Abstract
Histones form the protein core around which genomic DNA is wrapped in eukaryotic chromatin. Numerous genetic studies have established that the structure and transcriptional state of chromatin are closely related to histone post-translational modifications. Further elucidation of the precise mechanistic roles for individual histone modifications requires the ability to isolate and study homogeneously modified histones. However, the highly heterogeneous nature of histone modifications in vivo poses a significant challenge for such studies. Chemical tools that have enabled biochemical and biophysical studies of site-specifically modified histones are the focus of this minireview.
Keywords: Chromatin/Modification, Chromosomes/Histones, Enzymes, Histones/Modification, Peptides/Chemical Synthesis, Protein/Post-translational Modification, Protein Ligation, Semisynthesis
Introduction
Genomic DNA is stored as chromatin in the nuclei of eukaryotic cells. The fundamental repeating unit of chromatin is the mononucleosome. Each mononucleosome consists of ∼147 bp of double-stranded DNA wrapped around an octameric protein complex composed of two copies each of the four core histones, H2A, H2B, H3, and H4 (Fig. 1A) (1).
FIGURE 1.
Histones and their PTMs. A, mononucleosome structure with histone tails protruding from the core. This figure was generated from Protein Data Bank code 1KX5 with PyMOL. The colors used for individual histones are the same as described for B. B, schematic representation of histone tails and their modifications. Some modifications at the histone C terminus and globular core are also shown. Groups are indicated as follows: ac, acetyl; Cit, citrullyl; me, methyl; ph, phosphoryl; pr, propionyl; rib, ADP-ribosyl; and Ub, ubiquityl. This figure was adapted from Ref. 49.
The histones were first isolated and characterized in 1884 by Albrecht Kossel (2). However, it was not until 1950 that Stedman and Stedman (3) identified multiple forms of histones in the nuclei of cells and put forth the hypothesis that different cellular phenotypes in an organism may arise from the suppression of different genes by cell-specific histones. A decade later, in vitro experiments with cell-free systems demonstrated that histones were indeed inhibitory to DNA-templated RNA synthesis (4). This period also saw the discovery of histone acetylation by Phillips (5) and of histone lysine ϵ-N-methylation by Murray (6). As the protein synthesis inhibitor puromycin did not inhibit these histone modifications, Allfrey et al. (7) suggested that acetylation and methylation were PTMs2 of histones. On the basis of the observation that chemical acetylation of histones greatly reduced their inhibitory effect on RNA synthesis, they put forth the prescient hypothesis that small reversible PTMs of histones could switch RNA synthesis on or off at different loci along the chromosome.
Since these early studies, a large number of histone PTMs (Fig. 1B), along with the various proteins responsible for installing (writers), removing (erasers), and binding (readers) these PTMs, have been identified (8). It is now well established that both the position and chemical property of histone modifications dictate the structure of chromatin as well as its functions in transcription, replication, and DNA repair (9). This has led to the histone code hypothesis for epigenetic control of cellular events, whereby distinct histone modifications, on one or more tails, act sequentially or in combination to bring about distinct downstream events (9). Understanding the specific roles for histone modification, either individually or in combination with other modifications, and histone interactions with chromatin-associated proteins is key to understanding the mechanisms underlying epigenetic control of cellular activity. Chemistry provides a growing arsenal of tools to study the roles for histone PTMs, and these will be discussed below.
