Abstract
Heparan sulfate (HS) is involved in essential physiological and pathophysiological functions. HS is a highly sulfated polysaccharide consisting of glucuronic acid (or iduronic acid) linked to glucosamine carrying various sulfo groups. Biosynthesis of HS involves sulfotransferases and an epimerase. The HS C5-epimerase converts glucuronic acid to iduronic acid. The method for determining the activity has been cumbersome due to the use of a site-specifically 3H-labeled polysaccharide substrate. Here, we report a two-enzyme coupling assay to determine the activity of C5-epimerase. HS 2-O-sulfotransferase (2OST) transfers the sulfo group to the 2-OH-position of glucuronic or iduronic acid. Unlike the wild type protein, 2-O-sulfotransferase mutant (2OST Y94I) transfers sulfate to the iduronic acid but not to the glucuronic acid. Thus, 2OST Y94I cannot sulfate N-sulfated heparosan, a polysaccharide containing glucuronic acid. Incubating N-sulfated heparosan with C5-epimerase converts some of the glucuronic acid to iduronic acid, thus becoming a substrate for 2OST Y94I. The susceptibility of the C5-epimerase-treated N-sulfated heparosan to 2OST Y94I modification directly correlates to the amount of the activity of C5-epimerase, proving that this two-enzyme coupling system can be used to assay for C5-epimerase. The method was further used to determine the activities of various C5-epimerase mutants. Our approach will significantly reduce the complexity for assaying the activity of C5-epimerase and facilitate the structural and functional analysis of C5-epimerase.
Keywords: Carbohydrate/Biosynthesis, Carbohydrate/Polysaccharide, Enzymes/Sulfotransferase, Extracellular Matrix/Glycosaminoglycan, Extracellular Matrix/Glycosaminoglycans, Extracellular Matrix/Heparan Sulfate, Extracellular Matrix/Heparin, Glycoproteins/Biosynthesis
Introduction
Heparan sulfate (HS)2 is a highly sulfated linear polysaccharide carrying negative charges under physiological pH. HS interacts with a variety of proteins (1, 2) and is involved in numerous essential physiological and pathophysiological activities. These include developmental processes, nutritional metabolism, wound repair, blood coagulation and tumor metastasis, and assisting viral infections (3–6). Heparin, a widely used anticoagulant drug, is a special form of HS with higher iduronic acid content and sulfation levels. The specific interactions between HS/heparin and proteins are attributed to the location of glucuronic (GlcUA)/iduronic (IdoUA) acid residues and the sulfation patterns (7–9).
The biosynthesis of HS occurs in the Golgi apparatus. The biosynthetic pathway involves a series of specialized enzymes, including HS polymerases, an epimerase, and several sulfotransferases. HS is first synthesized as a polysaccharide containing repeating units of GlcUA and GlcNAc. The polysaccharide is then subjected to various enzyme-catalyzed modifications. The glucosaminyl N-deacetylase/N-sulfotransferase converts a GlcNAc unit to N-sulfoglucosamine. Following the N-sulfation step, C5-epimerase (C5-epi) transforms GlcUA units into IdoUA (Fig. 1A). Because of the reversibility of the reaction catalyzed by C5-epi, the resultant polysaccharide contains both GlcUA and IdoUA units. The resultant polysaccharide is then modified by the 2-O-sulfotransferase (2OST) (Fig. 1B), 6-O-sulfotransferase, and 3-O-sulfotransferase. Understanding the mechanism of action of HS biosynthetic enzymes will delineate the biosynthetic mechanism of HS and aid the development of enzyme-based synthesis of HS and heparin (10).
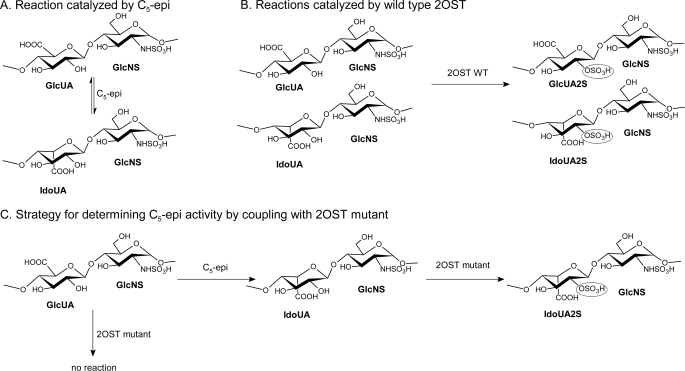
FIGURE 1.
Chemical reactions catalyzed by C5-epi and 2OST. A, HS C5-epi-catalyzed reaction. HS C5-epi catalyzes the conversion of a glucuronic acid unit in HS to iduronic acid and the reverse reaction, namely from the iduronic acid to glucuronic acid. B, 2OST WT-catalyzed reaction. 2OST WT catalyzes the 2-O-sulfation of both GlcUA and IdoUA units in the polysaccharide. C, strategy for determining C5-epi activity by coupling C5-epi and 2OST mutant. By coupling these two enzymes together, the IdoUA produced by the epimerase-catalyzed reaction is converted to IdoUA2S, which can be detected by the level of [35S]sulfate incorporation to the polysaccharide.
