Abstract
The homeodomain transcription factor Pitx2 and the T-box transcription factors are essential for organogenesis. Pitx2 and T-box genes are induced by growth factors and function as transcriptional activators or repressors. Gene expression analyses on abdominal tissue were used to identify seven of the T-box genes of the genome as Pitx2 target genes in the abdomen at embryonic day.10.5. Pitx2 activated Tbx4, Tbx15, and Mga and repressed Tbx1, Tbx2, Tbx5, and Tbx6 expression. As expected, activated genes showed reduced expression patterns, and repressed T-box genes showed increased expression patterns in the abdomen of Pitx2 mutants. Pitx2 occupied chromatin sites near all of these T-box genes. Co-occupancy by coactivators, corepressors, and histone acetylation at these sites was frequently Pitx2-dependent. Genes repressed by Pitx2 generally showed increased histone acetylation and decreased histone deacetylase (HDAC)/corepressor occupancy in Pitx2 mutants. The lower N-CoR, HDAC1, and HDAC3 occupancy observed at multiple sites along Tbx1 chromatin in mutants is consistent with the model that increased histone acetylation and gene expression of Tbx1 may result from a loss of recruitment of corepressors by Pitx2. Genes activated by Pitx2 showed less consistent patterns in chromatin analyses. Reduced H4 acetylation and increased HDAC1/nuclear receptor corepressor (N-CoR) occupancy at some Tbx4 sites were accompanied by increased H3 acetylation and reduced HDAC3 occupancy at the same or other more distal chromatin sites in mutants. Pitx2-dependent occupancy by corepressors resulted in alteration of the acetylation levels of several T-box genes, whereas Pitx2-dependent occupancy by coactivators was more site-localized. These studies will provide the basic scientific underpinning to understand abdominal wall syndromes.
Keywords: Chromatin Immunoprecipitation (ChiP), Coactivator Transcription, Development, Gene Transcription, Transcription Regulation, Transcription Target Genes
Introduction
Definitive endoderm and lateral plate mesoderm cells are formed and begin to migrate laterally between the ectoderm and primitive endoderm during early gastrulation. Together, these cells form the abdominal wall that begins to enclose the internal organs shortly after the mouse embryo turns. The abdominal wall at mouse embryonic day (E)3 9.5 is composed of lateral plate mesoderm-derived mesenchymal cells inserted between ectoderm- and endoderm-derived cell layers. Abdominal somites are beginning to extend ventrally into the lateral plate-derived mesenchyme to form abdominal wall muscle anlagen. Classic ventral body wall defects are characterized by a thin body wall, muscular dysplasia, and/or absence of midline fusion (1, 2).
Several sequence-specific DNA binding transcription factors (SSTFs) are involved in the pathogenesis of congenital body wall defects. These include the homeodomain transcription factor Pitx2, which is essential for organ formation and body wall closure (3–6). Pitx2 is a target of growth factor signaling pathways that mediate cell type-specific control of proliferation. Activation of the Wnt/β-catenin pathway results in the release of Pitx2-associated corepressors and mediates recruitment of specific coactivator complexes in myoblasts (7). T-box transcription factors are involved in induction of mesendoderm (8), subdivision of posterior mesoderm into rostral and caudal domains, organ patterning and formation (9, 10), and body wall development (11). T-box genes can be rapidly induced by growth factors (12), function as transcriptional activators or repressors (13–20), and interact with nucleosome assembly (21) or chromatin-modifying proteins (22).
Coactivators and corepressors mediate the regulatory actions of SSTFs by a series of exchanged cofactor complexes that execute enzymatic modifications of nucleosomes and chromatin and interact with the basal transcription apparatus. Coactivators and corepressors are components of complexes that exhibit diverse enzymatic activities. These typically involve the covalent modification of histone tails by adding or removing acetyl, methyl, phosphate, ADP-ribose, ubiquitin, or small ubiquitin-like modifier moieties (23). Acetylation of histones H3 and H4 counteracts the tendency of nucleosomal fibers to fold into higher order chromatin structures. Relaxed chromatin is more accessible to interacting proteins and active gene expression. Acetylation is mediated by a series of histone acetyl transferases including the CBP/p300 family. In contrast, histone deacetylases (HDACs) reduce gene expression by deacetylating lysine residues in the N-terminal tails of histone proteins, thereby encouraging chromatin compaction. HDAC1, HDAC2, and HDAC3 have also been found in both N-CoR and silencing mediator for retinoid or thyroid-hormone receptor (SMRT) complexes (24–26).
Pitx2 is first expressed in the ventrally located somatopleure at E8.5, which will become the abdominal wall, and follows myogenic expression in the trunk (27). Pitx2 plays a critical role in jaw development by regulating Tbx1 (28). It is known that several T-box genes are expressed in and play essential roles in body wall development (11). We therefore examined T-box expression in Pitx2 mutants during abdominal wall development. Seven T-box genes were identified and validated as Pitx2 target genes. The genomic non-coding regions surrounding all of these genes showed clustered and conserved Pitx2 binding motifs within and outside of conserved and ultraconserved regions. Chromatin immunoprecipitation (ChIP) assays with abdominal tissue were used to demonstrate Pitx2 occupancy at all T-box target genes in vivo and compare Pitx2 occupancy at several sites scattered throughout the Pitx2-repressed Tbx1 and Pitx2-activated Tbx4 genes. The Pitx2 dependence of co-occupancy by coactivators (CBP and PCAF), corepressors (N-CoR, HDAC1, and HDAC3), and histone acetylation was also examined at these sites. Pitx2-dependent changes in chromatin were consistent with changes in expression for almost all sites in Pitx2-repressed T-box genes. In contrast, chromatin changes in Pitx2-activated T-box genes showed a less predictable pattern. The data were consistent with the model that Pitx2 represses its target genes by recruiting corepressors and HDACs at numerous genomic locations within these genes (7). The opposing Pitx2-dependent chromatin effects observed at different genomic sites in Tbx4 suggest that Pitx2 activates its target genes by more site-localized, or indirect, mechanisms.
EXPERIMENTAL PROCEDURES
Mice
All research was conducted in compliance with the Public Health Service (PHS) Policy on the Humane Care and Use of Laboratory Animals, the United States Department of Agriculture (USDA) Animal Welfare Act and Regulations (66), and the United States Government Principles for the Utilization and Care of Vertebrate Animals Used in Research, Teaching and Testing. All research that involved the use of vertebrate animals was reviewed and approved by the Oregon State University Institutional Animal Care and Use Committee (IACUC). ICR Pitx2+/LacZ mouse line (6) was used. Pitx2+/LacZ mice were bred, and females were checked for the presence of vaginal plug (E0). Embryos were isolated at E10.5, and yolk sacs were used for genotyping.
RNA Preparation and Microarray Analysis
Total RNA was prepared using Qiagen RNeasy mini kit, labeled using Affymetrix one-step labeling, and used to probe the Affymetrix Mouse Genome 430a 2.0 array. Results were analyzed using FileMakerpro software (29, 30).
Whole Mount RNA in Situ Hybridization
Mouse embryos free of membranes were fixed with 4% paraformaldehyde, dehydrated, and rehydrated with a graduated MeOH in phosphate-buffered saline, 0.1% Tween series. Embryos were treated with 10 μg/ml protease K (Invitrogen) and processed as described previously (28). Stained samples were photographed with a Discovery V8 Zeiss microscope and Axiocam camera system.
Quantitative Real-time PCR (qPCR)
cDNA or immunoprecipitated (IP) DNA (25 ng) was analyzed by qPCR using SYBR Green I methodology. DNA was preincubated at 95 °C for 10 min followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 60 °C for 32 s, extension at 72 °C for 32 s, and a final elongation step of 2 min at 72 °C. The melting curves of all samples were routinely determined by melting curve analysis to ascertain that only the expected products had been generated. All samples were analyzed in triplicate and normalized by glyceraldehyde-3-phosphate dehydrogenase expression levels. Primers used are listed in supplemental Table 1.
ChIP
Abdominal wall tissue was dissected from E10.5 mice, cross-linked with 1% formaldehyde for 10 min at room temperature, and followed by the addition of glycine (125 mm) to quench formaldehyde. Tissue was washed twice with ice-cold phosphate-buffered saline and lysed in 200 μl of lysis buffer (20 mm Tris-HCl, pH 8.1, 1% SDS, 10 mm EDTA, 1× protease inhibitor mixture (Roche Applied Science)). Lysates were sonicated nine times for 10 s (40% output, Branson digital sonifier) to yield sheared DNA amplicons averaging less than 500 bp and clarified by centrifugation (13,000 rpm, 10 min, 4 °C). Each sample was split into input and IP. The material for IP was diluted 1:10 in dilution buffer (1% Triton X-100, 2 mm EDTA, 150 mm Tris-HCl, pH 8.1, protease inhibitor mixture). Preclearing was performed with 2 μg of salmon sperm DNA, 20 μl of goat IgGs, and 45 μl of protein G-Sepharose (50% slurry) for 2 h at 4 °C. Samples were incubated with 2 μg of goat anti-Pitx2 (C-16) polyclonal antibody (Santa Cruz Biotechnology, sc-8748 (28)), anti-HDAC1 (Santa Cruz Biotechnology, sc8410 (7, 31)), anti-HDAC3 (Bethyl Laboratories), anti-N-CoR (Santa Cruz Biotechnology; sc-8994 (7)), anti-CBP (Santa Cruz Biotechnology, sc-369 (7, 32)), anti-PCAF (Santa Cruz Biotechnology, sc-8999 (7, 33)), anti-acetylated H3 or H4 (Upstate Biotechnology) overnight at 4 °C followed by the addition of 45 μl of protein G-Sepharose for 1 h at 4 °C. Sepharose beads were washed sequentially with TSE1 (0.1% SDS, 1% Triton X-100, 20 mm Tris-HCl, pH 8.1, 2 mm EDTA, 150 mm NaCl), TSE2 (0.1% SDS, 1% Triton X-100, 20 mm Tris-HCl, pH 8.1, 2 mm EDTA, 500 mm NaCl), and TSE3 (0.25 m LiCl, 1% Nonidet P-40, 1% deoxycholate, 10 mm Tris-HCl, pH 8.1, 1 mm EDTA). Beads were washed with Tris-EDTA and eluted with 1% SDS, 0.1 m NaHCO3, and 0.3 m NaCl. RNase A was added, and the samples were incubated overnight at 65 °C. Following ethanol precipitation, the samples were resuspended (10 mm Tris-HCl, pH 7.5, 5 mm EDTA, 0.25% SDS) and treated with proteinase K for 2 h at 45 °C. DNA was purified with QIAquick columns (Qiagen) and analyzed in a 7500 real-time PCR system using SYBR Green I to evaluate the relative abundance of sequences in input and IP material. Primers used are listed in supplemental Table 1.
