Abstract
TDP-43 (43-kDa TAR DNA-binding protein) is a major constituent of ubiquitin-positive cytosolic aggregates present in neurons of patients with amyotrophic lateral sclerosis (ALS) and ubiquitin-positive fronto-temporal lobar degeneration (FTLD-U). Inherited mutations in TDP-43 have been linked to familial forms of ALS, indicating a key role for TDP-43 in disease pathogenesis. Here, we describe a Drosophila melanogaster model of TDP-43 proteinopathy. Expression of wild-type human TDP-43 protein in Drosophila motor neurons led to motor dysfunction and dramatic reduction of life span. Interestingly, coexpression of ubiquilin 1, a previously identified TDP-43-interacting protein with suspected functions in autophagy and proteasome targeting, reduced steady-state TDP-43 expression but enhanced the severity of TDP-43 phenotypes. Finally, ectopically expressed TDP-43 was largely localized to motor neuron nuclei, suggesting that expression of wild-type TDP-43 alone is detrimental even in the absence of cytosolic aggregation. Our findings demonstrate that TDP-43 exerts cell-autonomous neurotoxicity in Drosophila and further imply that dose-dependent alterations of TDP-43 nuclear function may underlie motor neuron death in ALS.
Keywords: Aging, Diseases/Neurodegeneration, Genetics/Drosophila, Neurobiology/Neuroscience, Proteases/Proteasomes, Protein/Folding, RNA/Processing
Introduction
Intracellular aggregates containing misfolded or improperly modified proteins are a hallmark of many neurodegenerative diseases (1). One such disease is amyotrophic lateral sclerosis (ALS),3 which targets spinal motor neurons that control voluntary movement. ALS carries a cumulative lifetime risk of 1 in 1,000 and is fatal, leading to respiratory failure within 3–5 years (2). Although extensively studied, the pathogenesis of ALS is not well understood, and consequently there are no effective treatments to slow the course of this disease.
The majority of ALS cases are sporadic, but ∼10% of cases are familial (fALS) and have a clear genetic cause (2). The most common genetic abnormalities leading to fALS are dominant mutations in superoxide dismutase 1 (SOD1), with over 100 mutations identified to date (3). SOD1 has therefore been extensively studied, and a mouse model of SOD1-induced ALS is the major tool used to test potential therapeutics (4). However, SOD1 mutations account for ∼2% of all ALS cases, and thus far, therapeutic strategies developed in SOD1 rodent models have not met with success in the clinical setting (5).
Recently, Neumann et al. (6) identified the 43-kDa TAR DNA-binding protein (TDP-43) as a common constituent of cytosolic inclusions in patients with both ALS and a related disorder, ubiquitin-positive fronto-temporal lobar degeneration (FTLD-U). TDP-43 is highly conserved across species, ubiquitously expressed, and localizes exclusively to the nucleus under normal conditions (7). Consistent with its nucleic acid binding ability, TDP-43 has been implicated in the regulation of gene transcription and mRNA splicing (8–10). In patients with ALS and FTLD-U, TDP-43 is mislocalized to the cytosol, where hyperphosphorylated, ubiquitylated, and cleaved forms have all been detected in insoluble aggregates (6, 11–13). Furthermore, a subset of fALS cases has been linked to dominant mutations in the gene encoding TDP-43, TARDBP, with at least 25 different mutations identified to date (14). Interestingly, TARDBP mutations have recently been found in sporadic ALS cases as well (15, 16). Combined with the occurrence of TDP-43 aggregates in ALS and FTLD-U patients even in the absence of such mutations, these data strongly suggest that these related conditions are caused by TDP-43 proteinopathy.
We recently identified ubiquilin 1 (UBQLN) as a TDP-43 binding partner (17). UBQLN is a ubiquitously expressed cytosolic protein that is believed to function primarily in targeting misfolded proteins to the proteasome for degradation (18) and has been linked to Huntington and Alzheimer disease (19, 20). When overexpressed in mammalian cells, TDP-43 and UBQLN colocalize in cytosolic aggregates that strongly overlap with LC3-positive autophagosomes (17). Combined with recent evidence suggesting a broad role for UBQLN in the unfolded protein response (UPR), these findings indicate that UBQLN may be involved in delivering TDP-43 to the proteasome and/or autophagosome for degradation (21). However, the contribution of UBQLN to TDP-43 proteostasis and toxicity in vivo is unknown.
