Abstract
Mismatch repair in Escherichia coli involves a number of proteins including MutL and UvrD. Eukaryotes also possess MutL homologues; however, no UvrD helicase homologues have been identified. The hyperthermophilic bacterium Aquifex aeolicus has a MutL protein (Aae MutL) that possesses a latent endonuclease activity similar to eukaryotic, but different from E. coli, MutL proteins. By sequence homology Aq793 is a member of the PcrA/UvrD/Rep helicase subfamily. We expressed Aae MutL and the putative A. aeolicus DNA helicase (Aq793) proteins in E. coli. Using synthetic oligonucleotide substrates, we observed that lower concentrations of Aq793 were required to unwind double-stranded DNA that had a 3′-poly(dT) overhang as compared with double-stranded DNA with a 5′-poly(dT) or lacking a poly(dT) tail. This unwinding activity was stimulated by adding Aae MutL with maximal stimulation observed at an ∼1.5:1 (MutL:Aq793) stoichiometric ratio. The enhancement of Aq793 helicase activity did not require the Aae MutL protein to retain endonuclease activity. Furthermore, the C-terminal 123 amino acid residues of Aae MutL were sufficient to stimulate Aq793 helicase activity, albeit at a significantly reduced efficacy. To the best of our knowledge this is the first time a human PMS2 homologue has been demonstrated to stimulate a PcrA/UvrD/Rep subfamily helicase, and this finding may further our understanding of the evolution of the mismatch repair pathway.
Keywords: DNA/Damage, DNA/Enzymes, DNA/Helicase, DNA/Protein Interaction, DNA/Repair, DNA/Uvr ABCD System, MutL, Helicase
Introduction
The mismatch repair (MMR)2 pathway corrects base mispairings that arise during DNA replication and errant recombination events in bacteria (1) and eukaryotes (2–4). MMR is therefore an important mechanism in the maintenance of genomic stability. The mechanism and major components of the Escherichia coli and human MMR pathways have been extensively studied (1, 5–7). In E. coli, MutS initiates MMR by binding to unpaired bases in duplex DNA. The mispair binding triggers an adenine nucleotide exchange in MutS and subsequent recruitment of MutL. The DNA-MutS-MutL complex stimulates the latent endonuclease activity of MutH that nicks the unmethylated strand at hemimethylated GATC (dam methylation) sites and thereby discriminates the daughter from the parental DNA strand. Interestingly, MutL is not only instrumental in activating the MutH endonuclease activity, it also stimulates UvrD helicase loading at the nick site, facilitating the displacement of the incorrect DNA strand. The displaced strand is finally hydrolyzed by exonucleases, and the ssDNA gap is stabilized by single-stranded binding protein. A complementary DNA strand is synthesized by polymerase III, and E. coli DNA ligase completes the repair.
One difference between E. coli and human mismatch repair is the absence of an identified MutH homologue in the latter organism. Furthermore, MutH homologues appear to be absent in a large number of bacteria and all eukaryotes that have been investigated. Modrich and co-workers (8, 9) determined that the endonuclease activity in human MMR resides in the MutLα protein itself instead of a separate MMR component. This may be a general trend in organisms that lack MutH homologues as exemplified by studies in yeast (10) and thermophilic bacteria (11).
MutL appears to stimulate UvrD helicase activity by increasing the UvrD affinity for DNA and thereby increasing its loading rate at a nick (12). E. coli MutL exists as a homodimer, has a weak ATPase activity, and has been reported to bind DNA (13). The monomeric MutL protein can be divided into three domains: the N-terminal LN40, the linker, and the C-terminal LC20 regions. The highly conserved N-terminal LN40 domain contains the ATPase motif of the ATPase/kinase superfamily members, GHLK (14). The LN40 domain is involved in ATP-dependent protein dimerization, perhaps causing the MutL protein to wrap around the DNA helix (15). Interestingly, the LN40 region of MutL homologues is relatively well conserved between organisms regardless of whether MutH is present or absent in that species.
The MutL C-terminal domain is not well conserved between organisms that possess or lack a MutH homologue. Human PMS2 (16), a subunit of human MutLα, has two notable motifs in the C-terminal domain that are absent in organisms that possess MutH. Mutations in the first motif, DQHAX2EX4E, were found to abolish the endonuclease activity of MutLα (8). Similar mutations in hPMS2 homologues in yeast and thermophilic bacteria confirmed the importance of an intact DQHAX2EX4E motif for nicking activity (10, 11, 17). The role of the second motif, CPHGRPI, is not well understood, although one study suggests that it is important in ATP responsiveness (11). MutL from the hyperthermophilic bacterium Aquifex aeolicus, a hPMS2 homologue, is known to possess endonuclease activity, and no MutH homologues have been discovered in this organism.