C5-epi is responsible for the synthesis of the IdoUA unit, a structurally flexible unit in HS polysaccharide. The flexibility contributes to the biological activities (11). For example, 2-O-sulfated iduronic acid (IdoUA2S) unit, the product of C5-epi and 2OST modifications, is required for binding to fibroblast growth factors (12, 13). Consequently, the HS containing the IdoUA2S unit displays the activity in stimulating cell proliferation via fibroblast growth factor and fibroblast growth factor receptor signaling pathways. The contribution of IdoUA or IdoUA2S to the binding affinity of antithrombin, a key regulating protein of the blood coagulation cascade, is more complex. The IdoUA2S unit is required for a pentasaccharide to bind to antithrombin (14). However, the fragments larger than a decasaccharide lacking IdoUA2S bind to antithrombin effectively (12). Only one isoform of C5-epi is present in the human genome. In vivo functions of C5-epi were uncovered from gene knock-out experiments. C5-epi knock-out mice are lethal, manifesting immature lung phenotype, abundant skeletal abnormalities, and kidney agenesis (15). It should be noted that C5-epi and 2OST reportedly form a complex in vivo, suggesting the intimate relationship between those two enzymes (16). As expected, the phenotypes displayed by C5-epi-knock-out mice are similar to those observed in 2OST-null mice.
One of our research goals is to understand the mechanism of action of C5-epi using a structural biology approach coupled with site-directed mutagenesis. The crystal structures of several HS sulfotransferases, including the N-sulfotransferase domain of N-deacetylase/N-sulfotransferase, three isoforms of 3-O-sulfotransferase, and 2OST, have been solved (17–21). We also demonstrated the feasibility to alter the substrate specificities based on the crystal structures (19, 20). Unlike sulfotransferases, the current method for measuring the activity of C5-epi is less straightforward. The method measures the release of 3H label from a site-specifically 3H-labeled polysaccharide (22). Although this method was successfully employed to purify the C5-epi (23), the preparation of the site-specifically 3H-labeled polysaccharide involves the use of a large amount of [3H]H2O, which cannot be readily prepared in a standard academic laboratory. Here, we report a two-enzyme coupled method to measure the activity of C5-epi using a 2OST mutant (Fig. 1C). The substrate used in this method is N-sulfoheparosan, a polysaccharide that contains only GlcUA. The mutant 2OST is engineered to specifically sulfate IdoUA. Therefore, the mutant 2OST does not react with N-sulfoheparosan unless the GlcUA unit is converted to an IdoUA unit by C5-epi (Fig. 1C). This method is able to screen the activity of C5-epi mutants in relatively large numbers. A disaccharide analysis of HS that was modified by C5-epi mutant/2OST confirmed the conclusions. Our method provides a unique application of engineered 2OST. This approach should enhance the study of the mechanism of action of C5-epi.
EXPERIMENTAL PROCEDURES
Preparation of Recombinant C5-epi
Human C5-epi was expressed in Origami-B DE3 cells (Novagen), which contain pGro7 (Takara, Japan) plasmid expressing chaperonin proteins GroEL and GroES of Escherichia coli as reported previously (24). Different N-terminally truncated forms of C5-epi were cloned into pMalc2x vector (New England BioLabs) to form maltose-binding protein (MBP)-epimerase fusion proteins. Several N-truncation points were prepared at Asp34, Glu53, Gln72, Asn92, Trp203, and Phe257. The transformed cells were grown in LB medium supplemented with 12.5 μg/ml tetracycline, 15 μg/ml kanamycin, 40 μg/ml chloramphenicol, and 50 μg/ml carbenicillin at 37 °C. The temperature was decreased to 22 °C when the A600 reached 0.4–0.8, followed by the addition of l-arabinose (1 mg/ml) and isopropyl β-d-thiogalactopyranoside (0.5 mm) to induce the expression of chaperone and C5-epi proteins, respectively. After shaking overnight, the cells were pelleted and lysed by sonication. The expressed proteins were then purified using an amylose-agarose (New England BioLabs) column following the protocol provided by the manufacturer. The protein product was analyzed by 10% SDS-PAGE stained with Coomassie Blue.
Preparation of Mutants for 2OST and C5-epi
The mutagenesis primers were purchased from Invitrogen. The expression of chicken 2OST was carried out as described previously (20). The preparation of 2-OST mutants followed the same procedures as reported (20). Human C5-epi mutant plasmids were prepared using the plasmid expressing MBP-C5-epi fusion protein as a template (24). This construct expresses the catalytic domain of C5-epi, which includes Glu53-Asn617. Unless otherwise specified, C5-epi WT is referred to MBP-C5-epi (Glu53-Asn617) fusion protein. The C5-epi mutants were expressed as described above. All of the mutant plasmids were confirmed by DNA sequencing (Genomic Analysis Core Facility, University of North Carolina). The expression levels of mutant proteins were assessed by SDS-PAGE by comparison with the wild type enzymes.
Preparation of N-Sulfoheparosan
Heparosan was isolated from the E. coli K5 strain, and the deacetylated heparosan was prepared following the method reported previously (25, 26). The deacetylated heparosan was incubated with N-sulfotransferase and the components for the PAPS (3′-phosphoadenosine 5′-phosphosulfate) regeneration system (12). The N-sulfoheparosan was recovered by a small diethylaminoethyl (DEAE)-Sephacel (40–160 μm; Sigma) column (27). The loaded sample was rinsed with a total of 3 ml of UPAS buffer (50 mm NaAcO, pH 5, 150 mm NaCl, 1 mm EDTA, 3 m urea, and 0.01% Triton X-100) four times, followed by washing with 3 ml of 250 mm NaCl with 0.001% Triton X-100. The polysaccharide was eluted with 1 ml of 1 m NaCl with 0.001% Triton X-100. To assess the level of N-sulfation, the N-sulfoheparosan (∼10 μg) was digested to disaccharides by a mixture of Flavobacterium heparinum heparin lyase I, II, and III (10 μg of each lyase) in 200 μl of 50 mm sodium phosphate buffer, pH 7.0, at 37 °C overnight (21, 28). The disaccharides were resolved by a C18 column under the reverse-phase ion pairing HPLC conditions as described below. We typically obtained a ratio of the N-sulfodisaccharide (ΔUA-N-sulfoglucosamine) and the N-acetylated disaccharide (ΔUA-GlcNAc) greater than 9, suggesting that the level of N-sulfation reached around 90% or more. The amount of the N-sulfoheparosan was also determined by the disaccharide analysis against a standard curve developed by the disaccharide standards (Seikagaku America).