Synthetic Peptide Strategies
The research groups of Allfrey and Merrifield first demonstrated the application of a synthetic peptide to address the substrate specificity of histone-deacetylating enzymes (HDACs) (10). A histone H4 peptide corresponding to residues 15–21 and 14C-acetylated at Lys16 was synthesized by automated solid-phase peptide synthesis. Whereas a chymotryptic peptide from 3H-acetylated H4 (residues 1–37) was deacetylated by HDACs purified from calf thymus, the heptameric synthetic peptide was not. This suggested that HDACs require a minimum sequence for activity in vitro but, importantly, not the entire H4 sequence. The authors then synthesized the longer H4 peptide (residues 1–37) uniformly 14C-acetylated at Lys12 and 3H-acetylated at Lys16 and subjected it to enzymatic deacetylation (11). Interestingly, the relative amounts of [14C]acetyl released from Lys12 and [3H]acetyl from Lys16 were equal throughout the time course of the HDAC assay, indicating that both acetylated residues were substrates for deacetylase activity. More recently, studies with varying degrees of acetylated H4 tail peptides (residues 1–36) have revealed the multivalent engagement of multiply acetylated lysines by two tandem bromodomains in the RNA polymerase II transcription factor D (TFIID) complex protein TAFII250 (12). Acetylated H4 tail peptides have also revealed an unprecedented cooperative binding of two acetyl groups at H4 Lys5 and Lys8 by a single bromodomain module (13). Biotinylated Lys4-methylated H3 tail peptides (residues 1–20) have also been employed to identify the proteins WDR5 and BPTF as readers for the subtly different H3 Lys4-dimethylated and Lys4-trimethylated states, respectively (14).
The generation of peptide libraries has allowed high-throughput screening of histone-protein interactions in a microarray format. Toward this goal, Bedford and co-workers (15) have designed a chromatin-associated domain array (CADOR) chip containing an array of immobilized glutathione S-transferase-tagged histone-binding domains, including tudor and MBT domains, bromodomains, and chromodomains. Binding experiments with fluorophore-tagged N-terminal peptides from H3 and H4 bearing varying sites and degrees of methylation revealed novel interactions with chromodomains and tudor and MBT domains from various chromatin-associated proteins. Rathert et al. (16) have utilized SPOT synthesis to generate arrays of as many as 420 mutant H3 tail peptides (residues 1–21) and tested the substrate specificity of the H3 Lys9 methyltransferase Dim-5 from Neurospora crassa. Results from these assays suggested an important role for Thr11 and Gly12 in the H3 tail in conferring specificity for Dim-5 activity and its discrimination against other lysines.
An impressive example of the combinatorial power of peptide synthesis was reported by Denu and co-workers (17), who developed a one-bead one-compound combinatorial library of 800 peptides bearing all possible permutations of the known modifications within the 21 N-terminal amino acids of histone H4. Peptide modifications included phosphorylation, acetylation, citrullination, and all possible methylation states of Lys and Arg. From the initial library, 512 members were used to elucidate the binding preferences of the double tudor domain of the human demethylase JMJD2A for the H4 tail. Interestingly, binding hits with various combinations of modifications revealed a rheostat-like continuum of binding affinities for human JMJD2A from 1 μm to 1 mm. Finally, a self-assembled monolayer for matrix-assisted laser desorption-ionization (SAMDI) mass spectrometric assay was applied by Gurard-Levin and Mrksich (18) to characterize the activity of HDAC8 for H4 tail peptides. Their results indicated the importance of both distal residues (residues 16–19) and those immediately adjacent to acetylated Lys12 for HDAC8 activity.
Amber Suppression Strategies
An understanding of the physiological roles for histone modifications requires the ability to study them in the context of nucleosomes and chromatin. Although peptide models are often sufficient for studies of binary protein interactions, they cannot address the effects of modifications on directing trans-tail histone modifications, on nucleosomal or higher order chromatin structure, and on chromatin-remodeling complexes.
Amber suppression mutagenesis of proteins with orthogonal pairs of amber suppressor tRNAs and their cognate aminoacyl-tRNA synthetases could be employed to incorporate modified amino acids into full-length histones. Toward this goal, Schultz and co-workers (19) have evolved a mutant Methanococcus jannaschii tyrosyl amber suppressor tRNA, TyrMjtRNACUA/tyrosyl-tRNA synthetase pair to site-specifically incorporate (Se)-phenylselenocysteine in response to the amber TAG codon in Escherichia coli (Fig. 2A). Oxidative elimination of phenylselenic acid yielded Dha (20), which underwent Michael addition with N-acetylated or N-methylated derivatives of 2-aminoethanethiol to produce the thiol-containing analogs of N-acetylated and N-methylated lysine (Fig. 2B). This methodology was employed to generate an analog of histone H3 acetylated at Lys9, which underwent phosphorylation at Ser10 by the Aurora B kinase. A potential limitation of this methodology may be the well established absence of diastereoselectivity in non-enzymatic thiol additions to Dha (20), although the local protein conformation may significantly influence the final diastereomeric ratios of the addition products (21).
