Abstract
The ability to genetically encode unnatural amino acids beyond the common 20 has allowed unprecedented control over the chemical structures of recombinantly expressed proteins. Orthogonal aminoacyl-tRNA synthetase/tRNA pairs have been used together with nonsense, rare, or 4-bp codons to incorporate >50 unnatural amino acids into proteins in Escherichia coli, Saccharomyces cerevisiae, Pichia pastoris, and mammalian cell lines. This has allowed the expression of proteins containing amino acids with novel side chains, including fluorophores, post-translational modifications, metal ion chelators, photocaged and photocross-linking moieties, uniquely reactive functional groups, and NMR, IR, and x-ray crystallographic probes.
Keywords: Amino Acid, Aminoacyl-tRNA Synthetase, Protein Chemical Modification, Protein Chemistry, Translation Control, Protein Engineering, Protein Evolution, Protein Translation, Unnatural Amino Acid
Early Methodology
With few exceptions, all organisms are restricted to the 20 common amino acid building blocks for the ribosomal biosynthesis of proteins. However, it is clear that proteins require a higher level of chemical complexity for many functions, as evidenced by the frequent use of post-translational modifications and the dependence of many enzymes on cofactors. Thus, the addition of new amino acid building blocks to the genetic code should further expand the range of functions available to proteins and provide powerful new tools for probing protein structure and function both inside and outside of living cells (1). Although recent advances in synthetic and semisynthetic methods have proven very useful for the incorporation of unnatural amino acids into proteins, they are generally limited by low yields and are technically cumbersome in the production of larger proteins. The use of the cellular biosynthetic machinery to introduce novel amino acids abrogates issues relating to scalability and protein size and simplifies the study of modified proteins in living cells.
To add a new amino acid to the genetic repertoire, a codon is needed that uniquely specifies that amino acid. The 20 canonical amino acids are encoded by 61 degenerate triplet codons, leaving the remaining three codons (TAG, amber; TAA, ochre; and TGA, opal) to serve as translational stops. Previously, we and others (2) used the redundancy of these “blank” stop codons, together with the ribosomal machinery, to site-specifically incorporate unnatural amino acids into proteins in vitro in response to the amber nonsense codon. This was accomplished by chemically aminoacylating a nonsense suppressor tRNA (tRNACUA, the amber suppressor tRNA) with the desired unnatural amino acid and adding the aminoacyl-tRNA to a cell-free transcription/translation system along with the gene of interest harboring a TAG mutation at the target site. In the decade that followed, this method was used to site-specifically incorporate a large number of unnatural amino acids with a wide variety of structures into proteins (3, 4). These amino acids were used to probe the roles of specific amino acid side chains and backbone groups in protein folding and stability, catalytic mechanisms, and biomolecular interactions. Later, this approach was extended to the incorporation of unnatural amino acids in cells by microinjecting chemically aminoacylated suppressor tRNACUAs into Xenopus oocytes along with the mutated mRNA (5). Although methodologies using chemically aminoacylated tRNAs are quite useful, they are limited to the production of small quantities of protein due to their stoichiometric nature and require microinjection or transfection into cells. To expand the utility of this methodology, we have engineered the translational machinery to directly incorporate unnatural amino acids into proteins in living cells.
General Considerations for the in Vivo Incorporation of Unnatural Amino Acids
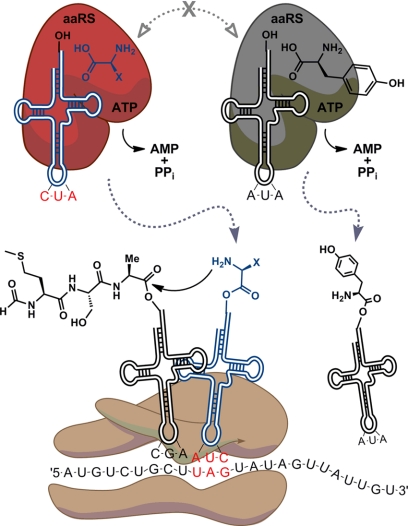
A number of new components must be integrated into the protein biosynthetic machinery to add new amino acids to the genetic repertoire. For example, selenocysteine is incorporated into proteins in response to the UGA nonsense codon by means of a unique seryl-tRNA, which is enzymatically converted to selenocysteine and then translated with an alternative elongation factor (SelB in bacteria) and mRNA SECIS (selenocysteine insertion sequence) element (6). Pyl2 is biosynthesized and subsequently incorporated in response to the UAG nonsense codon by a unique aaRS/tRNA pair (7). The minimal requirements to encode additional amino acids beyond the common 20 include a codon that does not code for one of the natural amino acids so that the unnatural amino acid can be uniquely incorporated into a protein at the desired site. In addition, a functional aaRS/tRNA pair is required that acts independently of the endogenous aminoacylation machinery of the cell (Fig. 1). Orthogonality must be maintained such that the suppressor tRNA is not a substrate for any endogenous aaRS, and the orthogonal aaRS does not aminoacylate any endogenous tRNAs. The active site of the aaRS must activate and aminoacylate its cognate tRNA with only the unnatural amino acid of interest and no endogenous host amino acids. Finally, the new amino acid must be efficiently transported into the cell (or biosynthesized by the cell), nontoxic, and stable to the metabolic enzymes of the cell.
