Abstract
Given the role of transcriptional misregulation in the pathogenesis of human disease, there is enormous interest in the development of molecules that exogenously control transcription in a defined manner. The past decade has seen many exciting advancements in the identification of molecules that mimic or inhibit the interactions between natural transcriptional activators and their binding partners. In this minireview, we focus on four activator·target protein complexes, highlighting recent advances as well as challenges in the field.
Keywords: Diseases/Cancer, DNA/Transcription, Gene/Regulation, Gene/Transcription, Protein/Motifs, Receptors/Nuclear, Transcription/p53, Transcription Coactivators
Introduction
Just as specific transcriptional patterns signify the differentiation of stem cells into individual tissues, unique transcriptional signatures are associated with every human disease either as a cause or as an effect. For example, >50% of human cancers exhibit loss-of-function mutations in the tumor suppressor and transcriptional activator p53 (1, 2). There is thus enormous interest in identifying exogenous agents that one can use to influence transcriptional events in a predefined manner. One attractive strategy is the design or discovery of molecules that reconstitute the function of a natural transcription factor that can then be used to directly impact the transcriptional state of predefined genes. A second, related approach is to identify molecules that mimic one aspect of the structure of a particular transcription factor and in doing so serve as competitive inhibitors of important binding interactions. For both of these strategies, transcriptional activators of the amphipathic class have served as the inspiration, and molecules that reconstitute one or more of the functions of these complex transcription factors have been outstanding mechanistic tools with considerable therapeutic promise. As a visible and rapidly evolving field, progress in this area is frequently reviewed (3–8). In this minireview, we focus on four examples of activator·target protein complexes to highlight recent successes as well as the significant challenges remaining in the field.
Transcriptional Activators
Transcriptional activators participate in a remarkably complex web of binding interactions as they regulate their cognate gene(s). After initiation of a signal transduction cascade, transcriptional activators are often post-translationally modified and, if not already in the nucleus, must translocate there. Once in the nucleus, activators localize to their cognate DNA-binding sites and recruit chromatin-remodeling enzymes; transcriptional activators utilize an additional series of interactions to stimulate the assembly of the preinitiation complex (RNA polymerase II holoenzyme and its associated cofactors) (9). Despite the need to interact with many different partners, transcriptional activation can be accomplished with only two domains: a DBD2 and a TAD. These domains can be present in a single polypeptide chain or associate through non-covalent interactions. As the name suggests, the DBD confers much of the gene-targeting specificity to the protein, localizing it to particular sites within genomic DNA. In contrast, it is the TAD that participates in many of the protein-protein interactions (and post-translational modifications) that dictate the timing and extent of transcription. The largest class of eukaryotic transcriptional activators is the amphipathic class, exemplified by TADs containing hydrophobic amino acids interspersed with polar residues. An ever-growing number of amphipathic activators are identified as aberrantly functioning in disease states (10, 11). For example, the transcription factor HIF-1α regulates a number of genes in cancer metastasis (12, 13). In another example, ESX (ESE/ELF3/ERT/Jen), an epithelium-specific transcription factor, regulates expression of ErbB2 (HER2), an oncogenic protein that is overexpressed in ∼30% of all breast cancers (11, 14, 15). Perhaps the most widely studied member of this class is p53, an amphipathic activator that is misregulated in >50% of all human cancers (1, 2). A wealth of mechanistic and structural information exists regarding the interactions between p53 and its various binding partners, and thus, p53 is a particularly useful example with which to discuss transcriptional intervention strategies.
The activity of transcriptional activators can be modulated through interference of DBD-DNA interactions or by TAD mimics that block binding interactions with protein-binding partners (Fig. 1). In the former case, several classes of non-natural DNA-targeting molecules have successfully been used to inhibit activator-stimulated transcription in both cell-free and cellular systems, and this area has been extensively reviewed (3–8). More difficult has been the discovery of molecules that competitively inhibit the TAD binding interactions, and it is the latest developments in this arena on which we will largely focus. A single TAD typically utilizes several distinct binding modes, ranging from binding interactions that are of high affinity and specificity to those that are more modest in strength and are not particularly specific. An example of the former case is the masking interaction, an intermolecular or, more rarely, intramolecular complex that blocks the TAD from activating transcription in a signal-responsive fashion. Using p53 as an example, the masking protein MDM2 (human DM2) binds to a region of the p53 TAD (15–29) with a Kd of 575 nm (16), thus rendering p53 transcriptionally inactive; MDM2 does not mask any other transcriptional activators. At the other end of the spectrum are the complexes formed between amphipathic TADs and coactivators within the transcriptional machinery; these tend to be micromolar in strength and poorly defined in terms of structure and specificity. Returning to p53, the same TAD sequence that interacts with MDM2 binds to the coactivator and the histone acetyltransferase CBP (17). In addition, TAD-binding motifs within CBP bind to multiple amphipathic activators. The CBP KIX domain, for example, interacts with at least 12 amphipathic TADs. There is also considerable evidence that TADs interact with multiple coactivators in the transcriptional machinery as part of transcription initiation, including chromatin-remodeling enzymes, various members of the Mediator complex, and the proteasome, among others (18–29). One might therefore anticipate that molecules that block activator function through inhibiting these interactions would need to exhibit a similar multipartner binding profile.