E. coli helicase II, also known as UvrD helicase, is a member of the PcrA/UvrD/Rep (PUR) helicase subfamily and is involved in the nucleotide excision repair (18) and the methyl-directed mismatch repair pathways (1). UvrD is a motor protein capable of unwinding dsDNA in a multi-step, ATP hydrolysis-driven reaction while traveling on one DNA strand in the 3′ to 5′ direction (12, 19, 20). It is also an effective translocase with a reported processivity on ssDNA of >2 kb (19). In the MMR pathway, UvrD has its unwinding activity stimulated by the E. coli mismatch repair protein MutL (21, 22) and by the UvrAB complex in the nucleotide excision repair pathway (23). In MMR, this stimulation is correlated with the DNA binding activity of MutL (12, 24). Although UvrD is an important component of the E. coli methyl-directed MMR pathway, UvrD homologues are not involved in the nick-directed MMR pathway of yeast and human.
E. coli UvrD shares ∼40% sequence similarity with another PUR helicase family member found in E. coli, the Rep protein. Furthermore, these proteins both form homodimers and display the same 3′ to 5′ polarity of DNA unwinding (25, 26). Their primary physiological roles, however, appear to be different with UvrD involved in methyl-directed mismatch repair, nucleotide excision repair, and recombinational processes and Rep playing a role in genomic DNA replication. There may be redundancy in function or exceptions in certain instances. For example, UvrD is required for the replication of certain plasmids in E. coli that utilize rolling circle replication (27).
Stimulation of helicases UvrD and Rep by MutL has been described for E. coli proteins (28); however, MutL has been reported unable to stimulate UvrD in the thermophilic bacteria Thermoanaerobacter tengcongensis and Bacillus stearothermophilus, suggesting that the stimulation of helicase activity in PUR subfamily members may be specific to mesophilic bacteria (29). The T. tengcongensis MutL (Tte MutL) and B. stearothermophilus MutL (Bst MutL) C-terminal domains are well conserved with those from organisms lacking the endonuclease MutH. Both MutL homologues have in the C-terminal domain the distinctive heavy metal binding motif, DQHAX2EX4E, and the CPHGRPI motif characteristic of hPMS2 family members.
A. aeolicus MutL (Aae MutL) also belongs to the hPMS2 family and carries a heavy metal binding motif responsible for its endonuclease activity. We have cloned and expressed in E. coli the putative ATP-dependent DNA helicase Aq793, Aae MutL, a truncated form of Aae MutL (Aae MutL-CTD) and an endonuclease-deficient mutant of Aae MutL (Aae MutL (E357K)). The PUR helicase subfamily homologue Aq793 was found to possess helicase activity in common with the homologues found in E. coli, B. stearothermophilus, and T. tengcongensis. Furthermore, we demonstrate for the first time that a hPMS2-like MutL can stimulate the unwinding activity of a PUR subfamily helicase. The C-terminal domain alone was found to be sufficient to stimulate the helicase activity of Aq793, although significantly higher protein concentrations were required.
MATERIALS AND METHODS
Protein Production
Recombinant wild-type and mutant A. aeolicus MutL proteins were expressed fused to a C-terminal Mxe GyrA Intein tag in the pTWIN1 vector (30) (New England Biolabs, Inc.), whereas the A. aeolicus helicase (Aq793) protein was expressed with a hexahistidine tag. Protein expression was performed in the E. coli Rosetta™ (DE3) strain (Novagen®, Madison, WI) (F− ompT hsdSB(rB− mB−) gal dcm (DE3) pRARE2 (CamR)) and purified with either chitin affinity chromatography or nickel-nitrilotriacetic acid affinity chromatography, respectively. MutL proteins were further purified using a Q column (GE Healthcare) and Aq793 with a heparin column. Both were eluted with a salt gradient from 0.05 to 0.3 m NaCl. Protein purity was assessed to be >95% by SDS-PAGE analysis.