FIGURE 1.
Orthogonal aaRS/tRNACUA pair and translational machinery. The orthogonal aaRS (red) aminoacylates the orthogonal tRNACUA (blue, red CUA) with an unnatural amino acid (blue X) and does not cross-react with the endogenous aaRS/tRNA pairs (shown is the TyrRS (gray)/tRNAAUA (black) pair). Aminoacyl-tRNACUA travels to the ribosome (brown), where it incorporates the unnatural amino acid in response to the UAG codon (red UAG).
Incorporation of Unnatural Amino Acids into Proteins in Escherichia coli
Initial efforts to encode unnatural amino acids in E. coli were aimed at evolving an endogenous aaRS/tRNA pair to be orthogonal in E. coli (8). However, misaminoacylation of native tRNAs by the evolved aaRS precluded its use for unnatural amino acid incorporation. To overcome this problem, in vitro studies identified orthogonal pairs from other organisms with distinct tRNA identity elements that do not interact with EcaaRS/EctRNA pairs but still function efficiently in translation. An engineered tyrosyl-tRNA synthetase and cognate nonsense suppressor tRNA evolved from the archaea Methanocaldococcus jannaschii (MjTyrRS/MjtRNATyr) was the first such orthogonal pair to be used to successfully incorporate an unnatural amino acid (9). Because TAG (amber stop codon) is the least used stop codon in the E. coli genome (93% of E. coli genes end with TAA or TGA), it was reassigned to the unnatural amino acid with the expectation that TAG suppression would have little impact on the E. coli native proteome. To recognize the TAG codon, the anticodon of MjtRNATyr was mutated to CUA to create an amber suppressor tRNA (MjtRNACUATyr).
To alter the specificity of MjTyrRS to recognize a desired unnatural amino acid, a large library of aaRS active-site mutants (≥108 mutants) was constructed and subjected to a double-sieve selection (9). In the first step, mutant aaRSs, together with orthogonal MjtRNACUATyr, are selected for their ability to suppress an amber mutation at a permissive site in the chloramphenicol acetyltransferase gene in the presence of an unnatural amino acid and chloramphenicol. Survivors of this positive selection encode MjTyrRS mutants that can aminoacylate MjtRNACUATyr with either the unnatural amino acid or an endogenous amino acid (10). To select against mutants that aminoacylate endogenous amino acids, a negative selection step was carried out (11). In this round, mutant aaRSs and cognate orthogonal MjtRNACUATyr are grown in the absence of unnatural amino acid; those pairs that can suppress three amber mutations at permissive sites in the toxic barnase gene with an endogenous amino acid are eliminated (9). Using this selection scheme, the MjTyrRS/MjtRNACUATyr pair has been engineered to incorporate >30 unnatural amino acids in E. coli with fidelities and efficiencies near those of natural ribosomal protein synthesis (1).
In addition to amber suppression by the MjTyrRS/MjtRNATyr pair, opal (TGA) and 4-base (AGGA) decoding pairs have been derived from a chimeric Methanobacterium thermoautotrophicum LeuRS/Halobacterium sp. NRC-1 suppressor tRNALeu pair (12). Chin and co-workers (13) also demonstrated that the aaRS/tRNACUA pairs that incorporate Pyl in Methanosarcina barkeri and Desulfitobacterium hafniense are orthogonal in E. coli and can be evolved to accept Pyl analogs as substrates. Our laboratory and others have recently evolved the Pyl pair from Methanosarcina mazei (MmPylRS/MmtRNACUAPyl) in E. coli to accept lysine derivatives (14, 15). Besides allowing the incorporation of multiple amino acids in a single protein, these new pairs should facilitate the evolution of new aaRSs specific for unnatural amino acids with increasing structural diversity.
