Abstract
At the biochemical level, motor proteins are enzymatic molecules that function by converting chemical energy into mechanical motion. The key element for energy transduction and a major unresolved question common for all motor proteins is the coordination between the chemical and conformational steps in ATP hydrolysis. Here we show time-lapse monitoring of an in vitro ATP hydrolysis reaction by the motor domain of a human Kinesin-5 protein (Eg5) using difference Fourier transform infrared spectroscopy and UV photolysis of caged ATP. In this first continuous observation of a biological reaction coordinate from substrate to product, direct spectral markers for two catalytic events are measured: proton abstraction from nucleophilic water by the catalytic base and formation of the inorganic phosphate leaving group. Simultaneous examination of conformational switching in Eg5, in parallel with catalytic steps, shows structural transitions in solution consistent with published crystal structures of the prehydrolytic and ADP-bound states. In addition, we detect structural modifications in the Eg5 motor domain during bond cleavage between the β- and γ-phosphates of ATP. This conclusion challenges mechanochemical models for motor proteins that utilize only two stages of the catalytic cycle to drive force and motion.
Keywords: ATPases, Biophysics, Enzyme Catalysis, Fourier Transform IR (FTIR), Kinesin, Chemical Dynamics, Structural Dynamics, Transition State
Introduction
Coupling between chemical events and conformational transitions is crucial for the catalytic function of proteins. Notable examples include molecular motors, signaling proteins, and membrane transporters, in which large scale conformational transitions are integral to the chemical steps of ATP hydrolysis (see Fig. 1a) to yield the products, ADP and inorganic phosphate, or to phosphorylation in which Pi is covalently attached to a protein and ADP is a leaving group. The complex nature of the phenomenon still renders it an outstanding challenge to establish cause-and-effect relationships inherent within mechanochemical coupling in molecular motors.
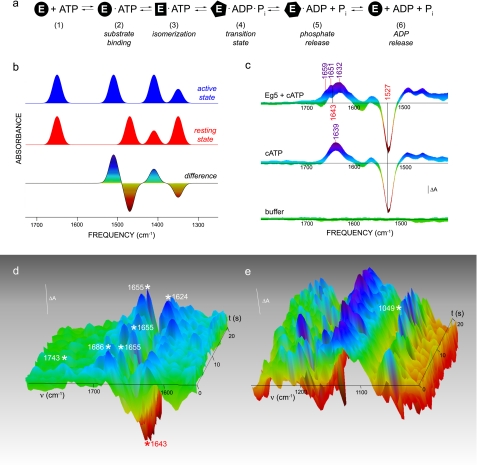
FIGURE 1.
Time-resolved IR spectroscopy of Eg5 catalysis in solution. a, schematic diagram of the Eg5 ATPase cycle. Free ATP (1) from caged ATP photolysis is bound to Eg5 (2), which undergoes conformational isomerization (3). The biochemical reaction proceeds through the transition state to form products (4). Sequential release of inorganic phosphate (5) and ADP (6) completes the catalytic cycle. b, Difference techniques applied to vibrational spectroscopy. When the sample is triggered to initiate catalysis, infrared modes in the protein can differ in frequency relative to the resting state. Positive (blue) or negative (red) lines in the difference data reflect either acquisition/loss of or chemical changes in functional groups upon reaction. c, three-dimensional representations of difference photolysis spectra for (i) wild-type Eg5 in the presence of caged ATP (Eg5 + cATP; n = 180), (ii) caged ATP alone (n = 40), and (iii) buffer (n = 38). The t = 0 s designates the laser flash applied to the sample. The scale bar represents 1 × 10−4 absorbance units. d and e, the amide I region (d) and the phosphate regions (e) of the time-resolved hydrolytic spectra. Alterations in absorbance are visualized by color, in which blue is the greatest amplitude and red is the lowest negative amplitude. The bar represents 0.5 × 10−4 absorbance units. All spectral data have 12-cm−1 resolution.
The motor protein kinesin (1) is a member of the P-type NTPase superfamily. Greater than 50 crystal structures have been solved for members of the kinesin protein family, arrested at various stages in the enzymatic cycle. However, important gaps in the chemistry of catalysis and the sequence of structural changes exist; in particular, there is little information on the unhydrolyzed ATP-bound state (substrate binding and isomerization, see Fig. 1a) and transition state of these proteins, which are short lived. As a result, open questions in kinesin catalysis include the chemical nature of the catalytic base, timing of proton abstraction from the nucleophilic water in relation with major conformational changes, the nature of the transition state, and understanding of which steps within ATP hydrolysis drive major structural transitions.