Sulfotransferase Activity of 2OST and Mutants
The activities of 2OST WT and mutants were tested using N-sulfoheparosan or completely desulfated N-sulfated (CDSNS) heparin as a substrate. N-Sulfoheparosan contains GlcUA, whereas CDSNS heparin (designated as fully de-O-sulfated heparin from the Neoparin catalogue) contains mainly IdoUA. To determine the reactivity of 2OST toward two different substrates, enzymes (0.6 μg) were incubated with 6.6 × 105 cpm of [35S]PAPS and 2.0 μg of N-sulfoheparosan or CDSNS heparin (5 μg) in 100 μl of a reaction buffer containing 50 mm MES, 10 mm MnCl2, 5 mm MgCl2, 0.1 mg/ml bovine serum albumin, and 1% Triton X-100 for 5 min at 37 °C. The reaction was stopped by adding 100 μl of UPAS. The 35S-labeled polysaccharide product was purified by a DEAE column. The amount of 35S radioactivity was determined by scintillation counting.
Coupling 2OST and C5-epi to Determine the Activity of C5-epi
Wild type C5-epi (1.5 μg) was incubated with 2 μg of N-sulfoheparosan in the buffer containing 50 mm MES (pH 7.0), Triton X-100, and 1 mm CaCl2 at 37 °C for 30 min. The mixture was heated at 100 °C for 5 min and then cooled down to room temperature. After centrifugation to discard the insoluble matter, 0.6 μg of 2OST Y94I, 0.01 mg of bovine serum albumin, and [35S]PAPS (5–7 × 105 cpm) were added to the reaction, followed by incubation at 37 °C for another 5 min. The resultant 35S-labeled polysaccharide was purified by DEAE chromatography. To determine the linear range of this assay, a dose-response curve was also generated using wild type C5-epi (0–3.3 μg) and 2OST Y94I mutant (0.6 μg). The activities of various C5-epi mutants (1.5 μg) were also tested using this protocol.
Disaccharide Analysis of the Polysaccharide Modified by C5-epi and 2OST WT
The 35S-labeled polysaccharides were prepared as described above except that 2OST WT was used. Here we scaled up the reaction volume to 3 ml. Briefly, After a 30-min incubation of N-sulfoheparosan with C5-epi or its mutants, the reaction was terminated by heating at 100 °C for 5 min. Then 2OST WT, bovine serum albumin, and [35S]PAPS were added to the supernatant after the reaction mixture was spun down and incubated at 37 °C for an additional 5 min. After DEAE column purification, dialysis against deionized water, and drying by speed vacuum, the polysaccharide products were subjected to nitrous acid degradation at pH 1.5 (29). The disaccharides were separated by a BioGel P-2 column, which was equilibrated with 0.1 m ammonium bicarbonate (27, 30). The resultant disaccharides were resolved on a C18 column (0.46 × 24 cm; Vydac) under the reverse-phase ion-pairing (RPIP)-HPLC conditions. Here, the column was eluted with a stepwise gradient of acetonitrile containing 0.5 mm H3PO4, 9.5 mm NH4H2PO4, and 1 mm tetrabutylammonium phosphate monobasic (Sigma). The column was washed with 5.2% acetonitrile for 30 min first, followed by 15 min of 9% acetonitrile elution and then 60 min of 11.7% acetonitrile elution. The identities of the disaccharides were determined by coeluting with authentic disaccharide standards (31).
RESULTS
Redirecting the Substrate Specificity of 2OST
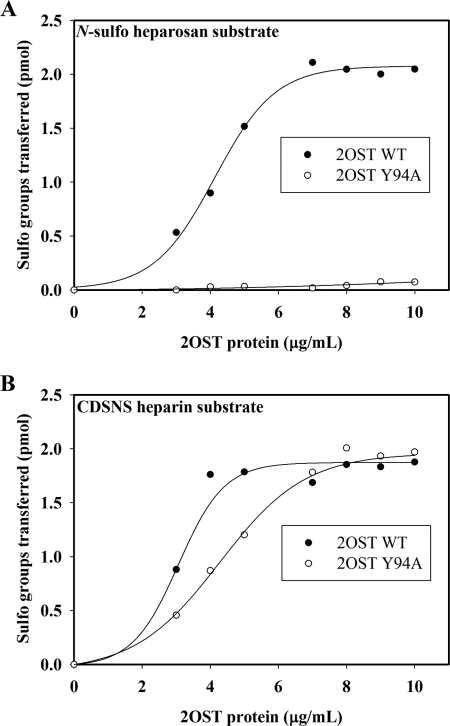
Wild type 2OST transfers the sulfate to the 2-OH position of either a GlcUA or an IdoUA unit as shown in Fig. 1B. Previously, introducing a single point mutation to the active site of 2OST altered the substrate specificity of the enzyme (20). The 2OST mutants, Y94A and H106A, selectively sulfate the IdoUA unit, whereas R189A selectively sulfates the GlcUA unit. To further confirm this finding, we compared the susceptibility of 2OST WT and 2OST Y94A to N-sulfoheparosan, a polysaccharide containing exclusively GlcUA across a range of concentrations of enzymes. Indeed, 2OST Y94A displayed merely a basal level of sulfation, whereas 2OST WT demonstrated excellent activity toward N-sulfoheparosan (Fig. 2A). In a separate experiment using CDSNS heparin as a substrate, a polysaccharide predominantly containing the IdoUA unit, both mutant and wild type proteins exhibited similar susceptibility, especially at higher enzyme concentrations (Fig. 2B). Taken together, our data suggest that the 2OST mutant could be used to detect the presence of IdoUA in the polysaccharide, leading to the possibility of assaying the activity of C5-epi using a two-enzyme coupling approach as depicted in Fig. 1C.