Incorporation of Unnatural Amino Acids into Proteins in Eukaryotes
Edwards and Schimmel (16) previously showed that the EcTyrRS/EctRNATyr pair is orthogonal in Saccharomyces cerevisiae, making it a prime candidate for unnatural amino acid incorporation in yeast. Yokoyama and co-workers (17) furthered this work with the semirational design of an EcTyrRS that aminoacylates a suppressor EctRNACUATyr with 3-iodotyrosine in cell-free systems. To create mutant EcTyrRS/EctRNACUATyr pairs that can be used to incorporate structurally diverse unnatural amino acids in living yeast cells, our laboratory developed a double-sieve selection scheme that is analogous to the system developed in E. coli. This system is based on suppression of two TAG mutations in the GAL4 transcriptional activator protein, which drives transcription of the HIS3 and URA3 genes (18). A library of EcTyrRS active-site mutants, together with the cognate EctRNACUATyr, are cotransformed with the selection markers into a S. cerevisiae histidine auxotroph. When grown in the absence of histidine and the presence of an unnatural amino acid, only those EcTyrRS mutants that can successfully charge EctRNACUATyr produce histidine and survive. To negatively select those mutants that accept an endogenous amino acid, 5-fluorootic acid is added to the medium in the absence of unnatural amino acid. Expression of URA3 under these conditions converts 5-fluorootic acid to the toxic product 5-fluorouracil and eliminates EcTyrRS mutants with activity for endogenous amino acids (19). Using this selection scheme, >20 unnatural amino acids have been added to the S. cerevisiae genetic code.
Because transformation efficiencies and slow doubling times limit the construction and selection of libraries in mammalian cells, methods have been developed to transfer aaRS/tRNA pairs evolved in yeast to mammalian cells. Yokoyama and co-workers (20) first demonstrated this approach using a semirationally engineered EcTyrRS. Because the EctRNACUATyr lacks the essential internal promoter sequences (A- and B-box identity elements) for transcription by pol III in higher eukaryotes, an amber suppressor tyrosyl-tRNA from Bacillus stearothermophilus (BstRNACUATyr) was paired with the EcTyrRS. Using this novel EcTyrRS/BstRNACUATyr pair, 3-iodotyrosine was incorporated in response to an amber codon. To increase the diversity of unnatural amino acids that can be used in mammalian cells, our laboratory developed a method of transferring EcTyrRSs evolved in yeast selection systems directly into mammalian cells (21). By using S. cerevisiae as a gateway, we have been able to incorporate ∼10 unnatural amino acids in Chinese hamster ovary cells, 293T cells, and primary cells using the EcTyrRS/BstRNACUATyr pair.
Alternate pairs for unnatural amino acid incorporation have been developed in eukaryotes as well. Evolution of the E. coli leucyl pair (EcLeuRS/EctRNACUALeu) in S. cerevisiae has allowed the incorporation of novel fluorogenic and photocaged amino acids (22). This pair has also been adapted to mammalian cells with good success (23). In addition, the MmPylRS/MmtRNACUAPyl pair has been used in mammalian cells to incorporate lysine analogs that may be useful in studying histone modifications (13, 15). This pair has the advantage of being orthogonal in both E. coli and higher eukaryotes, allowing aaRSs with novel specificities to be evolved in E. coli and subsequently transferred to mammalian cells.
