Abstract
Many of the neurodegenerative diseases that afflict people are caused by intracytoplasmic aggregate-prone proteins. These include Parkinson disease, tauopathies, and polyglutamine expansion diseases such as Huntington disease. In Mendelian forms of these diseases, the mutations generally confer toxic novel functions on the relevant proteins. Thus, one potential strategy for dealing with these mutant proteins is to enhance their degradation. This can be achieved by up-regulating macroautophagy, which we will henceforth call autophagy. In this minireview, we will consider the reasons why autophagy up-regulation may be a powerful strategy for these diseases. In addition, we will consider some of the drugs and associated signaling pathways that can be used to induce autophagy with these therapeutic aims in mind.
Keywords: Diseases/Amyloid, Metabolism, Signal Transduction/Protein Kinases/Calmodulin, Subcellular Organelles/Lysosomes, Tissue/Organ Systems/Brain, Toxins/Drugs/Xenobiotics/Drug Action, Autophagy, Drug Screen
Intracellular Protein Aggregation in Neurodegenerative Diseases
Intracellular protein misfolding and aggregation are features of many late-onset neurodegenerative diseases called proteinopathies. These include Alzheimer disease, Parkinson disease, tauopathies, and polyQ3 expansion diseases such as HD and various SCAs such as SCA3 (1, 2). Currently, there are no effective strategies to slow or prevent the neurodegeneration resulting from these diseases in humans.
All known polyQ mutant proteins form intracellular aggregates (inclusions) with amyloid-like structures in susceptible neurons (3). HD, the most prevalent of the nine polyQ expansion diseases, is caused by an abnormally expanded CAG trinucleotide repeat tract in the IT15 gene (>35 repeats). These repeats are translated into an elongated polyQ tract close to the N-terminal end of the huntingtin protein. Huntingtin is mainly cytosolic, but a small proportion is nuclear (4). In HD, intranuclear inclusions are seen in the rarer juvenile-onset cases, but extranuclear inclusions predominate in the more typical adult-onset cases. The causal role for inclusions in these diseases is debated because some have reported dissociations between cell death and inclusion formation (4, 5). Strong genetic and transgenic data argue that the primary consequence of the polyQ expansion mutations is to confer toxic gain of function on the mutant proteins (1, 2, 4, 6). Indeed, a gain-of-function mechanism appears to underlie most of the Mendelian disorders caused by aggregate-prone proteins, including tauopathies and other polyQ expansion disorders. This does not exclude that the gain-of-function toxicity in diseases like HD may be modulated to some degree by loss-of-function effects, although transgenic data suggest that such putative effects are likely to be small (7). Because the mutations causing many proteinopathies (e.g. polyQ diseases and tauopathies) confer novel toxic functions on the specific proteins and because disease severity frequently correlates with expression levels, it is important to understand the factors regulating the synthesis and clearance of these aggregate-prone proteins.
Autophagic Clearance of Intracytosolic Aggregate-prone Proteins
Our data suggest that accelerating the removal of toxic huntingtin fragments may be a tractable therapeutic strategy for HD (Fig. 1). We showed that the ubiquitin-proteasome and autophagy-lysosome pathways are the major routes for mutant huntingtin fragment clearance (8). Although the narrow proteasome barrel precludes entry of oligomers/aggregates of mutant huntingtin (or other aggregate-prone intracellular proteins), such substrates can be degraded efficiently by macroautophagy (which we will call autophagy).
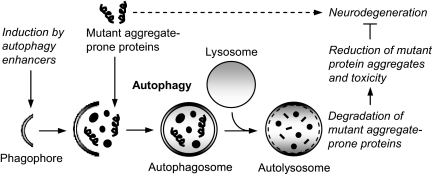
FIGURE 1.
Autophagy as a protective pathway for neurodegenerative diseases. Autophagy is a major degradation pathway for the clearance of various intracytosolic toxic aggregate-prone proteins associated with neurodegenerative diseases. Chemical induction of autophagy by autophagy enhancers triggers cellular signaling pathways, leading to formation of double-membrane cytoplasmic structures called phagophores. These structures elongate and engulf mutant aggregate-prone proteins along with portions of the cytoplasm to form autophagosomes. Autophagosomes then ultimately fuse with the lysosomes to form autolysosomes, where their contents are degraded by acidic lysosomal hydrolases. Enhancing autophagic clearance of these mutant aggregate-prone proteins results in reduction of mutant protein aggregates and toxicity, which is protective in several models of neurodegenerative diseases.
Autophagy involves the formation of double-membrane isolation structures called phagophores, which expand and engulf portions of the cytoplasm, forming double-membrane vesicles called autophagosomes (Fig. 1) (9, 10). Autophagosomes are formed randomly in the cytoplasm and are then trafficked along microtubules in a dynein-dependent fashion toward the microtubule-organizing center, where they fuse with lysosomes, forming autolysosomes, after which their contents are degraded (11, 12). The only known mammalian protein that specifically associates with the autophagosome membrane (as opposed to other vesicles) is MAP1 LC3 (microtubule-associated protein 1 light chain 3), which is post-translationally modified into cytosolic LC3-I, which conjugates with phosphatidylethanolamine upon autophagy induction to form autophagosome-associated LC3-II (13).