FIGURE 2.
The sulfotransferase activity of 2OST WT and Y94A mutant toward different polysaccharides. Different amounts of 2OST proteins (0–1.0 μg) were incubated with either 2.0 μg of N-sulfoheparosan (A) or 2.0 μg of CDSNS heparin (B) and 5.9 × 105 cpm of [35S]PAPS for 5 min at 37 °C. The sulfated polysaccharide was purified by DEAE resin and was determined by scintillation counting. The results for 2OST WT are shown by filled circles, and results for 2OST Y94A are shown in empty circles.
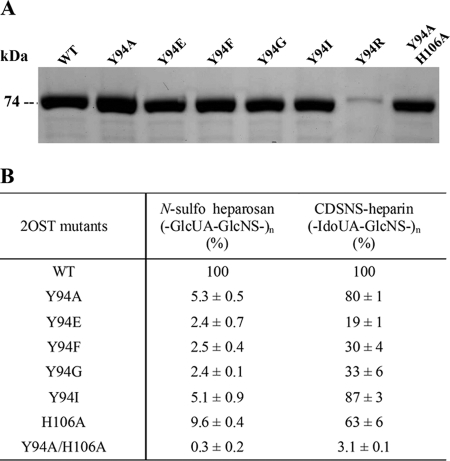
In an effort to further optimize the substrate selectivity of 2OST mutants, a series of single point mutations were introduced to replace Tyr94. The mutants were selected by replacing it with an acidic residue, such as Glu, or a basic residue, such as Arg. We also altered the size of the side chain by replacing it with either Phe or Gly. In addition, a double mutant, Y94A/H106A, was prepared based on the fact that 2OST H106A reportedly sulfates the IdoUA unit only (20). With the exception of 2OST Y94R, the mutants and the wild type proteins were all expressed successfully as an MBP fusion protein form exhibiting a molecular mass of 74 kDa as determined by the SDS-PAGE analysis (Fig. 3A). The sulfotransferase activity of the mutant proteins toward CDSNS heparin and N-sulfoheparosan was analyzed (Fig. 3B). Among the mutants, Y94A mutant retained most of the activity to CDSNS heparin (80%) while retaining only 5% of the activity to N-sulfoheparosan, consistent with previous findings (20). Similar results were obtained for the Y94I mutant, retaining activity at 87 and 5%, respectively. Substitution of tyrosine to phenylalanine (2OST Y94F) or glycine (2OST Y94G) also resulted in significant loss of sulfotransferase activity toward N-sulfoheparosan while maintaining substantial activity toward CDSNS heparin, suggesting that the size of side chain of the Tyr residue is not essential for the substrate selectivity. Similar results were obtained for the mutation of the tyrosine to negatively charged glutamate, although its sulfotransferase activity to CDSNS heparin decreased even further (19% of wild type protein). It should be noted that we do not know the contribution of a basic amino acid residue to the substrate selectivity because we failed to obtain a sufficient amount of 2OST Y94R to carry out the study. Unexpectedly, a double mutant, Y94A/H106A, lost nearly all activity toward both GlcUA and IdoUA units. Because substitution of Tyr94 with either alanine or isoleucine gave similar substrate selectivity to GlcUA and IdoUA units, we decided to choose 2OST Y94I for the subsequent study for assaying the activity of C5-epi.
FIGURE 3.
The expression and the sulfotransferase activity of 2OST mutants. The 2OST mutants were overexpressed in Origami-B cells using pMalc2x vector. The purified proteins were analyzed by 10% SDS-PAGE with 2OST WT (A). The sulfotransferase activities of these mutants were tested using either N-sulfoheparosan or CDSNS heparin as a substrate and are shown in percentage relative to wild type enzyme (B). The results are presented as average ± S.D., which was calculated based on three determinations.
Determination of the Activity of C5-epi Using Engineered 2OST
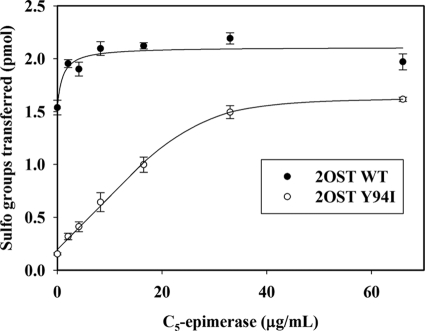
C5-epi catalyzes the reaction that converts a GlcUA unit of N-sulfoheparosan to an IdoUA unit (Fig. 1A). Because 2OST Y94I selectively sulfates the IdoUA, we employed the mutant protein to detect the level of IdoUA resulting from the action of C5-epi as illustrated in Fig. 1C. Indeed, incubation of N-sulfoheparosan with increasing amounts of C5-epi elevated the susceptibility to 2OST Y94I modification as determined by the amount of [35S]sulfo groups incorporated into the polysaccharide substrate (Fig. 4). A clear linear relationship (R2 = 0.99) between the quantity of C5-epi and the susceptibility to the 2OST Y94I modification was observed, suggesting that the method is valid to measure the activity of C5-epi. A control experiment using 2OST WT showed a high level of incorporation of [35S]sulfo groups to the N-sulfoheparosan in the absence of epimerase. Such result is anticipated because the wild type protein sulfates the GlcUA unit effectively. Further, a very narrow linear range of the 35S-labeled sulfated product in response to epimerase was observed, suggesting that 2OST WT enzyme is unsuitable for determining the activity of C5-epi using this method.