Expanded Genetic Code
Orthogonal pairs derived using the above selection schemes have been used to incorporate >50 unnatural amino acids in response to unassigned or reassigned codons. Unnatural amino acids with orthogonal chemical reactivities have enabled the site-specific modification of proteins through a diverse “toolkit” of chemistries (Fig. 2a). For example, a keto (1)-containing amino acid can be selectively modified with aminooxy groups, and an azide (2)- or alkyne (3)-containing amino acid can be selectively modified through 3 + 2 cycloaddition reactions (“click” chemistry) (24–26). These amino acids have been used to site-specifically derivatize proteins with polyethylene glycol molecules, sugars, oligonucleotides, fluorophores, peptides, and other synthetic moieties. In one example, fluorescence resonance energy transfer pairs were conjugated to T4 lysozyme and used to follow protein folding at single-molecule resolution (27). In another example, human growth hormone was expressed with a keto amino acid and polyethylene glycolylated through an oxime linkage on a kilogram scale to improve its pharmacological properties (and is currently in clinical trials) (24). A similar approach has been used to selectively conjugate toxins to antibodies to create novel targeted cancer therapies. The site-specific modification of proteins in this manner avoids the heterogeneous products generated by nonspecific electrophilic reagents and makes possible a form of “protein medicinal chemistry.” In addition to these chemistries, borono, iodo, olefinic, and aminophenyl residues have enabled selective modification reactions at the surface of proteins (28).
FIGURE 2.
Unnatural amino acids and their applications. a, orthogonal chemistries enable the site-specific modification of proteins with a wide variety of molecules. b, examples of unnatural amino acids described in text.
Unnatural amino acids with unique spectroscopic properties have facilitated structural studies of proteins (Fig. 2b). Applications include 1) the site-specific incorporation of 15N-, 13C-, or 19F-labeled residues (4 and 5) into fatty acid synthetase to use NMR to identify conformational changes that occur upon ligand binding (29); 2) the selective incorporation of a deuterated tyrosine (6) into dihydrofolate reductase to probe catalytic intermediates by infrared spectroscopy (this labeled residue was site-specifically incorporated into dihydrofolate reductase as a photocaged tyrosine whose caging group was removed with light) (30); 3) the site-specific introduction of an azide (2)-containing amino acid into rhodopsin to identify by Fourier transform infrared spectroscopy changes in specific residues that occur upon light activation (31); 4) the introduction of residues with altered redox potentials such as aminotyrosine (7) into ribonucleotide reductase to elucidate the mechanisms of electron transfer (32); 5) the site-specific incorporation of environmentally sensitive fluorogenic amino acids (8 and 9) as probes of local protein structure (for example, a hydroxycoumarin derivative (9) was used to probe the local unfolding of myoglobin, and a prodan derivative (8) was used to follow conformational changes that occur upon binding of glutamine to glutamine-binding protein and to label histones in living cells) (33, 34); and 6) incorporation of metal (10 and 11)-chelating or iodine (12)-containing amino acids to facilitate the solution of protein crystal structures (35). Metal-chelating amino acids may also be used for the de novo design of metalloproteins. For example, a catabolite activator protein mutant that site-specifically cleaves DNA has been engineered by introducing a redox-active Cu2+-chelating amino acid near the DNA backbone (36).
More recently, photochemically reactive amino acids have been used as probes of protein function in vitro and in cells. For example, photocaged cysteine (13), serine (14), lysine (15), and tyrosine (16) derivatives have been site-specifically incorporated into proteins to allow the photodynamic regulation of their activity (15, 37–39). In one example, the localization of the yeast transcription factor Pho4 was followed in living cells by selectively blocking its phosphorylation with a photocaged serine. Irradiation with low-energy blue light uncaged the serine and allowed nuclear export of the phosphorylated Pho4 to be followed in real time (39). To analyze biomolecular interactions in vivo, azide (2), benzophenone (17), and aziridine photocross-linking amino acids have been used to cross-link proteins to their interacting partners in the cell (40, 41). For example, this approach has been used to cross-link membrane protein ligands, such as G protein-coupled receptor peptide ligands, in S. cerevisiae (42). Finally, to selectively cleave proteins with light, o-nitrophenylalanine (18) has been genetically encoded, which results in the site-specific scission of the protein backbone through a light-induced radical mechanism (43).
Unnatural amino acid mutagenesis has also been used to selectively incorporate post-translationally modified amino acids into proteins, including sulfotyrosine (19) and methylated and acetylated lysine (44–46). In addition, a metabolically stable analog of phosphotyrosine (20) has been used to express a constitutively active human STAT1 (signal transducer and activator of transcription-1) protein and should be a general method for generating stable phosphoprotein mimetics (44). Unnatural amino acid mutagenesis can also be used to alter the polypeptide backbone. For example, to probe the effects of backbone hydrogen bonding on protein stability, an α-hydroxy acid (21) has been genetically encoded, which creates an ester bond in the polypeptide chain (47). This group can also be cleaved in vitro with mild base to allow the scarless purification of proteins. Finally, we have discovered that the site-specific incorporation of a single nitrophenylalanine (22) or nitrotyrosine into proteins can break immunological tolerance in mice (with or without adjuvant) (48). The incorporation of these immunogenic residues may allow a robust self-immune response to be raised against cancer-associated or weakly immunogenic antigens. This observation also raises the intriguing possibility that naturally occurring post-translational modifications may break tolerance and lead to autoimmune disease.