Helicase Assay
Three sets of synthetic double-stranded oligodeoxynucleotides were used to determine the helicase activity of Aq793 essentially as described previously (29). All of the dsDNAs were made of the same universal 23-mer top strand 5′-GCCCTGCTGCCGACCAACGAAGG-3′ annealed to 5′-ACCTTCGTTGGTCGGCAGCAGGGC-3′, 5′-(dT)40-ACCTTCGTTGGTCGGCAGCAGGGC-3′ or 5′-ACCTTCGTTGGTCGGCAGCAGGGC-(dT)40-3′ to form dsDNA substrates without a 40-nucleotide dT tail (dT40−), with a 5′ 40-nucleotide dT tail (5′-dT40), or with a 3′ 40-nucleotide dT tail (3′-dT40), respectively (see Fig. 1A). The top and bottom strands were annealed by mixing at a 1:1.5 (universal primer:variable primer) molar ratio in 1× ThermoPol reaction buffer (New England Biolabs, Inc.) and then heating to 95 °C followed by slow cooling to 25 °C. The substrates were labeled by strand extension using Klenow (exo−) (New England Biolabs, Inc.) containing [α-32P]dTTP and purified by size exclusion using Illustra™ MicroSpin G-25 columns (GE Healthcare). This previously reported protocol (25) adds a templated and a nontemplated [α-32P]dTTP to the 3′ ends of the oligonucleotide, permitting detection by autoradiography. The helicase reactions were performed by adding 0.02 nm radiolabeled duplex substrate in 1× Thermopol buffer and differing amounts of Aq793 in a 10-μl reaction volume. After a 2-min preincubation at 55 °C, an equal volume of 10 mm ATP in 1× Thermopol buffer was added to initiate the reaction (5 mm final ATP concentration). The reactions proceeded for 20 min and then were terminated by the addition of 5 μl of stop solution (0.25% bromphenol blue, 25% glycerol, 1% SDS, and 100 mm EDTA). A positive control was incubated for 15 min at 95 °C to completely denature the DNA duplexes and quenched by placing the reaction on ice. A negative control without helicase was performed in parallel. The samples were resolved on 20% TBE gels (Invitrogen) scanned with Typhon 9400® (GE Healthcare), and the bands were quantified using ImageQuant TL software (GE Healthcare).
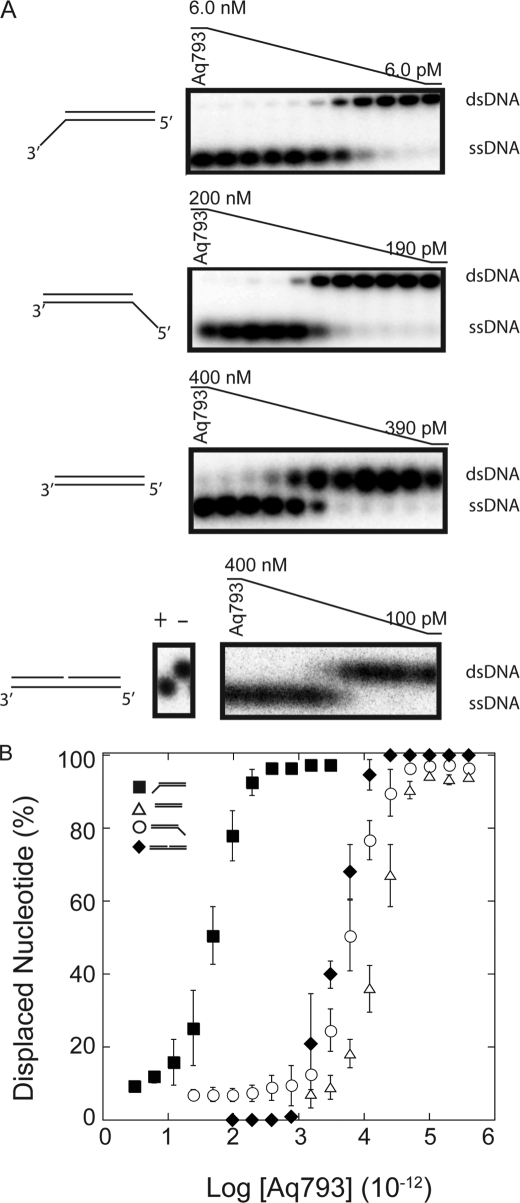
FIGURE 1.