FIGURE 2.
Amber suppression and cysteine-specific modification techniques. A, in vivo amber suppression methodology for incorporating PTMs into histones. 1, (Se)-phenylselenocysteine; 2, N-ϵ-Boc-N-ϵ-methyl-l-lysine; 3, N-ϵ-acetyl-l-lysine. B, conversion of 1 and 2 to modified lysine analogs after incorporation into histones. MeLys, methyllysine; AcLys, acetyllysine. C, in vitro cysteine modification strategy for generating MLAs. Br, bromo.
An alternate approach developed by Neumann et al. (22) utilized an evolved Methanosarcina barkeri pyrrolysyl-tRNA synthetase and its cognate amber suppressor, tRNACUA, to genetically incorporate N-ϵ-acetyllysine in response to the TAG codon in E. coli (Fig. 2A). This was employed to generate H3 acetylated at Lys56 that was incorporated into mononucleosomes and nucleosomal arrays. Förster resonance energy transfer-based experiments with fluorophore-labeled nucleosomes permitted the direct observation of DNA unwrapping. Acetylation was found to increase the extent of DNA unwrapping within the last turn of DNA on the nucleosome core by 7-fold. It also accelerated nucleosomal repositioning by remodeling complexes ∼20% over unmodified nucleosomes. However, acetylation did not affect the ATP-dependent H2A/H2B dimer transfer from mononucleosomes or higher order chromatin structure formation. Thus, the overall effects of H3 Lys56 acetylation on nucleosome structure and stability are fairly subtle and are manifest at the level of DNA breathing.
Recently, Chin and co-workers (23) have extended their methodology to incorporate N-ϵ-methyl-l-lysine into H3 at position 9, masked as the N-ϵ-Boc-N-ϵ-methyl-l-lysine derivative, which is an efficient substrate for the M. barkeri pyrrolysyl-tRNA synthetase/tRNACUA pair (Fig. 2A). Subsequent acidolytic deprotection of the Boc group generated H3 monomethylated at Lys9 (Fig. 2B), which was demonstrated to bind an anti-H3-K9me1 antibody and HP1 (heterochromatin protein 1).
Cysteine Modification Strategies
Cys is the most convenient amino acid for selective modification because of its highly nucleophilic side chain sulfhydryl group (pKa ∼ 8.5). Shokat and co-workers (24) have taken advantage of the unique reactivity of Cys to generate N-methylated aminoethylcysteine residues (Fig. 2C). Coupled with mutation of the single Cys in H3 (Cys110) to Ala, their methodology permitted the site-specific installation of MLAs in histone H3. Thiol-containing analogs of H3 methylated at Lys9 (H3-KC9me), Lys4, Lys36, and Lys79 and of H4 methylated at Lys20 were successfully recognized by methylation-specific antibodies. Furthermore, both an H3-KC9me2 peptide (residues 1–14) and nucleosome-associated full-length H3-KC9me2 were demonstrated to bind the heterochromatin-binding protein HP1α, which is known to bind H3-K9me2. Similar degrees of methylation of H3 Lys9 and the H3-KC9 analog by the methyltransferase SUV39H1 demonstrated the equivalence of MLAs in biochemical assays. The ease of access to MLAs in histones also permitted structural determination of mononucleosomes where both copies of the native histones were substituted with either H4-KC20me3 or H3-KC79me2 (25). In either case, methylation did not significantly affect mononucleosome structure. In solution, however, sedimentation velocity analysis indicated that 12-mer nucleosomal arrays reconstituted with octamers containing H4-KC20me3 had an enhanced propensity to form maximally folded and higher order structures with increasing Mg2+ concentrations compared with H3-KC79me2-containing or wild-type nucleosomal arrays. MLAs have also been applied in studies of DNA replication (26), recruitment of HDACs to histones (27), the propagation of repressive modifications (28), and cross-talk between modifications (29).
