Abstract
Glioblastoma multiforme is the most common and lethal primary brain cancer in adults. Tumor cells diffusely infiltrate the brain making focal surgical and radiation treatment challenging. The invasion of glioma cells into normal brain is facilitated by the activity of ion channels aiding dynamic regulation of cell volume. Recent studies have specifically implicated ClC-3, a voltage-gated chloride channel, in this process. However, the interaction between ClC-3 activity and cell movement is poorly understood. Here, we demonstrate that ClC-3 is highly expressed on the plasma membrane of human glioma cells where its activity is regulated through phosphorylation via Ca2+/calmodulin-dependent protein kinase II (CaMKII). Intracellular infusion of autoactivated CaMKII via patch pipette enhanced chloride currents 3-fold, and this regulation was inhibited by autocamtide-2 related inhibitory peptide, a CaMKII-specific inhibitor. CaMKII modulation of chloride currents was also lost upon stable small hairpin RNA knockdown of ClC-3 channels indicating a specific interaction of ClC-3 and CaMKII. In ClC-3-expressing cells, inhibition of CaMKII reduced glioma invasion to the same extent as direct inhibition of ClC-3. The importance of the molecular interaction of ClC-3 and CaMKII is further supported by our finding that CaMKII co-localizes and co-immunoprecipitates with ClC-3. ClC-3 and CaMKII also co-immunoprecipitate in tissue biopsies from patients diagnosed with grade IV glioblastoma. These tumor samples show 10-fold higher ClC-3 protein expression than nonmalignant brain. These data suggest that CaMKII is a molecular link translating intracellular calcium changes, which are intrinsically associated with glioma migration, to changes in ClC-3 conductance required for cell movement.
Keywords: Biophysics, Cell Migration, Chloride Channels, Ion Channels, Protein Phosphorylation
Introduction
Glioblastomas account for 60–70% of malignant gliomas (1) and are the most common and lethal type of primary malignant brain tumors among adults. The prognosis for glioblastoma patients is poor despite treatment consisting of surgical debulking, radiotherapy, and chemotherapy. A unique feature contributing to the disease aggressiveness is the ability of malignant glioma cells to actively migrate along the brain vasculature (2, 3) instead of passive metastasis through vascular circulation. To migrate along blood vessels through the narrow extracellular space of the brain, cells must be able to regulate their cell volume. We hypothesize that gliomas express ion channels that endow cells with an enhanced ability to extrude osmotically active ions, leading to the release of cytoplasmic water and cell shrinkage (4). Candidate channels providing the electrochemical driving force for ion movement in glioma cells are the Ca2+-activated potassium channel BK (5–7) and the voltage-gated chloride channel ClC-3 (8–10).
ClC-3 enhances migration of nasopharyngeal carcinoma cells (11), and pharmacological inhibition with NPPB2 demonstrates a requirement for chloride channels to support glioma invasion (8). Glioma cells express three members of the ClC family, ClC-2, -3, and -5 (10), and we have found that ClC-3 in particular is a critical regulator of cell volume changes associated with the cell cycle (9). Although a role for ClC-3 in vesicular and granule acidification (12–15) is established, recent evidence suggests that ClC-3 may also be involved in neuronal excitability (16), proliferation (17), and migration (18). Given the proposed roles for ClC-3, several groups have sought to understand regulation of this channel to provide further insight on its diverse functions. When ClC-3 was initially cloned and expressed, it was found to be inhibited by protein kinase C phosphorylation (19). Later characterization found that ClC-3 was inhibited by inositol 3,4,5,6-tetrakisphosphate (20) and activated by Ca2+/calmodulin-dependent protein kinase II (21, 22). CaMKII phosphorylated ClC-3 at Ser-109 (22) and led to a 22-fold increase in ClC-3 current in transfected tsA cells (21). This Ca2+-sensitive kinase may also play an important role in glioma biology, particularly because glioma cells require Ca2+ acting as a second messenger to support cell migration. Glioma cells show oscillatory changes in [Ca2+]i that correlate with cell migration (23), and this Ca2+ signal may be the consequence of AMPA-R activation (24). More specifically, gliomas express Ca2+-permeable AMPA-R, i.e. receptors that lack the GluR2 subunit, and mutations forming a Ca2+-impermeant channel retard glioma invasion in vivo (25). We hypothesize that Ca2+ acting via CaMKII leading to ClC-3 phosphorylation may be an important signaling event underlying glioma invasion.
Using a combination of biochemical and biophysical techniques, we found that CaMKII phosphorylates ClC-3 from human glioma cells, leading to an activation of native ClC-3 channels. Interestingly, we found that ClC-3 and CaMKII co-immunoprecipitate and that both proteins are necessary for glioma migration, furthering the importance of CaMKII-mediated phosphorylation of ClC-3. To extend our conclusions beyond cultured cells, we found that human biopsy tissue from grade IV glioblastoma patients expressed 10-fold more ClC-3 compared with normal brain and that ClC-3 from glioblastoma biopsy tissue is also associated with CaMKII. These data underscore the importance of understanding the role of ion channel regulation in glioma pathophysiology.
EXPERIMENTAL PROCEDURES
Cell Culture
D54 human glioma cells are a World Health Organization grade IV cell line derived from a glioblastoma and gifted to us by Dr. D. Bigner (Duke University, Durham, NC). Cells were passaged in Dulbecco's modified Eagle's medium/F-12 supplemented with 2 mm glutamine (Media Tech, University of Alabama at Birmingham Media Preparation Facility) and 7% fetal bovine serum (Hyclone, Logan, UT) and incubated at 37 °C and 10% CO2. All reagents were purchased from Sigma unless otherwise noted.