The objectives of this study were to develop a Drosophila model of TDP-43 proteinopathy and to determine the effect of UBQLN on TDP-43 toxicity. Expression of human TDP-43 in Drosophila motor neurons led to a dose-dependent reduction of life span. UBQLN coexpression reduced steady-state TDP-43 levels but unexpectedly increased the severity of TDP-43 phenotypes. Furthermore, TDP-43-dependent neurodegeneration occurred in the absence of aggregation. We propose that changes in gene expression and/or splicing due to alterations in TDP-43 nuclear gene dosage are responsible for pathologic motor neuron death in this model of ALS.
EXPERIMENTAL PROCEDURES
Fly Maintenance
Flies were maintained and all crosses were performed at 25 °C unless otherwise indicated. To create transgenic lines, human TDP-43 and UBQLN cDNAs were subcloned into the pUAST vector. Injection of plasmid into the w1118 strain was performed by Rainbow Technologies, Inc., and transformants were selected and balanced using standard methods. The lines generated were: UAS-TDP-43L1/CyO, UAS-TDP-43L2/TM3, UAS-TDP-43L3/TM3, UAS-UBQLNL1/CyO, and UAS-UBQLNL2/TM3. The GMR-Gal4 and D42-Gal4 driver lines were obtained from the Bloomington Drosophila Stock Center. The UBQLNRNAi line was generously provided by Dr. Ming Guo (22).
Antibodies and Protein Expression
The following antibodies were used in this study: α-TDP-43 (Proteintech), α-UBQLN (Zymed Laboratories Inc.), α-elav (Developmental Studies Hybridoma Bank), goat α-rabbit Alexa Fluor 488, and goat α-mouse Alexa Fluor 568 (Molecular Probes). To examine relative protein expression, transgenic lines were crossed to the GMR-Gal4 driver. Heads were homogenized in high salt lysis buffer (25 mm HEPES, pH 7.4, 300 mm NaCl, 1.5 mm MgCl2, 1 mm EGTA). Proteins were resolved by SDS-PAGE using standard methods, and Western blotting was then performed using α-TDP-43 (1:2000), α-UBQLN (1:1000), or α-elav (1:1000) antibodies. Quantification of Western blots was performed using ImageJ (23). For cellular fractionation, heads were first homogenized under low salt conditions (10 mm HEPES, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 0.5 mm dithiothreitol) and centrifuged at 20,000 × g to separate the cytosolic fraction from nuclear/insoluble material. The pellet was then treated with high salt buffer to lyse nuclei, and a second round of centrifugation at 20,000 × g separated the nuclear fraction from the insoluble fraction.
Microscopy
For immunostaining of Drosophila motor neurons, D42-Gal4-driven transgenic flies were dissected to isolate the thorax and abdomen. The tissue was incubated overnight in 4% paraformaldehyde, washed with phosphate-buffered saline (pH 7.4), and incubated overnight in 30% sucrose. Samples were placed in cryostat molds in OCT medium (Tissue-Tek) and frozen on dry ice. Blocks were cut in 7-μm sections on a Leica Cryocut 1800 cryostat. Sections were immunostained using standard methods. Antibody concentrations used were: α-TDP-43 (1:50), α-UBQLN (1:50), α-mouse-Alexa 568 (1:500), and α-rabbit-Alexa 488 (1:500). To ensure that sections were at the right depth to visualize the thoracic ganglia, this protocol was optimized using flies expressing green fluorescent protein in all neurons. Fluorescence microscopy was performed using a Zeiss Axiovert 200M fluorescence microscope. Transmission electron microscopy (TEM) of ommatidia was performed as described previously (24).
Survival Analysis
Flies were kept at 25 °C with no more than 15 flies per vial. Vials were changed every 3–5 days. Flies were considered dead when they no longer moved in response to tapping the side of the vial. For statistical analysis, a Cox regression was performed using the “survival” package available for the statistical program R (25, 26). All genotypes were included in the fit of the data. Statistical analysis found negligible differences between genders (data not shown); therefore, all data shown include both male and female flies.