Our broad aim in this work is to watch a biomolecule in real-time motion, and specifically, to observe coupling of chemistry and structural transitions in solution during catalysis. The experimental system is the human Eg5, a plus end-directed kinesin motor essential for chromosome segregation during cell division. Outside of its vital cellular role, this protein performs ATP hydrolysis with a slow kinetic rate in the absence of microtubules and thus provides an optimal model system for observing real-time catalysis at room temperature. Time-resolved vibrational spectroscopy has been established as an effective means to study enzymatic reactions. IR2 spectroscopy is capable of observing functional group chemistry under native conditions. Its Fourier transform implementation simultaneously measures all chemical moieties of the sample. Enzymatic reactions of Eg5 (2), distantly related NTPases (3, 4), and phosphotransferases (5, 6) have been monitored using IR methods as chemical reactions transform the substrate to an educt group and a product group, all of which have dissimilar infrared spectra.
Our prior steady-state work (2) showed physiochemical information on how individual amino acid residues participating in ATP hydrolysis are coupled to structural motions. Here we move toward real-time measurement of the kinesin catalytic cycle, acquiring IR data in the millisecond time regime (7). Spectroscopic signatures of conformational changes can be used to detect and define transient conformational states of a protein, an approach with a history in fluorescence and absorption spectroscopy. The advantage of IR spectroscopy is that a single experiment measures not only backbone conformation but also ligand chemistry simultaneously. Therefore, we can use this bona fide query to determine local chemical changes at the catalytic site and the structure of the Eg5 motor domain as a whole in a single experiment.
EXPERIMENTAL PROCEDURES
The motor domain of wild-type Eg5 monomer, composed of residues 1–367, was expressed in Escherichia coli BL21 CodonPlus(DE3)-RIL-competent cells (Stratagene) and purified as described (2). Steady-state rates of ATP hydrolysis were monitored by NADH-coupled assays (8) in microwell format. Kinetic rates reported are an average of three independent replicates.
FTIR measurements were acquired with previously described methods (2). A total of 20 μl/sample was prepared with a final concentration of 5 mg/ml Eg5 (0.12 mm), 3 mm NaCl, 20 mm HEPES, pH 7.5, 6.6 mm dithiothreitol, 0.1 mm EDTA, 1 mm MgCl2, and 2 mm caged ATP and sandwiched between two CaF2 windows. Parameters for the spectral acquisition on an IFS66v/S spectrometer (Bruker Optics, Billerica MA) with a water-cooled IR source and liquid nitrogen-cooled HgCdTe detector were: double-sided forward-backward acquisition mode, Happ-Genzel apodization function, 100-kHz mirror velocity, 12-cm−1 spectral resolution, 160-ms time resolution, 1 level of zero filling, and 2.5-mm aperture size. The time scale of caged ATP photolysis (∼10−5 s (9)) is an order of magnitude smaller than the experimental time resolution.
The background spectrum was collected for 500 scans to assess base-line stability, followed by 4 s of prephotolysis measurement. For photolysis, a single shot of an Nd:YAG Surelite (Continuum, Santa Clara, CA) laser with 355 nm excitation and a pulse duration of 6 ns was used. From our previous measurement (2) that ATP was released from 3–5% of caged ATP molecules by five laser flashes, we estimate that 1.2–2.0 μm substrate was liberated from the nitrophenylethyl moiety per laser flash. Twenty seconds of data acquisition followed (postphotolysis). After 17 min of relaxation, the whole sequence was repeated for the total of 8–20 times per sample (supplemental Fig. S1).
Spectra presented herein are difference data (see Fig. 1b), as defined by −log(I2/I1), where I2 and I1 refer to two single beam spectra of the same sample at different times. Difference spectra were analyzed using IGOR Pro (WaveMetrics, Lake Oswego, OR) by averaging normalized spectra. The numbers of spectra co-added for the samples were 40 and 180 for caged ATP and Eg5-caged ATP samples, respectively. Spectra of Eg5 alone and Eg5 with caged ATP in the absence of a photolytic trigger (data not shown) do not exhibit any detectable spectral features, and base-line drift over the time course of the experiment is negligible.