Immunoprecipitation and Immunoblotting
Nuclear extracts were prepared as described previously (34) from abdominal tissue of E10.5 Pitx2+/+ mice. 100 μg of nuclear extract were precleared with protein G-Sepharose (Amersham Biosciences) in Buffer IP150 (10 mm HEPES, pH 7.9, 10% glycerol, 1 mm EDTA, 150 mm NaCl, 0.1% Nonidet P-40) at 4 °C for 1 h to reduce nonspecific protein binding. After centrifugation, the precleared samples were incubated with 1 μg of antibodies at 4 °C overnight followed by the addition of protein G-Sepharose and incubation at 4 °C for 1 h. Sepharose beads were collected by centrifugation, washed three times with buffer IP350 (10 mm HEPES, pH 7.9, 10% glycerol, 1 mm EDTA, 350 mm NaCl, 0.1% Nonidet P-40) and once with IP150, and resuspended in denaturing sample buffer. Immune complexes were separated by SDS-PAGE and analyzed by Western blotting with appropriate antibodies.
RESULTS
Identification of T-box Genes Regulated by Pitx2 in Abdominal Tissue
Gene expression analysis was used to identify Pitx2 target genes during development. Abdominal walls of E10.5 mice were obtained by cutting across embryos behind the forelimb and in front of the hindlimb. The neural tube and obvious internal organs were removed. The abdominal tissue from which total RNA was extracted therefore consisted predominantly of abdominal body wall and included somites. Total RNA was prepared from the abdominal walls of three pools of wild type (Pitx2+/+, WT), three pools of heterozygote (Pitx2+/LacZ, HET), and three pools of mutant (Pitx2LacZ/LacZ, MUT) embryos. Probes prepared from these RNA were applied to nine Mouse Genome 430a 2.0 microarrays. The results from all nine arrays were normalized by RMA. The average expression value obtained from three biological replicates was compared between genotypes. Array results have been deposited for public access at ArrayExpress under the accession number E-MEXP-2332.
The arrays demonstrate that Pitx2 RNA levels decline significantly in the MUT isolates. Pitx2 expression was measured by two probe sets. Probe set 1424797-a-at, which was located within the 3′-non-translated region of Pitx2, produced a robust signal and showed 1.8-fold lower signal in MUT than WT or HET. A second Pitx2 probe set (1450482-a-at), which was located within the coding portion of the last exon, produced a 3-fold lower signal than the first probe and showed 4.7- and 4.3-fold lower signal in MUT than in WT or HET, respectively. Pitx2 levels were not significantly different in WT and HET arrays. Only 10 of ∼20,000 other genes showed higher -fold changes than Pitx2.
Pitx2 affected the expression of many other genes at lower levels. Scatter plot comparisons of individual arrays showed fewer changes in internal comparisons, between replicate arrays from the same genotype, than in cross comparisons, between individual arrays from different genotypes (data not shown). The number of regulated genes can be estimated by permutation fold-scanning analysis (28, 29). This non-parametric method counts probe set comparisons that fall above each -fold cutoff between conditions (cross comparisons) and within conditions (internal comparisons) and uses them to calculate the number of regulated genes as a function of -fold change or false discovery rate (FDR) (29, 30). An FDR of 8% was calculated for WT versus MUT comparisons at a 1.7-fold cutoff. There were 600 probe sets that change above this cutoff, corresponding to ∼300 genes. An FDR of 9% FDR was calculated for HET versus MUT comparisons at this -fold cutoff, and 550 probe sets changed above the cutoff. In contrast, a minimum FDR of 55% was calculated at a 1.4-fold cutoff for WT versus MUT comparisons, and less than 50 probe sets changed above this cutoff (data not shown).
Scatter plot comparisons between individual replicate arrays within conditions also showed that one of the three replicate arrays in each condition was different from the other two, which matched more closely. The outlier in each of the three conditions was the initial array that was produced for preliminary studies at a different time than the other two, suggesting a technical deviation. The latter two were therefore used to establish a list of putative T-box target genes (Table 1). T-box gene expression was consistently altered in all three replicates.
TABLE 1.
Microarray expression measurements on all T-box genes in the mouse genome
Genes in boldface were selected for further evaluation. *, difference not significant at p = 0.05 using Student's t test.
| MGI#a | WTb (Average ± S.D.) | MUTb (Average ± S.D.) | Fold Δ WT vs. MUTc (largest to smallest) | HETb (Average ± S.D.) | Fold Δ HET vs. MUTc (largest to smallest) | |
|---|---|---|---|---|---|---|
| Tbx1 | 98493 | 114 ± 4 | 143 ± 34 | 1.3* | 149 ± 1 | −1.0* |
| Tbx2 | 98494 | 413 ± 24 | 632 ± 34 | 1.5 | 463 ± 40 | 1.4 |
| Tbx3 | 98495 | 812 ± 27 | 975 ± 62 | 1.2, 1.1, −1.0*, −1.2* | 889 ± 75 | 1.1*, −1.0*, 1.2*, 1.1* |
| Tbx4 | 102556 | 1457 ± 120 | 411 ± 67 | −3.5, −3.2 | 879 ± 89 | −2.1, −2.4 |
| Tbx5 | 102541 | 355 ± 15 | 671 ± 101 | 1.9, 2.2 | 331 ± 4 | 2.0, 1.9 |
| Tbx6 | 102539 | 40 ± 1 | 37 ± 0.3 | −1.1 | 37 ± 4 | 1.0* |
| Tbx15 | 1277234 | 396 ± 91 | 129 ± 0.0 | −3.1, −3.3 | 64 ± 1 | 2.0, 1.4 |
| Tbx18 | 1923615 | 456 ± 33 | 393 ± 26 | −1.2*, 1.0* | 293 ± 3 | 1.3, 1.2 |
| Tbx19 | 1891158 | 49 ± 1 | 54 ± 2 | 1.1, 1.1* | 44 ± 2 | 1.2, 1.2* |
| Tbx20 | 1924493 | 424 ± 27 | 362 ± 13 | −1.2, −1.0*, −1.1* | 554 ± 37 | −1.5, −1.6, −1.1* |
| Tbx21 | 1888984 | 16 ± 2 | 15 ± 1 | −1.0* | 16 ± 0.4 | −1.0* |
| Tbx22 | 2389465 | 14 ± 1 | 14 ± 0.0 | 1.0* | 14 ± 0.4 | 1.0* |
| T | 98472 | 42 ± 7 | 36 ± 2 | −1.2* | 39 ± 3 | −1.1* |
| Tbr1 | 107404 | 11 ± 1 | 12 ± 0.0 | 1.0* | 12 ± 1 | −1.0* |
| Eomes | 1201683 | 40 ± 0.0 | 37 ± 2 | −1.1*, 1.0* | 38 ± 2 | −1.0*, −1.1* |
| Mga | 1352483 | 651 ± 38 | 419 ± 18 | −1.6, −1.3, −1.3*, 1.0*, −1.1* | 448 ± 23 | −1.1*, −1.2*, −1.1*, −1.1*, −1.2* |
a Mouse Genome Informatics identification number.
b Average of two biological replicates from the probe set with the highest average signal across all six arrays.
c -Fold changes computed by dividing the higher value by the lower and then assigning direction; Each -fold change is for a different probe set for the same gene; organized from highest to lowest average signal for that gene over all six arrays.
Previous studies have shown that Pitx2 regulates Tbx1 by direct interaction with its promoter during branchial arch development (30). Consequently, alteration of T-box gene expression was of particular interest to us in the abdominal wall context as well. All probe sets corresponding to T-box genes were therefore examined for Pitx2-dependent expression (Table 1). Tbx2, Tbx4, Tbx5, Tbx15, Tbx19, Tbx20, and Mga showed consistent, significant alterations of expression in different comparisons and/or with different probe sets. Six genes of the T-box family were selected as putative Pitx2 targets in the abdomen (Table 1, bold). Single probe sets for Tbx6 and Tbx1 showed little and/or insignificant change of expression in array data. These genes were selected for further analyses because of their known abdominal expression and interaction with Pitx2, respectively. Tbx1 is involved in craniofacial and cardiac development (35, 36). Tbx2 is involved in cardiac and limb development (37). Tbx4 is involved in allantois and hindlimb formation (38, 39). Tbx5 is involved in cardiac development and is required for forelimb formation (13, 40). Tbx15 is involved in skeletal development (41, 42). Mga is a dual specificity transcription factor that regulates both Max network and T-box genes with no known function yet during development (43). Tbx19 and Tbx20 were not selected for further evaluation because they showed very small changes in WT versus MUT comparisons, and they have predominantly been associated with pituitary (44) and cardiac development (45).
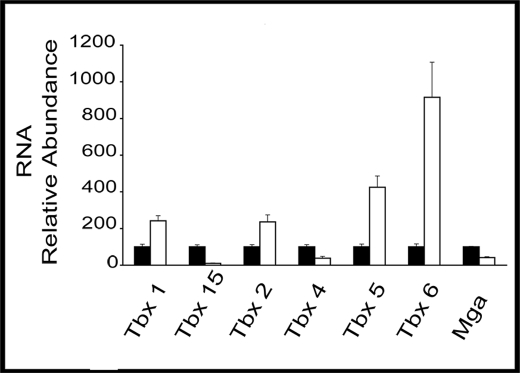
Microarray data were initially validated by qPCR on reverse-transcribed RNA from E10.5 abdominal tissue. RNA levels for selected putative T-box target genes were always altered in the same direction but sometimes to a different extent, as in microarray results. RNA was both from the same preparations used in array analyses and from fresh isolates. In triplicate analyses, RNA levels for Tbx15, Tbx4, and Mga were lower in MUT by 10-, 2.6-, and 2.4-fold, respectively (Fig. 1). Pitx2 therefore activated these three genes in the abdomen. Tbx1, Tbx2, and Tbx5 RNAs were higher in MUT by 2.4-, 2.3-, and 4.2-fold, respectively (Fig. 1). Additionally, qPCR showed that Tbx6, which showed no significant signal in the microarray data, produced 9-fold more RNA in mutants. Pitx2 therefore repressed these four genes in the abdomen.