RESULTS AND DISCUSSION
Overexpression of TDP-43 Is Toxic in the Fly Eye
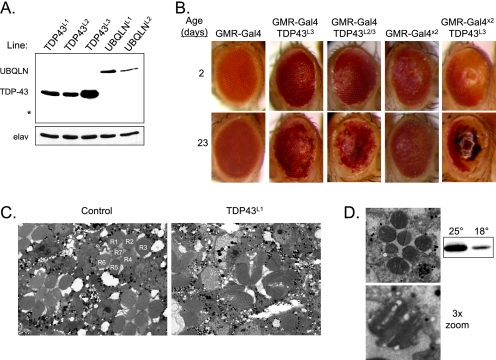
To test the consequences of the TDP-43/UBQLN interaction in vivo, we created transgenic Drosophila lines using cDNAs encoding either wild-type human TDP-43 or human UBQLN. Expression of these genes was under control of the upstream activating sequence (UAS) promoter, which is specifically activated by the yeast transcription factor Gal4. By crossing UAS-TDP-43 and UAS-UBQLN transgenic flies to driver lines that express Gal4 in a tissue-specific manner, TDP-43 and UBQLN protein were expressed only in the tissue of interest. Due to differences in genomic insertion sites, the lines expressed the transgenes to varying degrees. Of the three TDP-43 lines used in this study (designated TDP43L1, TDP43L2, and TDP43L3), TDP43L1 and TDP43L2 expressed the protein at similar levels, whereas TDP43L3 had more robust expression (Fig. 1A). Similarly, of the two UBQLN lines used (designated UBQLNL1 and UBQLNL2), UBQLNL1 expressed at higher levels.
FIGURE 1.
TDP-43 toxicity in the eye. A, Western blot analysis of transgenic protein expression in the TDP-43 and UBQLN lines used in this study, using the GMR-Gal4 driver. Each lane contains lysate from a single head. * indicates the expected size of TDP-43 cleavage products seen in cell culture (supplemental Fig. 2C). The neuronal marker elav is used as a loading control. B, external eye phenotype of control and TDP-43-expressing flies, using the indicated transgene combinations. Where possible, the same eye is shown for each genotype at age 2 and 23 days. C, TEM images of ommatidia from 7-day-old GMR-Gal4 and GMR-TDP43L1 flies; the individual photoreceptors of an intact ommatidium are labeled R1–R7. D, TEM image of an ommatidium from a GMR-TDP43L1 fly reared at 18 °C; a single rhabdomere is shown in the lower panel. Western blot (right) indicates relative TDP-43 expression at 18 °C as compared with 25 °C.
To determine whether human TDP-43 is toxic to Drosophila cells in vivo, we first expressed TDP-43 in the fly eye, using the GMR-Gal4 driver. Interestingly, lines TDP43L1 and TDP43L2 had no effect on external eye phenotype (data not shown). However, GMR-TDP43L3 flies exhibited depigmentation of the eye that increased with age (Fig. 1B). This is likely due to the death of retinal pigment cells. To further investigate TDP-43 toxicity, we used TEM to visualize the neuronal photoreceptors of eyes from GMR-TDP43L1 flies. In these flies, the regular pattern of seven rhabdomeres seen in control flies was severely disrupted (Fig. 1C), indicating that although no external phenotype was observed in these lines, TDP-43 is nevertheless toxic at the cellular level. The lack of a gross phenotype in lines TDP43L1 and TDP43L2 is therefore probably due to lower expression as compared with TDP43L3. These findings indicate that the toxicity of overexpressed TDP-43 in Drosophila is both dose-dependent and age-dependent.
We next sought to either increase or decrease TDP-43 expression to further test the hypothesis that toxicity is dose-dependent. To increase expression, we crossed GMR-TDP43L3 flies to either TDP43L2 or GMR-Gal4 flies; the former increases expression due to an increase in the number of TDP-43 transgenes, and the latter increases expression due to an increase in Gal4 driver expression. Both crosses yielded flies with increased depigmentation of the eye (Fig. 1B), with the most severe phenotype leading to almost no pigment and widespread necrosis. To decrease expression, we reared GMR-TDP43L1 flies at 18 °C instead of 25 °C, which reduces expression from the transgene due to heat shock elements in the promoter. In these flies, the normal structure of the ommatidia was restored; however, some degeneration was still present, as seen by the presence of vacuoles in the rhabdomeres (Fig. 1D). These data suggest that phenotypic severity is highly dependent on TDP-43 dose.