Estimates in the change of backbone structure and interaction (COBSI) index were calculated (10). Integration of amide I absorbance allows for a simple estimate of structural changes in the polypeptide backbone during a protein reaction; the integrated total protein absorbance is the averaged value between the resting and active states of the protein. Eg5 ATPase COBSI indices obtained were between 0.0074 and 0.0044 upon integration of the data in Fig. 1d and correction of active ATPase activity measured (via use of experimental measurement of ATP release per laser flash in our samples (2)).
RESULTS
Time-resolved Photolysis and Hydrolysis Spectra of Caged ATP·Eg5 Complexes
For this investigation, the catalytic reaction of Eg5 is initiated by photolytic cleavage of the caging group from 1-(2-nitrophenyl)ethyl adenosine 5′-triphosphate (caged ATP; supplemental Fig. S1). Laser flash photolysis of caged ATP·Eg5 complexes combined with FTIR monitoring with 160-ms time resolution yield two types of time-resolved difference spectra. The first is the photolysis difference spectra (Fig. 1c) in which positive lines originate from Eg5·ATP. In these data, a sequence of transient catalytic states can be tracked over time.
Also shown in Fig. 1c are control photolysis spectra of caged substrate alone and of buffer. The buffer spectra in Fig. 1c reflect the level of noise in our experiments throughout the entire spectral and temporal range. In both time-resolved spectra of samples with caged ATP, the negative band at 1527 cm−1, previously assigned to the disappearing NO2 group of the cage moiety (9), reflects the level of cage release by UV photolysis and serves as a quantifiable marker for normalizing data from different samples. When kinesin is present in the sample, there are clear amplitude changes and frequency shifts observable (Fig. 1c) when compared with the control caged ATP photolysis reaction. As Eg5 basal activity rates before and after the time-resolved experiment were 0.12 ± 0.02 and 0.11 ± 0.02 ADP/motors/s, respectively, our data support the conclusion that the kinesin motor domain was fully active throughout the experiment and comparable with other reports in the literature (11–14).
To isolate the chemical and structural alterations in Eg5 upon functional modulation between the active and inactive states of the motor domain, we focused on the second type of time-resolved data, termed hydrolytic difference spectra (Fig. 1, d and e), for which the spectra of caged ATP photolysis alone are subtracted from the spectra acquired from Eg5 in the presence of caged ATP (Fig. 1b). Positive lines in these data (Fig. 1, d and e, blue) are associated with protein modes from the Eg5·ATP collision complex, its isomerization state, and the transition state, as well as chemical modes of ADP and inorganic phosphate. Negative lines (Fig. 1, d and e, red) are associated with the Eg5·ADP and ATP contributions.
Spectral Identification of Key Chemical Transitions during ATP Hydrolysis
IR spectroscopy has been useful in determination of the structure of acids and bases in solution from characteristic vibrational marker modes. In these time-resolved hydrolytic spectra, we can examine the chemistry of the nucleophile, the base, and the product leaving groups during the course of the Eg5 enzymatic reaction. This allows us to distinguish between the mechanistic models of catalysis that are currently debated in the field of ATP hydrolysis by motor proteins.
Upon availability of free ATP substrate to Eg5 (t = 0 s), we observe a broad continuum infrared mode between 1800 and 1740 cm−1 (Fig. 1d). This broad band is a spectroscopic reporter of an organized water cluster undergoing protonation changes (15) during Eg5 catalysis in solution. A water molecule is the accepted nucleophile for ATP hydrolysis, and a recent crystallographic study of Eg5 kinesin (16) identified the catalytic base as a second water molecule in the active site. Thus, detection of this IR mode suggests that there is direct observation of the ordered nucleophile and base in the orthosteric site of Eg5 in real time.
Second, we observe an unambiguous spectroscopic marker that defines Pi product formation from the Eg5 ATPase reaction. From studies on model compounds, the PO2− and PO3− moieties are localized in the 1250–1090-cm−1 region, and the HPO42− modes are isolated between 1090 and 1000 cm−1. Appearing at ∼10 s, a positive 1049-cm−1 infrared line (Fig. 1e) is identified as a candidate HPO42− mode based on group frequency assignments (17). We note that this direct spectroscopic measurement of Pi formation from the ATPase reaction is at the anticipated time interval, based on the kinetic activity of our samples.
