Abstract
HSAN1 is an inherited neuropathy found to be associated with several missense mutations in the SPTLC1 subunit of serine palmitoyltransferase (SPT). SPT catalyzes the condensation of serine and palmitoyl-CoA, the initial step in the de novo synthesis of sphingolipids. Here we show that the HSAN1 mutations induce a shift in the substrate specificity of SPT, which leads to the formation of the two atypical deoxy-sphingoid bases (DSBs) 1-deoxy-sphinganine and 1-deoxymethyl-sphinganine. Both metabolites lack the C1 hydroxyl group of sphinganine and can therefore neither be converted to complex sphingolipids nor degraded. Consequently, they accumulate in the cell, as demonstrated in HEK293 cells overexpressing mutant SPTLC1 and lymphoblasts of HSAN1 patients. Elevated DSB levels were also found in the plasma of HSAN1 patients and confirmed in three groups of HSAN1 patients with different SPTLC1mutations. The DSBs show pronounced neurotoxic effects on neurite formation in cultured sensory neurons. The neurotoxicity co-occurs with a disturbed neurofilament structure in neurites when cultured in the presence of DSBs. Based on these observations, we conclude that HSAN1 is caused by a gain of function mutation, which results in the formation of two atypical and neurotoxic sphingolipid metabolites.
Keywords: Amino Acid, Lipid, Neurodegeneration, Neurological Diseases, Sphingolipid, Deoxysphingolipids, DoxSA, HSAN1, Neuropathie
Introduction
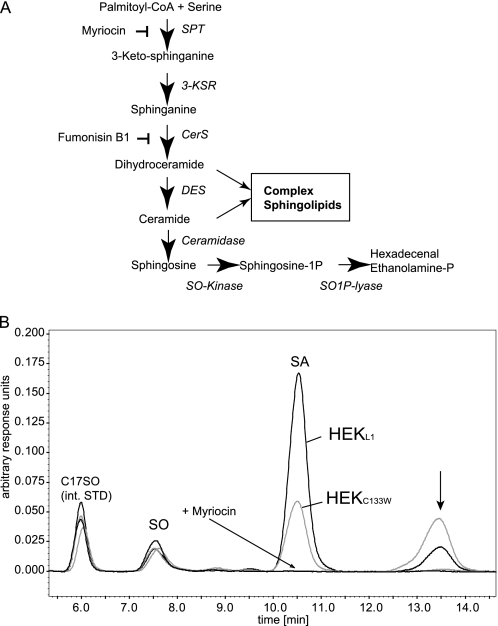
Hereditary sensory and autonomic neuropathy type 1 (HSN1 or HSAN1) is the most frequent HSAN2 subtype (1). It is an autosomal dominant condition, clinically characterized by loss of pain and temperature sensation in feet and hands, often accompanied by lancinating pain attacks, skin ulcers, and infections (2). Furthermore, degeneration of motor neurons frequently occurs accompanied by atrophy and weakness of distal limb muscles. Positional cloning identified mutations in the SPTLC1 gene (3, 4), which encodes the first of the three subunits of serine palmitoyltransferase (SPT) (5, 6). SPT catalyzes the pyridoxal phosphate-dependent condensation of l-serine and palmitoyl-coenzyme A, the first and rate-limiting step in the de novo synthesis of sphingolipids (7) (see Fig. 1A).
FIGURE 1.
A, de novo sphingolipid synthesis pathway. De novo ceramide synthesis involves several steps. SPT catalyzes the initial conjugation of palmitoyl-CoA with l-serine to form 3-keto-sphinganine, which is subsequently reduced to SA. SA is acetylated by ceramide synthase (CerS) and desaturated by ceramide desaturase (DES) to form ceramide. The degradation pathway starts with the deacetylation of ceramide by ceramidase. The formed SO is then phosphorylated by SO-kinase and finally degraded to hexadecenal and phosphoethanolamine by the action of the sphingosine-1-phospate lyase (SO1P lyase). B, HEK293 cells expressing the SPT-C133W mutant generate an unknown metabolite. HEK293 cells were transfected with either wild-type SPTLC1 or the SPTLC1-C133W mutant. De novo synthesis was blocked with FB1 for 24 h. This causes an accumulation of sphinganine but also of other potential SPT products. The accumulated sphingoid bases were extracted and analyzed by HPLC. We observed a significant accumulation of SA in HEKL1 cells (black). SA accumulation was lower in HEKC133W cells (gray), which reflects the reduced activity of the mutant. In parallel, we observed the appearance of a second, unknown peak (arrow). This peak appeared only in the presence of FB1 and was increased in cells expressing the C133W mutant. It was absent when SPT activity was blocked with myriocin. int. STD, internal standard.
Four missense mutations (C133W, C133Y, V144D, and G387A) in SPTLC1 have been reported in a total of 24 HSAN1 families. C133W is the most frequent mutation, found in 18 HSAN1 families (3, 4, 8–10), whereas C133Y was reported in three (3, 4, 11) families and V144D was reported in two families (3). The G387A mutation was described in one family (12) but is not disease-causing (13). Recently two further mutations in SPTLC1 were reported to be associated with HSAN1 (14). In vitro activity of the C133W and C133Y mutants is reduced in various cell types, including lymphocytes cultured from HSAN1 patients (15–17). HSAN1 is therefore commonly believed to be caused by a loss of SPT function. Following this reasoning, haploinsufficiency should be reflected in reduced total sphingolipid levels. This, however, could not be confirmed in HSAN1. Dawkins et al. (3) initially reported increased glucosylceramide levels in HSAN1 patients, whereas others found no changes in the lipid composition (16). Also, transgenic SPTLC1C133W mice showed significantly decreased SPT activity but no reduction in total sphingolipid levels (18). Nevertheless, these mice developed an age-dependent peripheral neuropathy with motor and sensory impairments (18). The hypothesis that HSAN1 is caused by haploinsufficiency is further challenged by the observation that heterozygous SPTLC1 and SPTLC2 knock-out mice, which have a significantly reduced SPT activity, do not develop neuropathic symptoms (19).
Here, we demonstrate that the HSAN1 mutations in SPT lead to a shift in substrate specificity of the enzyme, which then forms two atypical and neurotoxic deoxy-sphingoid bases (DSBs). Our findings therefore suggest that the pathological mechanism in HSAN1 is the accumulation of these neurotoxic metabolites rather than the reduced de novo sphingolipid synthesis. This gain of function concept is also consistent with the dominant mode of inheritance in HSAN1.