TDP-43 Expression in Motor Neurons Reduces Life Span
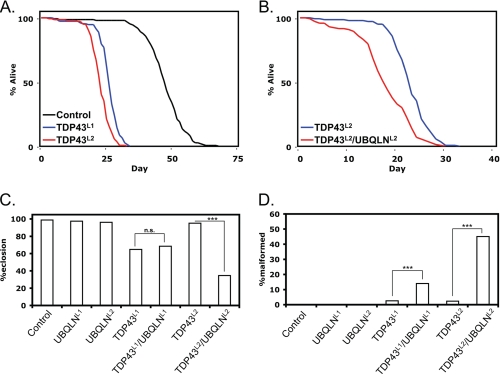
To more closely model ALS, we used a motor neuron-specific driver, D42-Gal4, to express TDP-43 exclusively in motor neurons. Interestingly, D42-TDP43L1 and D42-TDP43L2 lines were viable and phenotypically normal at eclosion. However, within 2–3 weeks, they displayed movement defects and ultimately paralysis, resulting in death within days of the onset of initial symptoms. Analysis of survival curves for D42-TDP43L1 and D42-TDP43L2 flies revealed that the TDP-43 transgenic lines survived approximately half as long as controls (Fig. 2A). Finally, no TDP43L3 flies survived past the second instar larval stage, further emphasizing the dose dependence of TDP-43 toxicity.
FIGURE 2.
UBQLN exacerbates TDP-43 motor neuron phenotypes. A, survival curves of D42-Gal4 control, D42-TDP43L1, and D42-TDP43L2 flies. B, survival curves of D42-TDP43L2 and D42-TDP43L2/UBQLNL2 flies. See supplemental Table 1 for statistical analysis. C, quantification of pupal eclosion defects seen with TDP-43 and UBQLN transgenic flies. >300 pupae were scored for each genotype; both a complete lack of eclosion and incomplete eclosion were counted as eclosion failure. n.s., not significant. D, quantification of wing defects seen with TDP-43 and UBQLN transgenic flies. >200 adult flies were scored for each genotype. ***, p < 0.001. p values were calculated using the chi square statistic.
As controls, we also examined the effect of UBQLN overexpression in motor neurons on survival. In addition to our two transgenic lines, we tested a third line (UBQLNRNAi) that knocks down the endogenous Drosophila UBQLN homolog in the tissue of interest via expression of a short hairpin RNA (22). As compared with control flies, D42-UBQLNL1 flies demonstrated slightly increased survival, D42-UBQLNL2 flies were no different, and D42-UBQLNRNAi demonstrated slightly reduced survival (supplemental Fig. 1A).
UBQLN Exacerbates TDP-43 Motor Neuron Phenotypes
We next sought to use the survival phenotype to determine the effect of UBQLN manipulation on TDP-43 toxicity in vivo. To do this, we created the following combinations of transgenes, all under the control of the D42 motor neuron-specific driver: TDP43L1/UBQLNL1, TDP43L1/UBQLNRNAi, and TDP43L2/UBQLNL2. The third combination allows for the simplest analysis because survival of D42-UBQLNL2 flies was similar to control flies (supplemental Fig. 1A). Surprisingly, D42-TDP43L2/UBQLNL2 flies had significantly reduced survival as compared with D42-TDP43L2 flies (Fig. 2B and supplemental Table 1). Similarly, D42-TDP43L1/UBQLNL1 flies also displayed a significant reduction in survival relative to that expected based on the TDP43L1 and UBQLNL1 survival curves (supplemental Fig. 1B and supplemental Table 1). Therefore, in both cases, UBQLN coexpression in motor neurons decreased the survival of TDP-43 flies. Finally, we found that D42-TDP43L1/UBQLNRNAi flies did not have significantly different survival relative to that expected based on the TDP43L1 and UBQLNRNAi survival curves (supplemental Fig. 1C and supplemental Table 1).