Recent studies have shown that constitutive autophagy may play a pivotal role in the clearance of normally occurring cellular misfolded proteins, as loss of basal autophagy by conditional knock-out of key autophagy genes, such as Atg5 and Atg7, in mouse brains resulted in a neurodegenerative phenotype and the formation of protein aggregates (14, 15). We have shown that mutant huntingtin fragments, expanded polyalanines tagged with GFP, and mutant forms of α-synuclein (associated with forms of Parkinson disease) are highly dependent on autophagy for their clearance in cell models (8, 16). The clearance of these mutant proteins is delayed by autophagy inhibitors like 3-methyladenine and bafilomycin A1 or by knockdown of autophagy genes, whereas autophagy induction with rapamycin enhances their clearance (8, 17, 18). Subsequently, other aggregate-prone proteins, such as Tau causing frontotemporal dementias, mutant ataxin-3 associated with SCA3, mutant SOD1 (superoxide dismutase 1) associated with ALS, and mutant prion proteins causing prion diseases, have been shown to be autophagy substrates (19–22). Although most of the wild-type counterparts of these mutant proteins are poor autophagy substrates, wild-type α-synuclein has been shown to be degraded by chaperone-mediated autophagy, a distinct lysosome pathway (23). Thus, up-regulating autophagy may be beneficial for the treatment of neurodegenerative diseases, and identification of autophagy enhancers could provide potential therapeutic candidates (Table 1) (24, 25).
TABLE 1.
List of autophagy enhancers for neurodegenerative diseases and their mode of action
| Autophagy enhancers | Mode of action | Refs. |
|---|---|---|
| Rapamycin, CCI-779, Glc, Glc-6-P, Torin1, perhexiline, niclosamide, rottlerin | Inhibit mTORC1 | 8, 20, 30, 31, 37, 39 |
| Lithium, L-690,330 | Inhibit IMPase and reduce inositol and IP3 levels; mTOR-independent | 41, 47 |
| Carbamazepine, sodium valproate | Reduce inositol and IP3 levels; mTOR-independent | 41, 48 |
| Verapamil, loperamide, amiodarone, nimodipine, nitrendipine, niguldipine, pimozide | Ca2+ channel blockers; reduce intracytosolic Ca2+ levels; mTOR-independent | 48, 54 |
| Calpastatin, calpeptin | Calpain inhibitors; mTOR-independent | 48 |
| Clonidine, rilmenidine | Imidazoline-1 receptor agonists; reduce cAMP levels; mTOR-independent | 48 |
| 2′,5′-Dideoxyadenosine | Adenylyl cyclase inhibitor; reduces cAMP levels; mTOR-independent | 48 |
| NF449 | Gαs inhibitor; mTOR-independent | 48 |
| Minoxidil | K+ATP channel opener; mTOR-independent | 48 |
| Penitrem A | Inhibits high conductance Ca2+-activated K+ channel; mTOR-independent | 54 |
| Fluspirilene, trifluoperazine | Dopamine antagonists; mTOR-independent | 54 |
| Trehalose | Unknown; mTOR-independent | 60 |
| SMER10, SMER18, SMER28, SMER analogs | Unknown; mTOR-independent | 50 |
Chemical Inhibitors of mTOR as Autophagy Inducers
At the time we did the first of these studies, the only drug that was known to induce autophagy that was in clinical use (for other indications) was rapamycin. Rapamycin is a highly specific inhibitor of mTOR. The mTOR pathway, which is essential for controlling cell growth, protein synthesis, ribosome biogenesis, nutrient metabolism, and autophagy, involves two functional complexes called mTORC1 and mTORC2 (26). In mammalian cells, rapamycin forms a complex with the immunophilin FKBP12 (FK506-binding protein of 12 kDa), which binds to mTORC1 and inhibits its activity (Fig. 2) (27). However, recent studies have shown that prolonged treatment with rapamycin can inhibit mTORC2 activity in certain mammalian cell types (28, 29). Recently, a selective ATP-competitive small molecule mTOR inhibitor called Torin1 has been found to induce autophagy to a much greater extent than rapamycin (30).
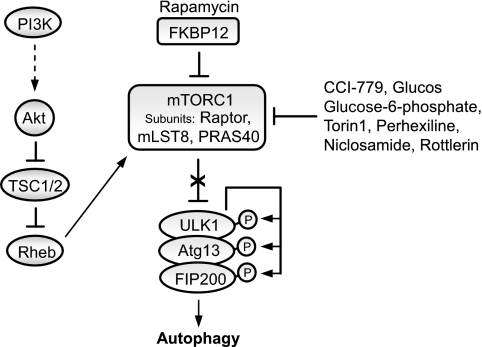
FIGURE 2.
Regulation of autophagy by the mTOR pathway. Autophagy is negatively regulated by mTOR, which is downstream in the phosphatidylinositol 3-kinase (PI3K) pathway. A diverse range of signals, such as growth factors and amino acids, regulates mTORC1 by inhibiting TSC1/2, thereby alleviating the inhibitory effect of TSC1/2 on Rheb, which subsequently activates mTORC1. Several kinases, such as Akt, signal to mTORC1 by phosphorylating TSC2 and inhibiting the activity of the TSC1/2 heterodimer. Rapamycin forms a complex with the immunophilin FKBP12, which inhibits the kinase activity of mTORC1. Inhibition of mTOR by rapamycin induces autophagy and enhances the clearance of mutant aggregate-prone proteins. The ULK1-Atg13-FIP200 complex acts as an integrator of the autophagy signals downstream of mTORC1. Under nutrient-rich conditions, mTORC1 suppresses autophagy by interacting with this complex and mediating phosphorylation-dependent inhibition of Atg13 and ULK1. Treatment with rapamycin dissociates mTOR from the complex, resulting in dephosphorylation-dependent activation of ULK1 and ULK1-mediated phosphorylations of Atg13, FIP200, and ULK1 itself, which triggers autophagy. Other chemical inhibitors of mTORC1 include CCI-779, glucose, glucose 6-phosphate, Torin1, perhexiline, niclosamide, and rottlerin. These may act directly or indirectly.