Aq793 helicase activity. DNA unwinding activity was assayed with serial dilutions of Aq793. The 20-μl unwinding reactions were incubated at 55 °C and contained 0.02 nm dsDNA substrate. The reactions were quenched after 20 min using stop solution and applied to a 20% TBE acrylamide gel as described under “Materials and Methods.” A, representative autoradiograms of TBE acrylamide gels. The DNA substrate is indicated to the left of the gel picture, and the expected mobility of dsDNA and ssDNA is shown to the right. The negative control (−) was performed using 0.02 nm of substrate incubated at 55 °C with 400 nm of Aq793 without ATP, and the positive control (+) was performed at 95 °C in the absence of Aq793. B, graph of data from TBE gel bands that were quantified using ImageQuant TL software. 3′-dT40 (solid squares), 5′-dT40 (open circles), dT40− (open triangles), and the nicked substrate (solid diamonds) are represented. The points represent the averages of three separate experiments. The error bars represent the standard deviation at each point.
Helicase Assay on Nicked Substrate
A set of double-stranded oligodeoxynucleotides containing a nick was used to determine the helicase activity of Aq793 at a nick. Three oligodeoxynucleotides were used to make the nicked DNA substrate: the bottom strand (B) 5′-GTCCACGGGCCCTCTCCGCCCTGCTGCCGATCAACGAAGGCGAGGCAGCGACCTCATTCCA-3′ and two top strands (T1) 5′-TGGAATGAGGTCGCTGCCTCGCCTTCGTTGA-3′ and (T2) 5′P-TCGGCAGCAGGGCGGAGAGGGCCCGTGGAC-3′. The T2 oligonucleotide was previously labeled by T4 polynucleotide kinase (New England Biolabs, Inc.) with [γ-32P]ATP and purified by size exclusion chromatography using Illustra™ MicroSpin G-25 columns (GE Healthcare). The three oligonucleotides were mixed at a ratio of 1.4:1.2:1 (T1:B:T2) in 1× ThermoPol reaction buffer (New England Biolabs, Inc.) and then heated to 95 °C followed by slow cooling to 25 °C.
Helicase Assays in the Presence of MutL
A titration of Aae MutL, Aae MutL (E357K), or Aae MutL-CTD was performed in a reaction mix containing 0.02 nm DNA substrate and 23.5 pm of Aq793 in 1× Thermopol buffer. After a 2-min equilibration at 55 °C the reactions were initiated by adding ATP to a final concentration of 5 mm. The reactions were stopped and resolved as described above. A titration of Aae MutL was performed in the same condition described above in the reaction mix containing 0.02 nm nicked DNA substrate and 1 nm Aq793 in 1× Thermopol buffer.
RESULTS
Aq793 Helicase Activity
The ability of Aq793 to unwind dsDNA templates was determined in vitro using synthetic oligonucleotide substrates as described under “Materials and Methods.” The assays were performed with two sets of dsDNA templates. One set used a common 24-mer single-stranded oligonucleotide that was annealed to one of three different complementary ssDNA strands to create a 3′ 40-nucleotide dT overhang (3′-dT40), a 5′ 40-nucleotide dT overhang (5′-dT40), or non-poly(dT)-tailed (dT40−) substrate. The templates were radiolabeled, and the assays were performed at 55 °C. The second set used a radiolabeled 30-mer and a nonlabeled 31-mer single-stranded oligonucleotide that was annealed to a 61-mer single-stranded oligonucleotide to create a nicked substrate. Likewise, the assay using the nicked substrate was performed at 55 °C. A titration of the helicase protein demonstrated that Aq793 could separate the DNA duplex into ssDNA constituents for all of the tailed substrates as well as the nicked substrate (Fig. 1A). A graphical representation of the TBE acrylamide gel data showed that 50 pm, 6 nm, and 40 nm of Aq793 was required to unwind 50% of the 3′-dT40, 5′-dT40, and dT40− substrates, respectively, under these conditions and that 4 nm of Aq793 was necessary to unwind 50% of the nicked DNA substrate (Fig. 1B). These data indicated that Aq793 unwinds DNA with the strongest preference for the substrate with a poly(dT) 3′ overhang.