FIGURE 4.
Dose response of C5-epi to the [35S]sulfation by 2OST Y94I and 2OST WT. Different amounts of C5-epi (0–66 μg/ml) were incubated with 2.0 μg of N-sulfoheparosan for 30 min at 37 °C. Then epimerization reactions were stopped by boiling for 5 min. After centrifugation, the supernatants were mixed with either 2OST WT or 2OST Y94I and [35S]PAPS (6.4 × 105 cpm), followed by incubation for 5 min at 37 °C. The sulfated polysaccharide was purified by DEAE resin and determined by scintillation counting. The results (average ± S.D.) obtained using 2OST WT are shown by filled circles, and 2OST Y94I results are shown by empty circles. The bars indicate S.E. of triplicates.
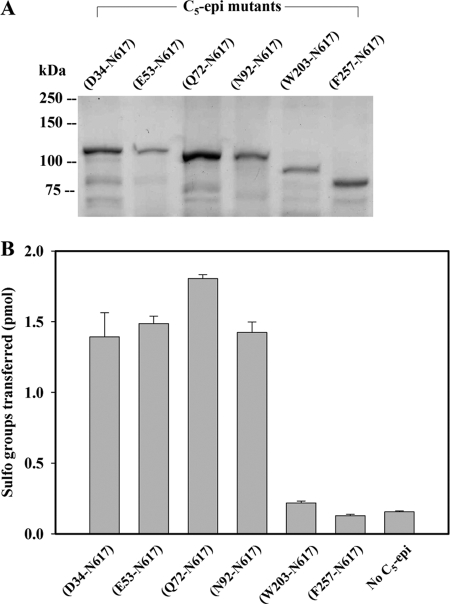
We then applied the newly developed assay to different C5-epi constructs, which differ in the sites of truncation from the N terminus. The human epimerase is a 617-amino acid protein that consists of three domains, cytoplasmic (Met1–Lys11), transmembrane (Thr12–Trp28), and catalytic domain (Asn29–Asn617) (see the UniProt data base on the World Wide Web). To further probe the catalytic domain of C5-epi, we prepared six different N-terminal truncated constructs in the format of MBP fusion proteins. To avoid the global structural changes in the protein, each truncation site was selected from a flexible loop based on the three-dimensional structural prediction using PredictProtein server (33). As expected, the size of the truncation mutants gradually decreased (Fig. 5A) as determined by SDS-PAGE, confirming the success in expressing the desired mutants. The mutant C5-epi (Asn92–Asn617) demonstrated epimerase activity similar to that of the three additional constructs, including C5-epi (Asp34–Asn617), C5-epi (Glu53–Asn617), and C5-epi (Gln72–Asn617). However, further truncated mutants C5-epi (Trp203–Asn617) and C5-epi (Phe257–Asn617) exhibited no epimerase activity (Fig. 5B). Our data suggest that the region of Asn92-Trp203 is important to the function of C5-epi.
FIGURE 5.
Activities of different truncations of C5-epi. The different truncations of human heparan sulfate C5 epimerase were overexpressed in Origami-B cells using the pMalc2x vector. A, purified proteins analyzed by SDS-PAGE. The values (average ± S.D.) for the activity measured by the developed assay (5.9 × 105 cpm of [35S]PAPS) are shown in B. The bars indicate S.E. of triplicates.
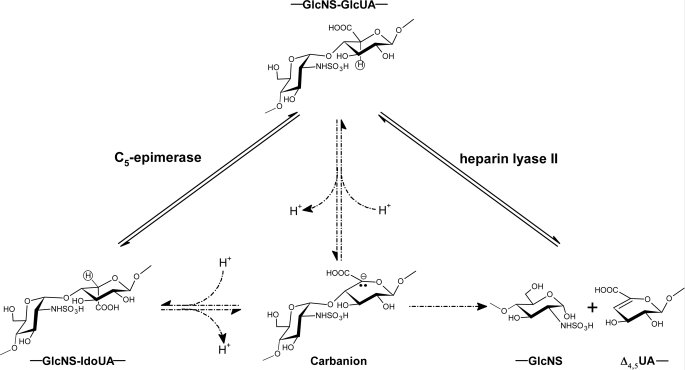
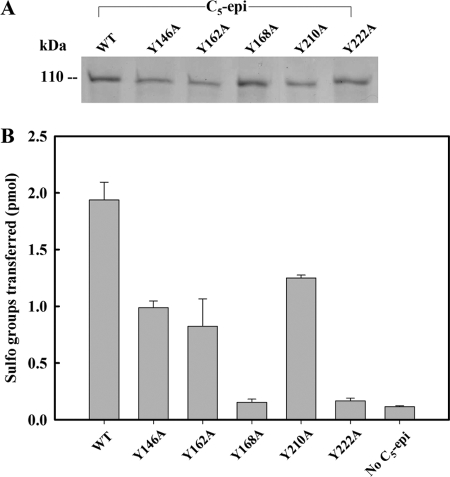
A tyrosine residue has been implicated to play an important role in performing the catalytic function of heparin lyases (34). It has been proposed that heparin lyase and C5-epi may share a similar transition state intermediate, a carbon anion at the C5-position of the GlcUA/IdoUA unit (Fig. 6) (34). Therefore, we speculated that some tyrosine residues could be essential for the activity of C5-epi. To this end, we mutated several selected tyrosine residues of C5-epi to alanine, including Tyr146, Tyr162, Tyr168, Tyr208, Tyr210, and Tyr222, and tested their activity in comparison with wild type enzyme using this assay. With the exception of C5-epi Y208A, all other mutants were expressed as analyzed by SDS-PAGE at a level comparable with that of the wild type protein (Fig. 7A). The activity measurement of C5-epi mutants revealed that the mutant proteins had different levels of activity. Both C5-epi Y168A and C5-epi Y222A mutants completely lost epimerase activity, suggesting a critical role of these two tyrosine residues in maintaining the activity of C5-epi (Fig. 7B). Other tyrosine mutants largely possess the activity, suggesting that these tyrosine residues are less crucial for the activity.