In addition to having reduced survival, D42-TDP-43 flies also exhibited eclosion defects (Fig. 2C). As well as complete eclosion failure, a number of flies died during eclosion with part of their bodies protruding from the pupal case (supplemental Fig. 1D). In both D42-TDP43L1 and D42-TDP43L1/UBQLNL1 pupae, the rate of eclosion was ∼65%. Interestingly, although D42-TDP43L2 pupae did not show an eclosion defect, D42-TDP43L2/UBQLNL2 pupae eclosed at a rate of only 35%. In this assay, therefore, UBQLN coexpression strongly exacerbates TDP-43 toxicity. Furthermore, given that only about one-third of D42-TDP43L2/UBQLNL2 pupae became adults, the reduced survival seen in this line likely underestimated the increase in toxicity seen with UBQLN coexpression. Finally, possibly as a result of difficult eclosion, some D42-TDP-43 flies had a shriveled wing phenotype (supplemental Fig. 1D). Both D42-TDP43L1/UBQLNL1 and D42-TDP43L2/UBQLNL2 flies had higher rates of wing malformation than the respective single TDP-43 transgenic lines, further emphasizing the increase in toxicity with both genes (Fig. 2D).
Additionally, we asked whether UBQLN had the same effect on TDP-43 phenotypes in mammalian cells. Coexpression of either full-length UBQLN or its ubiquitin-associated (UBA) domain, which was previously found to be responsible for TDP-43 binding (17), significantly increased TDP-43 toxicity in HeLa cells (supplemental Fig. 2A). Although this may be an artifact of overexpression, these findings suggest that UBQLN increases TDP-43 toxicity in both Drosophila and mammalian systems.
UBQLN Reduces TDP-43 Protein Levels and Does Not Coaggregate with TDP-43 in the Cytoplasm
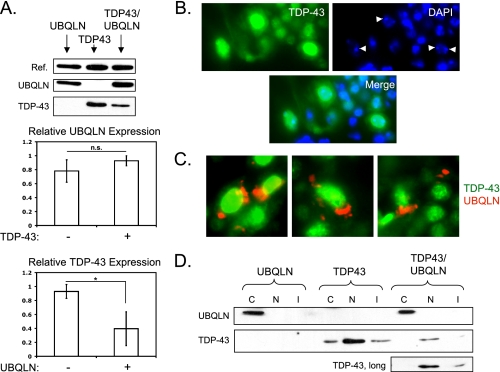
Our initial prediction was that UBQLN would attenuate TDP-43-dependent phenotypes by promoting clearance of misfolded and/or aggregated TDP-43 proteins; however, expression of UBQLN worsened motor neuron TDP-43 phenotypes. To test whether phenotypic exacerbation was due to increased TDP-43 protein upon UBQLN coexpression, we looked at the relative level of protein expression in flies expressing UBQLN, TDP-43, or both. We utilized the less toxic TDP43L2 line at a very early age to ensure that differences in protein levels were not due to different rates of cell loss. Interestingly, we found that although UBQLN levels were not changed by TDP-43 coexpression, TDP-43 levels were significantly reduced when coexpressed with UBQLN (Fig. 3A). This finding excludes increased TDP-43 expression as an explanation for worsened phenotypes in TDP-43/UBQLN transgenic flies. The results are, however, consistent with the hypothesis that UBQLN participates in the proteasomal and/or autophagosomal degradation of TDP-43.
FIGURE 3.
UBQLN reduces the expression of, but does not coaggregate with, TDP-43 in vivo. A, upper, protein expression levels in GMR-UBQLNL2 (left), GMR-TDP43L2 (center), and GMR-TDP43L2/UBQLNL2 (right) flies. The reference band (Ref.) is a nonspecific band detected with α-TDP-43 antibody. Middle, quantification of three experiments showing the relative ratio of UBQLN to the reference band with and without TDP-43 coexpression. Lower, quantification of three experiments showing the relative ratio of TDP-43 to the reference band with and without UBQLN coexpression. Error bars represent S.D. *, p < 0.05. p values were calculated using Student's t test. n.s., not significant. B, localization of TDP-43 in motor neurons of D42-TDP43L1 transgenic flies. Tissue sections were immunostained using α-TDP-43 (green) antibody and 4′,6-diamidino-2-phenylindole (DAPI) (blue). Arrowheads indicate condensed chromatin. C, localization of TDP-43 and UBQLN in motor neurons of D42-TDP43L1/UBQLNL1 transgenic flies. Tissue sections were immunostained using α-TDP-43 (green) and α-UBQLN (red) antibodies. D, Western blot analysis of TDP-43 and UBQLN localization in transgenic flies after cellular fractionation, using GMR-TDP43L1, GMR-UBQLNL1, and GMR-TDP43L1/UBQLNL1 flies. C = cytosolic; N = nuclear; I = insoluble.