FIGURE 1.
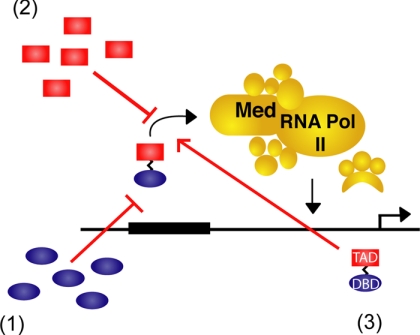
Strategies to modulate transcriptional activation. 1, interference of the endogenous activator DBD-DNA interaction using non-natural DBD mimics to inhibit transcriptional activation. 2, interference of the endogenous activator TAD-coactivator interaction using TAD mimics to inhibit transcription. TAD mimics can also be used to block activator TAD-masking protein interactions, thus stimulating transcriptional activation. 3, reconstitution of activator function using ATFs consisting of a non-natural TAD localized to the gene of interest using a DBD. Med, Mediator; RNA Pol II, RNA polymerase II.
Inhibition of Activator-Masking Protein Interactions
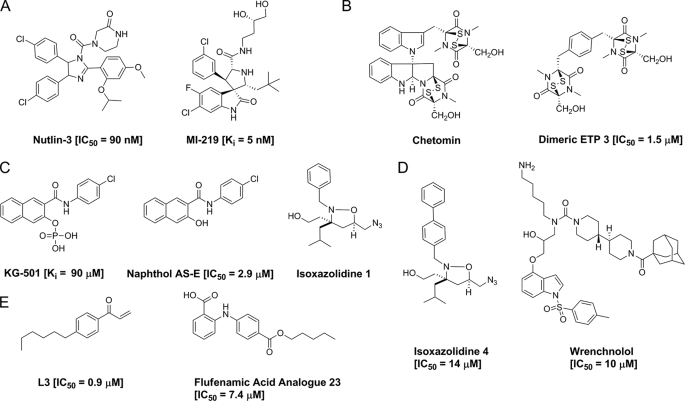
Perhaps the most well studied transcriptional activator-masking protein interaction is that of p53-MDM2. As mentioned previously, numerous human cancers have been linked to misregulated p53 function through either loss-of-function p53 mutations or overexpression of the p53-masking protein MDM2 (1, 2). Therefore, an exciting strategy for cancer therapeutics is the inhibition of the p53-MDM2 interaction for reactivation of the p53 pathway (30–32). High-resolution crystal structures of the N-terminal region of MDM2 complexed with short peptides of the N-terminal portion of p53 (residues 15–29) established that p53 binds as a short amphipathic helix in a deep hydrophobic pocket of MDM2, with four hydrophobic residues (Phe19, Leu22, Trp23, and Leu26) of p53 critical to this interaction, thus providing a framework for structure-based design of p53-MDM2 inhibitors (33, 34). Strategies such as library screening and structure-based de novo design have been employed in the discovery of MDM2 inhibitors, leading to a variety of inhibitor scaffolds, including chalcones, spiro-oxindoles, imidazoles, and benzodiazepines; these advances have been reviewed extensively (30–32). In particular, Nutlin-3a and MI-219 selectively inhibited the p53-MDM2 interaction and growth of cancer cell lines expressing wild-type p53 over those without wild-type p53. Furthermore, both compounds showed strong antitumor activity when administered orally in xenograft models of human cancers, with no apparent toxicity (Fig. 2A) (35–38). Recently, a Nutlin analog has entered into Phase I clinical trials.
FIGURE 2.
Small molecule transcriptional activator TAD mimics and NR inhibitors. A, p53 TAD mimic shown to inhibit p53-MDM2 interaction; B, TAD-CH1 domain (CBP/p300) inhibitors; C, TAD-KIX domain (CBP/p300) inhibitors; D, ESX mimics; E, inhibitors of thyroid receptors (L3) and ARs (23).
A perpetual challenge in the design of molecules that inhibit protein-protein interaction is that the broad interaction surface is difficult to entirely reconstitute with a drug-like small molecule. For this reason, the development of peptide-based inhibitors continues to be an active effort. β3-Peptides, for example, have been used by Schepartz and co-workers (39, 40) to recapitulate the side chain presentation of the p53 functional epitope and to inhibit MDM2. Recently, they improved upon one of the initial β3-peptide inhibitors, β53-8, using computational methods to identify β3-peptides with improved potency/selectivity, yielding β53-16, which has a Kd of 155 nm. Embedding cationic motifs within the β3-peptide improved cell permeability (41). The Verdine laboratory (42) successfully reactivated p53 activity in osteosarcoma cells using a “stapled” peptide in which an all-hydrocarbon cross-link was used to enforce the α-helical structure of a p53 TAD mimic. The stapled peptide showed dose-dependent inhibition of cell viability with an EC50 of 8.8 μm. As in the earlier example, the incorporation of cationic residues improved cell permeability.
