which Mg(II) and Ca(II) were added at physiological concentration to a previously divalent cation-free environment2. When only Mg(II) is present, no enzymatic activity occurs. This is to be put in relation with the F Xa (1P0S) structure where the ω-loop is not formed. When only Ca(II) is present, the enzymatic activity is signicantly higher, but can be enhanced by the addition of Mg(II). The three possible Mg(II) sites are constructed from three pairs of malonates provided by: site n° 5 Gla16 and 26 (Gla17 and 27 for F IXa), site n°7 Gla14 and 19 (Gla15 and 20 for FIXa) and site n°9 Gla25 and 29 (Gla26 and 30 for F IXa). Close examination of these sites reveals that they all share the same pattern in which the cation is coordinated by two groups of two oxygens residing in the same size of two almost parallel malonate groups. Such geometry recalls the optimized geometry of the two malonates bound to a cation complex. Because they are not perfectly tetrahedral, these three binding sites will be called “deformed tetrahedral” cation binding sites. Figure 6a and 6b show ELF pictures of such a “deformed tetrahedral” binding site occupied by a Mg(II) cation as extracted from site n° 9 of F IXa (1J34) and a Ca(II) cation as extracted from site n° 7 of F Xa (1IOD) respectively. From Table IV, RVS total energies gives us the preferrence of Mg(II) over Ca(II) within this type of sites by a difference of -109.4 kcal/mol for the optimized geometries, and -91.1 for the extracted ones. This is due to the excess of first order energies in favour of Mg(II), namely -89.4 and -76.5 kcal/mol respectively. This is consistent with Mg(II) not splitting its subvalence and thus involved in more electrostatic types of interactions. The presence of water molecules (two for Mg(II) and three for Ca(II)) within the coordination sphere of cations does not modify this conclusion (see Table V).
The four other sites, however, are less structured as Ca(II) cations are coordinated four to eight times, in both monodentate and bidentate mode, by several carboxylate groups belonging to different Gla residues, as well as by carbonyl groups of the backbone of Ala1 (Tyr1 for F IXa) and the side chain of Asn2. For example, in the site n° 6 of F IXa (1J34), Ca(II) is found eight times coordinated, by Tyr1, Gla21 in a monodentate mode, Gla7 and 17 in a bidentate mode and two water molecules (Figure 8b). The now familiar split of the subvalence of Ca(II) is observed. An ELF computation performed on this model, reveals that the net charge of the cation within the system is +1.67 instead of +2 in the fundamental state. This means that, up to one third of an electron was transferred from the ligands oxygen lone pairs to the Ca(II) cation, consistently with Ca(II) able to adapt its electronic density to its environment by splitting its subvalence, thus entering in more covalent types of interactions. This capacity of adaptation for Ca(II) explains why, when only present in a coagulation factor environment, all the binding sites are loaded with Ca(II) (F VIIa 1DAN, F IXa 1J35 and F Xa 1IOD).
Figure 8.
a Extracted geometry of binding site n° 5 of F VIIa (2A2Q) with Mg(II).
b Extracted geometry of binding site n° 6 of F IXa (1J34) with Ca(II).
Figure 8a shows a tetrahedral binding site loaded with a Mg(II) cation, that structure the ω-loop through a network of H-bonds established by highly polarized water molecules coordinated to the cation. Figure 8b shows direct interactions between a Ca(II) cation and some constitutive segments of the ω-loop. The subvalence pattern of the two cations stays consistent with the one observed in all the studied geometries, whatever the size of the system considered.
Therefore, with respect to the experimental results shown in figure 5 of ref. 2, a mechanism can be proposed for the activation of Vitamin K-dependent coagulation factors by Mg(II) and Ca(II) cations. Upon introducing first Mg(II) to a cation-free environment of an enzyme, one Mg(II) cation goes to each of the three “deformed tetrahedral” binding sites (n° 5, 7 and 9), completing its coordination sphere with two water molecules. At this stage, the enzyme is not active and the ω-loop is not structured (F Xa 1P0S). When Ca(II) is added, one Ca(II) cation goes to each of the four other sites (n° 3, 4, 6 and 8). This anchors Ala1 (or Tyr1 for F IXa) and Asn2 to the rest of the GLA domain47 and folds the peptide in this particular ω geometry through interactions involving Gla6 and 7 (Gla7 and 8 for F IXa) and the rest of the domain (F VIIa 2A2Q). However, prepared with the same mix of cations at physiological concentration39 as the one used to prepare F VIIa (2A2Q)41 for the crystallization process, F IXa (1J34) exhibits a Ca(II) cation in central site n° 5 instead of a Mg(II) cation. Could the occupation of the central binding site n° 5 by either a Mg(II) (2A2Q) or a Ca(II) (1J34) be explained by the topological difference of the two cations?