Conformational Switching during Kinesin ATP Hydrolysis
To concomitantly ascertain net changes in Eg5 secondary structure, we focus on the carbonyl stretching, or the amide I, vibration of the peptide backbone (18) between the 1690- and 1620-cm−1 region. In the amide region (Fig. 1d), four vibrational modes are transiently observed in our time-resolved hydrolysis spectra: three positive lines at 1686, 1655, and 1624 cm−1 and one negative line at 1643 cm−1.
Spectral components in the infrared amide I region can be assigned to specific forms of secondary structure elements. Assignments are made based on studies of proteins with known structures (19–24). The negative band at 1643 cm−1 is attributed to unordered structures in Eg5 kinesin. The 1655-cm−1 mode is assigned to α-helical structures, and the 1624-cm−1 mode is assigned to the low frequency component of β-sheets. The 1686-cm−1 mode may be ascribed to turns in the motor domain.
The time-resolved hydrolytic spectra report kinetic changes in the Eg5 structure during the catalytic cycle. Negative modes result from disappearing structures, and positive modes result from emerging structures. The negative 1643-cm−1 mode develops at 1.5 s after substrate availability and wanes at 13 s. These data show that there is a decrease in unordered structures at 1.5 s after ATP is introduced to Eg5, but there is a relaxation within the motor domain that results in the equivalent amount of unordered loops, to the inactive state, by 13 s. The 1686-cm−1 mode, identifying turn structures, emerges upon ATP release (t = 0 s) and persists until 14.5 s into the hydrolytic cycle. The α-helical 1655-cm−1 mode is visible in the hydrolysis spectra immediately upon ATP release through 3.8 s and then reappears from 8.7 to 13 s. The fourth mode at 1624 cm−1, visible between 10.5 and 13 s in the time-resolved hydrolytic data, provides direct evidence that alterations in Eg5 β-sheets occur after Pi formation.
DISCUSSION
Recent efforts in structural biology focus on dissecting the role of slow (millisecond) motions in enzyme catalysis as proteins proceed through multiple conformations serially and in parallel with catalysis. However, the typical and practical challenge is to isolate the conformational degrees of freedom with relevant impact on the chemical events in the protein. In this regard, the study of Eg5 kinesin with time-resolved IR spectroscopy provides an opportunity to probe the slow structural transitions that are tightly coupled with enzymatic chemistry.
The major accomplishment herein is the uninterrupted recording of transient events, or states, between the ATP-bound and ADP-bound states of Eg5 in solution. Continuous changes of molecular structure and chemistry are seldom observed, especially for large polyatomic molecules such as proteins. This failure limits our understanding of ATP hydrolysis to a level of an oversimplified reaction coordinate. Thus, our results provide insight regarding cause-and-effect among events of different scale and physicochemical nature that are typically absent in experiments.
Proton Abstraction from the Nucleophilic Water Can Be Observed in Real Time
The first chemical step in ATP hydrolysis is removal of a proton from the nucleophilic water by a neighboring catalytic base. In the Eg5·AMPPNP crystal structure (16), substrate binding and isomerization to the prehydrolysis catalytic state result in isolation of a two-water cluster in the active site: the nucleophilic water positioned for γ-phosphate attack and a second water that serves as a general base.
Vibrational spectroscopy can provide detailed information regarding 1) the chemical identity of and 2) the environment surrounding the proton-donating and proton-accepting molecules in this acid-base chemistry. The time-lapse experiment that examines Eg5 catalysis in real time provides independent, experimental verification that the chemical identities of the nucleophile and catalytic base in Eg5 kinesin are water molecules. Multiple waters have been theorized to be catalytically active in myosin (25), F1-ATPase (26, 27), and p21ras (28).
Although bulk water contributes free OH modes (>3500 cm−1) and the H–O–H scissoring modes, vibrational modes involving the central proton are crucial to understanding whether proton transfer occurs between water molecules. This requires examination of the 2100–1740-cm−1 region of O···H+···O fundamentals. Both calculation and experiment (29–31) characterize the broad IR continua between 1800 and 1740 cm−1 in our data (Fig. 1d) as a water cluster in which a proton is shared. Our Eg5·AMPPNP crystal structure (16) distinguishes assignment of the broad 1800–1740-cm−1 IR mode to a solvent-shared proton (Zundel cation, H5O2+) rather than a fully hydrated hydronium (Eigen cation, H9O3+).