The success to date in targeting the p53·MDM2 complex highlights the advantages of regulating the transcriptional activators through inhibiting masking interactions. Not surprisingly, a number of other activator-masking protein interactions have been identified as potential targets for the development of novel therapeutics, including the E2F·Rb complex, which is implicated in neuroblastomas and the AR-FOXO1 interaction (prostate cancer) (43, 44). Continued structural studies and advancements in improving cell permeability and specificity will lead to the next generation of inhibitors.
Disrupting Transcriptional Activator·Coactivator Complexes
Because transcriptional activators utilize binding interactions between their TADs and coactivators in the transcriptional machinery to initiate transcription, molecules that directly block the formation of these complexes would then function as transcriptional inhibitors. Thus, any TAD mimic, small molecule or peptide-based, should be able to competitively inhibit its natural counterpart. In practice, this simple idea has proven quite difficult to realize, with only a handful of TAD inhibitors reported in recent years (8). An enormous stumbling block has been difficulties in obtaining high-resolution structural information on the binding partners individually or in complex. Thus, it is not surprising that the most successful efforts in identifying TAD inhibitors are with activators that interact with the coactivator CBP/p300, a histone acetyltransferase that has been uniquely tractable for structural characterization.
Inhibiting TAD·CBP/p300 Complexes
The global coactivator CBP and its homolog p300 interact with >100 transcription factors, many of which have been implicated in human cancers; thus, CBP/p300 is a target of high interest for therapeutic intervention (25). CBP/p300 interacts with transcription factors through several distinct binding domains that each interact with multiple transcription factors (45). One such transcription factor, HIF-1α, regulates expression of hypoxia-adaptive related genes such as vascular endothelial growth factor, matrix metalloproteinases, and lysyl oxidase and contributes to cancer progression. HIF-1α interacts with the CH1 domain of CBP/p300; therefore, inhibition of HIF-1α transcriptional activity, through inhibition of the HIF-1α-CBP/p300 interaction, may lead to an effective strategy in combating cancer metastasis. Indeed, inhibiting the HIF-1α-p300 interaction via overexpression of the HIF-1α TAD in human tumor cells in a mouse xenograft model resulted in attenuated tumor growth (13). NMR structural analysis revealed that the HIF-1α TAD is significantly buried in the CH1 domain, utilizing extensive hydrophobic contacts with HIF-1α forming a clamp around the CH1 domain (12). The structural information will facilitate the optimization and refinement of lead inhibitors.
In 2004, high-throughput screening identified the natural product chetomin as an inhibitor of the HIF-1α-CBP/p300 interaction (46). Chetomin inhibited HIF-1α-mediated transcription in vitro and in a cellular context but showed necrosis, anemia, and leukocytosis in animal models. Recently, Olenyuk and co-workers (47) synthesized ETP-3, inspired by chetomin and designed to retain the 10-Å distance between the disulfide bridges found in chetomin (Fig. 2B). The disulfide bridges appear to be important for the biological activity of ETPs, as the corresponding ETP metabolic product diketopiperazine, which lacks disulfide bridges, did not bind to the p300 CH1 domain or exhibit effects on cell viability. In contrast, ETP-3 and chetomin bind to the p300 CH1 domain and alter its global fold, as observed using NMR and CD spectroscopy, suggesting that the altered structure prevents high-affinity binding to HIF-1α. Additionally, ETP-3 disrupted the HIF-1α·CBP/p300 complex in vitro via binding to the CH1 domain with an IC50 of 1.5 ± 0.2 μm and lower toxicity than chetomin as determined by cell viability assays. Furthermore, ETP-3 down-regulated HIF-1α target genes as evaluated by quantitative real-time PCR; however, global analysis of gene expression using microarrays showed that ETP-3 affected 403 genes, with 113 known to be directly controlled by HIF-1α. This is not surprising given the role of CBP/p300 as a master coactivator.
A second well characterized activator-binding motif within CBP/p300 is the KIX domain, a module that integrates signals of >12 transcriptional activators through at least two binding sites. Ground-breaking NMR spectroscopic work from the Wright and Lumb groups (48–53) have provided relatively high-resolution images of amphipathic TADs bound to this key target. Consistent with the prevailing model of TAD-coactivator interactions, the TADs of CREB, Myb, and MLL form amphipathic helices upon interacting with the KIX domain, with hydrophobic residues making the bulk of the contacts. The amenability of the KIX domain to NMR spectroscopic characterization has facilitated the discovery of transcriptional inhibitors. Using an NMR screening approach, Montminy and co-workers (54) identified a small molecule (KG-501) that bound to the KIX domain of CBP, inhibited the CREB-CBP interaction in vitro and in live cells (Fig. 2C), and down-regulated CREB target genes; however, KG-501 also inhibited other transcription factors such as NF-κB, highlighting the challenges of achieving small molecule modulators with the desired specificity. Overexpression of CBP has been linked to variety of human cancers, including breast cancer and prostate cancer (55, 56); therefore, inhibiting the CREB-CBP interaction via blocking of binding sites within the KIX domain with small molecule inhibitors may lead to novel anticancer agents. More recently, we described a group of amphipathic TAD mimics that interact with the MLL/Tat/c-Jun-binding site within this domain and competitively inhibit the activity of KIX domain-targeting activators (Fig. 2C) (57–59). Additionally, Li and Xiao (60) re-examined KG-501 and discovered that the dephosphorylated version of KG-501, naphthol AS-E, exhibited greater binding and a significantly lowered IC50 of 2.90 μm (Fig. 2C); they postulated that naphthol AS-E is the active form of KG-501.