EXPERIMENTAL PROCEDURES
Patients
Approval to perform this study was obtained from the National Hospital for Neurology and Neurosurgery and Institute of Neurology Joint Research Ethics Committee (Queen Square, London, UK) and the Institutional Review Board (Massachusetts General Hospital, Boston, MA). Plasma from blood samples was obtained after informed consent from affected individuals.
Cell Culture
HEK293 cells were obtained from the ATCC and cultured in Dulbecco's modified Eagle's medium (Sigma) with 10% fetal calf serum (Fisher Scientific FSA15-043) and penicillin/streptomycin (100 units per ml/0.1 mg per ml, Sigma). EBV-transformed lymphoblast cell lines were cultured in RPMI 1640 medium (Sigma) supplemented with 15% fetal calf serum (Fisher Scientific FSA15-043), penicillin/streptomycin (100 units per ml/0.1 mg per ml, Sigma), 2 mm l-glutamine, 1 mm sodium pyruvate, 0.075% sodium bicarbonate, and 1 mm HEPES.
Statistics
Significance levels were determined with an unpaired Student's t test using SigmaPlot version 10.0 (Systat Software, Inc.). In the legends for Figs. 2, 3, and 5, p values ≤ 0.05 were labeled with *, and p values ≤ 0.01 were labeled with **.
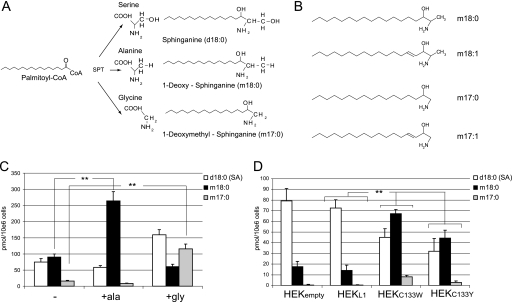
FIGURE 2.
A, products of the SPT reaction using serine, alanine, or glycine as substrates. The conjugation of palmitoyl-CoA with alanine and glycine leads to the formation of the two DSBs: m18:0 and m17:0. B, chemical structure of the DSBs. An abbreviated nomenclature for sphingoid bases is used in this work. The numbers of hydroxyls are designated by m (for mono-) and d (for di-) followed by the number of carbons. The second number indicates the double bonds. For example, d18:0 stands for sphinganine, and d18:1 stands for sphingosine. All shown metabolites were also found in the N-acetylated form. C, accumulation of m18:0 and m17:0 in HEKC133W cells after supplementing the culture medium with alanine or glycine. HEKC133W cells were cultured using either standard medium (−) or medium that was supplemented with 10 mm alanine (+ala) or 10 mm glycine (+gly). De novo synthesis was blocked with FB1 for 24 h, and the accumulated lipids were analyzed by LC-MS. D, accumulation of DSB in HEK cells expressing mutant forms of SPT. HEKempty, HEKL1, HEKC133W, and HEKC133Y cells were treated with FB1 for 24 h, and the extracted lipids were quantified by LC-MS. Error bars in C and D indicate S.E. p values ≤ 0.01 were labeled with **.
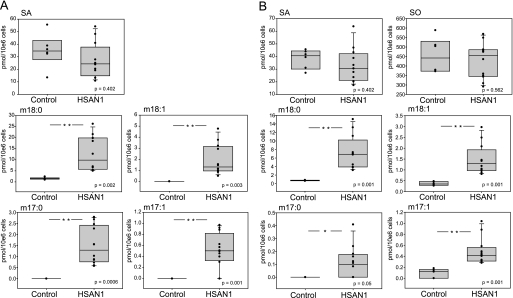
FIGURE 3.
Accumulation of DSBs in EBV-transformed lymphoblasts of HSAN1 patients. A, the de novo synthesis of DSBs was compared between EBV-transformed lymphoblasts from 12 HSAN1 patients (all C133W carriers) and six healthy controls. Lymphoblasts were cultured for 24 h in the presence of FB1, and lipids were extracted, base-hydrolyzed, and analyzed by LC-MS. SO was not detected in the lymphoblasts. B, sphingoid base levels in total lipid extracts of lymphoblasts from HSAN1 patients and healthy controls. Total cellular lipids were extracted and subjected to acid/base hydrolysis. The resulting free sphingoid bases were analyzed by LC-MS. No significant differences were seen in the total amounts of SA or SO, whereas the concentration of m17:0, m17:1, m18:0, and m18:1 was significantly higher in the HSAN1 lymphoblasts when compared with control cells. In A and B, p values ≤ 0.05 were labeled with *.
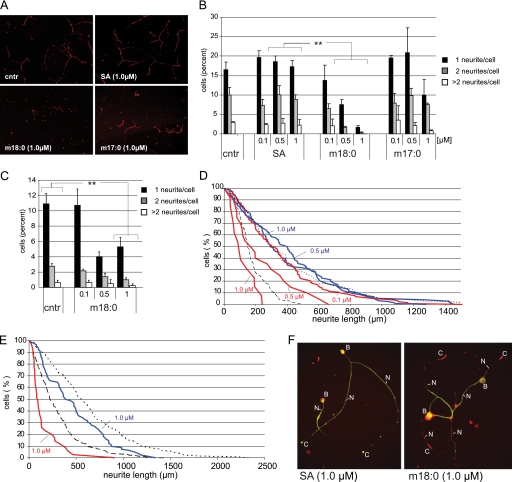
FIGURE 5.