Furthermore, we observe controlled proton transfer between the lytic water and the second water that serves as a catalytic base in the Eg5 active site. Typically, it is challenging to identify a hydrated proton at room temperature in aqueous conditions because the Zundel cation can assume broadly varying configurations. We conclude that the water molecules must be well ordered in the Eg5 active site upon ATP binding for the hydrated proton to be prominent. The large number of coordinations of Eg5 prehydrolytic state in a crystalline form (16) corroborates this deduction. Thus, orthosteric water molecules that serve as a proton transfer bridge are important in kinesins.
Time-resolved Hydrolytic Data Support an Associative Transition State for Eg5 Kinesin
In the classic representation of an associative ATPase mechanism, the nucleophilic attack by water at the phosphorus center occurs before the Pi leaving group departs; this results in a pentavalent phosphorane intermediate in the transition state. For a dissociative mechanism, the leaving group dissociates from the substrate prior to the nucleophilic attack; a metaphosphate intermediate state is involved.
Toward distinguishing the transition state and thus the mechanistic path, typical experiments are limited to the examination of geometric and electronic properties of ATP bound in the protein active site. Our hydrolytic data temporally resolve the formation of a H5O2+ intermediate (Fig. 1d, a proton being pulled off the nucleophilic water by the second water) well prior to the formation of the Pi leaving group (Fig. 1e). As a result, our data do not favor a dissociative mechanism for Eg5 ATP hydrolysis as the formation of this transition state requires that the Pi leaving group is formed prior to nucleophilic attack. Thus, our spectroscopic data favor an associative mechanism for kinesin ATP hydrolysis in solution. Unequivocal assignments regarding the bond order, electronic properties, or geometry of the catalytic transition state in these vibrational data await future isotopic labeling experiments.
Measured Structural Transitions Required for Formation of the Prehydrolytic State of Eg5 Are Consistent between IR and X-ray Crystallographic Studies
These infrared data (Fig. 1d) show the first direct observation of dynamic conformational switching caused by ATP binding and/or isomerization (Fig. 1a) in solution. Release of ATP from the caging group triggers initial gain of turn structures, quickly followed by the formation of α-helices, at the expense of unordered regions within the Eg5 motor domain.
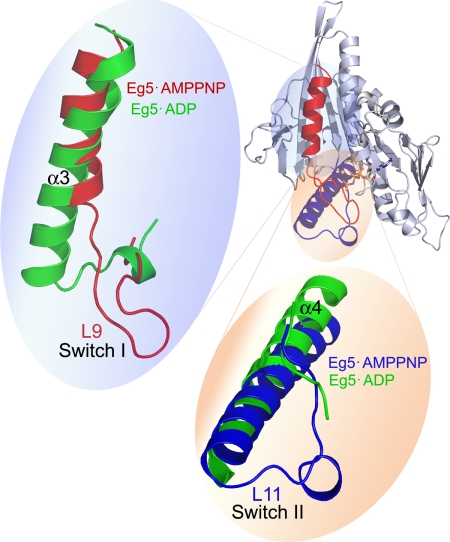
The conclusion that isomerization upon substrate binding involved conversion of disordered regions of Eg5 to turn and to helical structures is consistent with and complements the x-ray crystallographic data of Eg5. When ADP is bound to the Eg5 active site (32), the switch II region has unordered residues with a high local temperature factor (Fig. 2). In the Eg5·AMPPNP structure (16), the switch II region has an extended α4 helix, and the unordered residues, unobservable in the ADP-bound state, are resolved as a loop with a single helical turn. These x-ray structures also show loss of α3 helix and gain of turns in the switch I region (Fig. 2) in the prehydrolytic state of Eg5 when compared with the inactive state of the motor protein.
FIGURE 2.
Comparison of Eg5·ADP and Eg5·AMPPNP crystal structures. In the right inset is the ribbon representation of the Eg5·AMPPNP structure (PDB ID 3HQD (16)), with α3 and switch I highlighted in red and α4 and switch II in purple. Enlargements of these Eg5 regions provide overlays with the Eg5·ADP structure (PDB ID 1II6 (32)) in green. The structures were superpositioned using the P-loop residues.