Inhibiting TAD·Med23 Complexes
Beyond CBP/p300, there is very little coactivator structural information on or detailed characterization of the TAD· coactivator complexes, rendering the design of molecules or even effective screens quite challenging. One such complex is that between the Ras-linked coactivator Med23 (Sur2/DRIP130/CRSP130) and the TAD of the activator ESX. ESX is an epithelium-specific transcription factor that regulates expression of ErbB2 (HER2), an oncogenic protein that is overexpressed in ∼30% of all breast cancers (11, 14, 15). ESX interacts with multiple coactivators, presenting a significant challenge in the development of small molecule TAD mimics that exhibit a similar multipartner binding profile. Several lines of evidence suggest that the ESX·Med23 complex is an important contributor to ErbB2 expression (61), making it a target with great therapeutic potential. NMR studies of a minimal region of the ESX TAD (positions 129–145) suggested that it forms an amphipathic helix upon binding to Med23, and these data, in combination with mutagenesis studies, revealed that several hydrophobic residues (Trp138, Ile140, Leu142, Ile139, and Leu143) make the bulk of the contacts with the coactivator. On the basis of this information, Uesugi and co-workers (62) carried out a cell-based screen of a small molecule library biased toward indole-like compounds thought to mimic Trp138. Follow-up studies produced a second generation inhibitor named “wrenchnolol” (Fig. 2D) (63). In vitro studies demonstrated inhibition of the ESX·Med23 complex, and ErbB2-overexpressing breast cancer cells treated with wrenchnolol showed a reduction in ErbB2 expression and proliferation inhibition. Perhaps because of its large size and overall hydrophobicity, wrenchnolol exhibited limited nuclear distribution. More recently, a third generation version of this scaffold was reported, one with improved activity (3–4-fold greater than wrenchnolol) and affinity (2.6-fold greater than wrenchnolol) for Med23 (64). In an alternative approach, our group recently reported an isoxazolidine mimic of the ESX TAD that down-regulates ErbB2 expression in ErbB2-overexpressing cells (BT-474 and SKBR-3) at low micromolar concentrations (Fig. 2D) (81). Additionally, it attenuates cell proliferation of ErbB2-overexpressing cells, and gene expression analysis via quantitative real-time PCR showed down-regulation of erbB2 transcripts, consistent with lowered ErbB2 protein levels.
For both the ESX-Med23 inhibitors and the molecules targeting the KIX domain of CBP/p300, micromolar IC50 values for transcriptional inhibition are observed, despite considerable optimization in some examples. The origin of this moderate activity is not entirely clear. One contributing factor may be the nature of native TAD-coactivator interactions, interactions that often have Kd values in the micromolar range. Indeed, the reported dissociation constant for the minimal ESX TAD in complex with Med23 is 12 μm, suggesting that this surface may be difficult to recognize with high affinity. An additional challenge for all of the TAD-coactivator inhibitors is specificity. TAD-binding domains/surfaces within coactivators are often used by several activators, and thus, an inhibitor targeting a particular TAD-binding domain has the potential to interfere with more than one activator. For example, the KIX domain of CBP interacts with CREB in addition to c-Myb, c-Jun, and Stat1α, among others (25, 45). Defining the extent of this challenge awaits further genome-wide characterization of these and next-generation transcriptional inhibitors. Finally, although many other coactivators are of great therapeutic interest, the lack of structural data significantly hampers the identification of novel inhibitors.
Targeting a Third Domain: Nuclear Receptor·Coactivator Complexes
Although transcription can be minimally reconstituted with just a DBD and a TAD, many transcription factors have additional regulatory domains that also present attractive targets for the development of transcriptional modulators. Nowhere has this been more evident than with NRs. NRs contain an N-terminal amphipathic TAD (AF-1) and a C-terminal TAD (AF-2); AF-2 resides within a LBD that modulates AF-2 activity. Agonist binding induces a rearrangement of helix 12 of the LBD, creating a hydrophobic grove with which coactivators bind and activate transcription of specific genes. Thus, modulation of NR activity with exogenous molecules can be achieved by targeting AF-1, AF-2, or the LBD. AF-1 is unstructured in solution and closely resembles other amphipathic TADs; hence, to date, there are no reported small molecule regulators of AF-1. In contrast, AF-2 and the LBD are well defined and have been amenable to structural studies (65–67). Additionally, unlike the previously discussed activator-coactivator interactions, coactivators utilize a highly conserved and structurally well characterized α-helical LXXLL motif (NR box) to bind to AF-2 (68, 69). The wealth of structural information has resulted in many advancements in the development of NR regulators that either directly inhibit NR-coactivator interactions via targeting the hydrophobic groove with LXXLL helix mimics or allosterically inhibit NR-coactivator interactions by binding to the LBD within the ligand-binding pocket (70).