A, effect of SA, m18:0, and m17:0 on cultured DRG neurons. Dissociated sensory neurons were grown for 12 h in control medium before the lipids were added for another 24 h. The addition of SA (1 μm) had no effect on neurite number and length when compared with controls (cntr = BSA without lipids). In the presence of m18:0 (1 μm), the cells showed a greatly reduced number of neurites. The addition of m17:0 (1 μm) showed a similar but less pronounced effect. B, quantitative analysis of the formed neurites in the presence of SA, m18:0, and m17:0. The presence of SA had no significant effect on neurite outgrowth, whereas the presence of m18:0 clearly reduced the number of neurites in a dose-dependent manner. A similar effect was observed for m17:0 whereby the presence of m17:0 mainly affected cells with two and more neurites. C, effect of m18:0 on neurite formation in cultured motoneurons. Like sensory neurons, we observed a significant reduction of neurite numbers in the presence of m18:0. However, this reduction was less pronounced at higher m18:0 concentrations in motoneurons than in sensory neurons. Error bars in B and C indicate S.E., and p values ≤ 0.01 were labeled with **. D, distribution of neurite lengths in the presence of SA and deoxy-SA (m18:0) in sensory neurons. The distribution of neurite length was plotted as the percentage of neurons with neurites longer than a given length (y axis) versus neurite length (x axis) as introduced by Chang et al. (21). The length was measured as total neurite length elaborated per neuron. Neurons were cultured for 12 h prior to the addition of the lipids (dashed line). The presence of SA (1 and 0.5 μm, blue) showed no reduction in the length of the formed neurites when compared with the control (BSA alone, dotted line). The presence of m18:0 significantly reduced neurite length in a dose-dependent manner (red lines). No difference to the control was seen with 0.1 μm m18:0. In the presence of 0.5 and 1 μm m18:0, we observed a significant shortening of the neurites in a dose-dependent manner. At the highest m18:0 concentrations tested (1 μm), the neurites were even shorter than at the time the lipids were added (dashed line, neurite length prior to the addition of the lipids). E, the presence of m18:0 induced neurite retraction. Cells were cultured for 24 h prior to the addition of the lipids (dashed line). In the presence of SA (1 μm, blue line), the neurites continued growing, although the growth rate was a little diminished in comparison with the control (dotted line). In contrast, the addition of m18:0 (1 μm, red line) induced a significant retraction of the formed neurites. F, this effect was confirmed by immune fluorescence microscopy of sensory neurons that were cultured in the presence of SA or m18:0 (1 μm each, 24 h). The SA-treated cells looked healthy and showed a clear co-localization of actin (red) and neurofilament (green) over the whole length of the neurite (left panel). In contrast, m18:0-treated neurons showed a disruption of the neurite structure with a retraction of neurofilament and a disturbed actin-neurofilament interaction (right panel). B, cell body; N, neurite; C, growth cone.
DRG Neuron Cultures
Sensory neurons of dorsal root ganglia and motoneurons were cultured as single cells essentially as described previously (20). In brief, dorsal root ganglia (DRG) were dissected from chicken embryos at day 8 (motoneurons at day 6) and cultured for 12 h in serum-free medium before the lipids were added as a BSA complex at the concentrations indicated under “Results.” The lipid-BSA stock solution was prepared from a 50 mm lipid ethanol stock (sphinganine (SA); deoxy-SA (m18:0) and deoxymethyl-SA (m17:0) were obtained from Avanti Polar Lipids) by the dropwise dilution of the lipids into phosphate-buffered saline containing 0.4% BSA until a final concentration of 1 mm. After 24 h, cells were fixed by the addition of paraformaldehyde to the culture medium at a final concentration of 4%. Neurites were visualized by neurofilament staining with RMO270 (1:1500) and donkey anti-mouse IgG-Cy3 (1:500). Neurons were classified according to the number of neurites into four classes (no neurite, one neurite, two neurites, more than two neurites per cell). The total neurite length per cell was measured, and the distribution of neurite lengths was represented as percentage of neurons with a total neurite length longer than × (Chang plot) (21).
Cloning
SPTLC1 cDNA was cloned into a pcDNA3.1 expression vector and expressed in HEK293 cells as described previously (6). The C133W and C133Y mutation was introduced using site-directed mutagenesis as follows: C133fw, 5′-ggggacccagaggattttatggcacatttgatgttc-3′; C133Yfw, 5′-atggtcctcgaggattttatggcacatttgatgttc-3′; C133W+C133Yrv, 5′-aggtacccacgccatacttctttagagatgctaaagc-3′.
Fumonisin B1-dependent Accumulation of Sphingoid Bases
Fumonisin B1 (Sigma) was added to the medium of exponentially growing cells in a final concentration of 10 μg/ml. As a negative control, myriocin (10 μg/ml, Sigma) was added together with fumonisin B1. 24 h after fumonisin B1 addition, cells were washed twice with phosphate-buffered saline, harvested, and counted (Z2 Coulter Counter®, Beckman Coulter). Synthetic C17 sphingosine (Avanti Polar Lipids) was added to each sample as an internal extraction standard.
Lipid Extraction and Hydrolysis
Total lipids were extracted from cells or plasma and extracted according to the method of Riley et al. (22). The extracts were either base-hydrolyzed or acid- and base-hydrolyzed as stated under “Results.” Before extraction, cells were resuspended in 200 μl of phosphate-buffered saline, and plasma samples (human, 100 μl; mice, 50 μl) were filled up to a volume of 200 μl with phosphate-buffered saline. Briefly, lipids were extracted in 1 ml of extraction buffer 1 (2 volumes of methanol/1 volume of chloroform + 0.2 μl/ml C17 sphingosine (SO) (1 mm in EtOH)). 100 μl of ammonia (2 n) was added, and the lipids were extracted under constant agitation (1 h, 37 °C). Subsequently 0.5 ml of chloroform was added, and samples were centrifuged (12,000 × g, 5 min) to separate the organic from the water phase. The upper (water) phase was removed, and the lower phase was washed twice with 1 ml of alkaline water (1 ml of ammonia (2 m) in 100 ml of water) and dried under N2.
For acid hydrolysis, the dried lipids were resupended in 200 μl of methanolic HCl (1 n HCl/10 m water in methanol) and kept at 65 °C for 12–15 h. The solution was neutralized by the addition of 40 μl KOH (5 m) and subsequently subjected to base hydrolysis.
Base hydrolysis was performed as follows. 0.5 ml of extraction buffer (4 volumes of 0.125 m KOH in methanol + 1 volume of chloroform) was added and mixed. Subsequently, 0.5 ml of chloroform, 0.5 ml of alkaline water, and 100 μl of 2 m ammonia was added in this order. Liquid phases were separated by centrifugation (12,000 × g, 5 min). The upper phase was aspirated, and the lower phase was washed twice with alkaline water. Finally, the lipids were dried by evaporation of the chloroform phase under N2 and subjected to LC-MS analysis.
LC-MS
Extracted lipids were solubilized in 56.7% methanol, 33.3% ethanol, 10% water and derivatized with ortho-phthaldialdehyde (Sigma) (22). The lipids were separated on a C18 column (Uptispere 120 Å, 5 μm, 125 × 2 mm, Interchim, France) and analyzed by a serial arrangement of a fluorescence detector (HP1046A, Hewlett Packard) followed by an MS detector (LCMS-2010A, Shimadzu). Atmospheric pressure chemical ionization was used for ionization. Non-natural C17 sphingosine (Avanti Polar Lipids) was used as internal standard. Retention times were as follows: C17SO (internal standard), 6 min; SO, 7.5 min; m18:1, 9 min; m17:1, 10.5 min; SA, 10.5 min; m17:0, 13 min; m18:0, 13.5 min.
MS data were analyzed using LC-MS solution (Shimadzu) and MS Processor version 11 (Advanced Chemistry Development). The quantified lipids were normalized for cell number (per million cells) and the internal standard.