The addition of novel amino acids to the genetic code may also confer an advantage in the evolution of proteins with novel or enhanced functions. To begin to test this notion, phage-displayed antibody libraries that randomly incorporate unnatural amino acids into CDR3H (complementary-determining region 3 heavy chain) were subjected to in vitro selection experiments. Antibodies containing boronate (23) or sulfotyrosine (19) residues were found to outcompete natural antibodies in binding to acylic glucamine resins and the human immunodeficiency virus coat protein gp120, respectively (49, 50). Current protein evolution experiments are focused on harnessing the broad “chemical potential” of unnatural amino acids to overcome post-translational sequence constraints, introduce catalytic or structural metal ion-binding sites into proteins, or incorporate amino acids with reactive “chemical warheads” into peptides and antibodies to inhibit proteases or other therapeutically relevant enzymes (51).
An understanding of the molecular basis for unnatural amino acid recognition by the aaRS is critical to the evolution of aaRSs that can incorporate increasingly diverse chemical structures. Toward this end, we have solved the crystal structures of several substrate-bound MjTyrRS mutants (52). These structures show a significant degree of structural plasticity: both the side chains and the polypeptide backbone undergo significant conformational changes to create new hydrogen-bonding and hydrophobic interactions with the bound substrate (and remove those with the natural tyrosine substrate). These structures can also be used to iteratively generate new libraries to further expand the genetic repertoire.
Advances in the in Vivo Incorporation of Unnatural Amino Acids
Further increases in the efficiency of unnatural amino acid incorporation are leading to increased yields of mutagenized proteins. For example, several laboratories have overexpressed the suppressor tRNACUA gene in polycistronic constructs to improve the efficiency with which acylated tRNACUA can compete with release factors for the TAG codon (53, 54). In E. coli, this method increased the yields of mutant proteins by 20-fold. To increase yields in S. cerevisiae, Wang and Wang (55) expressed a single copy of EctRNACUA with an SNR52 promoter that contains the required identity elements for efficient pol III transcription. In a similar fashion, yields were increased in mammalian cells by expression of EctRNACUA from the pol III H1 promoter, which eliminates the need for the BstRNACUATyr (23). Optimization of suppressor tRNACUA sequences has also led to increased yields of mutant proteins. For example, in E. coli, mutant libraries focused on the T and acceptor stems of MjtRNACUATyr were passed through a double-sieve selection to identify tRNAs with up to 25-fold improved amber suppression efficiencies compared with wild-type MjtRNACUATyr (56). Protein yields were further increased in E. coli when an optimized MjtRNACUATyr sequence was coupled with inducible overexpression of the MjTyrRS (57). This system has been effective in standardizing unnatural amino acid incorporation in E. coli.
Mutations in host systems or alternate hosts have also been used to improve amber suppression efficiencies. For example, Chin and co-workers (58) have developed an orthogonal ribosome in E. coli (ribo-X) that increases suppression through a decreased functional interaction with release factor 1. To obtain higher yields (exceeding 150 mg/liters in shake flasks) of mutant proteins in a eukaryotic system, EcaaRS/EctRNACUA pairs evolved in S. cerevisiae have recently been transferred to the methylotrophic yeast Pichia pastoris (59). This new system has allowed the incorporation of unnatural amino acids into proteins that are poorly expressed in E. coli or S. cerevisiae hosts.
Multiple mutually orthogonal aaRS/tRNA pairs and additional reassigned codons are necessary for the expression of proteins containing multiple unnatural amino acids. To this end, ochre, opal, or 4-base codons (the latter are created by expanding the tRNA anticodon loop to complement a 4-base codon in the mRNA) have also been used to encode unnatural amino acids. In particular, the AGGA codon, together with an evolved Pyrococcus horikoshii lysyl-tRNA synthetase/tRNACUALys pair, has been used in conjunction with the MjTyrRS/MjtRNACUATyr pair to incorporate two unnatural amino acids into a single polypeptide expressed in E. coli (60).