One particularly exciting example is the recent identification of small molecule inhibitors of the TR and AR. A variety of diseases have been linked to aberrant TR and AR activity. For example, the AR has been shown to play a role in androgen-dependent prostate cancer, and anti-androgenic drugs have been used successfully against these cancers; however, these are not without significant side effects. Therefore, the development of novel small molecule modulators of NRs is of great therapeutic interest. A library screen for potential inhibitors of TR-coactivator interactions discovered a β-aminoketone, L3, which had an IC50 value of ∼0.9 μm for the TRβ isoform (Fig. 2E) (71). L3 acts irreversibly, exhibiting cell permeability and nearly full inhibition of transcription in a cell-based reporter assay at 4 μm. Structure-function analysis of this lead compound provided insight into properties contributing to potency, solubility, toxicity, and permeability, with the most potent inhibitors consisting of an electrophilic head, hydrophobic core, and a hydrophobic alkyl tail (72). Additionally, it appears that the hydrophobicity plays a role in the high selectivity (50-fold) of L3 for TRβ over TRα.
Similarly, library screening methods were also used to identify lead compounds that blocked AR transcriptional activity, which were then subjected to x-ray structural analysis. These studies identified FLF, a nonsteroidal anti-inflammatory drug, as an allosteric inhibitor of the AR (73). Interestingly, structural analysis revealed that FLF binds to a previously unidentified hydrophobic cleft in the AR, termed BF3, which appears to be necessary for AR function. Indeed, mutations at this site are involved in androgen-insensitivity syndromes (74). Interactions at BF3 abrogate binding of coactivators to the AR through an allosteric mechanism. Further derivatization of FLF produced analogs that antagonized the AR via binding at the hormone-binding site instead of at the BF3 site yet still inhibited AR transcriptional activity and AR target genes (Fig. 2E) (75).
The well defined nature of AF-2 and the LBD of NRs, in combination with the highly conserved LXXLL motif of NR coactivators, has greatly facilitated the identification of small molecule modulators of NR activity, in particular NR inhibitors. These exciting advancements may open the door to novel therapeutics and alternative strategies in the treatment of NR-related disorders such as prostate cancer and hyperthyroidism.
Putting It All Together: Activator Artificial Transcription Factors
Molecules that can reconstitute activator function are interesting as potential therapeutics aimed at replacing malfunctioning transcription factors. The predominant strategy for the development of ATFs has been to reconstitute activator function through replacement of either or both domains with synthetic or non-natural counterparts (Fig. 1) (8, 76, 77). Protein-based activator ATFs are at a more advanced development stage relative to ATFs constructed from one or more small molecule components, with, for example, designer zinc-finger ATFs in clinical trials (78). One of the primary reasons that small molecule ATFs have lagged behind is the considerable difficulty associated with recapitulating the multipartner binding profile of natural TADs with a small molecule, particularly given the many open questions surrounding activator-coactivator interactions. Despite this, there have been several advancements in ATFs in recent years. The Mapp laboratory reported the first small molecule TAD, an amphipathic isoxazolidine that was designed to generically mimic the amphipathic helices of natural TADs, and this and related molecules function in cell-free and cellular experiments; recent studies indicate that isoxazolidine 1 displays a multipartner binding profile analogous to that of natural TADs and in particular binds to the KIX domain of CBP using the same binding site utilized by MLL, Tat, and Tax (58, 59). A second class of TADs has emerged from the Kodadek laboratory (79, 80) through a screen of peptoids for binding to CBP, and from this, a number of peptoid TADs that function in cells have emerged. Uesugi and co-workers (64) were also able to up-regulate reporter and endogenous genes by targeting a wrenchnolol derivative to DNA. The next challenge is to be able to use the small molecule TADs to affect endogenous genes, a transition that would greatly increase their utility.
Future Directions
The past decade has seen many exciting advances in the development of synthetic modulators of transcriptional activation. Both screening and directed design strategies have been effective in identifying mimics of TADs that can be used as inhibitors or activators of transcription, depending on the context. However, the long-standing debate over the identity of functionally important activator-binding partners, coupled with limited structural data for activator-target protein complexes, continues to hinder the field. Advances in either of these areas will be enormously important for the discovery and characterization of transcriptional modulators with increased effectiveness and specificity.
Supplementary Material
This is the first of six articles in the “Chemical Biology Meets Biological Chemistry Minireview Series.” This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- DBD
- DNA-binding domain
- TAD
- transcriptional activation domain
- HIF-1α
- hypoxia-inducible factor 1α
- CREB
- cAMP-responsive element-binding protein
- CBP
- CREB-binding protein
- AR
- androgen receptor
- ETP
- epidithiodiketopiperazine
- MLL
- mixed lineage leukemia
- NR
- nuclear receptor
- LBD
- ligand-binding domain
- TR
- thyroid hormone receptor
- FLF
- flufenamic acid
- ATF
- artificial transcription factor.