FIGURE 6.
Proposed catalytic mechanisms of C5-epi and heparin lyase II (34). Both C5-epi-catalyzed (left) and heparin lyase II-catalyzed (right) reactions go through the same carbanion intermediate by abstracting the C5 proton. This carbanion intermediate can be reprotonated (left) to produce epimerized product or proceed to an elimination reaction by breaking up the glycosidic bond (right). Left, the C5 proton is shown in a circle, which can be 3H-labeled.
FIGURE 7.
Analysis of tyrosine mutants of C5-epi. The tyrosine mutants were prepared by site-directed mutagenesis using the plasmid containing a truncated form of human C5-epi (Glu53–Asn617) as template. The purified proteins were analyzed by SDS-PAGE, shown in A. The activities (average ± range) are shown in B. The bars indicate the range of assay results for epimerase mutants prepared from two different batches.
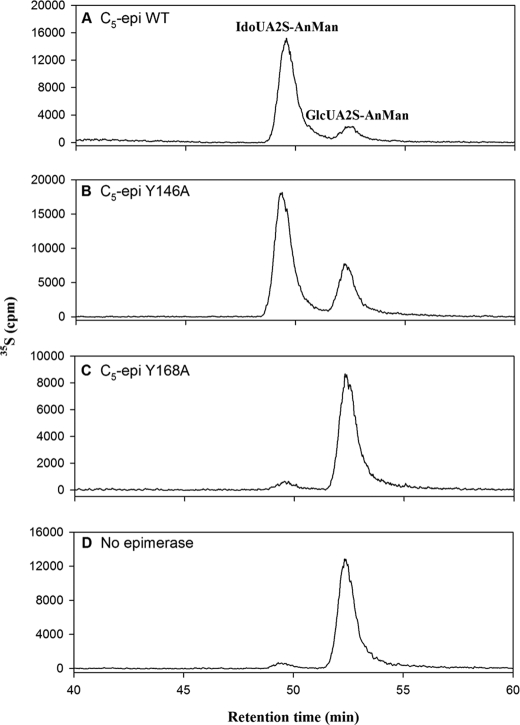
The activity levels of the different C5-epi tyrosine point mutants were also confirmed by disaccharide analysis of the 2-O-sulfated polysaccharide product using an established method (24). In this method, we reconstituted the biosynthesis of the polysaccharide containing IdoUA2S in vitro. The N-sulfoheparosan was incubated with various C5-epi point mutants, followed by the sulfation with 2OST WT. The resultant polysaccharide products were then subjected to nitrous acid degradation at pH 1.5 to yield 2-O-sulfated disaccharide products. The disaccharides were resolved by RPIP-HPLC to distinguish between IdoUA2S-AnMan and GlcUA2S-AnMan as described in the legend to supplemental Fig. S1. The presence of IdoUA2S-AnMan is indicative of active form C5-epi. As expected, when wild type C5-epi was present, IdoUA2S-AnMan was the major product (Fig. 8A). In the absence of C5-epi, only GlcUA2S-AnMan was detected (Fig. 8D). As the epimerase activity decreased (Y146A), the ratio of IdoUA2S and GlcUA2S dwindled (Fig. 8B). When the mutant (Y168A) lost all of the epimerase activity, no IdoUA2S-AnMan was found by the disaccharide analysis (Fig. 8C), just as in the case where C5-epi was absent (Fig. 8D). Additional data for the disaccharide analysis are shown in Table 1. Comparing the ratio of the two disaccharides and the sulfotransferase activity presented in Table 1, we concluded that the results of the disaccharide analysis are consistent with the analysis using the newly developed method.
FIGURE 8.
RPIP-HPLC chromatograms of the disaccharide analysis of the polysaccharides modified by various C5-epi mutants and 2OST WT. The polysaccharides were prepared by incubating with C5-epi and C5-epi point mutants followed by the [35S]sulfation using 2OST WT. The resultant disaccharides were resolved using RPIP-HPLC, as described under “Experimental Procedures.” A–D, chromatograms of the polysaccharide treated with C5-epi WT (A), C5-epi Y146A (B), C5-epi Y168A (C), and no C5-epi (D), respectively. It should be noted here that in C and D, the presence of the small peak at ∼49 min may be an artifact from nitrous acid degradation, and no IdoUA has been found in N-sulfoheparosan.
TABLE 1.
Summary of disaccharide analysis for C5-epi- and 2OST-modified HS
| C5-epi mutants | IdoUA2S-AnMan/GlcUA2S-AnMan | Relative C5-epi activitya |
|---|---|---|
| % | ||
| No C5-epi | ≤0.1 | |
| WT | 5.3 | 100 |
| Y146A | 2.2 | 40 |
| Y162A | 2.2 | 23 |
| Y168A | ≤0.1 | Inactive |
| Y210A | 6.2 | 71 |
| Y222A | ≤0.1 | Inactive |
a The relative activities were calculated from the data shown in Fig. 8.