c) Direct interactions with cations vs. interactions with cations through water
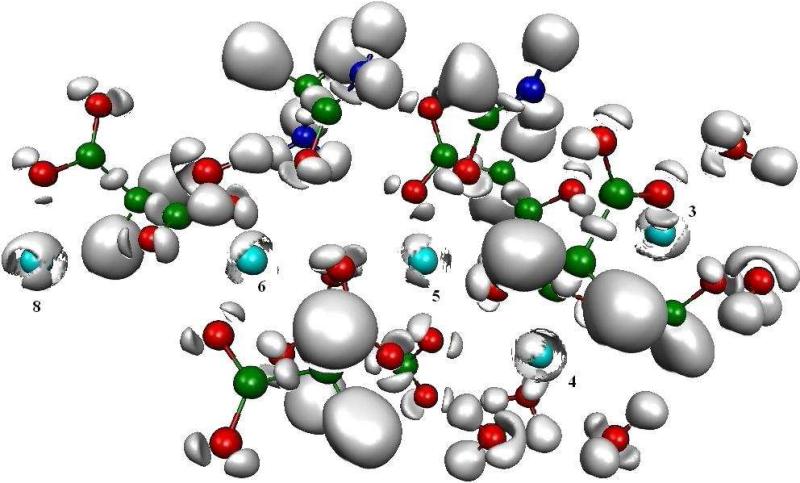
We consider here a system constituted by the five central Ca(II) binding sites extracted from the geometry of the GLA domain of F VIIa (1DAN) which is very similar to the one of F IXa (1J34). This complex was built from the backbone of Ala1, a formamide group representing the side chain of Asn2, two malonates for Gla6 and 7; Ala1, Asn2, Gla6 and Gla7 being part of the ω-loop, in addition with three malonates groups from Gla16, 20 and 26 and a formate given by Gla29 and the five Ca(II) ions. In addition, up to six water molecules were placed on the calcium ions according to their position in the X-ray structure. This system, which is globally neutral, is an X-ray crystal snapshot where hydrogens and waters have been added, focusing on the interactions between Ca(II) cations and their ligands. Such system is found in X-ray crystal structures 1DAN, 1J34, 1J35 and 1IOD in which cations directly interact with their environment. Figure 9 exhibits the ELF study on this system. This picture reveals the network of interactions between the subvalence basins of the cations and the lone pairs of the coordinating oxygens. The split of all Ca(II) subvalences indicates that these interactions are partially covalent because built from electronic exchanges between the cations and the lone pairs being present in their immediate environment. This results into a network of many-body interactions at the centre of the GLA domain. QTAIM values of M1 of each Ca(II), computed with the ELF function, confirm the intensity of the charge transfer. Ca(II) n° 3 and 5, being each seven-fold coordinated, have the lowest value of the series with a M1 of 0.041 D, and a net charge of +1.66 instead of +2, whereas Ca(II) n° 4 and 6, with six coordinations exhibit enhanced values, namely 0.060 D and +1.68, for M1 and the charge respectively. Ca(II) n° 8, being four times coordinated, shows values of M1 very close (0.166 D, +1.72) to the one found for the small bidentate formate-cation complex (0.160 D and +1.70 for M1 and the charge respectively, see Table 1). This indicates that the greater the coordination number, the less the polarization and therefore the residual charge.
Figure 9. Topological analysis of the five central Ca(II) binding sites of F VIIa (1DAN) GLA domain.