RESULTS
To test the hypothesis that a gain of function rather than a loss of function underlies the pathology in HSAN1, we generated HEK293 (HEK) cell lines expressing the SPTLC1 wild-type subunit (HEKL1), the SPTLC1 mutants C133W and C133Y (HEKC133W, HEKC133Y), or the vector alone (HEKempty). All constructs were expressed at similar levels without any sign of protein degradation (supplemental Fig. 1A). Overexpression of the SPTLC1 wild-type subunit had no influence on SPT activity (supplemental Fig. 1B), whereas cells expressing the HSAN1 show a 50% reduced SPT activity in vitro, as reported previously (15–17) (supplemental Fig. 1B).
Mutant Forms of SPT Generate an Atypical Metabolite
To detect possible side products of the SPT reaction, we blocked the sphingolipid de novo synthesis at the step of ceramide synthase (CerS) with fumonisin B1 (FB1) (Fig. 1A). The inhibition of ceramide synthase leads to a time-dependent accumulation of its substrate SA (supplemental Fig. 1C). Hence, it was assumed that potential side products of the SPT reaction would also accumulate under these conditions. The addition of FB1 showed no negative effect on cell survival within the given time window (supplemental Fig. 1C). After 24 h, the accumulated lipids were extracted, separated, and quantified by HPLC (22).
The addition of FB1 led to a significant accumulation of sphinganine (Fig. 1B), whereas no accumulation was seen when SPT activity was blocked with myriocin. SA generation was about 50% reduced in HEKC133W cells due to the lower activity of the mutant SPT. Concomitantly we observed the appearance of a second peak (Fig. 1B). The accumulation of this unknown metabolite was significantly higher in HEKC133W cells than in HEKL1 cells. The peak was also increased in HEKC133Y cells but was not observed when SPT activity was blocked with myriocin. This indicates that the unknown metabolite is a product of the SPT reaction.
HSAN1 Mutations Induce a Shift in SPT Substrate Specificity
For further characterization, we compared lipid extracts of FB1-treated HEKC133W cells with those of HEKempty and HEKL1 cells by LC-MS. The analysis revealed the presence of two distinct metabolites, which were merged and appeared as a single peak in the fluorescence chromatogram. The two metabolites had a mass to charge ratio (m/z) of 462.3 and 448.3, respectively (supplemental Fig. 1D). Hence, these metabolites were 16 and 30 Da smaller in mass than SA (m/z = 478.3). This corresponds to the masses of oxygen (16 Da) and a hydroxymethyl group (–CHOH; 30 Da), respectively. SA contains two hydroxyl groups, one at C3 that derives from the fatty acid moiety and a second at C1 that originates from the conjugated serine. Strikingly, the mass of serine differs from that of alanine and glycine by 16 and 30 Da, respectively. We therefore assumed that the two identified metabolites originated from the conjugation of palmitoyl-coenzyme A with alanine and glycine instead of serine. This reaction would result in two atypical sphingolipids, m18:0 and m17:0 (Fig. 2, A and B). A shared feature of these sphingoid bases is the lack of the C1 hydroxyl group, which assigns them to the class of DSBs.
To confirm this hypothesis, cultured HEKC133W cells were supplemented with either alanine (10 mm) or glycine (10 mm). Sphingolipid de novo synthesis was blocked with FB1 for 24 h, and the accumulated lipids were analyzed by LC-MS (Fig. 2C). The addition of alanine to the medium provoked an ∼3–4-fold increase of m18:0 without significantly influencing the m17:0 levels. Conversely, the addition of glycine induced a 10-fold increase of m17:0. The slight increase in SA, seen with the glycine-supplemented cells, is probably due to a partial, intracellular conversion of glycine to serine, which then, secondarily, stimulates the generation of SA (d18:0). This confirms that the HSAN1 mutations induce a shift in the substrate affinity of SPT from serine toward alanine and glycine.
Among the mutants, we observed a 4–5-fold higher accumulation of m18:0 in HEKC133W and HEKC133Y cells when compared with HEKempty or HEKL1 cells (Fig. 2D). This shows that the affinity toward alanine is greatly increased for the HSAN1 mutants. The differences between wild type and mutants were even more pronounced for the accumulation of 1-deoxymethyl-sphinganine (m17:0), which was only detected in the mutant but not in the wild type-expressing cells. HEKC133Y cells produced, in comparison with HEKC133W, about 25 and 60% lower levels of m18:0 and m17:0, respectively.
Interestingly, low levels of m18:0, but not of m17:0, were also detected in HEK wild-type and HEKL1 cells (Fig. 2D). This suggests that the wild-type SPT is also able to metabolize alanine, although to a much lesser extent than the HSAN1 mutants. The amount of accumulated m18:0 in HEK wild-type and HEKL1 cells was about 5-fold lower than the levels observed in HEKC133W or HEKC133Y cells.
Increased DSB Generation in Lymphoblasts of HSAN1 Patients
The overexpression of mutant SPTLC1 in the background of two wild-type alleles does not necessarily represent the situation in HSAN1 patients in which the mutant and wild-type alleles are present in equivalent doses. We therefore sought to confirm our findings by comparing EBV-transformed lymphoblast lines of HSAN1 patients and healthy controls. All HSAN1 cells were derived from C133W carriers. C133Y lymphoblasts were not available. In FB1-treated lymphoblasts from HSAN1 patients, but not in lymphoblasts from healthy controls, we observed a significant accumulation of m18:0 and m17:0 (Fig. 3A). SA levels were not significantly different between patient and control cells. No accumulation was seen when the cells were grown in the presence of myriocin. Although all HSAN1 cells showed consistently elevated DSB levels, the absolute accumulation of m17:0 and m18:0 varied considerably between the different patient lines. Because all cells were cultured under identical conditions, this heterogeneity either could reflect the variable genetic background of the patients or might be a consequence of the previous EBV transformation.