An alternate Cys-directed modification strategy developed by Davis and co-workers (30) is the oxidative elimination of Cys to Dha by the reagent O-mesitylenesulfonylhydroxylamine. Site-specific Cys mutagenesis in histones could in principle be coupled with this methodology to generate Dha, which would readily be converted to methylated and acetylated lysine analogs, similar to the methodology reported by Schultz and co-workers (19).
Native Chemical and Expressed Protein Ligation
The first report of EPL in 1998 by Muir et al. (31) marked a new avenue for employing chemistry to explore protein function. EPL extends the synthetic technique known as native chemical ligation (32), whereby two peptide halves, one bearing an N-terminal Cys and the other bearing a C-terminal thioester, are joined by a native amide bond. The peptide fragments can be obtained by solid-phase peptide synthesis employing either Boc or Fmoc (N-(9-fluorenyl)methoxycarbonyl) protecting group chemistry (33). Given the inevitable limitation of native chemical ligation by the length of the synthetic peptide halves, EPL significantly expands the range of proteins accessible for chemical modification. EPL employs an expressed protein in its C-terminal thioester form obtained by thiolysis of a C-terminally fused intein rather than by synthetic means. The expressed protein thioester undergoes trans-thioesterification when reacted with a second peptide/protein bearing an N-terminal Cys (Fig. 3). A subsequent S-to-N-acyl shift generates a native amide bond between the two halves, leading to the full-length target. EPL may also be reversed to enable the ligation of a C-terminal expressed protein half with an N-terminal synthetic peptide thioester. Most importantly, because any desired protein modification may be introduced in the synthetic half of the ligation partners, in principle, EPL permits the incorporation of any histone modification observed in Nature.
FIGURE 3.
EPL. Multiple strategies for incorporating synthetic peptides into full-length proteins. SPPS, solid-phase peptide synthesis; RSH, alkyl/aryl thiol.
Site-specific Histone Modification by EPL
Shogren-Knaak et al. (34) first reported the use of EPL to generate Xenopus laevis histone H3 bearing a pSer residue at position 10. Nucleosomal arrays reconstituted with H3 pSer10 were efficiently remodeled by the yeast SWI/SNF remodeling complex, demonstrating that semisynthetic H3 pSer10 did not drastically affect nucleosome structure. These arrays were also used to probe the substrate specificity of the histone acetyltransferase Gcn5 and revealed its different activities when presented with peptide versus nucleosomal substrates. This indicates the need for investigating the mechanisms of histone-modifying enzymes on intact nucleosomes rather than peptide substrates.
In another study, site-specifically Lys16-acetylated histone H4 was generated (35). This was incorporated into nucleosomal arrays, and its effect on chromatin compaction was determined by sedimentation velocity analysis during ultracentrifugation. The single acetylation of H4 Lys16 inhibited 30-nm fiber formation in nucleosomal arrays, similar to the absence of the histone tail. These results suggested a structural role for Lys16-acetylated H4 in establishing transcriptionally active euchromatic regions by decondensing chromatin.
EPL has permitted the generation of multiply modified histones with as many as three acetyl groups at Lys5, Lys8, and Lys12 in histone H4 and five acetyl groups at Lys4, Lys9, Lys14, Lys18, and Lys23 in H3 (36). An additional desulfurization step was added to these syntheses, converting the Cys introduced for ligation to a native Ala in the final step and rendering the ligation traceless. Histones are particularly amenable to chemical desulfurization because only a single Cys (Cys110 in H3) is present in the four core histones in higher eukaryotes, and Cys is altogether absent in yeast histones. Surprisingly, substituting wild-type H3 with semisynthetic pentaacetylated H3 did not interfere with RSF (remodeling and spacing factor) complex-mediated assembly of a chromatinized plasmid. Additionally, H3/H4 tetramers generated with pentaacetylated H3 were also demonstrated to be a substrate for the HDAC Sir1 and subsequently for the methyltransferase G9a.