Recently, Roberge and co-workers (31) reported a study in which they screened a library of 3500 chemicals with an automated cell-based assay to detect increases in autophagosome numbers. The screen identified four compounds (perhexiline, niclosamide, amiodarone, and rottlerin) that stimulated autophagy by inhibiting mTORC1 (but not mTORC2) signaling (Fig. 2). Rottlerin inhibited mTORC1 signaling via TSC2 (tuberous sclerosis complex 2), whereas the other drugs inhibited mTORC1 signaling in a TSC2-independent manner (31). Interestingly, three of the identified compounds (amiodarone, perhexiline, and niclosamide) are drugs already approved for other therapeutic indications, thereby reinforcing the rationale for targeting mTORC1 activity in diseases in which positive modulation of autophagy may be beneficial.
Recent studies have identified some of the molecular components in mammalian autophagy downstream of mTORC1. Rapamycin appears to regulate mammalian autophagy by inhibiting the mTOR-mediated phosphorylation of Atg13 and ULK1, which are involved in autophagosome formation. This leads to dephosphorylation-dependent activation of ULK1 (and ULK2) and ULK1-mediated phosphorylation of Atg13, FIP200, and ULK1 itself, which triggers autophagy (Fig. 2). Thus, the ULK1-Atg13-FIP200 complex appears to integrate the autophagy signals downstream of mTORC1 (32–34). However, it is not yet clear how phosphorylation of these proteins regulates their activities.
Subsequent studies have provided robust support for our assertions using genetic and chemical approaches and suggest that autophagy is important for clearance of mutant huntingtin fragments, at least as large as the first one-third of the protein as well as full-length mutant huntingtin, and that wild-type forms are far less dependent on autophagy for their clearance compared with the mutant forms (17, 18, 24, 35, 36). We showed that rapamycin attenuated mutant huntingtin fragment toxicity in cells and in transgenic Drosophila and mouse models of HD (37). The protective effects of rapamycin were blocked in flies expressing mutant polyalanines or expanded polyQ when the expression of different autophagy genes was reduced (20, 38), suggesting that the major benefits of this drug are autophagy-dependent and not mediated by alternative mechanisms such as impaired translation (at least in these in vivo settings). Our data in cell and fly models show that rapamycin-mediated autophagy up-regulation may be valuable for many other intracellular proteinopathies, including SCA3, and both mutant and wild-type Tau (20). Tau was of particular interest, as it is mutated in certain frontotemporal dementias, and wild-type Tau is the major component of the neurofibrillary tangles that are believed to contribute to pathology in sporadic Alzheimer disease (20). Furthermore, elevated intracellular glucose or glucose 6-phosphate also induces autophagy by inhibiting mTOR (Fig. 2) (39).
An additional benefit of autophagy up-regulation in these diseases is that it appears to protect cells against apoptotic insults (40). Thus, enhancing autophagy may have two beneficial effects in the context of neurodegenerative diseases. First, it enhances removal of the toxic aggregate-prone protein, and second, it protects cells from apoptosis.
The autosomal-dominant proteinopathies that are potentially amenable to autophagy up-regulation present an important opportunity for delaying the onset of disease. Most patients will have a positive family history, and thus, it is possible to identify most cases at risk of developing disease with a simple genetic test (4). Ideally, one would like to start treatment at the earliest possible age in such individuals to aim to delay the onset of disease. For instance, in HD, one would aim to delay onset from a median age of 40 until after normal life expectancy and thus effectively prevent the disease.
One issue that remains unresolved is whether long-term autophagy up-regulation may have deleterious effects. It is important to point out that our mouse studies involved rapamycin administration regimes that were pulsatile (37), and thus, it is very unlikely that autophagy was induced all the time; rather, autophagy would have been induced between periods of normal autophagy. Rapamycin is a drug designed for long-term use, and although it has some side effects in patients and mice due to mTOR inhibition, these do not appear to be mediated by autophagy. Although one may argue that the side effect profile of rapamycin is outweighed by its potential benefits in many of these devastating diseases, it would be desirable to identify compounds that are better tolerated because one may need to treat patients who are at risk for developing these diseases for decades. Hence, we have tried to identify drugs that act independently of mTOR (Table 1).
Chemical Inducers of mTOR-independent Autophagy
The first hint that there may be mTOR-independent pathways controlling autophagy was the discovery that intracellular IP3 levels negatively regulate autophagy (41). We have shown that autophagy can be induced by lowering intracellular inositol or IP3 levels independently of mTOR. Lithium and other mood-stabilizing agents used for treatment of bipolar disorder, such as carbamazepine and sodium valproate, enhances the clearance of autophagy substrates by reducing intracellular inositol levels (Fig. 3) (41, 42). The ability of lithium to induce autophagy is due to inhibition of IMPase, which prevents inositol recycling, leading to depletion of cellular inositol and inhibition of the phosphoinositol cycle (41, 43). Accordingly, the specific IMPase inhibitor L-690,330 mimics the effects of lithium on the clearance of autophagy substrates. Sodium valproate induces autophagy by inhibiting inositol synthesis and decreasing IP3 levels (41, 44). Consistent with a role of IP3 in autophagy, pharmacological inhibition of the IP3R by xestospongin B also induces autophagy (45). It was further shown that xestospongin B induces autophagy by disrupting the IP3R-beclin 1 complex, which can also be modulated by Bcl-2 levels (Fig. 3) (46).