This picture unravels the network of charge transfer based interactions between the five Ca(II) cations and the oxygen lone pairs borne by Gla residues of the two upper α-helices in one hand, and the oxygen lone pairs borne by Ala1 (backbone), Asn2 (side chain) and Gla6 and 7 of the lower ω-loop in the other hand. These interactions structure the ω-loop in its functional geometry. It can be noticed that Ca(II) cations show different patterns of their subvalence split according to their different numbers of coordinaton. The numbering os the Calcium ion binding sites in Fig. 4 is used.
In the GLA domain of F VIIa (2A2Q), the central Mg(II) is found six times coordinated: four times tetrahedrally by malonates of Gla 16 and 26 and two times by water molecules S209 and S363 (Figure 8a). The formamide moiety of Asn2, a formate group from Gla7 and a third water complete the network of H-bonds through which this central Mg(II) contributes to structure the ω-loop. As seen in Figure 8a, the central Mg(II) subvalence remains almost perfectly spherical, as in the bidentate formate-Mg(II) complex and in the two malonates-Mg(II) tetrahedral optimized geometry. Close examination of the RVS polarization contribution (Table V) reveals that, if the cation is as expected almost not polarized, the polarization of the three water molecules accounts for approximately 30% of the total polarization of -92.7 kcal/mol. Indeed, one of the water molecules is highly polarized and bears a Epol(RVS) value of -13.9 kcal/mol. Thus interactions between the cation and the ω-loop are established through highly polarized water molecules.
While the presence of Ca(II) is required2,3,48 in the central region for effective coagulation function, it has been demonstrated that the affinity of coagulation factors for, either cellular membranes45,49, Tissue Factors5 or anticoagulant agents36,40, is strongly enhanced when external sites are occupied by Mg(II)40. This supports our findings that these sites are better stabilized by Mg(II)37 rather than Ca(II). Therefore, the difference of the two geometries (1J34 and 2A2Q) resides in the fact that, in 2A2Q, the central site is occupied by a Mg(II) which is only able to complete its coordination shell with water, for which the residence time has been measured to be in the order of one micro second8. This explains the greater distance between the two α-helices and the ω-loop (2A2Q case), namely the interposition of a water layer. Therefore, one of the roles of the cations is to fix water, which in turn binds to the ω-loop through a network of H-bonds. By contrast, in FIXa 1J34, where all the central cation binding sites are occupied by Ca(II), interactions take place directly between metal cations and the lone pairs borne of its ligating oxygens. In this connection, it was recently demonstrated that water layers present either between separate domains of a protein or in between different proteins are in dynamic as short time exchanges with the solvation water present in the environment are observed50,51. Therefore, it is possible to make the hypothesis that F VIIa crystallized by Bajaj et al. (2A2Q) with interactions through water (ie: water mediated interactions), could be present free in the blood plasma since the latter is a highly hydrophilic environment. On the other hand, the GLA domain of F IXa crystallized by Shikamoto et al. (1J34), is found bound to a ligand (in this case a snake venom protein); this binding could impose the “direct interactions with cations” geometry. It is, then, perhaps reasonable to suggest that this is the geometry that inserts deeply into the cell membrane, the interior of which is a highly hydrophobic environment. Indeed, the insertion within the membrane may expel water molecules from the inserted part of the GLA domain, imposing a switch between “physics at play”: going from “through water structure” to “direct electronic interactions structure”.
Conclusion
In this contribution, we have used several theoretical tools to illustrate the relationship between the electronic structure of selected metal-cation complexes and the so-called hard and soft chemical behaviour. The ELF analysis allows ranking cations according to their topological signature, namely their ability to split their valence into sub-domains (subvalent basins). Hard cations will not exhibit such a capability as soft cations do so. The covalent character of the ligand-metal cation interaction is associated with a basin between the metal and the neighbouring heavy atoms. That way, such a covalent interaction can be directly affected by the cations environment and, as we have seen, an electrostatic interaction as in Mg(II) monodentate-like complexes can become covalent exhibiting an extra subvalent localization basin under specific stress conditions. This topological metal cation ranking has been shown to be relevant when compared to RVS and CSOV energy analysis as well as in good qualitative agreement with local Fukui Functions extracted from both QTAIM and ELF analysis.