TDP-43 is localized to cytosolic aggregates in motor neurons of ALS patients (6), and overexpression of UBQLN stimulated TDP-43 aggregation in mammalian cell lines (17). Thus, another plausible explanation for increased phenotypic severity in TDP-43/UBQLN transgenic flies is that UBQLN stimulates toxic TDP-43 aggregation in motor neurons. This idea was supported by the finding that TDP-43 and UBQLN colocalized to cytosolic aggregates when coexpressed in Drosophila S2 cells (supplemental Fig. 2B). To assess the localization patterns of TDP-43 and UBQLN in vivo, we isolated thoracic ganglia from D42-driven transgenic flies and stained cryosections of the tissue with α-TDP-43 and α-UBQLN antibodies. Somewhat unexpectedly, we found that TDP-43 was detected only in motor neuron nuclei, irrespective of UBQLN coexpression (Fig. 3, B and C). Although UBQLN formed distinct cytosolic structures, similar to those observed in cell culture, TDP-43 failed to strongly localize to the cytosol.
Cellular fractionation and Western blotting of lysates from GMR-TDP43 flies confirmed that the majority of TDP-43 localized to the nucleus in vivo, although a small amount was found in the cytosolic and insoluble fractions (Fig. 3D). Interestingly, coexpression of UBQLN reduced the amount of cytosolic TDP-43 such that it was undetectable even at higher exposures. This finding also suggests that UBQLN promotes the clearance of cytosolic TDP-43, even in the absence of coaggregation of the two proteins. In addition, although cleavage products of TDP-43 were clearly observed in extracts of transfected S2 cells (supplemental Fig. 2C), only the full-length form of TDP-43 was seen in vivo (Fig. 1A). These findings suggest that TDP-43 toxicity and reduction of life span do not require the cleavage or cytosolic aggregation of the protein in Drosophila motor neurons.
In this study, we described a Drosophila model for ALS in which the expression of wild-type human TDP-43 in neurons caused dose-dependent motor defects and severe reduction of life span. TDP-43-associated phenotypes ranged from photoreceptor degeneration and shortened life span in low expressing lines to gross disruption of eye architecture and larval lethality in high expressing lines. When combined with a recent report showing that deletion of the Drosophila TDP-43 gene causes a similar reduction of life span (27), our results suggest that motor neurons are exquisitely sensitive to the dose of TDP-43 and that too much or too little expression of the protein is deleterious.
Our data indicate that overexpression of wild-type TDP-43 in Drosophila motor neurons is sufficient for cell-autonomous toxicity in the absence of ALS-associated mutations or aggregation. The latter observation is congruent with a recent report that mutant TDP-43, when overexpressed in mice, causes neurodegeneration in the absence of cytosolic aggregation (28). Given that the majority of TDP-43 is found in the nucleus, we propose that one component of TDP-43 toxicity in this model could be due to changes in gene expression, splicing, or other processes regulated by TDP-43. Nuclear dysfunction due to TDP-43 expression is suggested by the observation that the nuclei of TDP-43-positive motor neurons contained abnormally condensed chromatin (Fig. 3B). Alterations in nuclear processes may arise due to the total dose of human and fly TDP-43 exceeding a certain threshold, above which the protein has a toxic gain-of-function. Alternatively, wild-type human TDP-43 may dominantly interfere with the endogenous Drosophila homolog, the functional knockdown of which has been found to alter expression of specific mRNAs (29) and result in reduced dendritic branching (30).
Importantly, we do see a small amount of TDP-43 in the cytosol, but the absence of aggregation suggests either that protein levels are not sufficiently high or that cytosolic TDP-43 is efficiently handled by the UPR and does not accumulate sufficiently to form insoluble aggregates. However, we cannot rule out the possibility that small aggregates, below the sensitivity of our assays, are present in Drosophila motor neurons; furthermore, the lack of aggregation in our model does not rule out a role for TDP-43 aggregation in ALS pathogenesis.