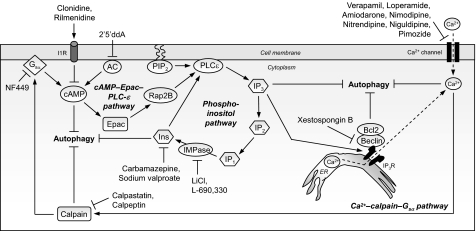
FIGURE 3.
Cyclical mTOR-independent autophagy pathway with multiple drug targets for neurodegenerative diseases. Shown is a cyclical mTOR-independent pathway regulating mammalian autophagy, comprising cAMP-Epac-PLCϵ-IP3 and Ca2+-calpain-Gαs pathways, which has multiple drug targets for neurodegenerative diseases. Intracellular cAMP levels are increased by adenylyl cyclase (AC) activity, thus activating Epac, which then activates the small G-protein Rap2B, thereby activating PLCϵ. PLCϵ mediates the production of IP3 from phosphatidylinositol 4,5-bisphosphate (PIP2), thereby increasing the levels of IP3 that binds to ER-resident IP3Rs, leading to release of Ca2+ from the ER stores. Intracytosolic Ca2+ levels are also increased by L-type Ca2+ channel agonists. Elevated intracytosolic Ca2+ activates calpains, which then cleave and activate Gαs. In turn, activation of Gαs increases adenylyl cyclase activity to elevate cAMP levels, thereby forming a loop. Activation of this pathway inhibits autophagy. Multiple drug targets acting at distinct stages in this pathway trigger autophagy, such as imidazoline-1 receptor (I1R) agonists (clonidine and rilmenidine) and the adenylyl cyclase inhibitor 2′,5′-dideoxyadenosine (2′5′ddA), which decrease cAMP levels; agents that lower inositol (Ins) and IP3 levels (carbamazepine and sodium valproate); IMPase inhibitors that also reduce inositol and IP3 levels (lithium and L-690,330); Ca2+ channel blockers (verapamil, loperamide, amiodarone, nimodipine, nitrendipine, niguldipine, and pimozide); calpain inhibitors (calpastatin and calpeptin); and the Gαs inhibitor NF449. Furthermore, inhibition of the IP3R by xestospongin B also induces autophagy by disrupting the IP3R-beclin 1 complex and, consequently, the Bcl-2-beclin 1 autophagy inhibitory complex. Enhancing autophagy through this mTOR-independent pathway is protective in various models of HD.
Lithium and sodium valproate reduced mutant huntingtin aggregation/toxicity in HD cell models and protected against neurodegeneration in Drosophila models of HD (41, 47, 48). Recently, lithium treatment in ALS patients and mouse models was found to increase survival and attenuate the disease progression (19). All of the ALS patients on lithium treatment for 15 months survived, but ∼30% of the control patients receiving riluzole died (19). Apart from the neuroprotective effects of lithium (49), this fascinating but preliminary result was attributed partly to autophagy up-regulation (19).
To identify novel pathways regulating autophagy that may be relevant to neurodegenerative diseases, we and others have performed chemical screens. Three major screens have been reported so far. The first of these screens started in the yeast Saccharomyces cerevisiae and aimed to identify small molecule chemical modifiers of the growth inhibitory effect of rapamycin (50). Of 50,729 compounds tested in a high-throughput screen, a number of small molecule inhibitors and enhancers (SMERs) of the cytostatic effects of rapamycin were identified and subsequently analyzed in a secondary screen in mammalian cells by analyzing the clearance of A53T mutant α-synuclein (a good autophagy substrate) in the absence of rapamycin as putative modulators of autophagy. We confirmed that SMER10, SMER18, and SMER28 were positive regulators of autophagy acting independently of rapamycin. These SMERs increased autophagosome synthesis and enhanced the clearance of model autophagy substrates such as A53T α-synuclein and mutant huntingtin fragments. Autophagy induced by SMERs was mTOR-independent. Furthermore, these SMERs were protective in a Drosophila model of HD. Further screening of the structural analogs of these three SMERs identified 18 additional small molecules that enhanced the clearance of aggregate-prone proteins (50).