REFERENCES
- 1.Hainaut P., Hollstein M. (2000) Adv. Cancer Res. 77, 81–137 [DOI] [PubMed] [Google Scholar]
- 2.Hollstein M., Sidransky D., Vogelstein B., Harris C. C. (1991) Science 253, 49–53 [DOI] [PubMed] [Google Scholar]
- 3.Arndt H. D. (2006) Angew. Chem. Int. Ed. Engl. 45, 4552–4560 [DOI] [PubMed] [Google Scholar]
- 4.Berg T. (2008) Curr. Opin. Chem. Biol. 12, 464–471 [DOI] [PubMed] [Google Scholar]
- 5.Dervan P. B. (2001) Bioorg. Med. Chem. 9, 2215–2235 [DOI] [PubMed] [Google Scholar]
- 6.Dervan P. B., Doss R. M., Marques M. A. (2005) Curr. Med. Chem. Anticancer Agents 5, 373–387 [DOI] [PubMed] [Google Scholar]
- 7.Dervan P. B., Edelson B. S. (2003) Curr. Opin. Struct. Biol. 13, 284–299 [DOI] [PubMed] [Google Scholar]
- 8.Majmudar C. Y., Mapp A. K. (2005) Curr. Opin. Chem. Biol. 9, 467–474 [DOI] [PubMed] [Google Scholar]
- 9.Ptashne M., Gann A. (2002) Genes & Signals, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 10.Darnell J. E., Jr. ( 2002) Nat. Rev. Cancer 2, 740– 749 [DOI] [PubMed] [Google Scholar]
- 11.Perou C. M., Sørlie T., Eisen M. B., van de Rijn M., Jeffrey S. S., Rees C. A., Pollack J. R., Ross D. T., Johnsen H., Akslen L. A., Fluge O., Pergamenschikov A., Williams C., Zhu S. X., Lønning P. E., Børresen-Dale A. L., Brown P. O., Botstein D. (2000) Nature 406, 747–752 [DOI] [PubMed] [Google Scholar]
- 12.Freedman S. J., Sun Z. Y., Poy F., Kung A. L., Livingston D. M., Wagner G., Eck M. J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5367–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kung A. L., Wang S., Klco J. M., Kaelin W. G., Livingston D. M. (2000) Nat. Med. 6, 1335–1340 [DOI] [PubMed] [Google Scholar]
- 14.Kristensen V. N., Sørlie T., Geisler J., Langerød A., Yoshimura N., Kåresen R., Harada N., Lønning P. E., Børresen-Dale A. L. (2005) Clin. Cancer Res. 11, 878s–883s [PubMed] [Google Scholar]
- 15.Pupa S. M., Tagliabue E., Ménard S., Anichini A. (2005) J. Cell. Physiol. 205, 10–18 [DOI] [PubMed] [Google Scholar]
- 16.Schon O., Friedler A., Bycroft M., Freund S. M., Fersht A. R. (2002) J. Mol. Biol. 323, 491–501 [DOI] [PubMed] [Google Scholar]
- 17.Lee C. W., Arai M., Martinez-Yamout M. A., Dyson H. J., Wright P. E. (2009) Biochemistry 48, 2115–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agalioti T., Lomvardas S., Parekh B., Yie J., Maniatis T., Thanos D. (2000) Cell 103, 667–678 [DOI] [PubMed] [Google Scholar]
- 19.Ard P. G., Chatterjee C., Kunjibettu S., Adside L. R., Gralinski L. E., McMahon S. B. (2002) Mol. Cell. Biol. 22, 5650–5661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Black J. C., Choi J. E., Lombardo S. R., Carey M. (2006) Mol. Cell 23, 809–818 [DOI] [PubMed] [Google Scholar]
- 21.Chan H. M., La Thangue N. B. (2001) J. Cell Sci. 114, 2363–2373 [DOI] [PubMed] [Google Scholar]
- 22.Chang C., Gonzalez F., Rothermel B., Sun L., Johnston S. A., Kodadek T. (2001) J. Biol. Chem. 276, 30956–30963 [DOI] [PubMed] [Google Scholar]
- 23.Fishburn J., Mohibullah N., Hahn S. (2005) Mol. Cell 18, 369–378 [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez F., Delahodde A., Kodadek T., Johnston S. A. (2002) Science 296, 548–550 [DOI] [PubMed] [Google Scholar]
- 25.Goodman R. H., Smolik S. (2000) Genes Dev. 14, 1553–1577 [PubMed] [Google Scholar]
- 26.Marr M. T., 2nd, Isogai Y., Wright K. J., Tjian R. (2006) Genes Dev. 20, 1458–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reeves W. M., Hahn S. (2005) Mol. Cell. Biol. 25, 9092–9102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roeder R. G. (2005) FEBS Lett. 579, 909–915 [DOI] [PubMed] [Google Scholar]
- 29.Yang F., Vought B. W., Satterlee J. S., Walker A. K., Jim Sun Z. Y., Watts J. L., DeBeaumont R., Saito R. M., Hyberts S. G., Yang S., Macol C., Iyer L., Tjian R., van den Heuvel S., Hart A. C., Wagner G., Näär A. M. (2006) Nature 442, 700–704 [DOI] [PubMed] [Google Scholar]
- 30.Hu C. Q., Hu Y. Z. (2008) Curr. Med. Chem. 15, 1720–1730 [DOI] [PubMed] [Google Scholar]
- 31.Shangary S., Wang S. (2009) Annu. Rev. Pharmacol. Toxicol. 49, 223–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vassilev L. T. (2005) J. Med. Chem. 48, 4491–4499 [DOI] [PubMed] [Google Scholar]