We hypothesized that overexpression of UBQLN, which shuttles ubiquitylated substrates to the proteasome and regulates autophagy, would phenotypically rescue TDP-43 transgenic flies. In support of this idea, UBQLN coexpression reduced TDP-43 protein levels in vivo. Surprisingly, however, coexpression of UBQLN significantly worsened TDP-43-associated phenotypes in motor neurons. Phenotypic worsening by UBQLN was not a consequence of additive toxic effects; in fact, UBQLN expression alone in motor neurons actually extended Drosophila life span (supplemental Fig. 1A). What, then, is the explanation for this paradox? Although definitive evidence is still lacking, we propose that UBQLN-TDP-43 complexes compete with endogenous UPR substrates for a limited number of proteasome and autophagosome binding sites. Endogenous UPR substrates may then accumulate and cause proteostatic stress that exacerbates TDP-43-dependent motor neuron degeneration. Importantly, there is precedent for a similar model in the literature. Korolchuk et al. (31) showed that overexpression of p62, a protein structurally and functionally related to UBQLN, led to inhibition of the UPR in the presence of high levels of misfolded huntingtin protein. Therefore, in addition to nuclear dysregulation, another component of TDP-43 toxicity may be due to inhibition of the UPR and other cytosolic processes, although the majority of the protein remains in the nucleus.
In conclusion, we have developed an in vivo model of TDP-43 toxicity that recapitulates aspects of motor neuron degeneration observed in ALS. Importantly, we found that aggregation and cleavage of the protein were not required for TDP-43 toxicity in Drosophila. This model will be a powerful tool for identifying drugs and genes that modify TDP-43-dependent neurodegeneration and that may have therapeutic benefit in ALS and related disorders.
Supplementary Material
Acknowledgments
We thank Satoshi Kinoshita and Ben August for help with tissue sectioning and Dr. Bryan Hanson for help with statistical analysis.
Note Added in Proof
Li et al. have recently reported that TDP-43 transgenic flies undergo neurodegeneration (Li, Y. et al. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3169–3174).
This work was supported, in whole or in part, by National Institutes of Health Grants CA124722 (to R. S. T.) and NS059001 (to D. A. W.). This work was also supported by grants from the American Cancer Society and a Shaw Scientist Award from the Greater Milwaukee Foundation (to R. S. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1, supplemental Experimental Procedures, and supplemental Figs. 1 and 2.
- ALS
- amyotrophic lateral sclerosis
- fALS
- familial ALS
- TDP-43
- 43-kDa TAR DNA-binding protein
- FTLD-U
- fronto-temporal lobar degeneration
- UBQLN
- ubiquilin 1
- UAS
- upstream activating sequence
- UPR
- unfolded protein response
- TEM
- transmission electron microscopy
- RNAi
- RNA interference.
REFERENCES
- 1.Winklhofer K. F., Tatzelt J., Haass C. (2008) EMBO J. 27, 336–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitsumoto H., Chad D. A., Pioro E. P. (1998) Amyotrophic Lateral Sclerosis, F.A. Davis, Philadelphia [Google Scholar]
- 3.Andersen P. M., Sims K. B., Xin W. W., Kiely R., O'Neill G., Ravits J., Pioro E., Harati Y., Brower R. D., Levine J. S., Heinicke H. U., Seltzer W., Boss M., Brown R. H., Jr. (2003) Amyotroph. Lateral Scler. Other Motor Neuron Disord. 4, 62–73 [DOI] [PubMed] [Google Scholar]
- 4.Gurney M. E. (1997) J. Neurol Sci. 152, Suppl. 1, S67–S73 [DOI] [PubMed] [Google Scholar]
- 5.Benatar M. (2007) Neurobiol. Dis. 26, 1–13 [DOI] [PubMed] [Google Scholar]
- 6.Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., Feiden W., Kretzschmar H. A., Trojanowski J. Q., Lee V. M. (2006) Science 314, 130–133 [DOI] [PubMed] [Google Scholar]
- 7.Wang H. Y., Wang I. F., Bose J., Shen C. K. (2004) Genomics 83, 130–139 [DOI] [PubMed] [Google Scholar]