DSB Accumulated in HSAN1 Lymphoblasts
The lack of the C1 hydroxyl group in the DSBs has consequences for the metabolism of these lipids. Due to the absence of the hydroxyl group, the DSBs cannot be converted to higher substituted sphingolipids, such as phospho- and glycosphingolipids, but are also not degraded by the classical pathway because this requires the formation of a phosphoester bond at C1 (Fig. 1A). Nevertheless, the DSBs serve as a substrate for ceramide synthase as shown previously by testing the effect of several synthetic sphingoid base analogs (23). The DSBs are N-acylated and also desaturated by ceramide desaturase (DES), which results in the formation of deoxy-ceramide and deoxymethyl-ceramide. The release of the N-linked acyl chain by ceramidase then results in the formation of deoxy-SO (m18:1) and deoxymethyl-SO (m17:1), respectively. To confirm the metabolic fate of the DSBs, we analyzed the sphingoid base composition in lipid extracts of HSAN1 lymphoblasts. For this, we subjected the extracted lipids to an acid and base hydrolysis. The acid hydrolysis specifically breaks the N-alkyl chain, whereas the basic hydrolysis leads to a release of the O-linked phosphoester or carbohydrate head group. A combined acid/base hydrolysis reveals, therefore, the total sphingoid base contents in a cell. The lipid extracts of HSAN1 lymphoblasts lines showed significantly higher DSB levels when compared with control cells (Fig. 3B). The levels of saturated 1-deoxy-sphinganine (deoxy-SA) and 1-deoxymethyl-sphinganine (deoxymethyl-SA) (m18:0 and m17:0) significantly increased, whereas SA (d18:0) and SO (d18:1) levels were not different between control and HSAN1 cells. This confirms earlier reports that the HSAN1 cells are able to maintain normal SA and SO levels despite the reduced SPT activity (16). Besides the saturated DSBs, the unsaturated DSBs (m18:1 and m17:1) were also significantly increased in the HSAN1 lymphoblast. Although standards for m18:1 and m17:1 are commercially not available, the metabolites were identified according to their mass difference of 2 Da and their correlation to their precursors m17:0 and m18:0 (supplemental Fig. 1E).
Among the d18 sphingoid bases, d18:1 (SO) was the most prominent. A d18:0 (SA) backbone was found in about 10% of the d18 sphingoid bases, which shows that the majority of the de novo generated SA is metabolized to ceramide. This is different for the DSBs. About 80% of the m18 bases in the lymphoblast were found in the saturated form (m18:0). In contrast, m17:0 and m17:1 were present in equal levels. The disproportion between m18:0 and m18:1 was also observed in HEKC133W cells, whereas m17 bases were only present as m17:1 (supplemental Fig. 2). The higher levels of saturated over unsaturated DSBs indicate that m18:0 is a less favorable substrate for the ceramide desaturase than SA (d18:0).
DSB Levels Are Elevated in the Plasma of HSAN1 Patients
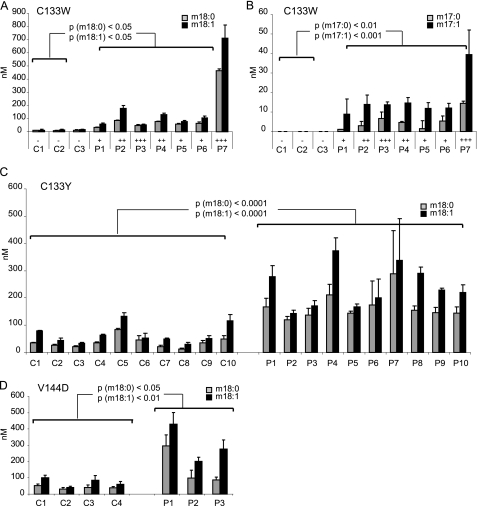
To confirm these in vitro results, we analyzed DSB levels in the plasma of three different sets of HSAN1 patients. The first set consisted of seven C133W carriers from three unrelated families and three unrelated non-carriers (Fig. 4, A and B). The second set included 10 carriers and 10 non-carriers from one single C133Y family (Fig. 4C). A third set consisted of three unrelated V144D carriers and four unrelated non-carriers (Fig. 4D). The plasma samples were subjected to acid/base hydrolysis, and the DSB levels were analyzed by LC-MS. For all C133W carriers, the levels of the m18 and m17 sphingoid bases were significantly elevated (Fig. 4, A and B). All HSAN1 samples (P1–7) showed elevated levels for m18:0 and m18:1 bases when compared with controls (C1–7). In most patients, m18:1 was more abundant than m18:0. Minor amounts of m18:0 and m18:1 were also detected in the plasma of control subjects, whereas m17:0 and m17:1 were only found in the patients. In plasma, we found generally higher proportions of unsaturated (m17:1 and m18:1) than saturated (m17:0 and m18:0) DSBs. The highest DSB levels were seen in patient 7, who also presented the most severe HSAN1 phenotype in this collective. One patient (patient 3) developed a severe form of the disease but had only intermediately elevated DSB levels. This outlier can possibly be explained by a history of alcohol abuse in this patient that could have accelerated the neuropathic symptoms. The other patients were moderately affected and showed moderately elevated DSB levels.
FIGURE 4.
DSB plasma levels in HSAN1 patients and healthy controls. A, total plasma lipids from seven HSAN1 patients (C133W carriers) and three unrelated healthy controls were extracted and subjected to acid and base hydrolysis. P1–7, plasma of HSAN1 patients; C1–7, control plasma. B, the m17 sphingoid bases were only detected in the plasma of HSAN1 patients. No m17 bases were found in control plasma. The symbols are: −, not affected; +, mildly affected, ++, moderately affected, +++, severely affected. C, comparison of m18:0 and m18:1 levels in affected (P1–10) and unaffected members (C1–10) of a C133Y family. All C133Y carriers showed significantly higher m18:0 and m18:1 levels when compared with family members who do not carry the mutation. No m17 sphingoid bases were detected in the plasma of the C133Y patients. D, m18:0 and m18:1 sphingoid bases were also significantly elevated in three carriers of a V144D mutation (P1–3) when compared with four non-carriers (C1–4). No m17 sphingoid bases were detected in the V144D carriers. Error bars indicate S.E.
The analysis of a C133Y family gave similar results (Fig. 4C), with the exception that in these patients, the m17 bases were below the detection limit. This indicates that the C133Y mutant has a lower affinity for glycine when compared with the C133W mutation, which is also reflected in a lower m17:0 generation in HEKC133Y cells when compared with HEKC133W cells (Fig. 2D). Significantly elevated m18:0 and m18:1 plasma levels were also found in patients with the V144D mutation (Fig. 4D). This shows that the V144D mutation, although located at a different site of the protein, induces the same shift in substrate specificity as the C133W/C133Y mutations. As for the C133Y mutation, the m17 sphingoid bases were below the detection limit in the plasma of the V144D patients.
The DSBs Impair Neurite Formation in Culture
To see whether the identified metabolites functionally affect the neuronal integrity, we tested their effect on cultured DRG neurons. DRGs were dissected, dissociated, and cultured in serum-free medium for 12 h prior to the addition of the lipids. After another 24 h of incubation, cells were fixed and stained for neurofilament to assess neurite outgrowth (Fig. 5A). The addition of SA (1 μm) had no obvious effect on neurites in comparison with control cells where no lipids where added (BSA only). The addition of m18:0 (1 μm), however, resulted in a remarkable reduction of neurite numbers (Fig. 5A). Also, the addition of m17:0 (1 μm) showed an effect on neurite formation, although this effect was less pronounced than with m18:0.