FIGURE 1.
T-box genes as Pitx2 targets in developing abdomen. qPCR of cDNA from E10.5 mouse abdominal tissue was used to measure relative RNA expression. Black and white bars represent WT and MUT samples, respectively. Samples were normalized to glyceraldehyde-3-phosphate dehydrogenase. Error bars represent three qPCR technical replicates.
Abdominal Expression Patterns of T-box Target Genes
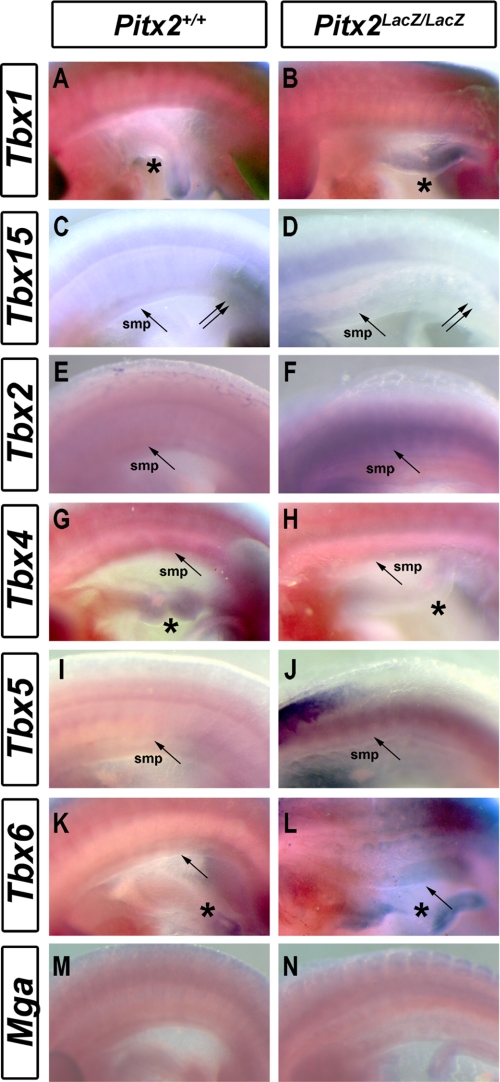
The T-box genes, identified as Pitx2 target genes, were examined by RNA whole mount in situ hybridization of E10.5 WT and MUT mice. Tbx1 was expressed in the foregut but not in the abdominal wall of WT animals (Fig. 2A) (46). However, MUT had a severely underdeveloped and hypocellular intestine, and the observed, elevated Tbx1 expression of the MUT may also be in the body wall as well as in the intestine (Fig. 2B, asterisk). This would be consistent with the idea that Tbx1+ splanchnopleure cells fail to colonize an underdeveloped gut in MUT. Tbx2 was strongly expressed in the somatopleure (Fig. 2D) forelimbs, hindlimbs, and the surrounded area in MUT. This expression was not observed in WT (Fig. 2C). Tbx5 expression was apparent in the MUT somatopleure (Fig. 2F, arrow) but not in WT embryos (Fig. 2E). Tbx6 expression was higher in the intestine (Fig. 2H, asterisk) and somatopleure of MUT (Fig. 2H, arrow). Tbx4 expression was lower in the intestine (Fig. 2J, asterisk), somatopleure (Fig. 2J, asterisk), and hindlimbs (data not shown) of MUT. Tbx15 was expressed in the margin of hindlimbs (Fig. 2K, double arrows) and somatopleure (Fig. 2K, arrow) in WT mice. It was not expressed in any of these tissues in the MUT (Fig. 2L). Mga is expressed before gastrulation (43), and no changes of expression pattern were detected in the abdomen at E10.5 (Fig. 2, M and N). The expression patterns of all selected T-box genes, except Mga, were altered in the area between the posterior margin of forelimb and the anterior margin of hindlimb in a manner consistent with the array and qPCR results. Thus, Pitx2 regulates expression of at least six T-box genes in the abdomen.
FIGURE 2.
Altered spatial expression patterns of T-box genes in Pitx2 mutants. A–N, whole mount RNA in situ for T-box genes in WT (A, C, E, G, I, K, and M) and MUT (B, D, F, H, J, L, and N) E10.5 mice. A and B, Tbx1 expression in the foregut (asterisks) was higher in mutants. B, Tbx1 expression was higher in mutants. C and D, Tbx2 expression was higher in the somatopleure and limb mesenchyme of mutants. E and F, Tbx5 expression was higher in somatopleure (arrow) and intestine (asterisk) of mutants. G and H, Tbx6 expression was higher in somatopleure (arrow) and intestine of mutants. I and J, Tbx4 expression was lower in somatopleure (arrow) and intestine (asterisk) of mutants. K and L, Tbx15 expression was lower in the hindlimb margin (double arrows) and somatopleure of mutants. M and N, no alteration of Mga expression pattern was apparent.
Selection of Test Amplicons in T-box Genes
The genomic sequences surrounding Pitx2-dependent T-box genes were searched for possible cis-regulatory modules that could mediate the observed Pitx2-dependent expression. Several methods were used to identify PCR amplicons of genomic sequences for subsequent ChIP analyses. The mouse genomic sequences (mouse build 37) encompassing each T-box transcripts were downloaded using the genomic representative sequences link in the mouse genome informatics (MGI) web site. Genomic sequences 20 kb upstream and downstream of the transcription unit were included in the download to give the genomic loci to be searched. Initially, these loci were searched for the optimal consensus Pitx2 binding motif TAATCY (47, 48). Several PCR amplicons were selected from the region 20 kb upstream of the MGI transcription start site by looking for clusters of these motifs or individual motifs clustered with bicoid motifs. A bicoid motif was defined as CAATCC, TGATCC, TATTCC, or AAATCC (49).
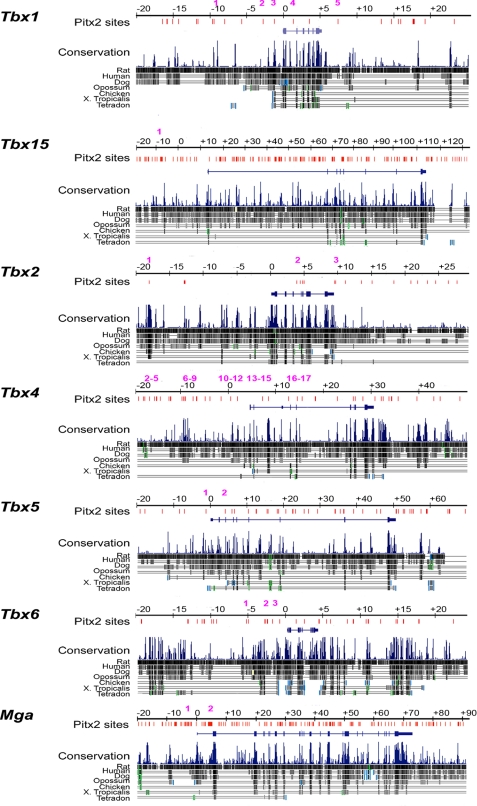
All other amplicons were selected by the following method. The sequence of each genomic locus was submitted to GenomeVISTA, selecting Mouse Feb 2006 as the base genome (mm8). Results were viewed as a VISTA track on the UCSC Genome Browser (mm8). The sites containing the TAATCY motif were identified. Each of these sites was evaluated for the number of species (of mouse, rat, human, dog, opossum, chicken, Xenopus, Tetraodon) in which the motif was conserved and the number of species in which the surrounding block was conserved (Fig. 3). Amplicons were designed around sites that were conserved in multiple species.
FIGURE 3.
Evolutionarily conserved Pitx2 binding sites in T-box genes. A comparative analysis of the Pitx2 consensus binding sites at all T-box Pitx2 target genes is shown. Intergenic and intronic consensus sequences 20 kb upstream and downstream of T-box genes were mapped against the aligned sequences. At Tbx1, evolutionarily conserved Pitx2 occupancy sites were identified at positions −9409 and −1335 relative to the transcriptional start site. At Tbx15, Pitx2 occupancy sites were identified in ultraconserved elements at positions −11,669, +21,323, +21,594, +26,749, +46,749, +46,603, +55,090, +55,102, and +55,651. An ultraconserved element is defined as sequences of >200 bp with 100% identity between human, mouse, and rat (60). Additional conserved Pitx2 occupancy sites were identified in positions +2106, +7634, +17,116, +17,163, +26,685, +32,604, +32,939, +61,418, +61,498, +61,604, +75,701, +90,837, and +117,266. In Tbx2, Pitx2 occupancy sites at positions −18,063 and +9149 were in ultraconserved sequences, with conserved sites found in −12,199 and +4500. In Tbx4, ultraconserved Pitx2 occupancy sites were found at positions −48,363 and +3932 with additional conserved sites at −13,589, −13,079, and −3945. In Tbx5, conserved Pitx2 binding sites were found in intronic sequences at positions +10,300, +12,639, +22,555, and +36,062. In Tbx6, conserved Pitx2 occupancy sites were found at positions −2542, −1728, and +14,914. In Mga, ultraconserved Pitx2 occupancy sites were found at +38,331, +38,453, and +67,530, and conserved sites were found at +4418, +25,169, +26,674, +41,112, +44,985, +61,088, +69,817, and +79,964. The multiple positions of consensus Pitx2 occupancy sites are indicated with red bars.
Pitx2 Occupies Genomic Sites in All T-box Target Genes
ChIP assays of E10.5 abdominal tissue from WT and MUT mice were performed to determine whether Pitx2 occupies identified sites in T-box loci in vivo. Formaldehyde cross-linked chromatin was sonicated, immunoprecipitated with an anti-Pitx2 antibody, and analyzed by qPCR. The signal from immunoprecipitated chromatin was represented as a percentage of the signal obtained from the chromatin put into the immunoprecipitate (% input (% 1P)). Test genomic amplicons were analyzed for each T-box gene to determine whether any enrichment of Pitx2 occupancy could be demonstrated in WT abdominal tissue (Table 2). At each T-box gene, Pitx2 occupancy was strongly enriched in the abdominal tissue of WT when compared with MUT mice. The crossing threshold Ct value in MUT was not significantly different from the no-antibody control. The E10.5 abdominal tissue preparations include a large fraction of Pitx2+ cells derived from the somatopleure, splanchnopleure, and dermomyotome. However, they also include a large fraction of Pitx2− cells derived from the sclerotome and other ectoderm-derived cells. Pitx2 occupancy indicated that it was physically present in the chromatin near the T-box target genes and that it may directly influence their expression at the transcriptional level.