The fidelity of the Eg5 secondary structural transitions measured between vibrational data in solution herein and static measurements in a crystalline lattice (16, 32) is remarkably high. Furthermore, quantitation of the number of residues involved in these structural changes is quite close between these techniques. COBSI estimates (10) of the maximum number of residues involved in these structural changes, measured by vibrational spectroscopy, are 5–14 residues. Using DSSP assignments in the Protein Data Bank, alterations in the peptide backbone of 16 Eg5 residues produce the net structural changes in the switch regions within Eg5 crystals (16, 32) noted above. These data demonstrate that these time-resolved infrared methods are an effective and quantitative probe of conformational switching in real time.
In Solution, Millisecond Conformational Transitions Are Observed in the Eg5 Enzymatic Transition State
An open question in understanding conformational switching of motor proteins is whether the transition state of ATP hydrolysis is capable of driving any major structural transitions. It is widely held that large scale structural transitions implicated in most “molecular machines” are directly coupled to the binding of ATP and release of the ADP leaving group. This tenet is borne out in the comparison of the Eg5·AMPPNP (16) and Eg5·ADP (32) crystal structures. In contrast, hydrolysis of ATP to form ADP·Pi only involves subtle electrostatic changes that may not drive major structural transitions; conformational changes associated with the transition state of ATP catalysis have not been experimentally observed by x-ray crystallography, fluorescence, or absorbance spectroscopy.
Conformational switching from the transition state through product formation can be followed in our hydrolytic FTIR spectra. Following the prehydrolysis state, there are two types of conformational changes (Fig. 1d): one that precedes and one that is coincident with product formation. After the formation of the prehydrolysis state, there are helical rearrangements in the Eg5 motor domain. First, loss of helical structure follows loss of hydrated proton signature (Fig. 1d). Second, a larger increase in α-helical content in the motor domain is measured at 8.7 s immediately prior to Pi formation. These structural changes are complemented by the inverse measure of turns in the motor domain. Therefore, we suggest that sequential interconversion between α-helices and turns occurs during the transition state toward bond cleavage between the β- and γ-phosphates of ATP (Fig. 1a). Lastly, the formation of inorganic phosphate induces structural alterations in α-helices, β-sheets, and turns until 13 s, at which point these structural changes cease.
This work establishes that IR spectroscopy can be used to refine and extend enzymatic information provided by x-ray crystallography. Here we supply novel evidence that the intermediate steps within the ATP hydrolysis reaction are also capable of driving major structural transitions. This conclusion suggests that mechanistic models of motor proteins may need to explore a greater number of conformational states, not only for mechanochemical function but also in development of allosteric drugs.
Supplementary Material
Acknowledgments
We are indebted to Edward J. Wojcik for instrumental planning, initial experiments involving this work, and comments on the manuscript. We thank Timothy Mitchison, Christine Fields, and Rebecca Ward in the Systems Biology Department at Harvard Medical School for support and encouragement, Ross Willis and R. Scott Neville for technical assistance, and Courtney Parke for thoughtful, intellectual discussions.
This work was supported by a grant from the Louisiana Board of Regents (to S. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- IR
- infrared
- FTIR
- Fourier transform IR
- AMPPNP
- 5′-adenylyl-β,γ-imidodiphosphate
- COBSI
- change of backbone structure and interaction.