The quantitative analysis confirmed these observations and revealed a dose-dependent effect of m18:0 on neurite number and length (Fig. 5B). About 30% of the control cells had one or more neurites per cell. This was unchanged when the cells were cultured in the presence of SA. In the presence of m18:0 (1 μm), however, the number of cells with one or more neurites was significantly diminished (Fig. 5B). The addition of m17:0 showed a milder effect and primarily reduced the number of cells with more than two neurites. Interestingly, the addition of m18:0 and m17:0 did not affect the number of viable neurons when compared with control (BSA only) or SA-treated cells. This shows that up to a concentration of 1 μm, the DSBs do not induce cell death in cultured sensory neurons. Rather, it seems that the DSBs specifically disturb neurite formation in these neurons.
When compared with sensory neurons, motoneurons are reported to be less affected in HSAN1. We therefore tested whether motoneurons are less sensitive to the DSBs (Fig. 5C). Also, motoneurons that were cultured in the presence of m18:0 showed an overall reduction in neurite numbers. However, this reduction was less pronounced than in sensory neurons (Fig. 5B). Especially at higher concentrations of m18:0 (1 μm), the proportion of cells with one or more neurites was clearly higher in moto- than in sensory neurons.
The analysis of neurite length showed a concentration-dependent effect of m18:0 on the length of the formed neurites (Fig. 5D). Low concentrations of m18:0 (0.1 μm) had no significant effect on neurite length in comparison with control conditions, e.g. no added lipids or 0.5–1 μm SA. Higher m18:0 concentrations (0.5 and 1 μm) resulted in a significant and dose-dependent reduction of neurite length. Remarkably, the highest tested concentration of m18:0 (1 μm) not only prevented further neurite growth but even seemed to result in a net reduction of neurite lengths when compared with the length of the neurites before adding the lipids (Fig. 5D, dashed line). We therefore repeated this experiment on neurons, which were precultured for 24 h before the lipids were added (Fig. 5E). In control and SA (1 μm)-treated cells, the neurites continued to grow over time, although the growth rate seemed to be slightly diminished in the presence of SA. In contrast, the addition of m18:0 resulted in a significant retraction of the already formed neurites with an increased percentage of neurites with less than 200 μm in length (Fig. 5E). The analysis of the neurites by immune fluorescence microscopy showed a co-localization of actin and neurofilament over the whole length of the neurites in neurons, which were cultured in the presence of SA (Fig. 5F). In contrast, the neurites of neurons that were cultured in the presence of m18:0 had a clearly disturbed cytoskeletal structure. The neurofilament staining was significantly shortened in these cells and only partly co-localized with the actin staining, whereas the actin signal was detected over the whole length of the neurites. This indicates that m18:0 influences the stability and dynamics of neurofilament formation.
In summary, our results demonstrate a neurotoxic effect of the alternative metabolites formed by the mutant forms of SPT. Thus, we conclude that the neuropathology in HSAN1 is due to the accumulation of these aberrant SPT products rather than haplotype insufficiency.
DISCUSSION
Various mutations in the SPTLC1 subunit of SPT cause HSAN1, a form of hereditary sensory and autonomic neuropathy. It has been repeatedly shown that these mutations result in reduced enzymatic activity. However, this reduced activity does not induce changes in total cellular sphingolipid levels (16). Also, our own analysis showed no significant differences between the total sphingolipid levels of lymphoblasts from HSAN1 patients and those from healthy controls (Fig. 2B). Neither were total sphingoid base levels different in plasma from HSAN1 patients when compared with healthy controls. This incongruous finding can be explained by the fact that cells can also generate ceramide by the degradation of sphingomyelin from external sources (24) and are, therefore, principally able to compensate for a reduced de novo ceramide synthesis. Furthermore, heterozygous SPTLC1 and SPTLC2 knock-out mice have significantly reduced SPT activity but do not develop neuropathic symptoms (19). Thus, a reduced SPT activity is not, by itself, a sufficient explanation for the pathogenesis in HSAN1.
In this study, we demonstrated that the C133W and the C133Y mutations, which are the most frequent HSAN1 mutations, cause a shift in the substrate specificity of SPT. We showed that mutated SPT is able to use alanine and, to a lesser extent, glycine as an alternative substrate. As a consequence, the two sphingolipid metabolites deoxy-SA (m18:0) and deoxymethyl-SA (m17:0) are formed, which both lack the hydroxyl group at C1. This prevents the further conversion of these metabolites to phospho- or glycosphingolipids but also impedes their degradation, which requires the phosphorylation at C1 and the subsequent breakdown by sphingosine-1-phospate lyase (SO1P lyase) (Fig. 1A). Consequently, these metabolites were found in high concentrations in those cells that express the mutant SPT. Elevated DSB levels were found in lymphoblasts of HSAN1 patients, and plasma DSB levels were significantly elevated in HSAN1 patients, who carry the C133W, the C133Y, or the V144D mutation.
Elevated DSB levels were also found in plasma of mice that are transgenic for the SPTLC1C133W mutant as reported recently (25). Interestingly, double transgenic mice, which concomitantly overexpress the C133W mutant together with wild-type SPTLC1, do not develop neurological symptoms (25). This correlates with significantly lower DSB plasma levels in these mice. However, DSB levels in the double transgenic mice are still higher than in wild-type, SPTLC1 wild type-overexpressing mice, or heterozygous SPTLC1 +/− knock-out mice, suggesting that DSBs can be tolerated up to a certain threshold level. Tissue analysis revealed the presence of highly elevated DSB levels in the sciatic nerves of the C133W mice but not of wild-type mice (25). No differences in the DSB levels were seen in brain tissue of C133W and wild-type mice. This shows that the DSBs accumulate specifically in peripheral nervous system but not in central nervous system tissue, which fully corresponds to the pattern of pathology in HSAN1 as the central nervous system is not affected in this disease.
Low amounts of m18 sphingoid bases, but not of m17 sphingoid bases, could also be detected in control plasma and non-HSAN1 cells. This also indicates that the wild-type SPT has a certain affinity toward alanine. Similar findings were reported recently by Riley and co-workers (26), which demonstrated the accumulation of deoxy-SA in FB1-treated LLC-PK1 cells. Structurally, deoxy-SA resembles the members of the fumonisin “B” series, whereas the glycine derivative shares a structural homology with the “C” series of fumonisins. These mycotoxins are formed in a variety of fungal species by the condensation of a polyketide chain to alanine or glycine, which is catalyzed by a monomeric SPT homologue (for a review, see Ref. 27).