TABLE 2.
Pitx2 occupancy at T-box genes
| Gene | Amplicon number | Amplicon location | % of IP WT | % of IP MUT | % of IP no Aba |
|---|---|---|---|---|---|
| Tbx1 | 0 | none (−3800, +10,013)b | <0.1 ± 0.01 | <0.1 ± 0.01 | <0.1 ± 0.01 |
| 1 | −9409 | 19.1 ± 1.3 | <0.1 ± 0.01 | <0.1 ± 0.01 | |
| 2 | −2711 | 26.3 ± 3.1 | <0.1 ± 0.01 | <0.1 ± 0.01 | |
| 3 | −1335 | 18.3 ± 1.6 | <0.1 ± 0.01 | <0.1 ± 0.01 | |
| 4 | +1691 | 7.2 ± 3.0 | <0.1 ± 0.01 | <0.1 ± 0.01 | |
| 5 | +7153 | 3.0 ± 0.5 | 0.2 ± 0.04 | <0.1 ± 0.01 | |
| Tbx15 | 1 | −7619, −7568 | 11.9 ± 0.3 | <0.1 ± 0.01 | <0.1 ± 0.01 |
| Tbx2 | 1 | −18,063 | 0.9 ± 0.01 | <0.1 ± 0.01 | <0.1 ± 0.01 |
| 2 | +4279, +4500 | 32.8 ± 0.9 | 0.8 ± 0.1 | 0.4 ± 0.2 | |
| 3 | +9149 | 1.1 ± 0.2 | 0.1 ± 0.01 | 0.1 ± 0.01 | |
| Tbx4 | 0 | none (+422)a | <0.1 ± 0.01 | <0.1 ± 0.01 | <0.1 ± 0.01 |
| 1 | −48,363 | 1.1 ± 0.2 | 0.1 ± 0.01 | 0.1 ± 0.01 | |
| 2 | −17,798, −17,671 | 1.40 ± 0.1 | <0.1 ± 0.01 | <0.1 ± 0.01 | |
| 3 | −15,585, −15,510, −15,482 | 2.6 ± 0.6 | <0.1 ± 0.01 | <0.1 ± 0.01 | |
| 4 | −13,589, −13,401 | 2.1 ± 0.3 | <0.1 ± 0.01 | <0.1 ± 0.01 | |
| 5 | −13,142, −13,079 | 0.7 ± 0.3 | <0.1 ± 0.01 | <0.1 ± 0.01 | |
| 6 | −10,654 | 0.1 ± 0.01 | <0.1 ± 0.01 | <0.1 ± 0.01 | |
| 7 | −9923, −9849 | 17.1 ± 4.9 | 0.3 ± 0.1 | 0.1 ± 0.1 | |
| 8 | −8167 | <0.1 ± 0.01 | <0.1 ± 0.01 | <0.1 ± 0.01 | |
| 9 | −7289 | <0.1 ± 0.01 | <0.1 ± 0.01 | <0.1 ± 0.01 | |
| 10 | −3945 | 25.8 ± 3.8 | <0.1 ± 0.01 | <0.1 ± 0.01 | |
| 11 | −3286 | 7.7 ± 0.2 | 0.1 ± 0.01 | <0.1 ± 0.01 | |
| 12 | −2000 | 7.4 ± 0.7 | 0.1 ± 0.01 | 0.02 ± 0.01 | |
| 13 | +3288 | 10.5 ± 1.2 | <0.1 ± 0.01 | <0.1 ± 0.01 | |
| 14 | +3932 | 7.3 ± 1.3 | <0.1 ± 0.01 | <0.1 ± 0.01 | |
| 15 | +8505 | 1.8 ± 0.3 | 0.2 ± 0.05 | 0.1 ± 0.01 | |
| 16 | +11,161 | 2.4 ± 0.5 | <0.1 ± 0.01 | <0.1 ± 0.01 | |
| 17 | +13,839, +13,865 | <0.1 ± 0.01 | <0.1 ± 0.01 | <0.1 ± 0.01 | |
| Tbx5 | 1 | −1424 | 30.7 ± 2.2 | 0.5 ± 0.2 | 0.2 ± 0.1 |
| 2 | +4347 | 0.6 ± 0.01 | <0.1 ± 0.01 | <0.1 ± 0.01 | |
| Tbx6 | 1 | −4917 | 10.6 ± 1.1 | 0.2 ± 0.05 | 0.1 ± 0.01 |
| 2 | −2542, −2437 | 12.4 ± 0.4 | 0.4 ± 0.1 | 0.4 ± 0.1 | |
| 3 | −1728 | 17.5 ± 3.4 | 0.5 ± 0.1 | 0.4 ± 0.2 | |
| Eomes | 1 | −1579 | <0.1 ± 0.01 | <0.1 ± 0.01 | <0.1 ± 0.01 |
| 2 | +8879 | <0.1 ± 0.01 | <0.1 ± 0.01 | <0.1 ± 0.01 | |
| 3 | +9518 | <0.1 ± 0.01 | <0.1 ± 0.01 | <0.1 ± 0.01 | |
| Mga | 1 | −3464, −3399, −3297 | <0.1 ± 0.01 | <0.1 ± 0.01 | <0.1 ± 0.01 |
| 2 | +4251, +4270, +4279 | 7.6 ± 0.9 | 0.1 ± 0.01 | <0.1 ± 0.01 | |
| +4353, +4418, +4423 |
a no Ab, no-antibody control.
b Sites without Pitx2 binding motifs.
Pitx2-dependent Recruitment of Coactivators to T-box Target Genes
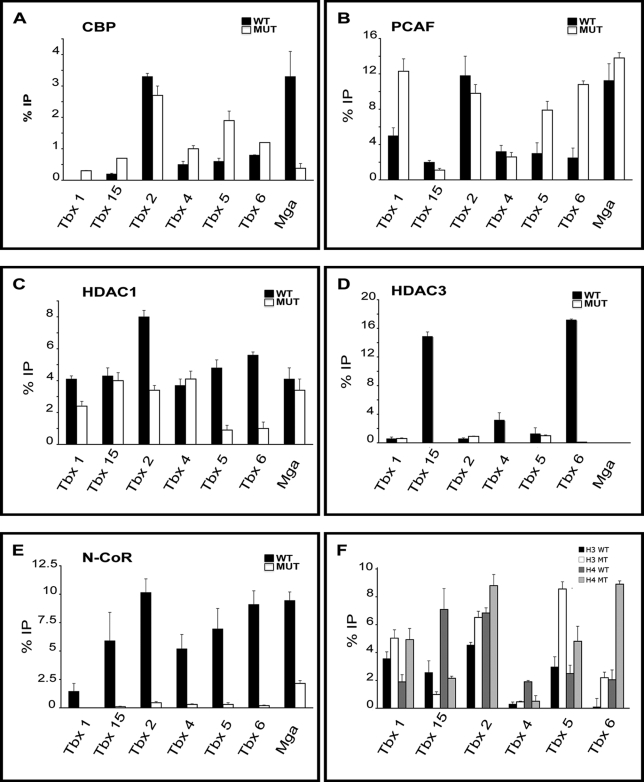
A large number of complexes have been identified that serve as coactivators and corepressors of transcription. SSTFs can recruit distinct combinations of these cofactors depending on cell type, promoter, DNA binding site, and the activity of different signaling pathways. Thus, we tested whether Pitx2 affected the recruitment of coactivators or corepressors to T-box target genes. ChIP assays were used to assess the role of Pitx2 in recruiting coactivators or corepressors to T-box genes in the abdominal tissue at E10.5 (Fig. 4). The recruitment of the coactivators CBP (Fig. 4A) and PCAF (Fig. 4B) and of the corepressors HDAC1 (Fig. 4C), HDAC3 (Fig. 4D), and N-CoR (Fig. 4E) was determined at the same seven genomic sites shown to be occupied by Pitx2.
FIGURE 4.
Pitx2-dependent occupancy of cofactors in T-box genes. ChIP assays for cofactor occupancy at Pitx2 sites on abdominal wall tissue from E10.5 WT and MUT mice in T-box genes are shown. Data were represented as %IP relative to input. WT samples are represented with black bars, and Pitx2LacZ/LacZ samples are represented with white bars. All experiments were performed in triplicates. A, CBP Pitx2-dependent occupancy at Tbx1 (not detectable in WT mice), Tbx15, Tbx4, Tbx5, Tbx6, Mga, and Tbx2 in MUT. Decreased CBP occupancy was detected at Tbx2 and Mga in MUT tissue. Increased CBP occupancy was detected at Tbx1, Tbx5, Tbx6, Tbx4, and Tbx15 in MUT tissue. B, PCAF Pitx2-dependent occupancy was strong at Tbx1, Tbx5, Tbx6, and Tbx15 and with no significant change at Tbx4 and at Mga in MUT tissue. C, HDAC1 Pitx2-dependent occupancy at the repressed T-box genes. Significant reduced occupancy HDAC1 was detected at Tbx1, Tbx2, Tbx5, and Tbx6 in MUT tissue. HDAC1 occupancy was not Pitx2-dependent at Tbx4, Tbx15, and Mga. D, HDAC3 Pitx2-dependent occupancy at Tbx15, Tbx4, and Tbx6 in WT. No occupancy was detected in MUT. E, N-CoR Pitx2-dependent occupancy at Tbx1, Tbx15, Tbx2, Tbx4, Tbx5, and Tbx6 in WT. No significant occupancy was detected in mutants, except Mga. F, acetylated histone H3 and H4 was higher at Tbx1, Tbx2, Tbx5, and Tbx6, whereas lower acetylated histone H3 and H4 was detected at Tbx15 and Tbx4 (only acetylated H4) in MUT. Error bars represent three qPCR technical replicates.