Our integrated methodological approach uncovered a clear relationship between the underlying metal electron structure and the biological activity of enzymes such as Vitamin K-dependent coagulation factors. The present approach could also be then extended to other biological systems involving metal cations. It is also important to point out that these results on metal cations-ligands, such as the dissymmetry between internal and external carboxylate oxygen lone pairs, could also provide useful informations for the design of new force fields7a,52. Moreover, applications to the problem of blood coagulation allowed us to address the Ca(II) vs. Mg(II) selectivity in their interaction with GLA domains. The two only X-ray structures, crystallized using the same Mg(II)/Ca(II) mix found in physiological conditions (1J34 and 2A2Q) have been investigated.
In the first (1J34), Ca(II) has been shown to be more covalently bonded to ligand than Mg(II) enabling the creation of a “direct charge transfer network” between its subvalence and the carboxylate oxygen lone pairs. We also showed that this concept could also uncover the distinct role of the two cation binding sites present in GLA domains. Being at the origin of the observed charge transfer network, calcium cations are mandatory in the central region to conserve the folding, whereas external binding sites are better stabilised by Mg(II) rather than Ca(II) in agreement with experiment thanks to strong electrostatic interactions with the environment.
In the second structure (2A2Q), in which some interactions between metal cations and Gla are established through the presence of crystallized water molecules, we have shown that magnesium is able to form “an indirect charge transfer network” with malonates through its interaction with two highly polarized water molecules that are responsible for a structure of the ω-loop similar to that observed in the first structure (1J34). To conclude, as the two crystal structures have been solved, they could be the two interconverting forms of a same system, each form with a different water requirement. In both cases, the ω-loop is present. This is a requirement for the active enzyme, but the underlying physics is not the same. In the first structure, the folding is mainly due to electronic effects: Ca(II) is preferred in the five central binding sites. By contrast, the presence of water molecules is able to reverse the direct electronic selectivity of cations (Mg(II) could be present if interacting with two water molecules). This clearly indicates that solvent dynamical effects could play a key role in the observed structure of the domain. The pathway between the two structures might be established through a series of molecular dynamics simulations. It also emphasizes again53 the importance of “discrete” water molecule to understand the stability of biological systems as the presence of a limited number of structured water molecules could be critical to obtain meaningful theoretical models.
Supplementary Material
Acknowledgments
The computations have been performed on the national IDRIS (F. 91403 Orsay, France), CRIHAN (F.76800 Saint-Etienne-du-Rouvray, France), and CINES (F. 34097 Montpellier, France) supercomputing centers. Some ELF computations have been run at the local CCRE centre at Université Pierre et Marie Curie, Univ Paris 6 (F. 75252 Paris CEDEX 05, France). Support from the French National Research Agency (ANR) on project LASIHMODo is acknowledged. This research was supported in part by the Intramural Research program of the NIH and NIEHS.