Interestingly, deoxy-SA (m18:0; also referred to as “Spisulosine” or “ES-285”) has been isolated from the arctic clam Spisula polynyma as an investigational marine anticancer drug (28). It induces cell death in various breast cancer cell lines with an LD50 value in the lower μm range (29). It was shown that deoxy-SA stimulates an atypical apoptosis pathway without affecting c-Jun N-terminal kinase (JNK), extracellular signal-regulated kinase (Erk), or Akt signaling by activating protein kinase Cζ (29, 30).
In our work, we demonstrated that the supplementation of the DSBs to neuronal DRG cultures is neurotoxic and interferes with the formation of the neurites in vitro. This effect was less pronounced for motoneurons, which is congruent with the pattern of pathology in HSAN1 where motoneurons are less affected. Interestingly, the presence of deoxy-SA not only impaired the number and length of newly formed neurites but also induced the retraction of already formed neurites. Immune fluorescence microscopy indicated a disruption of the neurite cytoskeletal structure in the presence of deoxy-SA, where we observed a retraction of neurofilament although actin was still present along the full length of the neurites (Fig. 5F).
The observed neurotoxic effect was clearly detectable for deoxy-SA but less pronounced for deoxymethyl-SA. The fact that deoxymethyl-SA was not detected in the plasma of C133Y and V144D patients indicates that the presence of deoxymethyl-SA is not essential for the pathogenesis of HSAN1. However, whether the m18:0 or one of its metabolic products, such as deoxy-dihydroceramide, deoxy-ceramide, or deoxy-SO, is primarily responsible for the observed neurotoxic effect needs to be addressed in a subsequent study.
The finding that elevated DSB levels are closely associated with HSAN1 also provides a new aspect in the diagnosis of HSAN1. Although HSAN1 is a rare disease, its clinical appearance can be very heterogeneous, and symptoms closely resemble the group of Charcot-Marie-Tooth (CMT) neuropathies. CMT is the most commonly inherited neurological disorder, with an incidence of 30–40 in 100,000. The lack of a blood biomarker for the diagnosis of CMT requires, classically, the verification of a clinically diagnosed CMT by sequencing. This often results in the extensive sequencing of many gene loci, especially in the case of a rare or spontaneous de novo mutation. The association of elevated plasma DSB levels with HSAN1 provides, for the first time, a blood marker that can be used, together with the clinical history, to confirm or exclude HSAN1 from other types of CMT. Furthermore, aberrations of SPT activity might be more frequently involved in peripheral neuropathies than is commonly believed. Very recently, two new mutations in the SPTLC1 gene were reported to be associated with a sensory neuropathy (14), and further SPT mutations might be identified in the future.
However, the exact pathomechanism by which the DSBs exert their neurotoxic activity is not yet fully clear. Our observations indicate that the DSBs act neurotoxically by disturbing neuronal cytoskeleton formation. In this context, it is interesting that deoxy-SA was reported earlier to impair cytoskeleton formation by interfering with the Rho signaling cascade (28). Members of the Rho GTPase family have been shown to regulate various aspects of intracellular actin and neurofilament dynamics, including cell polarization, axonal stabilization, and growth cone formation (31). It is therefore conceivable that a destabilization of axonal structures or an impairment of growth cone formation would lead to a progressive degeneration of axons as seen in HSAN1.
Another possibility could be that the DSBs have an inhibitory effect on one or several consecutive enzymes of the phospho- and glycosphingolipid pathway. A selective inhibition of one of these enzymes might reduce the levels of certain functional glycosphingolipids without significantly affecting total sphingolipid levels. An inhibitory effect on SO-kinase, for instance, would result in a reduction of sphingosine-1-phospate, lyase, which is a potent neurotrophic factor (32, 33). Nevertheless, the findings presented here provide conclusive evidence that HSAN1 is closely associated with elevated DSB levels and that DSB levels, at concentrations similar to those found in the plasma of HSAN1 patients, exert pronounced neurotoxic effects on cultured sensory neurons. This suggests that the same neurotoxic activity is also responsible for the atrophy of sensory neurons in HSAN1 patients.
Acknowledgments
We are grateful to the families for their essential help with this work. We also thank Ursula Gutteck for the technical help with the MS analysis and Diane McKenna-Yasek for the help in collecting patient samples. Part of this work was undertaken at the University College London Hospitals/University College London, which received a proportion of funding from the Department of Health National Institute for Health Research Biomedical Research Centres funding scheme.
This work was supported, in whole or in part, by National Institutes of Health Grant K08NS52550 (to F. E.) through the NINDS. This work was also supported by grants from the Deater Foundation (to R. H. B. and F. E.) and from the MRC (to M. M. R. and H. H.) and the Muscular Dystrophy Campaign (to M. M. R. and M. L.). The work undertaken at the University Hospital of Zurich and was supported by grants from the Hartmann Müller Foundation, the Herzog-Egli Foundation, the Olga Mayenfisch Foundation, and the Foundation for Scientific Research (University of Zürich) as well as the German Society for Clinical Chemistry and Laboratory Medicine (DGKL) and the European Commission (LSHM-CT-2006-037631).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- HSAN
- hereditary sensory and autonomic neuropathy
- SA
- sphinganine
- SO
- sphingosine
- deoxy-SA
- deoxy-sphinganine
- deoxymethyl-SA
- 1-deoxymethyl-sphinganine
- m18:0
- 1-deoxy-sphinganine
- m17:0
- 1-deoxymethyl-sphinganine
- d18:0
- SA
- d18:1
- SO
- SPT
- serine palmitoyltransferase
- DSB
- deoxy-sphingoid base
- EBV
- Epstein-Barr virus
- DRG
- dorsal root ganglia
- CMT
- Charcot-Marie-Tooth
- BSA
- bovine serum albumin
- FB1
- fumonisin B1
- LC-MS
- liquid chromatography-mass spectrometry
- HPLC
- high pressure liquid chromatography.