- 33.Chi S. W., Lee S. H., Kim D. H., Ahn M. J., Kim J. S., Woo J. Y., Torizawa T., Kainosho M., Han K. H. (2005) J. Biol. Chem. 280, 38795–38802 [DOI] [PubMed] [Google Scholar]
- 34.Kussie P. H., Gorina S., Marechal V., Elenbaas B., Moreau J., Levine A. J., Pavletich N. P. (1996) Science 274, 948–953 [DOI] [PubMed] [Google Scholar]
- 35.Sarek G., Kurki S., Enbäck J., Iotzova G., Haas J., Laakkonen P., Laiho M., Ojala P. M. (2007) J. Clin. Invest. 117, 1019–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shangary S., Qin D., McEachern D., Liu M., Miller R. S., Qiu S., Nikolovska-Coleska Z., Ding K., Wang G., Chen J., Bernard D., Zhang J., Lu Y., Gu Q., Shah R. B., Pienta K. J., Ling X., Kang S., Guo M., Sun Y., Yang D., Wang S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3933–3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tovar C., Rosinski J., Filipovic Z., Higgins B., Kolinsky K., Hilton H., Zhao X., Vu B. T., Qing W., Packman K., Myklebost O., Heimbrook D. C., Vassilev L. T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1888–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vassilev L. T., Vu B. T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., Fotouhi N., Liu E. A. (2004) Science 303, 844–848 [DOI] [PubMed] [Google Scholar]
- 39.Kritzer J. A., Lear J. D., Hodsdon M. E., Schepartz A. (2004) J. Am. Chem. Soc. 126, 9468–9469 [DOI] [PubMed] [Google Scholar]
- 40.Kritzer J. A., Luedtke N. W., Harker E. A., Schepartz A. (2005) J. Am. Chem. Soc. 127, 14584–14585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harker E. A., Schepartz A. (2009) ChemBioChem 10, 990–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernal F., Tyler A. F., Korsmeyer S. J., Walensky L. D., Verdine G. L. (2007) J. Am. Chem. Soc. 129, 2456–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeGregori J. (2002) Biochim. Biophys. Acta 1602, 131–150 [DOI] [PubMed] [Google Scholar]
- 44.Ma Q., Fu W., Li P., Nicosia S. V., Jenster G., Zhang X., Bai W. (2009) Mol. Endocrinol. 23, 213–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vo N., Goodman R. H. (2001) J. Biol. Chem. 276, 13505–13508 [DOI] [PubMed] [Google Scholar]
- 46.Kung A. L., Zabludoff S. D., France D. S., Freedman S. J., Tanner E. A., Vieira A., Cornell-Kennon S., Lee J., Wang B., Wang J., Memmert K., Naegeli H. U., Petersen F., Eck M. J., Bair K. W., Wood A. W., Livingston D. M. (2004) Cancer Cell 6, 33–43 [DOI] [PubMed] [Google Scholar]
- 47.Block K. M., Hui W., Szabó L. Z., Polaske N. W., Henchey L. K., Dubey R., Kushal S., László C. F., Makhoul J., Song Z., Meuillet E. J., Olenyuk B. (2009) J. Am. Chem. Soc. 131, 18078–18088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campbell K. M., Lumb K. J. (2002) Biochemistry 41, 13956–13964 [DOI] [PubMed] [Google Scholar]
- 49.Lee C. W., Arai M., Martinez-Yamout M. A., Dyson H. J., Wright P. E. (2009) Biochemistry 48, 2115–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Radhakrishnan I., Pérez-Alvarado G. C., Parker D., Dyson H. J., Montminy M. R., Wright P. E. (1997) Cell 91, 741–752 [DOI] [PubMed] [Google Scholar]
- 51.Radhakrishnan I., Pérez-Alvarado G. C., Parker D., Dyson H. J., Montminy M. R., Wright P. E. (1999) J. Mol. Biol. 287, 859–865 [DOI] [PubMed] [Google Scholar]
- 52.Sugase K., Dyson H. J., Wright P. E. (2007) Nature 447, 1021–1025 [DOI] [PubMed] [Google Scholar]
- 53.Zor T., De Guzman R. N., Dyson H. J., Wright P. E. (2004) J. Mol. Biol. 337, 521–534 [DOI] [PubMed] [Google Scholar]
- 54.Best J. L., Amezcua C. A., Mayr B., Flechner L., Murawsky C. M., Emerson B., Zor T., Gardner K. H., Montminy M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17622–17627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chhabra A., Fernando H., Watkins G., Mansel R. E., Jiang W. G. (2007) Oncol. Rep. 18, 953–958 [PubMed] [Google Scholar]
- 56.Wu D., Zhau H. E., Huang W. C., Iqbal S., Habib F. K., Sartor O., Cvitanovic L., Marshall F. F., Xu Z., Chung L. W. (2007) Oncogene 26, 5070–5077 [DOI] [PubMed] [Google Scholar]