Footnotes
Supporting Information Available
S1: Performance of the B3LYP functional
S2: Effects of PCM solvation over the topology of two bidentate formate-cation complexes.
S3: CSOV computations (DFT level). This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Furie B, Furie BC. Cell. 1988;53:505–518. doi: 10.1016/0092-8674(88)90567-3. [DOI] [PubMed] [Google Scholar]
- 2.Prendergast FG, Mann KG. J. Biol. Chem. 1977;252:840–850. [PubMed] [Google Scholar]
- 3.van den Besselaar AMHP. Blood Coagulation & Fibrinolysis. 2002;13:19–23. doi: 10.1097/00001721-200201000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Wang SX, Hur E, Sousa CA, Brinen L, Slivka EJ, Fletterick RJ. Biochemistry. 2003;42:7959–7966. doi: 10.1021/bi027320a. [DOI] [PubMed] [Google Scholar]
- 5.Persson E, Ostergaard A. J. Tromb. Haemost. 2007;5:1977–1978. doi: 10.1111/j.1538-7836.2007.02661.x. [DOI] [PubMed] [Google Scholar]
- 6.a Lightstone FC, Schwegler E, Hood RQ, Gygi F, Galli G. Chem. Phys. Lett. 2001;343:549–555. [Google Scholar]; b Naor MM, Van Nostrand K, Dellago C. Chem. Phys. Lett. 2003;369:159–164. [Google Scholar]; c Bako I, Hutter J, Palinkas G. J. Chem. Phys. 2002;117:9838–9843. [Google Scholar]; d Lightstone FC, Schwegler E, Allesch M, Gygi F, Galli G. Chem. Phys. Chem. 2005;6:1745–1749. doi: 10.1002/cphc.200500053. [DOI] [PubMed] [Google Scholar]
- 7.a Piquemal J-P, Perera L, Cisneros GA, Ren P, Pedersen LG, Darden TA. J. Chem. Phys. 2006;125:054511–1-7. doi: 10.1063/1.2234774. [DOI] [PubMed] [Google Scholar]; b Babu CS, Lim C. J. Phys. Chem. A. 2006;110:691–699. doi: 10.1021/jp054177x. [DOI] [PubMed] [Google Scholar]
- 8.Neely J, Connick R. J. Am. Chem. Soc. 1970;92:3476–3478. [Google Scholar]
- 9.a Bader RFW. Atoms In Molecules: A Quantum Theory. Oxford University Press; Oxford, U. K.: 1990. [Google Scholar]; b Matta CF, Boyd RJ. The Quantum Theory of Atoms in Molecules: From Solid State to DNA and Drug Design. Wiley-VCH; Weinheim, Germany: 2007. [Google Scholar]; c Popelier PLA. Atoms In Molecules: An Introduction. Prentice-Hall; Harlow, U. K.: 2000. [Google Scholar]
- 10.a Becke AD, Edgecombe KE. J. Chem. Phys. 1990;92:5397–5403. [Google Scholar]; b Silvi B, Savin A. Nature (London) 1994;371:683–686. [Google Scholar]
- 11.a Stevens WJ, Fink W. Chem. Phys. Letts. 1987;139:15–22. [Google Scholar]; b Bagus PS, Illas F. J. Chem. Phys. 1992;96:8962–8970. [Google Scholar]
- 12.Silvi B, Fourré I, Alikani ME. Chemie. 2005;136:855–879. [Google Scholar]
- 13.Silvi B. J. Phys. Chem. A. 2003;107:3081–3085. [Google Scholar]
- 14.Martín Pendás A, Francisco E, Blanco MA. Chem. Phys. Lett. 2008;454:396–403. [Google Scholar]
- 15.a Gillespie RJ, Popelier PLA. Chemical Bonding and Molecular Geometry. Oxford University Press; Oxford U. K.: 2001. [Google Scholar]; b Gillespie RJ, Robinson EA. J. Comput. Chem. 2007;34:396–407. doi: 10.1002/jcc.20545. [DOI] [PubMed] [Google Scholar]
- 16.Gillespie R, J., Noury S, Pilme J, Silvi B. Inorg. Chem. 2004;43:3248–3256. doi: 10.1021/ic0354015. [DOI] [PubMed] [Google Scholar]
- 17.Piquemal J-P, Pilmé J, Parisel O, Gérard H, Fourré I, Bergès J, Gourlaouen C, de la Lande A, van Severen MC, Silvi B. Int. J. Quant. Chem. 2008;108:1951–1969. [Google Scholar]