Effect of Aae MutL on Aq793 Helicase Unwinding Activity
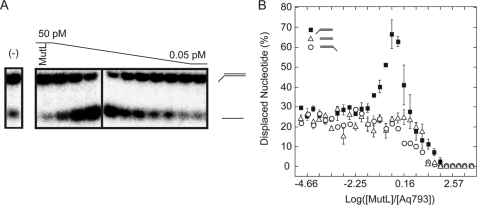
E. coli MutL has been reported to stimulate E. coli UvrD and Rep helicase activity (12). To investigate whether a similar effect occurs with the homologous A. aeolicus proteins, the helicase activity of Aq793 was assayed in the presence of differing amounts of Aae MutL. Conditions were chosen such that the helicase alone unwound ca. 20% of the respective substrate in 20 min as described under “Materials and Methods.” After 20 min the reactions were quenched by the addition of stop solution and applied to an acrylamide gel, and the products were visualized by autoradiography (Fig. 2A). The percentage of ssDNA formed was plotted against the Aae MutL:Aq793 mole ratio in the reaction. In the absence of Aae MutL, the helicase unwinding activity resulted in ca. 20% displaced ssDNA, as expected. Aq793 helicase activity was significantly stimulated by MutL near stoichiometric ratios of the proteins (Fig. 2B), with maximum stimulation occurring at 1.5:1 MutL:Aq793. No enhanced Aq793 activity was detected at low amounts of MutL; however, at high Aae MutL concentrations, the helicase activity was inhibited (Fig. 2B). Interestingly, MutL-enhanced Aq793 unwinding activity was not observed with the 5′-dT40 and dT40− deoxyoligonucleotide substrates (Fig. 2B).
FIGURE 2.
Aae MutL stimulation of Aq793 helicase activity. Serial dilutions of Aae MutL were incubated with a fixed concentration of Aq793 (23.5 pm) as described under “Materials and Methods.” The reactions were performed for 20 min at 55 °C and contained 0.02 nm duplex DNA substrate. The samples were resolved on a 20% TBE acrylamide gel and visualized by autoradiography. A, a representative TBE acrylamide gel of the reaction containing 3′-dT40 duplex DNA substrate. The expected mobility of dsDNA and ssDNA are indicated to the right of the picture. The negative control (−) contained Aq793 helicase but lacked MutL. B, graphical representation of the TBE acrylamide gel data. The gel bands were quantified using ImageQuant TL software, and the individual points represent the averages of three separate experiments, and the error bars show the standard deviation at each point. 3′-dT40 (solid squares), 5′-dT40 (open circles), and dT40− (open triangles) DNA substrates are shown.
Effect of Aae MutL Mutants on Aq793 Helicase Activity
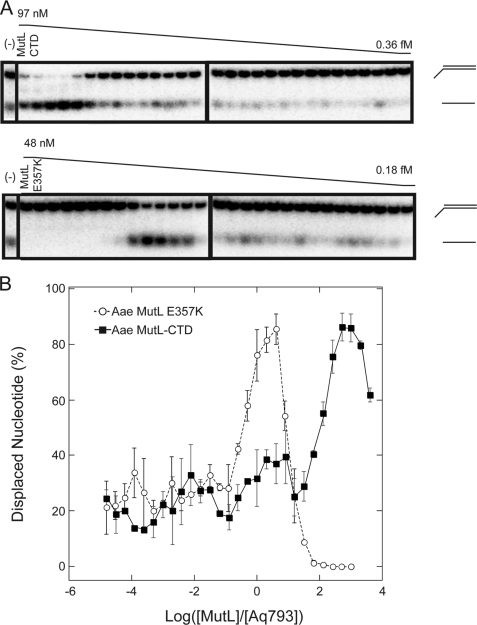
The effect that endonuclease-deficient and truncated forms of Aae MutL had on Aq793 unwinding activity was determined using the same methodology as described for the wild-type Aae MutL. Aae MutL (E357K) is an endonuclease-deficient form of the enzyme (8). It stimulated the helicase activity of Aq793 when the two proteins were present at nearly equimolar ratios (Fig. 3A) similar to the wild-type MutL protein. Also, in common with the wild-type enzyme, Aae MutL (E357K) at high molar ratios inhibited the Aq793 helicase activity (Fig. 3).
FIGURE 3.
Aae MutL (E357K) and Aae MutL-CTD stimulate Aq793 helicase activity. Aq793 was assayed for unwinding activity in the presence of serial dilutions of Aae MutL (E357K) and Aae MutL-CTD. The reactions were performed as described in the legend to Fig. 2 and under “Materials and Methods.” A, representative TBE acrylamide gel of the titration of Aae MutL-CTD (top gel picture) and Aae MutL (E357K) (bottom gel picture) mutants. Negative controls (−) contained Aq793 protein but not the respective MutL mutant. B, graphical representation of the effect of Aae MutL-CTD (solid squares) and Aae MutL (E357K) (open circles) on the oligonucleotide displacement activity of Aq793. Gel bands were quantified using ImageQuant TL software, and individual points represent the averages of three separate experiments.