In an attempt to identify novel potential therapeutic agents capable of inducing mTOR-independent autophagy, we screened a library of 253 compounds, comprising FDA-approved drugs and pharmacologically active compounds belonging to different classes with known biological activities, by analyzing their effects on the clearance of known autophagy substrates, such as the A30P and A53T α-synuclein mutants, in a stable inducible PC12 cell line. From the primary screen, we identified several mTOR-independent autophagy enhancers, such as L-type Ca2+ channel antagonists (verapamil, loperamide, amiodarone, nimodipine, and nitrendipine), a K+ATP channel opener (minoxidil), and a Gi-signaling activator (clonidine), whose mechanisms of action were linked in a cyclical manner (Fig. 3) (48). L-type Ca2+ channel antagonists, as well as the K+ATP channel opener, prevent the influx of Ca2+ and decrease intracytosolic Ca2+ levels, leading to inhibition of Ca2+-dependent cysteine proteases called calpains and induction of autophagy. This is consistent with an earlier finding that raised intracytosolic Ca2+ levels impair autophagy (51). Pharmacological inhibition of calpain by calpastatin or calpeptin or calpain knockdown also induces autophagy. Conversely, calpain activation inhibits autophagy by cleaving and activating the α-subunit of heterotrimeric G-proteins (Gαs), resulting in adenylyl cyclase activation and, consequently, cAMP production (48, 52). Reduction in cAMP levels by inhibitors of Gαs (NF449) or adenylyl cyclase (2′,5′-dideoxyadenosine) induces autophagy. Likewise, imidazoline-1 receptor agonists (clonidine and rilmenidine) that also reduce cAMP levels trigger autophagy (48). On the other hand, increased cAMP activates Epac, which in turn activates the small G-protein Rap2B, leading to activation of PLCϵ and, consequently, increased IP3 generation by PLCϵ-mediated hydrolysis of phosphatidylinositol 4,5-bisphosphate (53). Consistent with our previous finding that generation of IP3 inhibits autophagy, inhibition of the cAMP-Epac-Rap2B-PLCϵ-IP3 pathway activates autophagy (48). Regulation of autophagy by intracellular IP3 levels is most likely dependent on it being a signal for ER Ca2+ release, as elevated cytosolic IP3 levels bind the ER-resident IP3Rs to mobilize the ER Ca2+ stores and increase cytosolic Ca2+, which has autophagy inhibitory effects (43, 48). This creates an elaborate mTOR-independent autophagy pathway where Ca2+-calpain-Gαs signaling is linked to cAMP-Epac-PLCϵ-IP3 in a potential cyclical fashion. Among the various autophagy enhancers identified in this screen that reduced huntingtin aggregation/toxicity in HD cell models, verapamil and clonidine were shown to protect against neurodegeneration in HD fly models, and calpastatin, 2′,5′-dideoxyadenosine, verapamil, and clonidine were protective in an HD zebrafish model (48).
Some of the L-type Ca2+ channel antagonists identified in our screen as autophagy inducers were also reported by an independent high-throughput image-based screen with 480 bioactive compounds in which the number of GFP-LC3 vesicles was measured as a readout (54). This screen yielded eight compounds that trigger mTOR-independent autophagy and reduce expanded polyQ aggregates. These include fluspirilene and trifluoperazine (dopamine antagonists); pimozide, niguldipine, amiodarone, and loperamide (Ca2+ channel blockers); and penitrem A (inhibitor of high conductance Ca2+-activated K+ channels), which provide a number of potential therapeutic candidates for neurodegenerative disorders, as most of these compounds were FDA-approved drugs (Fig. 3) (54). Note that amiodarone, identified both in this and our screens, was classified as an mTOR-independent autophagy inducer (48, 54). This contrasts with the recent screen by Roberge and co-workers (31), in which this compound was shown to inhibit mTORC1 at 10–50-fold higher concentrations compared with where it can induce autophagy without inhibiting mTOR (48, 54).
The negative regulation of autophagy by intracytosolic Ca2+ levels was first suggested by Seglen and co-workers (51), consistent with the data from our recent screen (Fig. 3) (48). Autophagy was inhibited with agents that increase intracytosolic Ca2+ levels such as thapsigargin (an ER Ca2+/Mg2+-ATPase inhibitor that releases Ca2+ from ER stores) and ionomycin (a Ca2+ ionophore that releases Ca2+ from intracellular stores) (51). Interestingly, thapsigargin blocks autophagic flux at two stages of the pathway. It increases LC3-II levels and vesicle numbers by impairing autophagosome-lysosome fusion and also reduces autophagosome synthesis by activating calpains (48). Although another study reported an autophagy-inducing effect of thapsigargin, this measured only GFP-LC3 dots rather than autophagic flux, and our data suggested that these GFP-LC3 dots would increase due to impaired autophagosome-lysosome fusion: LC3 vesicle numbers can increase if there is either induction of autophagosome synthesis (increased autophagic flux) or inhibition of autophagosome-lysosome fusion (decreased flux) (55, 56). Furthermore, calcium phosphate precipitates were shown to induce autophagy at early time points but blocked autophagosome-lysosome fusion after longer exposures (57, 58). However, the compartments where these precipitates act in the context of autophagy are not known. Thus, these studies suggest complex roles for Ca2+ in autophagy.
Another chemical screen to identify inhibitors of polyQ-mediated protein aggregation in vitro identified trehalose (a disaccharide) as an inhibitor of mutant huntingtin aggregation, which reduced toxicity in HD cell models and attenuated disease pathology in a mouse model of HD (59). This protective effect of trehalose was suggested to be mediated by its ability to act as a chemical chaperone through its binding to the polyQ-expanded mutant huntingtin and influencing its protein folding and aggregation. However, we have shown that trehalose enhances the autophagy pathway independently of mTOR, thereby increasing the clearance of mutant aggregate-prone proteins (60). Additionally, trehalose protected against pro-apoptotic insults via autophagy (60). The myriad of protective properties of trehalose acting as an autophagy inducer and chemical chaperone, coupled with its lack of toxicity, may be of benefit in the treatment of neurodegenerative disorders.