CBP and PCAF have histone acetyl transferase activity. If Pitx2 recruited these proteins to the three Pitx2-activated target genes (Tbx4, Tbx15, Mga), then one would expect higher occupancy of these proteins in WT than in MUT. CBP occupancy at the Pitx2-occupied amplicons was higher in WT for only one of the three activated target genes (Mga; Fig. 4A). The amplicons in the other two activated target genes showed significantly lower CBP occupancy in WT (Tbx4 and Tbx15; Fig. 4A). PCAF occupancy at all three activated genes was not significantly dependent on Pitx2. The levels of Pitx2 and CBP occupancy at the Mga in WT were 3 and 6%, respectively. Both Pitx2 and CBP occupancy drops below 0.5% in MUT, consistent with the model that Pitx2 binds to the Mga and helps recruit CBP.
If Pitx2 occupancy prevented recruitment of coactivators to genomic sites in the four repressed target genes, then one would expect lower occupancy of coactivators at these sites in WT. Clearly, the Tbx1, Tbx5, and Tbx6 amplicons have significantly lower occupancy of both CBP and PCAF in WT when compared with MUT (Fig. 4, A and B). However, the Tbx2 test amplicon had significantly less CBP recruitment in MUT, and PCAF recruitment was not Pitx2-dependent (Fig. 4, A and B). The recruitment of the coactivators CBP and PCAF was not Pitx2-dependent across all test amplicons but may contribute to the transcriptional regulation of a subset of T-box genes.
Pitx2-dependent Occupancy by Corepressors to T-box Target Genes
Corepressors can reduce transcription in a chromatin region they occupy by recruiting histone deacetylase activity. If Pitx2 recruits corepressors to the sites it occupies on repressed T-box genes, then one would expect to see higher corepressor occupancy at these amplicons in WT tissue. HDAC1 occupancy at T-box genes in WT tissue was significantly higher in all four repressed target genes (Tbx1, Tbx2, Tbx5, Tbx6; Fig. 4C) and unaffected in all three activated target genes (Tbx4, Tbx15, Mga; Fig. 4C). The data were therefore consistent with Pitx2-dependent recruitment of HDAC1 at the repressed genes. HDAC3 occupancy was detected at three of the four repressed T-box genes, but it was not Pitx2-dependent (Tbx1, Tbx2, Tbx5; Fig. 4D). However, HDAC3 occupancy was not detected in MUT at test amplicons at Tbx15, Tbx4, Tbx6, and Mga (Fig. 4D). In the three activated target genes (Tbx4, Tbx15, Mga), all or nearly all of the Pitx2-occupied sites could also be occupied by N-CoR. N-CoR occupancy declined in all seven amplicons in MUT (Fig. 4E). The data were consistent with the model that Pitx2 recruits N-CoR to these sites in a subset of Pitx2-expressing abdominal cells. However, lower N-CoR occupancy in MUT at sites near activated target genes is inconsistent with the lower expression levels of these genes in MUT tissue.
Pitx2 Alters Histone Acetylation of T-box Genes In Vivo
The acetylation of histones H3 and H4 at each of the T-box gene amplicons was measured to determine whether Pitx2-dependent changes in gene expression were correlated with expected changes in histone acetylation (Fig. 4F). The Pitx2-dependent occupancy of HDACs at Pitx2-repressed loci suggested that histones at these loci would be less acetylated in the presence than in the absence of Pitx2. Immunoprecipitations with anti-ac-H3 or anti-ac-H4 antibodies demonstrated increased histone H3 and histone H4 acetylation in MUT at test amplicons of Pitx2-repressed genes. Thus, increased expression of Pitx2-repressed genes in MUT was clearly associated with increased histone acetylation. Similar tests on amplicons associated with the Pitx2-activated genes also showed the expected correlation. Both histones were less acetylated in the Tbx15 amplicon, and only H4 was less acetylated in the Tbx4 amplicon. No acetylation was detected in the Mga amplicon (data not shown).
These data suggest that Pitx2 is involved in regulating the acetylation state of chromatin in T-box genes in the abdominal wall. This is likely due to the recruitment of corepressor complexes including HDACs at Tbx1, Tbx2, Tbx5, and Tbx6, which were repressed by Pitx2. One gene that was activated by Pitx2, Tbx15, showed greater histone H3 and H4 acetylation in WT. This was surprising because WT tissue showed higher or equal occupancy by N-CoR, lower occupancy by CBP, and only modestly high occupancy by PCAF, suggesting that other factors are involved. The Tbx15 gene locus contains many potential Pitx2 sites, so it is possible that this is a result of the sites chosen for analysis. Tbx4 showed higher H4 acetylation and similar H3 acetylation in WT tissue. As with Tbx15, this was difficult to reconcile with the similar or higher corepressor HDAC occupancy and the similar or lower coactivator/histone acetyl transferase occupancy at this locus. Alternatively, in the absence of Pitx2, untested corepressors may increase occupancy at these genes, or contributions from other Pitx2 sites can be an indirect effect.
Pitx2 Interacts with Corepressor Complexes
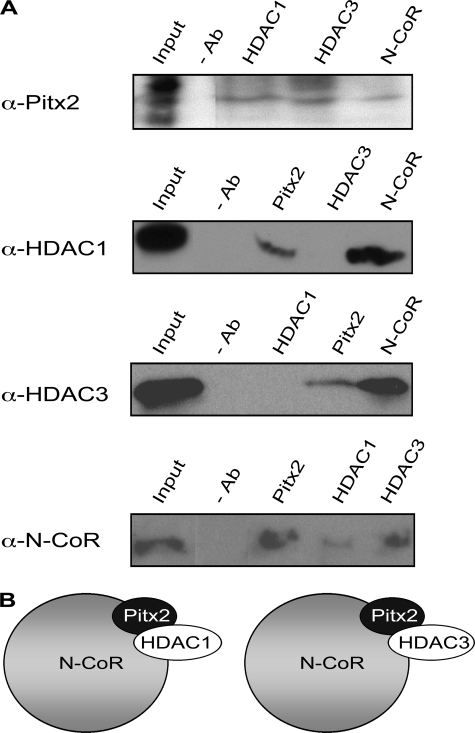
The co-occupancy of Pitx2 and corepressors at DNA sequences near Pitx2-repressed T-box genes suggested that Pitx2 interacts directly with these molecules. Reciprocal IP experiments were used as a second method to test whether Pitx2 coexisted in complexes with corepressors. Nuclear extracts were prepared from WT abdominal tissue harvested at E10.5. Specific antibodies against Pitx2, HDAC1, HDAC3, N-CoR, or normal IgG (without antibody) were used to precipitate complexes containing these proteins. Precipitated complexes were separated in denaturing gels and subjected to Western blot analysis to identify the proteins in the complexes. Pitx2 was detected in HDAC1-, HDAC3-, or N-CoR-containing complexes (Fig. 5A) and was not detected in CBP- or PCAF-containing complexes (data not shown). HDAC1 and HDAC3 were found in complexes precipitated with Pitx2 or N-CoR antibodies. N-CoR was detected in IP complexes with Pitx2, HDAC1, and HDAC3 antibodies. These data indicate that Pitx2 is found in N-CoR complexes with either HDAC1 or HDAC3 (Fig. 5B). Such Pitx2/corepressor complexes may repress T-box genes. Loss of Pitx2 in MUT would lead to a loss of these complexes and could cause the higher expression observed in MUT.
FIGURE 5.
Physical interactions of Pitx2 and cofactors. A, nuclear extracts prepared from the abdominal wall tissue of E10.5 WT tissue were immunoprecipitated with antibodies (Ab) against Pitx2, HDAC1, HDAC3, and N-CoR and hybridized with the same antibodies. 10% input and immunoprecipitates without specific antibodies were used as controls. Pitx2 interacted with HDAC1, HDAC3, and N-CoR. HDAC1 interacted with Pitx2 and N-CoR. HDAC3 interacted with Pitx2 and N-CoR. N-CoR interacted with Pitx2, HDAC1, and HDAC3. B, diagrammatic representation of the Pitx2 cofactorial complexes. Pitx2 was in a complex with N-CoR and HDAC1 and in a different complex with N-CoR and HDAC3.
Chromatin Analyses along the Repressed Tbx1 Locus
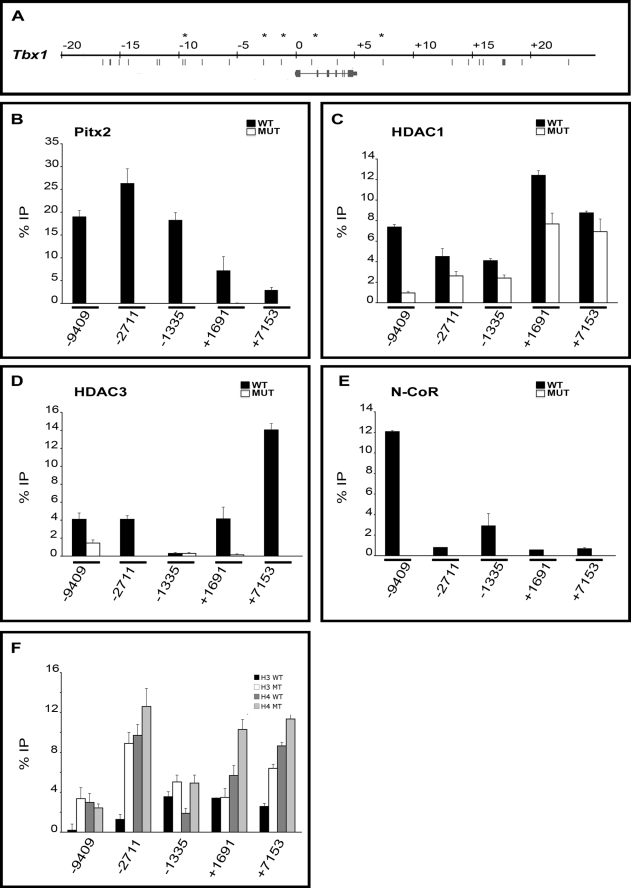
The analyses described above use a single site to probe Pitx2, corepressor, and coactivator occupancy in each Pitx2-regulated T-box gene. The fact that occupancy of Pitx2 was observed at each selected site suggests either that the site selection was very good or that Pitx2 occupancy is a property that can generally be demonstrated in conserved sites that contain consensus DNA binding motifs for Pitx2. Pitx2-dependent occupancy by corepressors was therefore tested at four other genomic sites around the repressed Tbx1 gene in addition to the position −1335 examined above (Fig. 6A).