The C-terminal 123 amino acid residues of Aae MutL, similar to the so-called LC20 domain of E. coli, was also able to stimulate Aq793 activity; however, maximum stimulation required an ∼1000-fold molar excess of Aae MutL-CTD over Aq793 (Fig. 3B). Very high excess of Aae MutL-CTD over Aq793 showed a trend toward less stimulation of helicase activity.
Effect of Aae MutL on Aq793 Helicase Activity on a Nicked Substrate
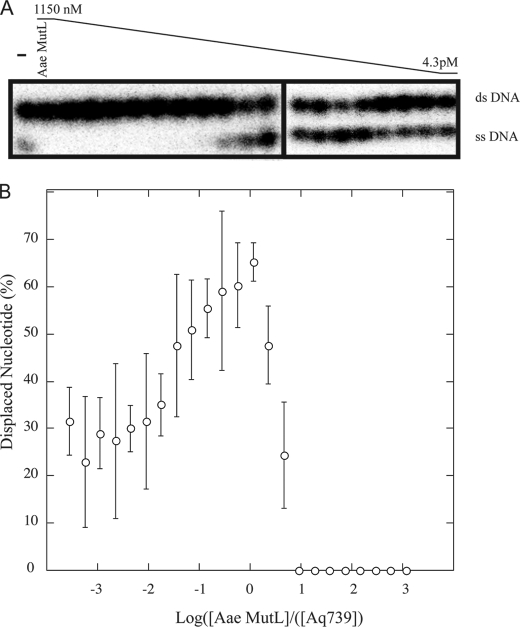
MutL has been reported to stimulate UvrD helicase activity during mismatch repair. This requires that MutL stimulate the UvrD helicase activity at a nick. To determine whether Aae MutL stimulated Aq793 activity at a nick, the Aq793 helicase activity was assayed on the nicked substrate in the presence of differing concentrations of Aae MutL. Conditions were optimized so that ∼10% of the nicked substrate was unwound in the presence of Aq793 alone as described under “Materials and Methods.” The reactions were quenched by the addition of stop solution and applied to a polyacrylamide gel. The reaction products were visualized by autoradiography (Fig. 4A), and the percentage of ssDNA was plotted against the Aae MutL:Aq793 mole ratio in the reaction (Fig. 4B). In the absence of Aae MutL, the helicase unwinding activity resulted in ca. 10% displaced ssDNA, as expected. Aq793 helicase activity was significantly stimulated by MutL near stoichiometric ratios of the proteins, with maximal stimulation occurring at a 1:1 MutL:Aq793 stoichiometric ratio. At high Aae MutL concentrations, the helicase activity was inhibited as observed with the other dsDNA substrates.
FIGURE 4.
Aae MutL stimulation of Aq793 helicase activity on a nicked substrate. Serial dilutions of Aae MutL were incubated with a fixed concentration of Aq793 (1 nm) as described under “Materials and Methods.” The reactions were performed for 20 min at 55 °C and contained 0.02 nm duplex DNA substrate. The samples were resolved on a 20% TBE acrylamide gel and visualized by autoradiography. A, a representative TBE acrylamide gel of the reaction containing nicked DNA substrate. The expected mobility of dsDNA and ssDNA are indicated to the right of the picture. The negative control (−) contained Aq793 helicase but lacked MutL. B, a graphical representation of the TBE acrylamide gel data. The gel bands were quantified using ImageQuant TL software. The individual points represent the averages of three separate experiments, and the error bars show the standard deviation at each point.
DISCUSSION
The E. coli UvrD helicase is involved in many DNA metabolic pathways. One of these pathways is the MMR pathway that ensures DNA replication fidelity and genomic integrity. In E. coli, MutL has been reported to stimulate UvrD helicase activity, most likely by increasing the rate of UvrD loading onto the DNA substrate (12). Eukaryotic MutL proteins have not been reported to stimulate a homologous helicase. Known mismatch repair pathways appear to use one of two methods to discriminate the daughter DNA strand from the parental strand. In E. coli MMR is methyl-directed, whereas in eukaryotes and many bacteria it appears to be nick-directed. The presence or absence of a MutH homologue may be diagnostic for which mechanism is utilized. Furthermore, the C-terminal domains of E. coli MutL homologues are significantly different from Aae MutL homologues, such as human PMS2 (8–10). It is also known that the LC20 domain of E. coli MutL is instrumental in binding and stimulating E. coli UvrD (22). Therefore, it is possible that the MutL proteins from organisms that lack MutH are not involved in UvrD helicase stimulation. This idea is supported by the previous publication that Tte and Bst MutL proteins, both hPMS2 homologues, did not stimulate UvrD helicase activity (29).