Additive Effects of mTOR-dependent and mTOR-independent Autophagy Pathways
The existence of mTOR-dependent and mTOR-independent pathways regulating autophagy allows the combined use of different perturbations to increase the autophagic clearance of aggregate-prone proteins. For instance, although lithium induces autophagy in an mTOR-independent manner by inhibiting IMPase, it also inhibits GSK-3β, which activates mTOR (47, 61). This mTOR activation acts to partially inhibit the autophagy-inducing effects of lithium action via IMPase inhibition (47). We have shown that treatment with rapamycin impedes the GSK-3β-dependent activation of mTOR that occurs with the simultaneous treatment with lithium, thereby eliminating the undesirable effects on autophagy resulting from mTOR activation. Combinatorial treatment with rapamycin and lithium enables greater autophagic clearance of mutant huntingtin in HD cell models and exerts a greater protection against the neurodegeneration in HD fly models compared with either treatment alone. Consequently, lithium treatment in an HD fly model with a heterozygous TOR mutation rescued against neurodegeneration to a greater extent than the heterozygous TOR mutation alone (47). Moreover, this strategy may also benefit from the cytoprotective effects of GSK-3β inhibition occurring as a result of lithium treatment due to activation of the β-catenin-T-cell factor pathway, which may serve as an additional protective effect in the context of neurodegenerative diseases in which there are secondary apoptotic insults (47, 62).
We have further shown that simultaneous treatment with rapamycin and other mTOR-independent autophagy inducers, such as trehalose, calpastatin, and the SMERs, results in a greater up-regulation of autophagy than the single treatments alone (48, 50, 60). The combined therapy approach may minimize the side effects arising from these treatments by lowering the required doses of each compound, thereby providing a safer strategy for long-term treatments. Therefore, the use of combination treatment with lower doses not only provides additive mechanisms for enhancing autophagy but also may abrogate undesirable effects resulting from the perturbations of these signaling pathways on their own.
Future Prospects of Autophagy as a Therapeutic Strategy
Recent advances in the field of autophagy have implicated its role in various physiological and pathological conditions such as development, longevity, cancer, and infectious and cardiovascular diseases (10). Indeed, autophagy is a critical pathway that regulates the clearance of diverse intracellular pathogens (63). We have shown that two of the autophagy-inducing SMERs could reduce the number of viable intracellular mycobacteria in primary macrophages in an autophagy-dependent manner, providing evidence for their benefit in infectious diseases as well (64). Thus, chemical inducers of autophagy offer great potential for future studies and could also be utilized in various contexts outside neurodegeneration, either in the treatment of disease conditions in which autophagy serves as a protective pathway or in the investigation of signaling pathways regulating autophagy.
Supplementary Material
This work was supported in part by the Medical Research Council, European Union Framework VI (EUROSCA), and the National Institute for Health Research Biomedical Research Centre at Addenbrooke's Hospital. This is the sixth of six articles in the “Chemical Biology Meets Biological Chemistry Minireview Series.” This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- polyQ
- polyglutamine
- HD
- Huntington disease
- SCA
- spinocerebellar ataxia
- GFP
- green fluorescent protein
- ALS
- amyotrophic lateral sclerosis
- mTOR
- mammalian target of rapamycin
- IP3
- inositol 1,4,5-trisphosphate
- IMPase
- inositol monophosphatase
- IP3R
- IP3 receptor
- SMER
- small molecule enhancer of rapamycin
- PLCϵ
- phospholipase Cϵ
- ER
- endoplasmic reticulum
- GSK-3β
- glycogen synthase kinase-3β.
REFERENCES
- 1.Rubinsztein D. C. (2006) Nature 443, 780–786 [DOI] [PubMed] [Google Scholar]
- 2.Ross C. A., Poirier M. A. (2004) Nat. Med. 10, (suppl.) S10–S17 [DOI] [PubMed] [Google Scholar]
- 3.Zoghbi H. Y., Orr H. T. (2000) Annu. Rev. Neurosci. 23, 217–247 [DOI] [PubMed] [Google Scholar]
- 4.Imarisio S., Carmichael J., Korolchuk V., Chen C. W., Saiki S., Rose C., Krishna G., Davies J. E., Ttofi E., Underwood B. R., Rubinsztein D. C. (2008) Biochem. J. 412, 191–209 [DOI] [PubMed] [Google Scholar]
- 5.Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S. (2004) Nature 431, 805–810 [DOI] [PubMed] [Google Scholar]
- 6.Rubinsztein D. C. (2002) Trends Genet. 18, 202–209 [DOI] [PubMed] [Google Scholar]