Perspective
Ongoing efforts by several laboratories are focused on the evolution of new orthogonal pairs, the encoding of additional unnatural amino acids, and the transition of these systems to other unicellular and multicellular organisms. The ability to encode more complex amino acids will likely require additional orthogonal aaRS/tRNA pairs, structure-based active-site libraries, and more complex selection schemes, including the stepwise selection of novel aaRSs (e.g. the bipyridylalanine-specific aaRS was evolved from a library created from a biphenylalanine-specific aaRS (61)). Of particular interest to our laboratory is the expansion of this methodology to Caenorhabditis elegans and mice. The evolution of new aaRS/tRNA pairs may also allow the in vivo synthesis of biopolymers with unnatural backbones (e.g. β-peptides). To “free up” additional codons, the degeneracy of the E. coli or yeast genome may be reduced by deleting rare codons in a synthetically produced genome. Additional experiments in our laboratory are aimed at the in vivo evolution and selection of functional peptides, proteins, or whole organisms harboring unnatural amino acids. Evolution experiments with unnatural amino acids may answer questions regarding the fitness of the modern genetic code. Finally, the application of unnatural amino acid methodology to protein therapeutics such as bispecific antibodies, immunotoxins, and vaccines is certain to have an impact on medicine.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM062159 and Division of Materials Sciences Award DE-FG03-00ER46051 from the United States Department of Energy. This is the second of six articles in the “Chemical Biology Meets Biological Chemistry Minireview Series.” This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- Pyl
- pyrrolysine
- aaRS
- aminoacyl-tRNA synthetase
- TyrRS
- tyrosyl-tRNA synthetase
- LeuRS
- leucyl-tRNA synthetase
- PylRS
- pyrrolysyl-tRNA synthetase
- pol III
- RNA polymerase III
- Ec
- E. coli
- Mj
- M. jannaschii
- Mm
- M. mazei
- Bs
- B. stearothermophilus.
REFERENCES
- 1.Wang L., Xie J., Schultz P. G. (2006) Annu. Rev. Biophys. Biomol. Struct. 35, 225–249 [DOI] [PubMed] [Google Scholar]
- 2.Noren C. J., Anthony-Cahill S. J., Griffith M. C., Schultz P. G. (1989) Science 244, 182–188 [DOI] [PubMed] [Google Scholar]
- 3.Mendel D., Cornish V. W., Schultz P. G. (1995) Annu. Rev. Biophys. Biomol. Struct. 24, 435–462 [DOI] [PubMed] [Google Scholar]
- 4.Martin A. B., Schultz P. G. (1999) Trends Cell Biol. 9, M24–M28 [PubMed] [Google Scholar]
- 5.Nowak M. W., Kearney P. C., Sampson J. R., Saks M. E., Labarca C. G., Silverman S. K., Zhong W., Thorson J., Abelson J. N., Davidson N., Schultz P. G., Dougherty D. A., Lester H. A. (1995) Science 268, 439–442 [DOI] [PubMed] [Google Scholar]
- 6.Leinfelder W., Zehelein E., Mandrand-Berthelot M. A., Böck A. (1988) Nature 331, 723–725 [DOI] [PubMed] [Google Scholar]
- 7.Srinivasan G., James C. M., Krzycki J. A. (2002) Science 296, 1459–1462 [DOI] [PubMed] [Google Scholar]
- 8.Liu D. R., Magliery T. J., Pastrnak M., Schultz P. G. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 10092–10097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L., Brock A., Herberich B., Schultz P. G. (2001) Science 292, 498–500 [DOI] [PubMed] [Google Scholar]
- 10.Pastrnak M., Magliery T. J., Schultz P. G. (2000) Helv. Chim. Acta 83, 2277–2286 [Google Scholar]
- 11.Wang L., Schultz P. G. (2001) Chem. Biol. 8, 883–890 [DOI] [PubMed] [Google Scholar]
- 12.Anderson J. C., Schultz P. G. (2003) Biochemistry 42, 9598–9608 [DOI] [PubMed] [Google Scholar]