In contrast to the above idea, the work described herein demonstrates that a bacterial hPMS2 homologue can stimulate the helicase activity of UvrD homologues. The stimulation increased with increasing ratios of Aae MutL:Aq793 with the maximum helicase enhancement at a ratio of ∼1:1. At higher stoichiometries the helicase activity was eventually inhibited, probably because a large excess of Aae MutL in solution, not productively bound to DNA, was competing with the DNA substrate for Aq793 binding. Interestingly, the Aae MutL-CTD domain alone was able to stimulate Aq793 helicase activity. Although the LC20 of E. coli MutL was reported to contribute important contacts to UvrD binding and helicase stimulation, this region alone was not found to significantly enhance E. coli UvrD unwinding activity (22). The reason for this difference is not known, but it should be noted that the Aq793 stimulation required 1000-fold higher Aae MutL-CTD:Aq793 ratios than for the full-length Aae MutL protein.
We demonstrated for the first time that the putative helicase, Aq793, was able to unwind DNA. It displayed a substantial preference for unwinding a DNA substrate with a 3′ poly(dT) overhang as compared with a 5′ overhang or a non-poly(dT) tailed DNA. This property is shared by E. coli UvrD and reinforces that Aq793 is a 3′-5′ helicase (25). The Aae MutL stimulation of Aq793 activity was only observed when substrates possessed a 3′-poly(dT) overhang or at a nick. The Aae MutL may be binding to the ssDNA regions and loading the Aq793, at which point the helicase moves in a 3′-5′ direction. This would obviously increase the rate of unwinding a substrate with a 3′ overhang, but loading the helicase on a 5′ overhang would simply result in the helicase translocating along the ssDNA region without causing substrate unwinding. Despite the fact that no stimulation was observed with the substrates other than with 3′-dT40, at high Aae MutL:Aq793 ratios, helicase activity was inhibited probably by the mechanism discussed above.
In the annotated genome of A. aeolicus, Aq793 is listed as Rep helicase. However, this has not been experimentally verified, and the PUR subfamily helicases are difficult to differentiate based on sequence alone. Interestingly, we show that Aae MutL was able to stimulate the helicase activity of Aq793 on a nicked substrate. This would be expected if Aq793 were the A. aeolicus UvrD homologue. This study focused on whether Aae MutL could act to stimulate a PUR family helicase, something that had not been previously reported to occur in an organism that lacks an obvious MutH homologue. More detailed characterization is needed to ascertain whether Aq793 is actually a UvrD homologue.
E. coli MutL has been reported to stimulate both UvrD and Rep helicases. Homologues of human MutLα have not been reported to stimulate the PUR subfamily of helicases. Aae MutL is a hPMS2 homologue and has been demonstrated to possess a latent endonuclease activity (11) similar to the human protein. Eukaryotic MutL proteins are not known to stimulate the activity of a helicase such as UvrD. However, despite the sequence and biochemical similarities of hPMS2 and Aae MutL, the latter protein stimulates Aq793 helicase activity in a manner very similar to the interaction of E. coli MutL with E. coli UvrD. The work herein indicates that MMR, in at least some bacteria that lack MutH homologues, is an intriguing blend of the better studied MMR mechanisms exemplified by E. coli and Homo sapiens. Furthermore, A. aeolicus is in the phylum Aquificae, which some argue contains some of the earliest eubacterial species (31), and only one ATP-dependent DNA helicase homologue was identified in A.aeolicus. Therefore, the MMR pathway in A. aeolicus may have occurred very early in evolution.
In conclusion, we demonstrate for the first time that a human PMS2 homologue can stimulate the activity of a ATP-dependent DNA helicase. Furthermore, the C-terminal domain of Aae MutL is sufficient to promote this stimulation. A. aeolicus lacks MutH and appears to use a mismatch repair pathway that has similarities and differences with both E. coli and human MMR. It will be exciting to see whether this is a general trend in bacteria that lack MutH.
Footnotes
- MMR
- mismatch repair
- PUR
- PcrA/UvrD/Rep
- ds
- double-stranded
- ss
- single-stranded
- h
- human.