- 7.Van Raamsdonk J. M., Pearson J., Rogers D. A., Bissada N., Vogl A. W., Hayden M. R., Leavitt B. R. (2005) Hum. Mol. Genet. 14, 1379–1392 [DOI] [PubMed] [Google Scholar]
- 8.Ravikumar B., Duden R., Rubinsztein D. C. (2002) Hum. Mol. Genet. 11, 1107–1117 [DOI] [PubMed] [Google Scholar]
- 9.Ravikumar B., Futter M., Jahreiss L., Korolchuk V. I., Lichtenberg M., Luo S., Massey D. C., Menzies F. M., Narayanan U., Renna M., Jimenez-Sanchez M., Sarkar S., Underwood B., Winslow A., Rubinsztein D. C. (2009) J. Cell Sci. 122, 1707–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008) Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jahreiss L., Menzies F. M., Rubinsztein D. C. (2008) Traffic 9, 574–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ravikumar B., Acevedo-Arozena A., Imarisio S., Berger Z., Vacher C., O'Kane C. J., Brown S. D., Rubinsztein D. C. (2005) Nat. Genet. 37, 771–776 [DOI] [PubMed] [Google Scholar]
- 13.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Mizushima N. (2006) Nature 441, 885–889 [DOI] [PubMed] [Google Scholar]
- 15.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. (2006) Nature 441, 880–884 [DOI] [PubMed] [Google Scholar]
- 16.Webb J. L., Ravikumar B., Atkins J., Skepper J. N., Rubinsztein D. C. (2003) J. Biol. Chem. 278, 25009–25013 [DOI] [PubMed] [Google Scholar]
- 17.Shibata M., Lu T., Furuya T., Degterev A., Mizushima N., Yoshimori T., MacDonald M., Yankner B., Yuan J. (2006) J. Biol. Chem. 281, 14474–14485 [DOI] [PubMed] [Google Scholar]
- 18.Iwata A., Christianson J. C., Bucci M., Ellerby L. M., Nukina N., Forno L. S., Kopito R. R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13135–13140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fornai F., Longone P., Cafaro L., Kastsiuchenka O., Ferrucci M., Manca M. L., Lazzeri G., Spalloni A., Bellio N., Lenzi P., Modugno N., Siciliano G., Isidoro C., Murri L., Ruggieri S., Paparelli A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2052–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berger Z., Ravikumar B., Menzies F. M., Oroz L. G., Underwood B. R., Pangalos M. N., Schmitt I., Wullner U., Evert B. O., O'Kane C. J., Rubinsztein D. C. (2006) Hum. Mol. Genet. 15, 433–442 [DOI] [PubMed] [Google Scholar]
- 21.Aguib Y., Heiseke A., Gilch S., Riemer C., Baier M., Schätzl H. M., Ertmer A. (2009) Autophagy 5, 361–369 [DOI] [PubMed] [Google Scholar]
- 22.Heiseke A., Aguib Y., Riemer C., Baier M., Schätzl H. M. (2009) J. Neurochem. 109, 25–34 [DOI] [PubMed] [Google Scholar]
- 23.Cuervo A. M., Stefanis L., Fredenburg R., Lansbury P. T., Sulzer D. (2004) Science 305, 1292–1295 [DOI] [PubMed] [Google Scholar]
- 24.Sarkar S., Ravikumar B., Floto R. A., Rubinsztein D. C. (2009) Cell Death Differ. 16, 46–56 [DOI] [PubMed] [Google Scholar]
- 25.Sarkar S., Ravikumar B., Rubinsztein D. C. (2009) Methods Enzymol. 453, 83–110 [DOI] [PubMed] [Google Scholar]
- 26.Guertin D. A., Sabatini D. M. (2009) Sci. Signal. 2, pe24. [DOI] [PubMed] [Google Scholar]
- 27.Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 28.Sarbassov D. D., Ali S. M., Sengupta S., Sheen J. H., Hsu P. P., Bagley A. F., Markhard A. L., Sabatini D. M. (2006) Mol. Cell 22, 159–168 [DOI] [PubMed] [Google Scholar]
- 29.Zeng Z., Sarbassov D. D., Samudio I. J., Yee K. W., Munsell M. F., Jackson C. E., Giles F. J., Sabatini D. M., Andreeff M., Konopleva M. (2007) Blood 109, 3509–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thoreen C. C., Kang S. A., Chang J. W., Liu Q., Zhang J., Gao Y., Reichling L. J., Sim T., Sabatini D. M., Gray N. S. (2009) J. Biol. Chem. 284, 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balgi A. D., Fonseca B. D., Donohue E., Tsang T. C., Lajoie P., Proud C. G., Nabi I. R., Roberge M. (2009) PloS One 4, e7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ganley I. G., Lam D. H., Wang J., Ding X., Chen S., Jiang X. (2009) J. Biol. Chem. 284, 12297–12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N., Guan J. L., Oshiro N., Mizushima N. (2009) Mol. Biol. Cell 20, 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung C. H., Jun C. B., Ro S. H., Kim Y. M., Otto N. M., Cao J., Kundu M., Kim D. H. (2009) Mol. Biol. Cell 20, 1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin Z. H., Wang Y., Kegel K. B., Kazantsev A., Apostol B. L., Thompson L. M., Yoder J., Aronin N., DiFiglia M. (2003) Hum. Mol. Genet. 12, 3231–3244 [DOI] [PubMed] [Google Scholar]
- 36.Rubinsztein D. C., Gestwicki J. E., Murphy L. O., Klionsky D. J. (2007) Nat. Rev. Drug Discov. 6, 304–312 [DOI] [PubMed] [Google Scholar]
- 37.Ravikumar B., Vacher C., Berger Z., Davies J. E., Luo S., Oroz L. G., Scaravilli F., Easton D. F., Duden R., O'Kane C. J., Rubinsztein D. C. (2004) Nat. Genet. 36, 585–595 [DOI] [PubMed] [Google Scholar]