Owen-Hughes and co-workers (37) employed semisynthetically tetraacetylated derivatives of H3 (Lys9, Lys14, Lys18, and Lys23) and H4 (Lys5, Lys8, Lys12, and Lys16) to generate specifically modified chromatin templates to study the interaction of various yeast remodeling complexes with differentially acetylated nucleosomes. It was shown that the ATP-dependent remodeling enzyme complex RSC (remodel the structure of chromatin) preferentially remodeled chromatin containing tetraacetylated H3, but not H4, ∼16-fold faster than unmodified chromatin. Kinetic analysis revealed this to be due to a 3-fold lower Km for the tetraacetylated nucleosomes and that a single acetylation at H3 Lys14 contributed most to this effect. On the other hand, tetraacetylation of H4 inhibited nucleosome remodeling by the Isw2 enzyme by ∼1.5-fold relative to unmodified nucleosomes. This was due to a reduction in the rate of ATP hydrolysis (kcat) by Isw2. These experiments categorically demonstrated that histone modifications affect nucleosome remodeling through distinct pathways for different remodeling enzymes.
EPL has also enabled investigations of the effects of acetylation near the nucleosome dyad pseudo-symmetry axis, where key histone-DNA contacts occur (38). Results from nucleosome competitive reconstitution experiments revealed that acetylation at H3 Lys115 near the dyad axis reduced DNA binding significantly more than acetylation at H3 Lys122. However, mononucleosomes acetylated at H3 Lys122 underwent thermal repositioning about twice as fast as those acetylated at Lys115. These results suggest that the different sites of acetylation near the nucleosome dyad may have different physiological consequences in vivo, such as their effect on genome positioning and nucleosome assembly/disassembly. Another interesting result from this study was the observation that the Lys-to-Gln mutation that is commonly employed to mimic lysine acetylation in vivo showed significant differences from acetyllysine in in vitro competitive reconstitution assays.
Our own laboratory has reported several advances toward the semisynthesis of histones for biochemical studies. Chiang et al. (39) have demonstrated the utility of a thiol-protected 2-hydroxy-3-mercaptopropionic acid linker (40) and a diaminobenzoic acid linker (41) to synthesize a histone H2B N-terminal peptide thioester containing pSer at position 14 as well as acetyllysines at positions 5, 11, 12, and 15. These synthetic strategies avoided epimerization at the peptide C terminus, which is known to occur during the solution-phase activation of side chain-protected peptides (42). Furthermore, a mild radical-based desulfurization methodology specific for cysteine (43) was employed to render the ligation product traceless. The phosphorylated and polyacetylated full-length H2B was probed with a commercial H2B pSer14-specific antibody. Surprisingly, the presence of multiple acetylations in the H2B tail prevented the recognition of pSer14 by this antibody. This result indicates a limitation of antibody-based ChIP (chromatin immunoprecipitation)-on-chip experiments for whole genome analysis because histones are often decorated with multiple modifications in vivo that may interfere with the detection of specific modifications. However, the acetyl groups did not interfere with phosphorylation of Ser14 by the human MST1 (mammalian sterile twenty-like 1) kinase, suggesting an unusual mechanism for H2B recognition by this enzyme.
One of the most dramatic PTMs of proteins is their conjugation with the small protein ubiquitin. Ubiquitylation is undertaken by a family of E1–E3 ligases that activate the C terminus of ubiquitin and catalyze its condensation with specific lysine side chain ϵ-amines in target proteins (44). Unlike its typical role in proteasome-assisted degradation, the ubiquitylation of histones is associated with DNA damage repair and both transcription elongation and repression (45). Ubiquitylation of H2B occurs at Lys123 in yeast (Lys120 in humans) and has been linked to transcription elongation and trans-tail methylation of H3 Lys4 and Lys79 through genetic studies (45). However, the small fraction (1–2%) of uH2B and the heterogeneity of histone modifications in vivo posed a serious challenge in the isolation of uH2B for biochemical studies aimed at identifying its mechanistic role in H3 methylation.