FIGURE 6.
Cofactor occupancy and acetylation state of Tbx1. A, diagram of 45.3 kb of Tbx1 locus. Asterisks indicate the primer set used for the ChIP assays. B–F, ChIP assays of abdominal wall tissue from WT and MUT mice at E10.5 detected differential Pitx2 occupancy. B, Pitx2 occupancy was detected at positions −2711, −9409, −1335, +1691, and +7153. C, Pitx2-dependent HDAC1 occupancy was detected at −9409, −2711, −1335, +1691, and +7153. D, Pitx2-dependent HDAC3 occupancy was lower at −9409, −2711, +1691, and +7153. E, Pitx2-dependent N-CoR occupancy was mostly enriched at −9409 and weaker at −2711, −1335, +1691, and +7153. F, differential levels of acetylation in Pitx2 amplicon detected by immunoprecipitation using specific antibodies for acetylated H3 and H4. Histone acetylation was higher at positions −9409, −2711, −1335, +1691, and +7153 in MUT for the most of the amplicons tested. All experiments were performed in triplicate. Error bars represent three qPCR technical replicates.
ChIP assays showed that Pitx2 occupied both intergenic and intronic regions of Tbx1 (Fig. 6B). There was no enrichment for any amplicon in MUT tissue. No Pitx2 enrichment was detected at a test amplicon to a region containing no Pitx2 sites (data not shown). It appeared that Pitx2 occupancy was higher in the upstream intergenic region and decreased significantly in the intronic and downstream intergenic region. HDAC1 occupancy decreased in MUT at all sites tested (Fig. 6C). Pitx2 dependence of HDAC1 occupancy was 4-fold higher at the position −9409 than at any of the other four amplicons. HDAC1 occupancy was greatest at the +1691 and +7153 amplicon, which showed the lowest Pitx2 occupancy. HDAC occupancy depended in part on Pitx2 at all amplicons and did so at some sites more strongly than at others.
HDAC3 occupancy showed a far greater variation in level and Pitx2 dependence than HDAC1. All amplicons, except one (−1335), showed higher HDAC occupancy in WT than MUT. This further confirms the correlation between corepressor occupancy in WT and Pitx2-repressed gene expression than was described above. The extent of Pitx2-dependent HDAC3 occupancy seems to complement that of HDAC1. The −9409 amplicon showed the lowest -fold change in HDAC3 occupancy and the highest -fold change in the HDAC1 occupancy. Conversely, the −2711, +1691, and +7153 amplicons showed less Pitx2 dependence in HDAC1 occupancy and far more Pitx2 dependence in HDAC3 occupancy. N-CoR occupancy at all amplicons was Pitx2-dependent with no occupancy detected in mutants. N-CoR occupancy was higher at −9409, the amplicon that had the highest Pitx2 dependence in HDAC1 occupancy (Fig. 6E).
Acetylation of histone H3 and/or H4 increased at all five amplicons in MUT (Fig. 6F). The −9409 amplicon showed Pitx2 dependence for H3 but not for H4 acetylation. The converse was true at the +1691 amplicon. The lack of acetylation of either H4 or H3 at these amplicons appeared to be compensated by robust increases in the acetylation of the other histones. Clearly, increased acetylation of H3 and H4 occurred in mutant tissue in all regions of the Pitx2-repressed genes.
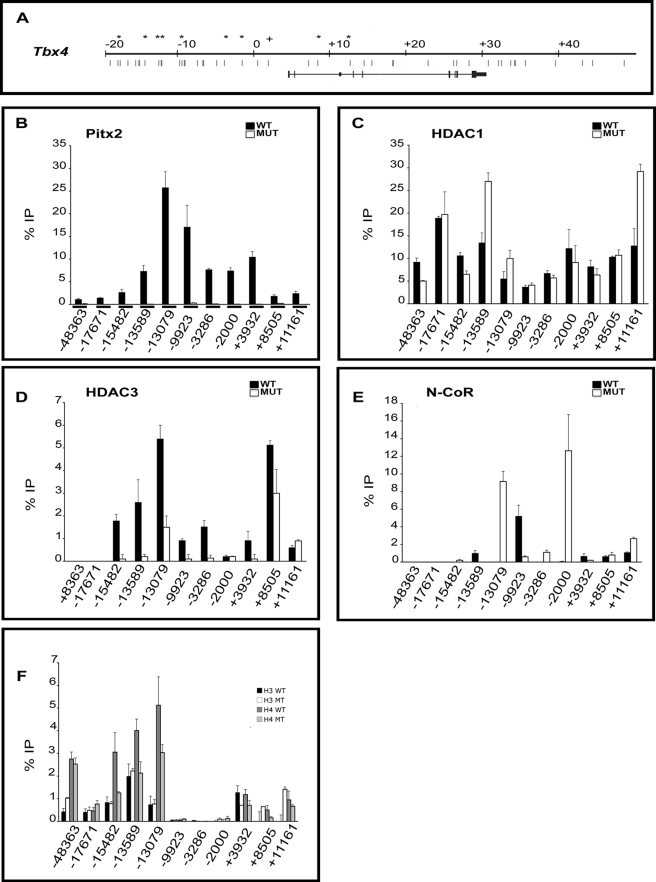
Chromatin Analyses along the Activated Tbx4 Locus
Similar studies were performed for Tbx4, a gene activated by Pitx2. 10 amplicons containing Pitx2 binding sites throughout the non-coding region of the Tbx4 gene were analyzed together with the −9923 amplicon analyzed above (Fig. 7A). The Pitx2 binding site at position −48,363 was within an ultraconserved region just downstream of the Tbx2 gene. Tbx2 and Tbx4 are neighboring genes. ChIP assays demonstrated that Pitx2 occupied numerous amplicons in WT tissue. However, Pitx2 occupancy was not detected at amplicons containing Pitx2 sites at positions −10,654 and +13,839 (data not shown), confirming that Pitx2 is not present at all Pitx2 elements. No occupancy was detected on any amplicons in MUT tissue. A control amplicon in a region with no Pitx2 binding sites found no enrichment above background in WT or MUT (data not shown). As with Tbx1, Pitx2 occupancy appeared to be greatest in amplicons located in the upstream intragenic region (Fig. 7B).
FIGURE 7.
Cofactor occupancy and acetylation state of Tbx4. A, diagram of 70 kb of the Tbx4 locus. Asterisks indicate the primer set used for the ChIP assays. Position −48,363 was within an ultraconserved region just downstream of Tbx2. Position −17,671 encompasses two Pitx2 sites, −17,798 and −1,7671. Position −15,482 encompasses three Pitx2 sites, −15,585, −15,510, and −15,482. Position −13,589 encompasses two Pitx2 amplicons at −13,589 (conserved across five species (mouse, rat, human, dog, and opossum)) and −13,401. Position −13,079, conserved across five species, encompasses two Pitx2 sites, −13,142 and −13,079. Position −9923 encompasses two non-conserved Pitx2 amplicons, −9923 and −9849. Positions −3286 and −2000 encompass single non-conserved Pitx2 amplicons. Position +3932 encompasses a single conserved Pitx2 amplicon in the 5′-untranslated region across seven species (mouse, rat, human, dog, opossum, chicken, and Xenopus tropicalis). Positions +8205 and +11,161 encompass non-conserved single intronic Pitx2 sites. B–F, ChIP assays of abdominal tissue from WT and MUT tissue detected differential Pitx2 occupancy. B, Pitx2 occupancy was detected at positions −48,363, −17,671, −15,482, −13,589, −13,079, −9923, −3286, −2000, +3932, +8505, and +11,161 in WT. C, Pitx2-dependent HDAC1 occupancy was lower at positions −48,363 and −15,482, −2000, whereas it was higher at −13,589, −13,079, and +11,161 in mutants. D, Pitx2-dependent HDAC3 occupancy was lower at positions −15,482, −13,589, −13,079, −9923, −3286, +3932, and +8505 and higher at +11,161 in mutants. E, Pitx2-dependent N-CoR occupancy was detected at −13,589, −9923, +3932, −15,482, −13,079, −3286, −2000, and +11,161 in mutants. F, Differential levels of acetylation in Pitx2 amplicons were detected by immunoprecipitation using specific antibodies for acetylated histone H3 and H4. Increased H3 histone acetylation was observed at −48,363, +8505, and +11,161 in MUT. Levels of acetylated histone H3 were lower at +3932, whereas levels of acetylated histone H4 were lower at −15,482, −13,589, −13,079, +3932, and +8505 in mutants. All experiments were performed in triplicate. Error bars represent three qPCR technical replicates.
HDAC1 occupancy could also be detected at all sites but was dependent on Pitx2 at only 5 of 11 amplicons (Fig. 7C). Pitx2 knock-out resulted in the increased occupancy in three proximal amplicons and decreased occupancy in two distal amplicons. HDAC1 occupancy was not correlated with Pitx2 occupancy at different sites. Moreover, the Pitx2 dependence of HDAC1 occupancy was also not correlated to Pitx2 occupancy.
HDAC3 occupancy was also strongly Pitx2-dependent but could not be detected on two amplicons. HDAC3 occupancy was lower in MUT at most amplicons. This would be expected to increase expression in MUT. Tbx4 expression decreases in MUT. Increased corepressor occupancy in MUT tissue was only sporadically observed for different corepressors at different amplicons. HDAC1 occupancy was increased at two amplicons (−13,589, +11,161), HDAC3 was increased only at +11,161, and N-CoR was increased at three amplicons (13,079, −2000, +11,161). Thus, corepressor occupancy of amplicons at the Pitx2-activated locus (Tbx4) was less consistent with changes in gene expression, less consistent across the locus, and less Pitx2-dependent than at the Pitx2-repressed locus (Tbx1).
Histone acetylation also did not change in as consistent fashion along the entire locus in the Pitx2-activated gene as in the Pitx2-repressed gene. H3 acetylation decreased in one amplicon and increased at in two amplicons in MUT tissue. H4 acetylation decreased at five amplicons. However, the hypoacetylation of chromatin in MUT was more consistent closer to the transcriptional start site. Tbx4 is downstream of Tbx2, a Pitx2-repressed gene, which may contribute to the inconsistent acetylation pattern in chromatin regions upstream of Tbx4. Histone acetylation was extremely low in amplicons surrounding the upstream end of the transcription unit.