- 38.Pandey U. B., Nie Z., Batlevi Y., McCray B. A., Ritson G. P., Nedelsky N. B., Schwartz S. L., DiProspero N. A., Knight M. A., Schuldiner O., Padmanabhan R., Hild M., Berry D. L., Garza D., Hubbert C. C., Yao T. P., Baehrecke E. H., Taylor J. P. (2007) Nature 447, 859–863 [DOI] [PubMed] [Google Scholar]
- 39.Ravikumar B., Stewart A., Kita H., Kato K., Duden R., Rubinsztein D. C. (2003) Hum. Mol. Genet. 12, 985–994 [DOI] [PubMed] [Google Scholar]
- 40.Ravikumar B., Berger Z., Vacher C., O'Kane C. J., Rubinsztein D. C. (2006) Hum. Mol. Genet. 15, 1209–1216 [DOI] [PubMed] [Google Scholar]
- 41.Sarkar S., Floto R. A., Berger Z., Imarisio S., Cordenier A., Pasco M., Cook L. J., Rubinsztein D. C. (2005) J. Cell Biol. 170, 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams R. S., Cheng L., Mudge A. W., Harwood A. J. (2002) Nature 417, 292–295 [DOI] [PubMed] [Google Scholar]
- 43.Berridge M. J. (1993) Nature 361, 315–325 [DOI] [PubMed] [Google Scholar]
- 44.Shaltiel G., Shamir A., Shapiro J., Ding D., Dalton E., Bialer M., Harwood A. J., Belmaker R. H., Greenberg M. L., Agam G. (2004) Biol. Psychiatry 56, 868–874 [DOI] [PubMed] [Google Scholar]
- 45.Criollo A., Maiuri M. C., Tasdemir E., Vitale I., Fiebig A. A., Andrews D., Molgó J., Díaz J., Lavandero S., Harper F., Pierron G., di Stefano D., Rizzuto R., Szabadkai G., Kroemer G. (2007) Cell Death Differ. 14, 1029–1039 [DOI] [PubMed] [Google Scholar]
- 46.Vicencio J. M., Ortiz C., Criollo A., Jones A. W., Kepp O., Galluzzi L., Joza N., Vitale I., Morselli E., Tailler M., Castedo M., Maiuri M. C., Molgó J., Szabadkai G., Lavandero S., Kroemer G. (2009) Cell Death Differ. 16, 1006–1017 [DOI] [PubMed] [Google Scholar]
- 47.Sarkar S., Krishna G., Imarisio S., Saiki S., O'Kane C. J., Rubinsztein D. C. (2008) Hum. Mol. Genet. 17, 170–178 [DOI] [PubMed] [Google Scholar]
- 48.Williams A., Sarkar S., Cuddon P., Ttofi E. K., Saiki S., Siddiqi F. H., Jahreiss L., Fleming A., Pask D., Goldsmith P., O'Kane C. J., Floto R. A., Rubinsztein D. C. (2008) Nat. Chem. Biol. 4, 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rowe M. K., Chuang D. M. (2004) Expert Rev. Mol. Med. 6, 1–18 [DOI] [PubMed] [Google Scholar]
- 50.Sarkar S., Perlstein E. O., Imarisio S., Pineau S., Cordenier A., Maglathlin R. L., Webster J. A., Lewis T. A., O'Kane C. J., Schreiber S. L., Rubinsztein D. C. (2007) Nat. Chem. Biol. 3, 331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gordon P. B., Holen I., Fosse M., Røtnes J. S., Seglen P. O. (1993) J. Biol. Chem. 268, 26107–26112 [PubMed] [Google Scholar]
- 52.Sato-Kusubata K., Yajima Y., Kawashima S. (2000) Biochem. J. 347, 733–740 [PMC free article] [PubMed] [Google Scholar]
- 53.Kelley G. G., Reks S. E., Ondrako J. M., Smrcka A. V. (2001) EMBO J. 20, 743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang L., Yu J., Pan H., Hu P., Hao Y., Cai W., Zhu H., Yu A. D., Xie X., Ma D., Yuan J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19023–19028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rubinsztein D. C., Cuervo A. M., Ravikumar B., Sarkar S., Korolchuk V., Kaushik S., Klionsky D. J. (2009) Autophagy 5, 585–589 [DOI] [PubMed] [Google Scholar]
- 56.Høyer-Hansen M., Bastholm L., Szyniarowski P., Campanella M., Szabadkai G., Farkas T., Bianchi K., Fehrenbacher N., Elling F., Rizzuto R., Mathiasen I. S., Jäättelä M. (2007) Mol. Cell 25, 193–205 [DOI] [PubMed] [Google Scholar]
- 57.Gao W., Ding W. X., Stolz D. B., Yin X. M. (2008) Autophagy 4, 754–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sarkar S., Korolchuk V., Renna M., Winslow A., Rubinsztein D. C. (2009) Autophagy 5, 307–313 [DOI] [PubMed] [Google Scholar]
- 59.Tanaka M., Machida Y., Niu S., Ikeda T., Jana N. R., Doi H., Kurosawa M., Nekooki M., Nukina N. (2004) Nat. Med. 10, 148–154 [DOI] [PubMed] [Google Scholar]
- 60.Sarkar S., Davies J. E., Huang Z., Tunnacliffe A., Rubinsztein D. C. (2007) J. Biol. Chem. 282, 5641–5652 [DOI] [PubMed] [Google Scholar]
- 61.Inoki K., Ouyang H., Zhu T., Lindvall C., Wang Y., Zhang X., Yang Q., Bennett C., Harada Y., Stankunas K., Wang C. Y., He X., MacDougald O. A., You M., Williams B. O., Guan K. L. (2006) Cell 126, 955–968 [DOI] [PubMed] [Google Scholar]
- 62.Carmichael J., Sugars K. L., Bao Y. P., Rubinsztein D. C. (2002) J. Biol. Chem. 277, 33791–33798 [DOI] [PubMed] [Google Scholar]
- 63.Deretic V., Levine B. (2009) Cell Host Microbe 5, 527–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Floto R. A., Sarkar S., Perlstein E. O., Kampmann B., Schreiber S. L., Rubinsztein D. C. (2007) Autophagy 3, 620–622 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.