- 18.a Pilmé J, Piquemal J-P. J. Comput. Chem. 2008;29:1440–1449. doi: 10.1002/jcc.20904. [DOI] [PubMed] [Google Scholar]; b Fukui K, Yonezawa T, Shingu H. J. Chem. Phys. 1952;20:722–725. [Google Scholar]; b Fukui K, Yonezawa T, Nagata C, Shingu H. J. Chem. Phys. 1954;22:1433–1442. [Google Scholar]; c Woodward RB, Hoffmann R. The Conservation of Orbital Symmetry. Chemie; Weinheim, Germany: 1970. [Google Scholar]; d Parr RG, Yang W. Density Functional Theory of Atoms and Molecules. Oxford University Press; New York, U. S. A.: 1989. [Google Scholar]; e Morell C, Grand A, Toro-Labbé A. J. Phys. Chem. A. 2005;109:205–212. doi: 10.1021/jp046577a. [DOI] [PubMed] [Google Scholar]; f Morell C, Grand A, Toro-Labbé A. Chem. Phys. Lett. 2006;425:342–346. [Google Scholar]; g Cioslowski J, Martinov M, Mixon ST. J. Phys. Chem. 1993;97:10948–10951. [Google Scholar]; h Contreras RR, Fuentealba P, Galván M, Pérez P. Chem. Phys. Lett. 1999;304:405–413. [Google Scholar]; i Bulat FA, Chamorro E, Fuentealba P, Toro-Labbé A. J. Phys. Chem. A. 2004;108:342–349. [Google Scholar]; j Tiznado W, Chamorro E, Contreras R, Fuentealba P. J. Phys. Chem. A. 2005;109:3220–3224. doi: 10.1021/jp0450787. [DOI] [PubMed] [Google Scholar]
- 19.a Lee C, Yang W, Parr RG. Phys. Rev. B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]; b Becke AD. J. Chem. Phys. 1993;98:5648–5652. [Google Scholar]; c Burda JV, Šponer J, Hobza PJ. J. Phys. Chem. 1996;100:7250–7256. [Google Scholar]; d Russo N, Toscano M, Grand A. J. Phys. Chem. A. 2003;107:11533–11538. [Google Scholar]
- 20.Jaguar 6.5. Schrodinger Inc.; Portland, OR: 2005. [Google Scholar]
- 21.Hay PJ, Wadt WR. J. Chem. Phys. 1985;82:299–310. [Google Scholar]
- 22.Krishnan R, Binkley JS, Seeger R, Pople JA. J. Chem. Phys. 1980;72:650–654. [Google Scholar]
- 23.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JJA, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03, Revision C.02. Gaussian Inc.; Wallingford, CT: 2007. [Google Scholar]
- 24.Noury S, Krokidis X, Fuster F, Silvi B. J. Comput. Chem. 1999;23:597–604. [Google Scholar]; the modified TopMod90 program (named TopChem) is available upon request. See the following website for details: Available at: http://www.lct.jussieu.fr/pagesperso/pilme.(01/15/2010)
- 25.Piquemal J-P, Marquez A, Parisel O, Giessner-Prettre C. J. Comput. Chem. 2005;26:1052–1062. doi: 10.1002/jcc.20242. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH, Koseki S, Matsunaga N, Nguyen KA, Su S, Windus TL, Dupuis M, Montgomery JA., Jr J. Comput. Chem. 1993;14:1347–1363. [Google Scholar]
- 27.Ratcliffe JV, Furie B, Furie BC. J. Biol. Chem. 1993;268:24339–24345. [PubMed] [Google Scholar]
- 28.de Courcy B, Gresh N, Piquemal J-P. Interdiscip. Sci. Comput. Life Sci. 2009;1:55–60. doi: 10.1007/s12539-008-0027-0. [DOI] [PubMed] [Google Scholar]
- 29.Parr RG, Pearson RG. J. Am. Chem. Soc. 1983;105:7512–7516. [Google Scholar]
- 30.Xu X, Zhang L, Shen D, Wu H, Peng L, Li J. J. Biolo. Inorg. Chem. 2009;14:559–571. doi: 10.1007/s00775-009-0470-3. [DOI] [PubMed] [Google Scholar]
- 31.Perera L, Foley C, Darden TA, Stafford D, Mather T, Esmon CT, Pedersen LG. Biophysical Journal. 2000;79:2925–2643. doi: 10.1016/S0006-3495(00)76530-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffman M. J. Thromb. Thrombolysis. 2003;16:17–20. doi: 10.1023/B:THRO.0000014588.95061.28. [DOI] [PubMed] [Google Scholar]
- 33.Roberts HR, Hoffman M, Monroe DM. Semin. Thromb. Hemost. 2006;32(Suppl. 1):32–38. doi: 10.1055/s-2006-939552. [DOI] [PubMed] [Google Scholar]