- 8.Ou S. H., Wu F., Harrich D., García-Martínez L. F., Gaynor R. B. (1995) J. Virol. 69, 3584–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buratti E., Baralle F. E. (2001) J. Biol. Chem. 276, 36337–36343 [DOI] [PubMed] [Google Scholar]
- 10.Ayala Y. M., Misteli T., Baralle F. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3785–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hasegawa M., Arai T., Nonaka T., Kametani F., Yoshida M., Hashizume Y., Beach T. G., Buratti E., Baralle F., Morita M., Nakano I., Oda T., Tsuchiya K., Akiyama H. (2008) Ann. Neurol 64, 60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inukai Y., Nonaka T., Arai T., Yoshida M., Hashizume Y., Beach T. G., Buratti E., Baralle F. E., Akiyama H., Hisanaga S., Hasegawa M. (2008) FEBS Lett. 582, 2899–2904 [DOI] [PubMed] [Google Scholar]
- 13.Igaz L. M., Kwong L. K., Xu Y., Truax A. C., Uryu K., Neumann M., Clark C. M., Elman L. B., Miller B. L., Grossman M., McCluskey L. F., Trojanowski J. Q., Lee V. M. (2008) Am. J. Pathol. 173, 182–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banks G. T., Kuta A., Isaacs A. M., Fisher E. M. (2008) Mamm. Genome 19, 299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sreedharan J., Blair I. P., Tripathi V. B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J. C., Williams K. L., Buratti E., Baralle F., de Belleroche J., Mitchell J. D., Leigh P. N., Al-Chalabi A., Miller C. C., Nicholson G., Shaw C. E. (2008) Science 319, 1668–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kabashi E., Valdmanis P. N., Dion P., Spiegelman D., McConkey B. J., Vande Velde C., Bouchard J. P., Lacomblez L., Pochigaeva K., Salachas F., Pradat P. F., Camu W., Meininger V., Dupre N., Rouleau G. A. (2008) Nat. Genet. 40, 572–574 [DOI] [PubMed] [Google Scholar]
- 17.Kim S. H., Shi Y., Hanson K. A., Williams L. M., Sakasai R., Bowler M. J., Tibbetts R. S. (2009) J. Biol. Chem. 284, 8083–8092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ko H. S., Uehara T., Tsuruma K., Nomura Y. (2004) FEBS Lett. 566, 110–114 [DOI] [PubMed] [Google Scholar]
- 19.Doi H., Mitsui K., Kurosawa M., Machida Y., Kuroiwa Y., Nukina N. (2004) FEBS Lett. 571, 171–176 [DOI] [PubMed] [Google Scholar]
- 20.Mah A. L., Perry G., Smith M. A., Monteiro M. J. (2000) J. Cell Biol. 151, 847–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.N'Diaye E. N., Kajihara K. K., Hsieh I., Morisaki H., Debnath J., Brown E. J. (2009) EMBO Rep. 10, 173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganguly A., Feldman R. M., Guo M. (2008) Hum. Mol. Genet. 17, 293–302 [DOI] [PubMed] [Google Scholar]
- 23.Rasband W. S. (2009) ImageJ, U. S. National Institutes of Health, Bethesda, MD [Google Scholar]
- 24.Rimkus S. A., Katzenberger R. J., Trinh A. T., Dodson G. E., Tibbetts R. S., Wassarman D. A. (2008) Genes Dev. 22, 1205–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.R Development Core Team (2009) R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 26.Therneau T., Lumley T. (2009) survival, R package, Version 2.35–4, Mayo Clinic, Rochester, MN [Google Scholar]
- 27.Feiguin F., Godena V. K., Romano G., D'Ambrogio A., Klima R., Baralle F. E. (2009) FEBS Lett. 583, 1586–1592 [DOI] [PubMed] [Google Scholar]
- 28.Wegorzewska I., Bell S., Cairns N. J., Miller T. M., Baloh R. H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18809–18814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fiesel F. C., Voigt A., Weber S. S., Van den Haute C., Waldenmaier A., Görner K., Walter M., Anderson M. L., Kern J. V., Rasse T. M., Schmidt T., Springer W., Kirchner R., Bonin M., Neumann M., Baekelandt V., Alunni-Fabbroni M., Schulz J. B., Kahle P. J. (2010) EMBO J. 29, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Y., Ferris J., Gao F. B. (2009) Mol. Brain 2, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korolchuk V. I., Mansilla A., Menzies F. M., Rubinsztein D. C. (2009) Mol. Cell 33, 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.