DISCUSSION
Pitx2 is a bicoid-related homeodomain transcription factor that plays a critical role in the development of multiple organs, including heart, lung, intestine, pituitary gland, tooth, muscles, and body wall closure (3–6). Although it is obvious that Pitx2 is required for proper organ and tissue formation, the role and mechanisms by which Pitx2 influences body wall formation and closure are unclear. In the present studies, we provide evidence indicating that Pitx2 acts upstream of T-box gene family members by occupying specific sites on their promoters. Occupancy by coactivators and corepressors occur by a Pitx2-dependent mechanism and results in modification of their histone acetylation status.
The most striking phenotype of the Pitx2 knock-out mice is the open body wall and the externalization of the thoracic and abdominal viscera during embryonic development (5, 6). Pitx2 is expressed in the lateral plate mesoderm and somatopleure (27), tissues that contribute to body wall formation. Thus, Pitx2 is likely placed temporally upstream in gene regulatory networks involved in this developmental process, and there was a need to identify its target genes. Gene expression analysis was used to identify the genes that are regulated by Pitx2 during body wall formation. One of the families regulated by Pitx2 was the evolutionarily conserved family of T-box genes. T-box genes are essential for patterning embryonic mesoderm and forming organs. Tbx2 superfamily members Tbx2, Tbx4, and Tbx5 and the Tbx1 superfamily member Tbx15 are expressed in the body wall and in the anterior and posterior margins of the developing limbs (11). Expression of all four genes was altered in the absence of Pitx2. Tbx1, Tbx2, and Tbx5 expression was up-regulated in mutant embryos (Figs. 1A and 2). The differential gene regulation of T-box genes by Pitx2 in different tissues during development could be explained by changes in the transcriptional machinery in time and space. These experiments were performed at E10.5 when Pitx2 MUT already exhibit gross morphological defects. T-box gene expression is widespread during development. T-box genes are expressed in all stages of development from the oocyte to the adult. Pitx2 expression overlaps in the developing embryo with T-box genes from as early as E8.0. Future experiments should characterize the role of Pitx2 expression on T-box gene expression at other stages in development.
Multiple sites occupied by Pitx2 have been identified in the upstream genomic sequences and intronic sequences of T-box genes. Evolutionary alignment of T-box genes confirmed the presence of evolutionarily conserved Pitx2 binding motifs (Fig. 3). The identification of multiple sites that matched the optimal DNA binding sequence of Pitx2 suggests that Pitx2 directly binds to T-box genes and may regulate their expression. This was confirmed with in vivo ChIP assays, where Pitx2 was found to occupy its optimal sites (Table 2). To fully characterize Pitx2 occupancy at the T-box gene loci, genome-wide ChIP-sequence would need to be used due to the number of potential Pitx2 binding sites at these loci. The exact role of Pitx2 occupancy in regulated transcription needs to be further characterized due to possible indirect effects from the altered expression or activity of other genes.
Regulated exchange of HDAC1/β-catenin converts Pitx2 from repressor to activator for cell cycle control genes, such as cyclin D2 (7). Pitx2-dependent HDAC1 occupancy was associated with decreased acetylation of histone H3 and H4 and repression of Tbx1, Tbx2, Tbx5, and Tbx6 (Figs. 5–7). However, coactivators CBP and PCAF and/or corepressors HDAC3 and Pitx2-dependent N-CoR occupancy did not universally correspond to activation or repression of T-box genes. Corepressor occupancy in Pitx2-activated genes was unaffected or affected in the direction opposite of what was expected at some sites, suggesting that although Pitx2 alters the occupancy of these cofactors, different complexes may occupy different Pitx2 sites. Due to the number of Pitx2 sites at T-box loci, the occupancy of coactivators and corepressors throughout the loci needs to be characterized. The coactivators and corepressors targeted to these sites in the presence and absence of Pitx2 are likely due to nearby other transcription factor binding sites. In addition, intronic Pitx2 binding sites can regulate gene transcription (50). Pitx2 interacts with multiple coactivator and corepressor complexes (7). Pitx2 was physically interacting with N-CoR and either HDAC1 or HDAC3 to occupy its T-box target genes. The interaction of Pitx2 with N-CoR, HDAC1, and HDAC3 is one mechanism by which corepressors occupy Pitx2 targets (Fig. 5). Pitx2-dependent HDAC1, HDAC3, and N-CoR occupancy was detected at multiple sites in Tbx1 (Fig. 6). These data suggest that decreased histone acetylation results in Tbx1 repression due to occupancy of Pitx2-dependent corepressor complexes. Recent studies also indicated that Pitx2 induces acetylation of histone H4 at the promoter of structural maintenance of chromosomes (SMC) proteins differentiation marker genes through exchange of HDAC2 and HDAC5 with p300 (51). In branchial arch mesoderm, Pitx2 activates Tbx1 (28), whereas repressing Tbx1 in the abdominal wall, further suggesting that Pitx2 can switch from an activator to a repressor due to interaction with different complexes in a tissue-specific manner.
In the absence of Pitx2, N-CoR and HDAC1 occupancy increased at several amplicons in the Tbx4 locus. Pitx2 has been shown to interact with several transcription factors such as Lef-1, β-catenin, and Pit-1 (7, 52, 53). Lef-1 can also act as a repressor in the absence of Wnt signaling by interacting with Groucho and HDACs (54), whereas β-catenin interacts with PCAF and CBP (55, 56). Pitx2 occupied amplicons in close proximity to the lymphoid enhancer factor/T-cell factor (Lef-1/TCF) and Pit-1 binding sites at Tbx4 and Tbx15, and our recent studies have shown that Lef-1 occupancy was lower in MUT (data not shown). Thus, Pitx2 may interact with Lef-1, β-catenin, Pit-1, and coactivators in Pitx2-activated genes.
The genomic sequence upstream of the Tbx4 transcriptional start site was hypoacetylated in MUT. Tbx4 was activated, and Tbx2 was repressed by Pitx2 despite their proximity in the genome. Tbx4 and Tbx2 are closely positioned on chromosome 11. Analysis of the Tbx2 gene showed increased acetyl-H3 and acetyl-H4 in the absence of Pitx2. Analysis of the amount of acetyl-H3 and acetyl-H4 in the Tbx4 showed that within the intergenic region between Tbx2 and Tbx4, there was a gradual shift from more acetylated histone H3 in the chromatin near Tbx2 to hypoacetylated histone H4 near Tbx4, suggesting that Pitx2 can regulate chromatin structure in a highly localized manner (Fig. 7). Histone modifications due to the presence of coactivators and corepressors are only one type of chromatin remodeling. Further studies will be necessary to determine whether other chromatin modifications are Pitx2-dependent at the T-box gene loci. These modifications can act alone or in concert, in a context-dependent manner, to facilitate or repress chromatin-mediated processes because they can influence one another (57).
Gene expression profiling of abdominal tissue and whole mount in situ hybridization at E10.5 indicated that several signaling pathways and transcription factors that regulate the expression of T-box genes are Pitx2-dependent. Pitx2 was acting as a repressor of Wnt10a, Wnt6, Dkk1, and Axin2 and as an activator on the Wnt5b, Tcf12, and adenomatous polyposis coli (7). The Wnt/β-catenin pathway regulates the expression of several T-box genes (58–60). Expression of several bone morphogenetic proteins (BMPs) was Pitx2-dependent, and BMP2 and BMP4 regulate Tbx2, Tbx3, and Tbx4 expression (10, 60). Additionally, T-box genes can regulate their own expression and the expression of other T-box genes through T-box binding elements. Tbx20 negatively regulates the expression of Tbx2 (10, 61, 63). Pitx1 negatively regulates Tbx1 expression in dental epithelium (64) and activates Tbx4 expression in the limb (65). Pitx1 expression was Pitx2-dependent in the abdominal tissue and dental epithelium (data not shown). These pathways and transcription factors may also contribute to Pitx2-dependent expression of T-box genes in the abdomen at E10.5.
We conclude that T-box gene expression depends on the Pitx2-dependent occupancy of coactivators and corepressors in the abdominal wall and that this reveals one of the pathways by which Pitx2 regulates development. However, Pitx2 is part of a complex gene network that can also regulate the expression of T-box genes via signaling pathways and other interactions.
Acknowledgments
We thank J. Menino for animal husbandry, A. M. Girard for microarray processing, and Drs. M. Leid and T. Filtz for stimulating discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant AR054406 (to C. K.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- E
- embryonic day
- ChIP
- chromatin immunoprecipitation
- IP
- immunoprecipitated
- HDAC
- histone deacetylase
- qPCR
- quantitative PCR
- SSTF
- sequence-specific DNA binding transcription factors
- CBP
- CREB-binding protein
- CREB
- cAMP-response element-binding protein
- PCAF
- p300/CBP-associated factor
- WT
- wild type
- HET
- heterozygote
- MUT
- mutant
- FDR
- false discovery rate
- BMP
- bone morphogenetic protein
- N-CoR
- nuclear repressor corepressor.