REFERENCES
- 1.Vale R. D. (1996) J. Cell Biol. 135, 291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wojcik E. J., Dalrymple N. A., Alford S. R., Walker R. A., Kim S. (2004) Biochemistry 43, 9939–9949 [DOI] [PubMed] [Google Scholar]
- 3.Du X., Frei H., Kim S. H. (2000) J. Biol. Chem. 275, 8492–8500 [DOI] [PubMed] [Google Scholar]
- 4.Allin C., Gerwert K. (2001) Biochemistry 40, 3037–3046 [DOI] [PubMed] [Google Scholar]
- 5.Liu M., Barth A. (2002) Biopolymers. 67, 267–270 [DOI] [PubMed] [Google Scholar]
- 6.White E. M., Holland A. R., MacDonald G. (2008) Biochemistry 47, 84–91 [DOI] [PubMed] [Google Scholar]
- 7.Braiman M. S., Ahl P. L., Rothschild K. J. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 5221–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Learman S. S., Kim C. D., Stevens N. S., Kim S., Wojcik E. J., Walker R. A. (2009) Biochemistry 48, 1754–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cepus V., Ulbrich C., Allin C., Troullier A., Gerwert K. (1998) Methods Enzymol. 291, 223–245 [DOI] [PubMed] [Google Scholar]
- 10.Barth A., von Germar F., Kreutz W., Mäntele W. (1996) J. Biol. Chem. 271, 30637–30646 [DOI] [PubMed] [Google Scholar]
- 11.Cochran J. C., Gatial J. E., 3rd, Kapoor T. M., Gilbert S. P. (2005) J. Biol. Chem. 280, 12658–12667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maliga Z., Kapoor T. M., Mitchison T. J. (2002) Chem. Biol. 9, 989–996 [DOI] [PubMed] [Google Scholar]
- 13.DeBonis S., Simorre J. P., Crevel I., Lebeau L., Skoufias D. A., Blangy A., Ebel C., Gans P., Cross R., Hackney D. D., Wade R. H., Kozielski F. (2003) Biochemistry 42, 338–349 [DOI] [PubMed] [Google Scholar]
- 14.Yan Y., Sardana V., Xu B., Homnick C., Halczenko W., Buser C. A., Schaber M., Hartman G. D., Huber H. E., Kuo L. C. (2004) J. Mol. Biol. 335, 547–554 [DOI] [PubMed] [Google Scholar]
- 15.Asmis K. R., Pivonka N. L., Santambrogio G., Brümmer M., Kaposta C., Neumark D. M., Wöste L. (2003) Science 299, 1375–1377 [DOI] [PubMed] [Google Scholar]
- 16.Parke C. L., Wojcik E. J., Kim S., Worthylake D. K. (2010) J. Biol. Chem. 285, 5859–5867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng H., Nikolic-Hughes I., Wang J. H., Deng H., O'Brien P. J., Wu L., Zhang Z. Y., Herschlag D., Callender R. (2002) J. Am. Chem. Soc. 124, 11295–11306 [DOI] [PubMed] [Google Scholar]
- 18.Krimm S., Bandekar J. (1986) in Advances in Protein Chemistry (Anfinsen C. B., Edsall J. T., Richards F. M. eds), pp 181–364, Academic Press, New York [Google Scholar]
- 19.Surewicz W. K., Mantsch H. H., Chapman D. (1993) Biochemistry 32, 389–394 [DOI] [PubMed] [Google Scholar]
- 20.Jackson M., Mantsch H. H. (1995) Crit. Rev. Biochem. Mol. Biol. 30, 95–120 [DOI] [PubMed] [Google Scholar]
- 21.Goormaghtigh E., Cabiaux V., Ruysschaert J. M. (1994) Subcell. Biochem. 23, 329–362 [DOI] [PubMed] [Google Scholar]
- 22.Prestrelski S. J., Byler D. M., Thompson M. P. (1991) Int. J. Peptide Protein Res. 37, 508–512 [PubMed] [Google Scholar]
- 23.Barth A., Zscherp C. (2002) Q. Rev. Biophys. 35, 369–430 [DOI] [PubMed] [Google Scholar]
- 24.Fabian H., Schultz C. P. (2000) in Encyclopedia of Analytical Chemistry (Meyers R. A. ed) pp 5779–5803, John Wiley & Sons Ltd., Chichester, UK [Google Scholar]
- 25.Li G., Cui Q. (2004) J. Phys. Chem. B. 108, 3342–3357 [DOI] [PubMed] [Google Scholar]
- 26.Ahmad Z., Senior A. E. (2004) J. Biol. Chem. 279, 46057–46064 [DOI] [PubMed] [Google Scholar]
- 27.Dittrich M., Hayashi S., Schulten K. (2004) Biophys. J. 87, 2954–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheidig A. J., Burmester C., Goody R. S. (1999) Structure 7, 1311–1324 [DOI] [PubMed] [Google Scholar]
- 29.Garczarek F., Gerwert K. (2006) Nature 439, 109–112 [DOI] [PubMed] [Google Scholar]
- 30.Mathias G., Marx D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6980–6985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baer M., Mathias G., Kuo I. F., Tobias D. J., Mundy C. J., Marx D. (2008) Chemphyschem 9, 2703–2707 [DOI] [PubMed] [Google Scholar]
- 32.Turner J., Anderson R., Guo J., Beraud C., Fletterick R., Sakowicz R. (2001) J. Biol. Chem. 276, 25496–25502 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.