DISCUSSION
In this study, we developed a sulfotransferase-based assay for measuring the HS C5-epi activity by coupling C5-epi with a 2OST mutant that specifically sulfates the IdoUA unit. C5-epi is a crucial enzyme in the HS biosynthetic pathway to prepare the HS with different biological functions. Campbell et al. (22) reported a method to measure the epimerase activity by determining the released tritium, using a site-specific 3H-labeled polysaccharide substrate. In this method, the 3H label was introduced at the C5-position of the GlcUA/IdoUA unit of CDSNS heparin (22) (Fig. 6). The C5-epi de-[3H]protonates at the C5-position of a GlcUA or IdoUA unit and thus dissociates the 3H label from the polysaccharide. Measuring the amount of 3H label released from the polysaccharide, one can determine the activity of C5-epi. Although the method was sufficiently robust to guide the purification of C5-epi from bovine liver (22), a large amount of [3H]H2O was used to prepare the 3H-labeled polysaccharide. Here, we bypass the need for the 3H-labeled polysaccharide for assaying C5-epi using a 2OST mutant with altered substrate specificity. This method demonstrated the linearity of the incorporation of 35S-sulfo groups versus the amount of C5-epimerase and can be efficiently utilized to screen various C5-epi mutants rapidly. Further, the results are validated with another method involving disaccharide analysis.
We attempted to expand the substrate selectivity of 2OST mutants toward the IdoUA unit by substituting Tyr94 with different types of amino acid residues. By comparing the susceptibility of various mutants to N-sulfoheparosan and CDSNS heparin, we found that 2OST Y94A or 2OST Y94I have comparable substrate selectivity and have the best substrate selectivity so far. The mechanistic role of the isoleucine (or alanine) residue in selecting IdoUA versus GlcUA unit could perhaps be revealed with the cocrystal structure of 2OST and an appropriate oligosaccharide substrate.
The similarity in catalytic transition state between heparin lyases and C5-epi has been noted because both enzymes involve deprotonation at the C5-position to yield a carbanion intermediate (Fig. 6). Such similarity may result from structural homology, at least to some extent (35). A recent study on the dermatan sulfate epimerase, an enzyme that catalyzes the conversion of the GlcUA to the IdoUA unit of chondroitin sulfate/dermatan sulfate, provides some evidence to support the idea (32). Three residues, His202, Tyr257, and His406, are found in the active site of the heparin lyase II from Pedobacter heparinus and may play catalytic roles based on the crystal structure (34). Corresponding histidine (His205 and His450) and tyrosine (Tyr261) were identified in dermatan sulfate epimerase by structural modeling, and mutation of these residues resulted in the complete loss of epimerase activity. Furthermore, the predicted three-dimensional structure of dermatan sulfate epimerase is very similar to the crystal structure of the heparin lyase II (34), suggesting that the two enzymes are evolutionarily related. Although no obvious amino acid sequence homology between C5-epi and heparin lyases was observed, we speculate that the tyrosine residue may also play important catalytic roles in C5-epi due to the fact that both heparin lyase II and C5-epi utilize a similar substrate and yield a carbanion intermediate (Fig. 6). We selected several tyrosine residues to investigate their roles in catalysis. A total of 31 Tyr residues are present in human C5-epi. Our attention was focused on those tyrosine residues located between Asn92 and Val235 based on the truncated mutational analysis and amino acid conservation among species. We postulated that some of the Tyr residues between Asn92 and Trp203 might contribute to activity because C5-epi Asn92–Asn617 is active, whereas C5-epi Trp203–Asn617 is inactive. In addition, we found that Glu105–Val235 is the most conserved region in the protein (73% identity) among different species, including zebrafish, platypus, mice, and humans. Indeed, mutation of tyrosine residues Tyr168 and Tyr222 of C5-epi abolished the activity, suggesting their possible roles in the catalytic function of C5-epi. Due to the inability to assess the overall structure of mutants, we could not rule out the possibility of the indirect effect of the tyrosine residues on the catalytic functions of C5-epi. Further experimental and structural evidence is required to elucidate the precise roles of these residues.
Coupling C5-epi with 2OST was previously described to qualitatively determine the presence of the activity of C5-epi, although a wild type 2OST was employed in this method (24). Because wild type 2OST sulfates both GlcUA and IdoUA, disaccharide analysis is required to confirm the activity of C5-epi by identifying the presence of a disaccharide-carrying IdoUA2S unit (as illustrated in supplemental Fig. S1). Unfortunately, the method is less quantitative, and the procedure is long due to the involvement of the disaccharide analysis. Our new method takes advantage of an engineered 2OST with unique substrate specificity, which sulfates only the IdoUA unit. The method is fast, sensitive, and quantitative. This simple and efficient assay will enhance the study of the structural and functional relationship of C5-epi.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants AI50050 and HL094463. This work is also supported by American Heart Association, MidAtlantic Affiliate, Grant-in-aid 0855424E.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- HS
- heparan sulfate
- C5-epi
- glucuronyl C5-epimerase
- CDSNS
- completely de-O-sulfated N-sulfated
- MBP
- maltose-binding protein
- GlcUA
- glucuronic acid
- IdoUA
- iduronic acid
- 2OST
- 2-O-sulfotransferase
- IdoUA2S
- 2-O-sulfoiduronic acid
- WT
- wild type
- DEAE
- diethylaminoethyl
- MES
- 4-morpholineethanesulfonic acid
- RPIP
- reverse-phase ion-pairing
- HPLC
- high pressure liquid chromatography
- PAPS
- 3′-phosphoadenosine 5′-phosphosulfate.