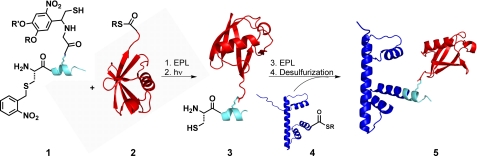
We have developed several semisynthetic approaches for the site-specific ubiquitylation of histones. McGinty et al. (46) have reported an EPL strategy to access uH2B that employs traceless peptide ubiquitylation (47). In this synthetic scheme (Fig. 4), the H2B protein was divided into a short synthetic C-terminal fragment (1) and recombinant N-terminal thioester (4). In the first step, ubiquitylation of the H2B C-terminal peptide (1) was accomplished with a photolytically removable ligation auxiliary that was coupled to the side chain of the residue corresponding to Lys120 in full-length H2B. The ligation auxiliary acted as an N-terminal cysteine surrogate and facilitated EPL with a recombinant ubiquitin thioester lacking its C-terminal residue Gly76 (2). Upon ligation, photolysis of the auxiliary with UV irradiation yielded native uH2B peptide (3) and simultaneously released a photoprotected Cys at the N terminus of the H2B peptide. In a second ligation step, the ubiquitylated peptide was reacted with the recombinant H2B thioester (4) to generate full-length uH2B(A117C). In a final step, the Cys was desulfurized to yield native uH2B (5). Biochemical assays of chemically ubiquitylated mononucleosomes with the human histone lysine methyltransferase DOT1L revealed that ubiquitylation directly stimulated intranucleosomal methylation of H3 Lys79 (46). This was the first direct biochemical evidence of cross-talk between PTMs on different histones.
FIGURE 4.
Semisynthesis of uH2B. A two-step ligation strategy followed by a final desulfurization step yielded native uH2B (5).
Very recently, we reported an alternate methodology for histone ubiquitylation that bypasses the need for the synthetically challenging ligation auxiliary (48). This was achieved by incorporating the mutation G76A at the ubiquitin C terminus, which allowed ligation with cysteine both at the lysine side chain and within H2B. Avoiding ligation onto a sterically hindered secondary amine, which was unavoidable in the ligation auxiliary approach, led to significantly reduced reaction times and higher yields. Furthermore, substitution of UV irradiation with chemical unmasking of the second cysteine greatly facilitated sample handling. Simultaneous desulfurization of both cysteines yielded the u(G76A)H2B protein, which was recognized by a uH2B-specific antibody and by the ubiquitin-specific hydrolase UCHL3. u(G76A)H2B also stimulated robust methylation at H3 Lys79 by human DOT1L, thus demonstrating similarity to native uH2B in a nucleosomal context. Subsequent kinetic and structure-activity relationship analyses with u(G76A)H2B have revealed a non-canonical role for ubiquitin in the enhancement of the chemical step of H3 Lys79 methylation. In particular, the hydrophobic patch on the surface of ubiquitin centered around Ile44, which forms critical interactions with most helical ubiquitin-binding domains, was found to be non-essential for stimulation of human DOT1L activity. Mutagenic studies aimed at identifying the specific surface residues of ubiquitin involved in human DOT1L stimulation are currently under way in our laboratory.
Conclusions and Future Directions
Beginning with their discovery >40 years ago, histone modifications have been shown to play critical roles in directing key cellular events such as transcription activation, gene silencing, DNA damage repair, and DNA replication. Several semisynthetic methodologies for the generation of homogeneously modified histones have been developed, and these have led to investigations of the mechanistic roles for individual histone modifications in these processes. Advances in protein chemistry have made accessible synthetically challenging histone modifications, such as those found in the globular core domains of mononucleosomes and those involving large proteins such as ubiquitin and SUMO (small ubiquitin-like modifier). In the future, experiments with semisynthetic histones in our own laboratories will be aimed at the level of testing the histone code hypothesis in nucleosomal arrays bearing controlled modifications for studies of gene transcription, replication, and repair.