REFERENCES
- 1.Brewer S., Williams T. (2004) Dev. Biol. 267, 399–417 [DOI] [PubMed] [Google Scholar]
- 2.Brewer S., Williams T. (2004) Bioessays 26, 1307–1321 [DOI] [PubMed] [Google Scholar]
- 3.Lu M. F., Pressman C., Dyer R., Johnson R. L., Martin J. F. (1999) Nature 401, 276–278 [DOI] [PubMed] [Google Scholar]
- 4.Kitamura K., Miura H., Miyagawa-Tomita S., Yanazawa M., Katoh-Fukui Y., Suzuki R., Ohuchi H., Suehiro A., Motegi Y., Nakahara Y., Kondo S., Yokoyama M. (1999) Development 126, 5749–5758 [DOI] [PubMed] [Google Scholar]
- 5.Gage P. J., Suh H., Camper S. A. (1999) Development 126, 4643–4651 [DOI] [PubMed] [Google Scholar]
- 6.Lin C. R., Kioussi C., O'Connell S., Briata P., Szeto D., Liu F., Izpisúa-Belmonte J. C., Rosenfeld M. G. (1999) Nature 401, 279–282 [DOI] [PubMed] [Google Scholar]
- 7.Kioussi C., Briata P., Baek S. H., Rose D. W., Hamblet N. S., Herman T., Ohgi K. A., Lin C., Gleiberman A., Wang J., Brault V., Ruiz-Lozano P., Nguyen H. D., Kemler R., Glass C. K., Wynshaw-Boris A., Rosenfeld M. G. (2002) Cell 111, 673–685 [DOI] [PubMed] [Google Scholar]
- 8.Kimelman D., Griffin K. J. (2000) Curr. Opin. Genet. Dev. 10, 350–356 [DOI] [PubMed] [Google Scholar]
- 9.Tada M., Smith J. C. (2001) Dev Growth Differ. 43, 1–11 [DOI] [PubMed] [Google Scholar]
- 10.Naiche L. A., Harrelson Z., Kelly R. G., Papaioannou V. E. (2005) Annu. Rev. Genet. 39, 219–239 [DOI] [PubMed] [Google Scholar]
- 11.Chapman D. L., Garvey N., Hancock S., Alexiou M., Agulnik S. I., Gibson-Brown J. J., Cebra-Thomas J., Bollag R. J., Silver L. M., Papaioannou V. E. (1996) Dev. Dyn. 206, 379–390 [DOI] [PubMed] [Google Scholar]
- 12.O'Reilly M. A., Smith J. C., Cunliffe V. (1995) Development 121, 1351–1359 [DOI] [PubMed] [Google Scholar]
- 13.Bruneau B. G., Nemer G., Schmitt J. P., Charron F., Robitaille L., Caron S., Conner D. A., Gessler M., Nemer M., Seidman C. E., Seidman J. G. (2001) Cell 106, 709–721 [DOI] [PubMed] [Google Scholar]
- 14.Garg V., Kathiriya I. S., Barnes R., Schluterman M. K., King I. N., Butler C. A., Rothrock C. R., Eapen R. S., Hirayama-Yamada K., Joo K., Matsuoka R., Cohen J. C., Srivastava D. (2003) Nature 424, 443–447 [DOI] [PubMed] [Google Scholar]
- 15.Habets P. E., Moorman A. F., Clout D. E., van Roon M. A., Lingbeek M., van Lohuizen M., Campione M., Christoffels V. M. (2002) Genes Dev. 16, 1234–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiroi Y., Kudoh S., Monzen K., Ikeda Y., Yazaki Y., Nagai R., Komuro I. (2001) Nat. Genet. 28, 276–280 [DOI] [PubMed] [Google Scholar]
- 17.Krause A., Zacharias W., Camarata T., Linkhart B., Law E., Lischke A., Miljan E., Simon H. G. (2004) Dev. Biol. 273, 106–120 [DOI] [PubMed] [Google Scholar]
- 18.Lamolet B., Pulichino A. M., Lamonerie T., Gauthier Y., Brue T., Enjalbert A., Drouin J. (2001) Cell 104, 849–859 [DOI] [PubMed] [Google Scholar]
- 19.Maira M., Couture C., Le Martelot G., Pulichino A. M., Bilodeau S., Drouin J. (2003) J. Biol. Chem. 278, 46523–46532 [DOI] [PubMed] [Google Scholar]
- 20.Stennard F. A., Costa M. W., Elliott D. A., Rankin S., Haast S. J., Lai D., McDonald L. P., Niederreither K., Dolle P., Bruneau B. G., Zorn A. M., Harvey R. P. (2003) Dev. Biol. 262, 206–224 [DOI] [PubMed] [Google Scholar]
- 21.Wang G. S., Hong C. J., Yen T. Y., Huang H. Y., Ou Y., Huang T. N., Jung W. G., Kuo T. Y., Sheng M., Wang T. F., Hsueh Y. P. (2004) Neuron 42, 113–128 [DOI] [PubMed] [Google Scholar]
- 22.Lickert H., Takeuchi J. K., Von Both I., Walls J. R., McAuliffe F., Adamson S. L., Henkelman R. M., Wrana J. L., Rossant J., Bruneau B. G. (2004) Nature 432, 107–112 [DOI] [PubMed] [Google Scholar]
- 23.Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 24.Jepsen K., Rosenfeld M. G. (2002) J. Cell Sci. 115, 689–698 [DOI] [PubMed] [Google Scholar]
- 25.Karagianni P., Wong J. (2007) Oncogene 26, 5439–5449 [DOI] [PubMed] [Google Scholar]
- 26.Jones P. L., Shi Y. B. (2003) Curr. Top. Microbiol. Immunol. 274, 237–268 [DOI] [PubMed] [Google Scholar]
- 27.Shih H. P., Gross M. K., Kioussi C. (2007) Gene Expr. Patterns 7, 441–451 [DOI] [PubMed] [Google Scholar]
- 28.Shih H. P., Gross M. K., Kioussi C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5907–5912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kioussi C., Gross M. K. (2008) PLoS One 3, e2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kioussi C., Shih H. P., Loflin J., Gross M. K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18621–18626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiper-Bergeron N., Wu D., Pope L., Schild-Poulter C., Haché R. J. (2003) EMBO J. 22, 2135–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y., Wolf L. V., Cvekl A. (2007) J. Mol. Biol. 369, 917–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edelstein L. C., Lagos L., Simmons M., Tirumalai H., Gélinas C. (2003) Mol. Cell. Biol. 23, 2749–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mengus G., May M., Jacq X., Staub A., Tora L., Chambon P., Davidson I. (1995) EMBO J. 14, 1520–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jerome L. A., Papaioannou V. E. (2001) Nat. Genet. 27, 286–291 [DOI] [PubMed] [Google Scholar]
- 36.Arnold J. S., Werling U., Braunstein E. M., Liao J., Nowotschin S., Edelmann W., Hebert J. M., Morrow B. E. (2006) Development 133, 977–987 [DOI] [PubMed] [Google Scholar]
- 37.Harrelson Z., Kelly R. G., Goldin S. N., Gibson-Brown J. J., Bollag R. J., Silver L. M., Papaioannou V. E. (2004) Development 131, 5041–5052 [DOI] [PubMed] [Google Scholar]
- 38.Naiche L. A., Papaioannou V. E. (2007) Development 134, 93–103 [DOI] [PubMed] [Google Scholar]
- 39.Naiche L. A., Papaioannou V. E. (2003) Development 130, 2681–2693 [DOI] [PubMed] [Google Scholar]
- 40.Agarwal P., Wylie J. N., Galceran J., Arkhitko O., Li C., Deng C., Grosschedl R., Bruneau B. G. (2003) Development 130, 623–633 [DOI] [PubMed] [Google Scholar]
- 41.Candille S. I., Van Raamsdonk C. D., Chen C., Kuijper S., Chen-Tsai Y., Russ A., Meijlink F., Barsh G. S. (2004) PLoS Biol. 2, E3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh M. K., Petry M., Haenig B., Lescher B., Leitges M., Kispert A. (2005) Mech. Dev. 122, 131–144 [DOI] [PubMed] [Google Scholar]
- 43.Hurlin P. J., Steingrìmsson E., Copeland N. G., Jenkins N. A., Eisenman R. N. (1999) EMBO J. 18, 7019–7028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J., Lin C., Gleiberman A., Ohgi K. A., Herman T., Huang H. P., Tsai M. J., Rosenfeld M. G. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8674–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kraus F., Haenig B., Kispert A. (2001) Mech. Dev. 100, 87–91 [DOI] [PubMed] [Google Scholar]
- 46.Choi M., Klingensmith J. (2009) PLoS Genet. 5, e1000395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amendt B. A., Sutherland L. B., Semina E. V., Russo A. F. (1998) J. Biol. Chem. 273, 20066–20072 [DOI] [PubMed] [Google Scholar]
- 48.Wilson D. S., Sheng G., Jun S., Desplan C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6886–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vadlamudi U., Espinoza H. M., Ganga M., Martin D. M., Liu X., Engelhardt J. F., Amendt B. A. (2005) J. Cell Sci. 118, 1129–1137 [DOI] [PubMed] [Google Scholar]
- 50.Shima Y., Zubair M., Komatsu T., Oka S., Yokoyama C., Tachibana T., Hjalt T. A., Drouin J., Morohashi K. (2008) Mol. Endocrinol. 22, 1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shang Y., Yoshida T., Amendt B. A., Martin J. F., Owens G. K. (2008) J. Cell Biol. 181, 461–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Amen M., Liu X., Vadlamudi U., Elizondo G., Diamond E., Engelhardt J. F., Amendt B. A. (2007) Mol. Cell. Biol. 27, 7560–7573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amendt B. A., Sutherland L. B., Russo A. F. (1999) Mol. Cell. Biol. 19, 7001–7010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arce L., Pate K. T., Waterman M. L. (2009) BMC Cancer 9, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ge X., Jin Q., Zhang F., Yan T., Zhai Q. (2009) Mol. Biol. Cell 20, 419–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hecht A., Vleminckx K., Stemmler M. P., van Roy F., Kemler R. (2000) EMBO J. 19, 1839–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suganuma T., Workman J. L. (2008) Cell 135, 604–607 [DOI] [PubMed] [Google Scholar]
- 58.Wardle F. C., Smith J. C. (2006) Semin. Cell Dev. Biol. 17, 99–109 [DOI] [PubMed] [Google Scholar]
- 59.Wardle F. C., Papaioannou V. E. (2008) Curr. Opin. Genet. Dev. 18, 418–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.White J. A., Heasman J. (2008) J. Exp. Zool. B. Mol. Dev. Evol. 310, 73–84 [DOI] [PubMed] [Google Scholar]
- 61.Behesti H., Holt J. K., Sowden J. C. (2006) BMC Dev. Biol. 6, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deleted in proof
- 63.Singh R., Horsthuis T., Farin H. F., Grieskamp T., Norden J., Petry M., Wakker V., Moorman A. F., Christoffels V. M., Kispert A. (2009) Circ. Res. 105, 442–452 [DOI] [PubMed] [Google Scholar]
- 64.Mitsiadis T. A., Tucker A. S., De Bari C., Cobourne M. T., Rice D. P. (2008) Dev. Biol. 320, 39–48 [DOI] [PubMed] [Google Scholar]
- 65.DeLaurier A., Schweitzer R., Logan M. (2006) Dev. Biol. 299, 22–34 [DOI] [PubMed] [Google Scholar]
- 66.United States Department of Agriculture (2009) Animal Welfare Act and Regulations, 9CFR [Google Scholar]