- 13.Neumann H., Peak-Chew S. Y., Chin J. W. (2008) Nat. Chem. Biol. 4, 232–234 [DOI] [PubMed] [Google Scholar]
- 14.Mukai T., Kobayashi T., Hino N., Yanagisawa T., Sakamoto K., Yokoyama S. (2008) Biochem. Biophys. Res. Commun. 371, 818–822 [DOI] [PubMed] [Google Scholar]
- 15.Chen P. R., Groff D., Guo J., Ou W., Cellitti S., Geierstanger B. H., Schultz P. G. (2009) Angew. Chem. Int. Ed. Engl. 48, 4052–4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards H., Schimmel P. (1990) Mol. Cell. Biol. 10, 1633–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiga D., Sakamoto K., Kodama K., Kigawa T., Matsuda T., Yabuki T., Shirouzu M., Harada Y., Nakayama H., Takio K., Hasegawa Y., Endo Y., Hirao I., Yokoyama S. (2002) Proc. Natl. Acad. Sci. U. S. A. 99, 9715–9720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chin J. W., Cropp T. A., Chu S., Meggers E., Schultz P. G. (2003) Chem. Biol. 10, 511–519 [DOI] [PubMed] [Google Scholar]
- 19.Chin J. W., Cropp T. A., Anderson J. C., Mukherji M., Zhang Z., Schultz P. G. (2003) Science 301, 964–967 [DOI] [PubMed] [Google Scholar]
- 20.Sakamoto K., Hayashi A., Sakamoto A., Kiga D., Nakayama H., Soma A., Kobayashi T., Kitabatake M., Takio K., Saito K., Shirouzu M., Hirao I., Yokoyama S. (2002) Nucleic Acids Res. 30, 4692–4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu W., Brock A., Chen S., Chen S., Schultz P. G. (2007) Nat. Methods 4, 239–244 [DOI] [PubMed] [Google Scholar]
- 22.Wu N., Deiters A., Cropp T. A., King D., Schultz P. G. (2004) J. Am. Chem. Soc. 126, 14306–14307 [DOI] [PubMed] [Google Scholar]
- 23.Wang W., Takimoto J. K., Louie G. V., Baiga T. J., Noel J. P., Lee K. F., Slesinger P. A., Wang L. (2007) Nat. Neurosci. 10, 1063–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L., Zhang Z., Brock A., Schultz P. G. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chin J. W., Santoro S. W., Martin A. B., King D. S., Wang L., Schultz P. G. (2002) J. Am. Chem. Soc. 124, 9026–9027 [DOI] [PubMed] [Google Scholar]
- 26.Deiters A., Schultz P. G. (2005) Bioorg. Med. Chem. Lett. 15, 1521–1524 [DOI] [PubMed] [Google Scholar]
- 27.Brustad E. M., Lemke E. A., Schultz P. G., Deniz A. A. (2008) J. Am. Chem. Soc. 130, 17664–17665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brustad E., Bushey M. L., Lee J. W., Groff D., Liu W., Schultz P. G. (2008) Angew. Chem. Int. Ed. Engl. 47, 8220–8223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cellitti S. E., Jones D. H., Lagpacan L., Hao X., Zhang Q., Hu H., Brittain S. M., Brinker A., Caldwell J., Bursulaya B., Spraggon G., Brock A., Ryu Y., Uno T., Schultz P. G., Geierstanger B. H. (2008) J. Am. Chem. Soc. 130, 9268–9281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groff D., Thielges M. C., Cellitti S., Schultz P. G., Romesberg F. E. (2009) Angew. Chem. Int. Ed. Engl. 48, 3478–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye S., Huber T., Vogel R., Sakmar T. P. (2009) Nat. Chem. Biol. 5, 397–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seyedsayamdost M. R., Xie J., Chan C. T., Schultz P. G., Stubbe J. (2007) J. Am. Chem. Soc. 129, 15060–15071 [DOI] [PubMed] [Google Scholar]
- 33.Wang J., Xie J., Schultz P. G. (2006) J. Am. Chem. Soc. 128, 8738–8739 [DOI] [PubMed] [Google Scholar]
- 34.Lee H. S., Guo J., Lemke E. A., Dimla R. D., Schultz P. G. (2009) J. Am. Chem. Soc. 131, 12921–12923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee H. S., Spraggon G., Schultz P. G., Wang F. (2009) J. Am. Chem. Soc. 131, 2481–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee H. S., Schultz P. G. (2008) J. Am. Chem. Soc. 130, 13194–13195 [DOI] [PubMed] [Google Scholar]