REFERENCES
- 1.Kreuger J., Spillmann D., Li J. P., Lindahl U. (2006) J. Cell Biol. 174, 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Capila I., Linhardt R. J. (2002) Angew. Chem. Int. Ed. Engl. 41, 391–412 [DOI] [PubMed] [Google Scholar]
- 3.Bishop J. R., Schuksz M., Esko J. D. (2007) Nature 446, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 4.Liu J., Pedersen L. C. (2007) Appl. Microbiol. Biotechnol. 74, 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J., Thorp S. C. (2002) Med. Res. Rev. 22, 1–25 [DOI] [PubMed] [Google Scholar]
- 6.Sasisekharan R., Shriver Z., Venkataraman G., Narayanasami U. (2002) Nat. Rev. Cancer 2, 521–528 [DOI] [PubMed] [Google Scholar]
- 7.Gama C. I., Tully S. E., Sotogaku N., Clark P. M., Rawat M., Vaidehi N., Goddard W. A., 3rd, Nishi A., Hsieh-Wilson L. C. (2006) Nat. Chem. Biol. 2, 467–473 [DOI] [PubMed] [Google Scholar]
- 8.Habuchi H., Habuchi O., Kimata K. (2004) Glycoconj. J. 21, 47–52 [DOI] [PubMed] [Google Scholar]
- 9.Nakato H., Kimata K. (2002) Biochim. Biophys. Acta 1573, 312–318 [DOI] [PubMed] [Google Scholar]
- 10.Peterson S. P., Frick A., Liu J. (2009) Nat. Prod. Rep. 26, 610–627 [DOI] [PubMed] [Google Scholar]
- 11.Raman R., Venkataraman G., Ernst S., Sasisekharan V., Sasisekharan R. (2003) Proc. Natl. Acad. Sci. 100, 2357–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J., Jones C. L., Liu J. (2007) Chem. Biol. 14, 986–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia J., Maccarana M., Zhang X., Bespalov M., Lindahl U., Li J. P. (2009) J. Biol. Chem. 284, 15942–15950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das S. K., Mallet J. M., Esnault J., Driguez P. A., Duchaussoy P., Sizun P., Herault J. P., Herbert J. M., Petitou M., Sinaÿ P. (2001) Chemistry 7, 4821–4834 [DOI] [PubMed] [Google Scholar]
- 15.Li J. P., Gong F., Hagner-McWhirter A., Forsberg E., Abrink M., Kisilevsky R., Zhang X., Lindahl U. (2003) J. Biol. Chem. 278, 28363–28366 [DOI] [PubMed] [Google Scholar]
- 16.Pinhal M. A., Smith B., Olson S., Aikawa J., Kimata K., Esko J. D. (2001) Proc. Natl. Acad. Sci. 98, 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kakuta Y., Li L., Pedersen L. C., Pedersen L. G., Negishi M. (2003) Biochem. Soc. Trans. 31, 331–334 [DOI] [PubMed] [Google Scholar]
- 18.Edavettal S. C., Lee K. A., Negishi M., Linhardt R. J., Liu J., Pedersen L. C. (2004) J. Biol. Chem. 279, 25789–25797 [DOI] [PubMed] [Google Scholar]
- 19.Xu D., Moon A. F., Song D., Pedersen L. C., Liu J. (2008) Nat. Chem. Biol. 4, 200–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bethea H. N., Xu D., Liu J., Pedersen L. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18724–18729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moon A. F., Edavettal S. C., Krahn J. M., Munoz E. M., Negishi M., Linhardt R. J., Liu J., Pedersen L. C. (2004) J. Biol. Chem. 279, 45185–45193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell P., Hannesson H. H., Sandbäck D., Rodén L., Lindahl U., Li J. P. (1994) J. Biol. Chem. 269, 26953–26958 [PubMed] [Google Scholar]
- 23.Li J., Hagner-McWhirter A., Kjellén L., Palgi J., Jalkanen M., Lindahl U. (1997) J. Biol. Chem. 272, 28158–28163 [DOI] [PubMed] [Google Scholar]
- 24.Muñoz E., Xu D., Avci F., Kemp M., Liu J., Linhardt R. J. (2006) Biochem. Biophys. Res. Commun. 339, 597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindahl U., Li J. P., Kusche-Gullberg M., Salmivirta M., Alaranta S., Veromaa T., Emeis J., Roberts I., Taylor C., Oreste P., Zoppetti G., Naggi A., Torri G., Casu B. (2005) J. Med. Chem. 48, 349–352 [DOI] [PubMed] [Google Scholar]
- 26.Vann W. F., Schmidt M. A., Jann B., Jann K. (1981) Eur. J. Biochem. 116, 359–364 [DOI] [PubMed] [Google Scholar]
- 27.Xia G., Chen J., Tiwari V., Ju W., Li J. P., Malmstrom A., Shukla D., Liu J. (2002) J. Biol. Chem. 277, 37912–37919 [DOI] [PubMed] [Google Scholar]
- 28.Duncan M. B., Liu M., Fox C., Liu J. (2006) Biochem. Biophys. Res. Commun. 339, 1232–1237 [DOI] [PubMed] [Google Scholar]
- 29.Shively J. E., Conrad H. E. (1976) Biochemistry 15, 3932–3942 [DOI] [PubMed] [Google Scholar]
- 30.Chen J., Duncan M. B., Carrick K., Pope R. M., Liu J. (2003) Glycobiology 13, 785–794 [DOI] [PubMed] [Google Scholar]
- 31.Chen J., Liu J. (2005) Biochim. Biophys. Acta 1725, 190–200 [DOI] [PubMed] [Google Scholar]
- 32.Pacheco B., Maccarana M., Goodlett D. R., Malmström A., Malmström L. (2009) J. Biol. Chem. 284, 1741–1747 [DOI] [PubMed] [Google Scholar]
- 33.Rost B., Yachdav G., Liu J. (2004) Nucleic Acids Res. 32, W321–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaya D., Tocilj A., Li Y., Myette J., Venkataraman G., Sasisekharan R., Cygler M. (2006) J. Biol. Chem. 281, 15525–15535 [DOI] [PubMed] [Google Scholar]
- 35.Shriver Z., Hu Y., Pojasek K., Sasisekharan R. (1998) J. Biol. Chem. 273, 22904–22912 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.