REFERENCES
- 1.Modrich P., Lahue R. (1996) Annu. Rev. Biochem. 65, 101–133 [DOI] [PubMed] [Google Scholar]
- 2.Schofield M. J., Hsieh P. (2003) Annu. Rev. Microbiol. 57, 579–608 [DOI] [PubMed] [Google Scholar]
- 3.Harfe B. D., Jinks-Robertson S. (2000) Annu. Rev. Genet. 34, 359–399 [DOI] [PubMed] [Google Scholar]
- 4.Kolodner R. (1996) Genes Dev. 10, 1433–1442 [DOI] [PubMed] [Google Scholar]
- 5.Kunkel T. A., Erie D. A. (2005) Annu. Rev. Biochem. 74, 681–710 [DOI] [PubMed] [Google Scholar]
- 6.Iyer R. R., Pluciennik A., Burdett V., Modrich P. L. (2006) Chem. Rev. 106, 302–323 [DOI] [PubMed] [Google Scholar]
- 7.Lahue R. S., Modrich P. (1988) Mutat. Res. 198, 37–43 [DOI] [PubMed] [Google Scholar]
- 8.Kadyrov F. A., Dzantiev L., Constantin N., Modrich P. (2006) Cell 126, 297–308 [DOI] [PubMed] [Google Scholar]
- 9.Jiricny J. (2006) Cell 126, 239–241 [DOI] [PubMed] [Google Scholar]
- 10.Kadyrov F. A., Holmes S. F., Arana M. E., Lukianova O. A., O'Donnell M., Kunkel T. A., Modrich P. (2007) J. Biol. Chem. 282, 37181–37190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukui K., Nishida M., Nakagawa N., Masui R., Kuramitsu S. (2008) J. Biol. Chem. 283, 12136–12145 [DOI] [PubMed] [Google Scholar]
- 12.Matson S. W., Robertson A. B. (2006) Nucleic Acids Res. 34, 4089–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ban C., Yang W. (1998) Cell 95, 541–552 [DOI] [PubMed] [Google Scholar]
- 14.Dutta R., Inouye M. (2000) Trends Biochem. Sci. 25, 24–28 [DOI] [PubMed] [Google Scholar]
- 15.Ban C., Junop M., Yang W. (1999) Cell 97, 85–97 [DOI] [PubMed] [Google Scholar]
- 16.Kosinski J., Plotz G., Guarné A., Bujnicki J. M., Friedhoff P. (2008) J. Mol. Biol. 382, 610–627 [DOI] [PubMed] [Google Scholar]
- 17.Mauris J., Evans T. C. (2009) PLoS One 4, e7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sancar A. (1994) Science 266, 1954–1956 [DOI] [PubMed] [Google Scholar]
- 19.Tomko E. J., Fischer C. J., Niedziela-Majka A., Lohman T. M. (2007) Mol. Cell 26, 335–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lohman T. M., Bjornson K. P. (1996) Annu. Rev. Biochem. 65, 169–214 [DOI] [PubMed] [Google Scholar]
- 21.Lahue R. S., Au K. G., Modrich P. (1989) Science 245, 160–164 [DOI] [PubMed] [Google Scholar]
- 22.Guarné A., Ramon-Maiques S., Wolff E. M., Ghirlando R., Hu X., Miller J. H., Yang W. (2004) EMBO J. 23, 4134–4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atkinson J., Guy C. P., Cadman C. J., Moolenaar G. F., Goosen N., McGlynn P. (2009) J. Biol. Chem. 284, 9612–9623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robertson A. B., Pattishall S. R., Gibbons E. A., Matson S. W. (2006) J. Biol. Chem. 281, 19949–19959 [DOI] [PubMed] [Google Scholar]
- 25.Matson S. W. (1986) J. Biol. Chem. 261, 10169–10175 [PubMed] [Google Scholar]
- 26.Yarranton G. T., Gefter M. L. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 1658–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruand C., Ehrlich S. D. (2000) Mol. Microbiol. 35, 204–210 [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi M., Dao V., Modrich P. (1998) J. Biol. Chem. 273, 9197–9201 [DOI] [PubMed] [Google Scholar]
- 29.An L., Tang W., Ranalli T. A., Kim H. J., Wytiaz J., Kong H. (2005) J. Biol. Chem. 280, 28952–28958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans T. C., Jr., Xu M. Q. (1999) Biopolymers 51, 333–342 [DOI] [PubMed] [Google Scholar]
- 31.Horiike T., Miyata D., Hamada K., Saruhashi S., Shinozawa T., Kumar S., Chakraborty R., Komiyama T., Tateno Y. (2009) Gene 429, 59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]