This is the third of six articles in the “Chemical Biology Meets Biological Chemistry Minireview Series.” This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- PTM
- post-translational modification
- HDAC
- histone deacetylase
- MBT
- malignant brain tumor
- Dha
- dehydroalanine
- Boc
- t-butoxycarbonyl
- MLA
- methyllysine analog
- EPL
- expressed protein ligation
- pSer
- phosphoserine
- uH2B
- ubiquitylated H2B.
REFERENCES
- 1.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 2.Kossel A. (1928) The Protamines and Histones, 1st Ed., Longmans, Green & Co., London [Google Scholar]
- 3.Stedman E., Stedman E. (1950) Nature 166, 780–781 [DOI] [PubMed] [Google Scholar]
- 4.Butler J. A. (1965) Nature 207, 1041–1042 [DOI] [PubMed] [Google Scholar]
- 5.Phillips D. M. (1963) Biochem. J. 87, 258–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murray K. (1964) Biochemistry 3, 10–15 [DOI] [PubMed] [Google Scholar]
- 7.Allfrey V. G., Faulkner R., Mirsky A. E. (1964) Proc. Natl. Acad. Sci. U.S.A. 51, 786–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 9.Allis C. D., Jenuwein T., Reinberg D., Caparros M.-L. (2007) Epigenetics, 1st Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 10.Krieger D. E., Levine R., Merrifield R. B., Vidali G., Allfrey V. G. (1974) J. Biol. Chem. 249, 332–334 [PubMed] [Google Scholar]
- 11.Krieger D., Vidali G., Erickson B., Allfrey V., Merrifield R. (1979) Bioorg. Chem. 8, 409–427 [Google Scholar]
- 12.Jacobson R. H., Ladurner A. G., King D. S., Tjian R. (2000) Science 288, 1422–1425 [DOI] [PubMed] [Google Scholar]
- 13.Morinière J., Rousseaux S., Steuerwald U., Soler-López M., Curtet S., Vitte A. L., Govin J., Gaucher J., Sadoul K., Hart D. J., Krijgsveld J., Khochbin S., Müller C. W., Petosa C. (2009) Nature 461, 664–668 [DOI] [PubMed] [Google Scholar]
- 14.Ruthenburg A. J., Allis C. D., Wysocka J. (2007) Mol. Cell 25, 15–30 [DOI] [PubMed] [Google Scholar]
- 15.Kim J., Daniel J., Espejo A., Lake A., Krishna M., Xia L., Zhang Y., Bedford M. T. (2006) EMBO Rep. 7, 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rathert P., Zhang X., Freund C., Cheng X., Jeltsch A. (2008) Chem. Biol. 15, 5–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garske A. L., Craciun G., Denu J. M. (2008) Biochemistry 47, 8094–8102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurard-Levin Z. A., Mrksich M. (2008) Biochemistry 47, 6242–6250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo J., Wang J., Lee J. S., Schultz P. G. (2008) Angew. Chem. Int. Ed. Engl. 47, 6399–6401 [DOI] [PubMed] [Google Scholar]
- 20.Zhu Y., van der Donk W. A. (2001) Org. Lett. 3, 1189–1192 [DOI] [PubMed] [Google Scholar]
- 21.Schmidt U., Ohler E. (1976) Angew. Chem. Int. Ed. Engl. 15, 42–42 [DOI] [PubMed] [Google Scholar]
- 22.Neumann H., Hancock S. M., Buning R., Routh A., Chapman L., Somers J., Owen-Hughes T., van Noort J., Rhodes D., Chin J. W. (2009) Mol. Cell 36, 153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen D. P., Garcia Alai M. M., Kapadnis P. B., Neumann H., Chin J. W. (2009) J. Am. Chem. Soc. 131, 14194–14195 [DOI] [PubMed] [Google Scholar]