- 34.Mc Donald JF, Shah AM, Schwalbe RA, Kisiel W, Dahlbäck B, Nelsestuen GL. Biochemistry. 1997;36:5120–5127. doi: 10.1021/bi9626160. [DOI] [PubMed] [Google Scholar]
- 35.Soriano-Garcia M, Padmanabhan K, de Vos AM, Tulinsky A. Biochemistry. 1992;31:2554–2566. doi: 10.1021/bi00124a016. [DOI] [PubMed] [Google Scholar]
- 36.Gopinath SCB, Shikamoto Y, Mizuno H, Kumar PKR. Biochem J. 2007;405:351–357. doi: 10.1042/BJ20061737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sekiya F, Yamashita T, Atoda H, Komiyama Y, Morita T. J. Biol. Chem. 1995;270:14325–14331. doi: 10.1074/jbc.270.24.14325. [DOI] [PubMed] [Google Scholar]
- 38.Banner DW, D'Arcy A, Chene C, Winkler FK, Guha A, Konigsberg WH, Nemerson Y, Kirchhofer D. Nature (London) 1996;380:41–46. doi: 10.1038/380041a0. [DOI] [PubMed] [Google Scholar]
- 39.Shikamoto Y, Morita T, Fujimoto Z, Mizuno H. J. Biol. Chem. 2003;278:24090–24094. doi: 10.1074/jbc.M300650200. [DOI] [PubMed] [Google Scholar]
- 40.Mizuno H, Fujimoto Z, Atoda H, Morita T. Proc. Natl. Acad. Sci. USA. 2001;98:7230–7234. doi: 10.1073/pnas.131179698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bajaj SP, Schmidt AE, Agah S, Bajaj MS, Padmanabhan K. J. Biol. Chem. 2006;281:24873–24888. doi: 10.1074/jbc.M509971200. [DOI] [PubMed] [Google Scholar]
- 42.Falls LA, Furie BC, Jacobs M, Furie B, Rigby AC. J. Biol. Chem. 2001;276:23895–23902. doi: 10.1074/jbc.M008332200. [DOI] [PubMed] [Google Scholar]
- 43.Ohbuko YZ, Tajkhorshid E. Structure. 2008;16:72–81. doi: 10.1016/j.str.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 44.Sunnerhagen M, Forsén S, Hoffrén A-M, Drakenberg T, Teleman O, Stenflo J. Nature Structural Biology. 1995;2:504–509. doi: 10.1038/nsb0695-504. [DOI] [PubMed] [Google Scholar]
- 45.Taboureau O, Olsen OH. Eur. Biophys. J. 2007;36:133–144. doi: 10.1007/s00249-006-0114-2. [DOI] [PubMed] [Google Scholar]
- 46.Davis CH, Deerfield D, II, Stafford DW, Pedersen LG. J. Phys. Chem. A. 2007;111:7257–7261. doi: 10.1021/jp068564y. [DOI] [PubMed] [Google Scholar]
- 47.Huang M, Furie BC, Furie B. J. Biol. Chem. 2004;279:14338–14346. doi: 10.1074/jbc.M314011200. [DOI] [PubMed] [Google Scholar]
- 48.Huang M, Rigby AC, Morelli X, Grant MA, Huang G, Furie B, Seaton B, Furie BC. Nature Structural Biology. 2003;10:751–756. doi: 10.1038/nsb971. [DOI] [PubMed] [Google Scholar]
- 49.Sekiya F, Yoshida M, Yamashita T, Morita T. J. Biol. Chem. 1996;271:8541–8544. doi: 10.1074/jbc.271.15.8541. [DOI] [PubMed] [Google Scholar]
- 50.Lin J, Balabin IA, Beratan DN. Science. 2005;310:1311–1313. doi: 10.1126/science.1118316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de La Lande A, Marti S, Parisel O, Moliner V. J. Am. Chem. Soc. 2007;129:11700–11707. doi: 10.1021/ja070329l. [DOI] [PubMed] [Google Scholar]
- 52.a Gresh N, Cisneros GA, Darden TA, Piquemal J-P. J. Chem. Theory Comput. 2007;3:1960–1986. doi: 10.1021/ct700134r. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Piquemal J-P, Chevreau H, Gresh N. J. Chem. Theory Comput. 2007;3:824–837. doi: 10.1021/ct7000182. [DOI] [PubMed] [Google Scholar]; c Piquemal J-P, Cisneros GA, Reinhardt P, Gresh N, Darden TA. J. Chem. Phys. 2006;124:104101–1-12. doi: 10.1063/1.2173256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Courcy B, Piquemal J-P, Garbay C, Gresh N. J. Am. Chem. Soc. 2010 doi: 10.1021/ja9059156. online, DOI:10.1021/ja9059156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.