- 37.Deiters A., Groff D., Ryu Y., Xie J., Schultz P. G. (2006) Angew. Chem. Int. Ed. Engl. 45, 2728–2731 [DOI] [PubMed] [Google Scholar]
- 38.Bose M., Groff D., Xie J., Brustad E., Schultz P. G. (2006) J. Am. Chem. Soc. 128, 388–389 [DOI] [PubMed] [Google Scholar]
- 39.Lemke E. A., Summerer D., Geierstanger B. H., Brittain S. M., Schultz P. G. (2007) Nat. Chem. Biol. 3, 769–772 [DOI] [PubMed] [Google Scholar]
- 40.Hino N., Okazaki Y., Kobayashi T., Hayashi A., Sakamoto K., Yokoyama S. (2005) Nat. Methods 2, 201–206 [DOI] [PubMed] [Google Scholar]
- 41.Chin J. W., Martin A. B., King D. S., Wang L., Schultz P. G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11020–11024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang L. Y., Umanah G., Hauser M., Son C., Arshava B., Naider F., Becker J. M. (2008) Biochemistry 47, 5638–5648 [DOI] [PubMed] [Google Scholar]
- 43.Peters F. B., Brock A., Wang J., Schultz P. G. (2009) Chem. Biol. 16, 148–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie J., Supekova L., Schultz P. G. (2007) ACS Chem. Biol. 2, 474–478 [DOI] [PubMed] [Google Scholar]
- 45.Liu C. C., Schultz P. G. (2006) Nat. Biotechnol. 24, 1436–1440 [DOI] [PubMed] [Google Scholar]
- 46.Guo J., Wang J., Lee J. S., Schultz P. G. (2008) Angew. Chem. Int. Ed. Engl. 47, 6399–6401 [DOI] [PubMed] [Google Scholar]
- 47.Guo J., Wang J., Anderson J. C., Schultz P. G. (2008) Angew. Chem. Int. Ed. Engl. 47, 722–725 [DOI] [PubMed] [Google Scholar]
- 48.Grünewald J., Hunt G. S., Dong L., Niessen F., Wen B. G., Tsao M. L., Perera R., Kang M., Laffitte B. A., Azarian S., Ruf W., Nasoff M., Lerner R. A., Schultz P. G., Smider V. V. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4337–4342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu C. C., Mack A. V., Tsao M. L., Mills J. H., Lee H. S., Choe H., Farzan M., Schultz P. G., Smider V. V. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17688–17693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu C. C., Mack A. V., Brustad E. M., Mills J. H., Groff D., Smider V. V., Schultz P. G. (2009) J. Am. Chem. Soc. 131, 9616–9617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu C. C., Schultz P. G. (2010) Annu. Rev. Biochem. 79, in press [DOI] [PubMed] [Google Scholar]
- 52.Turner J. M., Graziano J., Spraggon G., Schultz P. G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6483–6488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen S., Schultz P. G., Brock A. (2007) J. Mol. Biol. 371, 112–122 [DOI] [PubMed] [Google Scholar]
- 54.Ryu Y., Schultz P. G. (2006) Nat. Methods 3, 263–265 [DOI] [PubMed] [Google Scholar]
- 55.Wang Q., Wang L. (2008) J. Am. Chem. Soc. 130, 6066–6067 [DOI] [PubMed] [Google Scholar]
- 56.Guo J., Melancon C. E., 3rd, Lee H. S., Groff D., Schultz P. G. (2010) Angew. Chem. Int. Ed. Engl. 48, 9148–9151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young T. S., Ahmad I., Yin J. A., Schultz P. G. (2010) J. Mol. Biol. 395, 361–374 [DOI] [PubMed] [Google Scholar]
- 58.Wang K., Neumann H., Peak-Chew S. Y., Chin J. W. (2007) Nat. Biotechnol. 25, 770–777 [DOI] [PubMed] [Google Scholar]
- 59.Young T. S., Ahmad I., Brock A., Schultz P. G. (2009) Biochemistry 48, 2643–2653 [DOI] [PubMed] [Google Scholar]
- 60.Anderson J. C., Wu N., Santoro S. W., Lakshman V., King D. S., Schultz P. G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7566–7571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie J., Liu W., Schultz P. G. (2007) Angew. Chem. Int. Ed. Engl. 46, 9239–9242 [DOI] [PubMed] [Google Scholar]