- 24.Simon M. D., Chu F., Racki L., de la Cruz C. C., Burlingame A. L., Panning B., Narlikar G. J., Shokat K. M. (2007) Cell 128, 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu X., Simon M. D., Chodaparambil J. V., Hansen J. C., Shokat K. M., Luger K. (2008) Nat. Struct. Mol. Biol. 15, 1122–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Francis N. J., Follmer N. E., Simon M. D., Aghia G., Butler J. D. (2009) Cell 137, 110–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B., Jackson J., Simon M. D., Fleharty B., Gogol M., Seidel C., Workman J. L., Shilatifard A. (2009) J. Biol. Chem. 284, 7970–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margueron R., Justin N., Ohno K., Sharpe M. L., Son J., Drury W. J., 3rd, Voigt P., Martin S. R., Taylor W. R., De Marco V., Pirrotta V., Reinberg D., Gamblin S. J. (2009) Nature 461, 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hung T., Binda O., Champagne K. S., Kuo A. J., Johnson K., Chang H. Y., Simon M. D., Kutateladze T. G., Gozani O. (2009) Mol. Cell 33, 248–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernardes G. J., Chalker J. M., Errey J. C., Davis B. G. (2008) J. Am. Chem. Soc. 130, 5052–5053 [DOI] [PubMed] [Google Scholar]
- 31.Muir T. W., Sondhi D., Cole P. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6705–6710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dawson P. E., Muir T. W., Clark-Lewis I., Kent S. B. (1994) Science 266, 776–779 [DOI] [PubMed] [Google Scholar]
- 33.Muralidharan V., Muir T. W. (2006) Nat. Methods 3, 429–438 [DOI] [PubMed] [Google Scholar]
- 34.Shogren-Knaak M. A., Fry C. J., Peterson C. L. (2003) J. Biol. Chem. 278, 15744–15748 [DOI] [PubMed] [Google Scholar]
- 35.Shogren-Knaak M., Ishii H., Sun J. M., Pazin M. J., Davie J. R., Peterson C. L. (2006) Science 311, 844–847 [DOI] [PubMed] [Google Scholar]
- 36.He S., Bauman D., Davis J. S., Loyola A., Nishioka K., Gronlund J. L., Reinberg D., Meng F., Kelleher N., McCafferty D. G. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12033–12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferreira H., Flaus A., Owen-Hughes T. (2007) J. Mol. Biol. 374, 563–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manohar M., Mooney A. M., North J. A., Nakkula R. J., Picking J. W., Edon A., Fishel R., Poirier M. G., Ottesen J. J. (2009) J. Biol. Chem. 284, 23312–23321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiang K. P., Jensen M. S., McGinty R. K., Muir T. W. (2009) ChemBioChem 10, 2182–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.George E. A., Novick R. P., Muir T. W. (2008) J. Am. Chem. Soc. 130, 4914–4924 [DOI] [PubMed] [Google Scholar]
- 41.Blanco-Canosa J. B., Dawson P. E. (2008) Angew. Chem. Int. Ed. Engl. 47, 6851–6855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodman M., McGahren W. J. (1967) Tetrahedron 23, 2031–2050 [DOI] [PubMed] [Google Scholar]
- 43.Wan Q., Danishefsky S. J. (2007) Angew. Chem. Int. Ed. Engl. 46, 9248–9252 [DOI] [PubMed] [Google Scholar]
- 44.Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 45.Weake V. M., Workman J. L. (2008) Mol. Cell 29, 653–663 [DOI] [PubMed] [Google Scholar]
- 46.McGinty R., Kim J., Chatterjee C., Roeder R., Muir T. W. (2008) Nature 453, 812–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chatterjee C., McGinty R. K., Pellois J. P., Muir T. W. (2007) Angew. Chem. Int. Ed. Engl. 46, 2814–2818 [DOI] [PubMed] [Google Scholar]
- 48.McGinty R., Kohn M., Chatterjee C., Chiang K., Pratt M., Muir T. W. (2009) ACS Chem. Biol. 4, 958–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhaumik S. R., Smith E., Shilatifard A. (2007) Nat. Struct. Mol. Biol. 14, 1008–1016 [DOI] [PubMed] [Google Scholar]