REFERENCES
- 1.Auer-Grumbach M., Mauko B., Auer-Grumbach P., Pieber T. R. (2006) Neuromolecular. Med. 8, 147–158 [DOI] [PubMed] [Google Scholar]
- 2.Auer-Grumbach M. (2008) Orphanet. J. Rare Dis. 3, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawkins J. L., Hulme D. J., Brahmbhatt S. B., Auer-Grumbach M., Nicholson G. A. (2001) Nat. Genet. 27, 309–312 [DOI] [PubMed] [Google Scholar]
- 4.Bejaoui K., Wu C., Scheffler M. D., Haan G., Ashby P., Wu L., de Jong P., Brown R. H., Jr. (2001) Nat. Genet. 27, 261–262 [DOI] [PubMed] [Google Scholar]
- 5.Hanada K., Hara T., Fukasawa M., Yamaji A., Umeda M., Nishijima M. (1998) J. Biol. Chem. 273, 33787–33794 [DOI] [PubMed] [Google Scholar]
- 6.Hornemann T., Richard S., Rütti M. F., Wei Y., von Eckardstein A. (2006) J. Biol. Chem. 281, 37275–37281 [DOI] [PubMed] [Google Scholar]
- 7.Hanada K. (2003) Biochim. Biophys. Acta 1632, 16–30 [DOI] [PubMed] [Google Scholar]
- 8.Bi H., Gao Y., Yao S., Dong M., Headley A. P., Yuan Y. (2007) Neuropathology 27, 429–433 [DOI] [PubMed] [Google Scholar]
- 9.Klein C. J., Wu Y., Kruckeberg K. E., Hebbring S. J., Anderson S. A., Cunningham J. M., Dyck P. J., Klein D. M., Thibodeau S. N., Dyck P. J. (2005) J. Neurol. Neurosurg. Psychiatry 76, 1022–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houlden H., King R., Blake J., Groves M., Love S., Woodward C., Hammans S., Nicoll J., Lennox G., O'Donovan D. G., Gabriel C., Thomas P. K., Reilly M. M. (2006) Brain 129, 411–425 [DOI] [PubMed] [Google Scholar]
- 11.Geraldes R., de Carvalho M., Santos-Bento M., Nicholson G. (2004) J. Neurol. Sci. 227, 35–38 [DOI] [PubMed] [Google Scholar]
- 12.Verhoeven K., Coen K., De Vriendt E., Jacobs A., Van Gerwen V., Smouts I., Pou-Serradell A., Martin J. J., Timmerman V., De Jonghe P. (2004) Neurology 62, 1001–1002 [DOI] [PubMed] [Google Scholar]
- 13.Hornemann T., Penno A., Richard S., Nicholson G., van Dijk F. S., Rotthier A., Timmerman V., von Eckardstein A. (2009) Neurogenetics 10, 135–143 [DOI] [PubMed] [Google Scholar]
- 14.Rotthier A., Baets J., De Vriendt E, Jacobs A., Auer-Grumbach M., Lévy N., Bonello-Palot N., Kilic S. S., Weis J., Nascimento A., Swinkels M., Kruyt M. C., Jordanova A., De Jonghe P., Timmerman V. (2009) Brain 132, 2699–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bejaoui K., Uchida Y., Yasuda S., Ho M., Nishijima M., Brown R. H., Jr., Holleran W. M., Hanada K. (2002) J. Clin. Invest. 110, 1301–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dedov V. N., Dedova I. V., Merrill A. H., Jr., Nicholson G. A. (2004) Biochim. Biophys. Acta 1688, 168–175 [DOI] [PubMed] [Google Scholar]
- 17.Gable K., Han G., Monaghan E., Bacikova D., Natarajan M., Williams R., Dunn T. M. (2002) J. Biol. Chem. 277, 10194–10200 [DOI] [PubMed] [Google Scholar]
- 18.McCampbell A., Truong D., Broom D. C., Allchorne A., Gable K., Cutler R. G., Mattson M. P., Woolf C. J., Frosch M. P., Harmon J. M., Dunn T. M., Brown R. H., Jr. (2005) Hum. Mol. Genet. 14, 3507–3521 [DOI] [PubMed] [Google Scholar]
- 19.Hojjati M. R., Li Z., Jiang X. C. (2005) Biochim. Biophys. Acta 1737, 44–51 [DOI] [PubMed] [Google Scholar]
- 20.Stoeckli E. T., Ziegler U., Bleiker A. J., Groscurth P., Sonderegger P. (1996) Dev. Biol. 177, 15–29 [DOI] [PubMed] [Google Scholar]
- 21.Chang S., Rathjen F. G., Raper J. A. (1987) J. Cell Biol. 104, 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riley R. T., Norred W. P., Wang E., Merrill A. H. (1999) Nat. Toxins 7, 407–414 [DOI] [PubMed] [Google Scholar]
- 23.Humpf H. U., Schmelz E. M., Meredith F. I., Vesper H., Vales T. R., Wang E., Menaldino D. S., Liotta D. C., Merrill A. H., Jr. (1998) J. Biol. Chem. 273, 19060–19064 [DOI] [PubMed] [Google Scholar]
- 24.Tani M., Ito M., Igarashi Y. (2007) Cell. Signal. 19, 229–237 [DOI] [PubMed] [Google Scholar]
- 25.Eichler F. S., Hornemann T., McCampbell A., Kuljis D., Penno A., Vardeh D., Tamrazian E., Garofalo K., Lee H. J., Kini L., Selig M., Frosch M., Gable K., von Eckardstein A., Woolf C. J., Guan G., Harmon J. M., Dunn T. M., Brown R. H., Jr. (2009) J. Neurosci. 29, 14646–14651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zitomer N. C., Mitchell T., Voss K. A., Bondy G. S., Pruett S. T., Garnier-Amblard E. C., Liebeskind L. S., Park H., Wang E., Sullards M. C., Merrill A. H., Jr., Riley R. T. (2009) J. Biol. Chem. 284, 4786–4795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du L., Zhu X., Gerber R., Huffman J., Lou L., Jorgenson J., Yu F., Zaleta-Rivera K., Wang Q. (2008) J. Ind. Microbiol. Biotechnol. 35, 455–464 [DOI] [PubMed] [Google Scholar]
- 28.Cuadros R., Montejo de Garcini E., Wandosell F., Faircloth G., Fernández-Sousa J. M., Avila J. (2000) Cancer Lett. 152, 23–29 [DOI] [PubMed] [Google Scholar]
- 29.Salcedo M., Cuevas C., Alonso J. L., Otero G., Faircloth G., Fernandez-Sousa J. M., Avila J., Wandosell F. (2007) Apoptosis 12, 395–409 [DOI] [PubMed] [Google Scholar]
- 30.Sánchez A. M., Malagarie-Cazenave S., Olea N., Vara D., Cuevas C., Díaz-Laviada I. (2008) Eur. J. Pharmacol. 584, 237–245 [DOI] [PubMed] [Google Scholar]
- 31.Heasman S. J., Ridley A. J. (2008) Nat. Rev. Mol. Cell Biol. 9, 690–701 [DOI] [PubMed] [Google Scholar]
- 32.Milstien S., Gude D., Spiegel S. (2007) Acta Paediatr. Suppl. 96, 40–43 [DOI] [PubMed] [Google Scholar]
- 33.Okada T., Kajimoto T., Jahangeer S., Nakamura S. (2009) Cell. Signal. 21, 7–13 [DOI] [PubMed] [Google Scholar]