- 57.Buhrlage S. J., Bates C. A., Rowe S. P., Minter A. R., Brennan B. B., Majmudar C. Y., Wemmer D. E., Al-Hashimi H., Mapp A. K. (2009) ACS Chem. Biol. 4, 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Minter A. R., Brennan B. B., Mapp A. K. (2004) J. Am. Chem. Soc. 126, 10504–10505 [DOI] [PubMed] [Google Scholar]
- 59.Rowe S. P., Casey R. J., Brennan B. B., Buhrlage S. J., Mapp A. K. (2007) J. Am. Chem. Soc. 129, 10654–10655 [DOI] [PubMed] [Google Scholar]
- 60.Li B. X., Xiao X. (2009) ChemBioChem 10, 2721–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Asada S., Choi Y., Yamada M., Wang S. C., Hung M. C., Qin J., Uesugi M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12747–12752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Asada S., Choi Y., Uesugi M. (2003) J. Am. Chem. Soc. 125, 4992–4993 [DOI] [PubMed] [Google Scholar]
- 63.Shimogawa H., Kwon Y., Mao Q., Kawazoe Y., Choi Y., Asada S., Kigoshi H., Uesugi M. (2004) J. Am. Chem. Soc. 126, 3461–3471 [DOI] [PubMed] [Google Scholar]
- 64.Jung D., Shimogawa H., Kwon Y., Mao Q., Sato S., Kamisuki S., Kigoshi H., Uesugi M. (2009) J. Am. Chem. Soc. 131, 4774–4782 [DOI] [PubMed] [Google Scholar]
- 65.Darimont B. D., Wagner R. L., Apriletti J. W., Stallcup M. R., Kushner P. J., Baxter J. D., Fletterick R. J., Yamamoto K. R. (1998) Genes Dev. 12, 3343–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nolte R. T., Wisely G. B., Westin S., Cobb J. E., Lambert M. H., Kurokawa R., Rosenfeld M. G., Willson T. M., Glass C. K., Milburn M. V. (1998) Nature 395, 137–143 [DOI] [PubMed] [Google Scholar]
- 67.Shiau A. K., Barstad D., Loria P. M., Cheng L., Kushner P. J., Agard D. A., Greene G. L. (1998) Cell 95, 927–937 [DOI] [PubMed] [Google Scholar]
- 68.Chang C., Norris J. D., Grøn H., Paige L. A., Hamilton P. T., Kenan D. J., Fowlkes D., McDonnell D. P. (1999) Mol. Cell. Biol. 19, 8226–8239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heery D. M., Kalkhoven E., Hoare S., Parker M. G. (1997) Nature 387, 733–736 [DOI] [PubMed] [Google Scholar]
- 70.Chen T. (2008) Curr. Opin. Chem. Biol. 12, 418–426 [DOI] [PubMed] [Google Scholar]
- 71.Arnold L. A., Estébanez-Perpiñá E., Togashi M., Jouravel N., Shelat A., McReynolds A. C., Mar E., Nguyen P., Baxter J. D., Fletterick R. J., Webb P., Guy R. K. (2005) J. Biol. Chem. 280, 43048–43055 [DOI] [PubMed] [Google Scholar]
- 72.Arnold L. A., Kosinski A., Estébanez-Perpiñá E., Fletterick R. J., Guy R. K. (2007) J. Med. Chem. 50, 5269–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Estébanez-Perpiñá E., Arnold L. A., Nguyen P., Rodrigues E. D., Mar E., Bateman R., Pallai P., Shokat K. M., Baxter J. D., Guy R. K., Webb P., Fletterick R. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16074–16079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McPhaul M. J. (2002) Mol. Cell. Endocrinol. 198, 61–67 [DOI] [PubMed] [Google Scholar]
- 75.Feau C., Arnold L. A., Kosinski A., Zhu F., Connelly M., Guy R. K. (2009) ACS Chem. Biol. 4, 834–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ansari A. Z., Mapp A. K. (2002) Curr. Opin. Chem. Biol. 6, 765–772 [DOI] [PubMed] [Google Scholar]
- 77.Mapp A. K., Ansari A. Z. (2007) ACS Chem. Biol. 2, 62–75 [DOI] [PubMed] [Google Scholar]
- 78.Reik A., Zhou Y., Collingwood T. N., Warfe L., Bartsevich V., Kong Y., Henning K. A., Fallentine B. K., Zhang L., Zhong X., Jouvenot Y., Jamieson A. C., Rebar E. J., Case C. C., Korman A., Li X. Y., Black A., King D. J., Gregory P. D. (2007) Biotechnol. Bioeng. 97, 1180–1189 [DOI] [PubMed] [Google Scholar]
- 79.Xiao X., Yu P., Lim H. S., Sikder D., Kodadek T. (2007) J. Comb. Chem. 9, 592–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xiao X., Yu P., Lim H. S., Sikder D., Kodadek T. (2007) Angew. Chem. Int. Ed. Engl. 46, 2865–2868 [DOI] [PubMed] [Google Scholar]
- 81.Lee L. W., Taylor C. E., Desaulniers J. P., Zhang M., Højfeldt J. W., Pan Q., Mapp A. K. (2009) Bioorg. Med. Chem. Lett. 19, 6233–6236 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.