Immunocytochemistry
Cells were cultured on round 12-mm glass coverslips (Macalaster Bicknell, New Haven, CT) in a 24-well plate for 2–4 days, washed with phosphate-buffered saline, and fixed with 4% paraformaldehyde for 10 min. Cells were then blocked and permeabilized for 30 min at room temperature with phosphate-buffered saline containing 10% normal goat serum and 0.3% Triton X-100. After incubation in primary antibodies overnight at 4 °C, cells were washed with a 1:3 dilution of the blocking buffer in phosphate-buffered saline and incubated in secondary antibodies for 1 h at room temperature. After further washing with the diluted blocking buffer, cells were incubated with 4′,6-diamidino-2-phenylindole (DAPI) at 1:2000 for 5 min at room temperature. Cells were then washed, and coverslips were mounted onto 3 × 1-inch × 1-mm glass slides (Fisher) with Fluoromount (Sigma) and stored at −20 °C.
We immunolabeled ClC-3 with a rabbit polyclonal anti-ClC-3 antibody targeted against residues 592–661 of ClC-3 (lot no. AN-06, Alomone Labs, Jerusalem, Israel) used at 1:250. CaMKII was labeled with a mouse monoclonal anti-CaMKII antibody (Abcam, Cambridge, MA) used at 1:250. The following secondary antibodies obtained from Invitrogen were used at 1:500: goat anti-rabbit Alexa 488 and goat anti-mouse Alexa 546. Phalloidin conjugated to Alexa 546 (Invitrogen) was used at 1:50. Fluorescent images were acquired with Slidebook software (Intelligent Imagining Innovations) using a Hamamatsu IEEE1394 digital CCD camera mounted on an Olympus IX81 motorized inverted microscope with an Olympus disk scanning unit to remove out-of-focus light. Using a 60× oil immersion lens (numerical aperture, 1.42) with digital zooms of individual cells, 20 images were taken at 0.5-μm steps totaling 10-μm image stacks through the center of the cells. Alexa 488 fluorescence was imaged with a fluorescein isothiocyanate filter set (excitation, 482 ± 17 nm; emission, 536 ± 20 nm), and Alexa 546 fluorescence was imaged with a TRITC filter set (excitation, 543 ± 22 nm; emission, 593 ± 20 nm), and DAPI fluorescence was imaged with a DAPI filter set (excitation, 387 ± 5.5 nm; emission, 447 ± 30 nm) (Semrock). Images were interpolated with Slidebook software and displayed in “three-view.”
Immunoprecipitation
Whole-cell lysates of D54 human glioma cells were collected in 500 μl of RIPA buffer (150 mm NaCl, 50 mm Tris-HCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS) supplemented with 1:100 protease inhibitor mixture and 1:100 phosphatase inhibitor mixture. Protein concentrations were obtained using a NanoDrop 1000 spectrophotometer (Thermo Scientific). Protein A (for CaMKII pulldown) or protein G (for ClC-3 pulldown) magnetic beads (Millipore, Billerica, MA) were washed two times in RIPA buffer and equilibrated in RIPA buffer for 2 h on ice. Lysates were pre-cleared with 50 μl of beads rotating for 1 h at 4 °C. After the pre-clear, 6 μg of precipitating antibody was added to the lysate and rotated for 30 min at 4 °C. Polyclonal rabbit anti-ClC-3 (Alpha Diagnostic International, San Antonio, TX) and monoclonal rabbit anti-CaMKII (Abcam, Cambridge, MA) were the precipitating antibodies. 100 μl of beads were then added and rotated overnight at 4 °C. The next day, beads were washed three times with RIPA. Bound protein was eluted from beads with 40 μl of 0.2 m glycine, pH 3, and then neutralized with 5 μl of 1 m Tris base. Blots were run as described below.
For the CaMKII kinase experiments, beads with bound ClC-3 were split into two tubes of equal volume. CaMKII was activated similar to the manufacturer's directions (P6060, New England Biolabs, Ipswich, MA). Briefly, 1 μl of CaMKII (500,000 units/ml) was diluted in 1× CaMKII reaction buffer, 200 μm ATP, 1.2 μm calmodulin, and 2 mm CaCl2. This mixture was incubated at 37 °C for 10 min. After this pre-activation, beads with bound ClC-3 were incubated with the CaMKII reaction mixture at 37 °C for 40 min. Beads were then washed two times with RIPA buffer, and protein was eluted as described above.
Electrophysiology
Whole-cell patch clamp recording was done on D54 human glioma cells after 1–3 days in culture using standard recording techniques (26). Patch pipettes, with resistances of 3–5 megohms, were pulled from thin-walled borosilicate glass (TW150F-4, World Precision Instruments, Sarasota, FL) with an upright puller (PP-830, Narashige Instruments, Tokyo, Japan). Recordings were done with a MultiClamp 700B amplifier (Molecular Devices) and digitized on line at 10 kHz and low pass filtered at 2 kHz using a Digidata 1322A digitizer (Axon Instruments). pClamp 9.0 (Axon Instruments) software was used to acquire and store the data onto a personal computer. Series resistance was compensated to 80%, and cells in which series resistance was >12 megohms were omitted because of poor voltage clamping. Standard pipette solution (140 mm KCl, 1 mm MgCl2, 0.2 mm CaCl2, 10 mm EGTA, 10 mm Hepes sodium salt) was adjusted to 302–304 mosm, pH 7.2, with Tris base. Standard bath solution (130 mm NaCl, 5 mm KCl, 1 mm CaCl2, 10.55 mm glucose, 32.5 mm Hepes acid) was adjusted to 308–312 mosm and pH 7.4 with NaOH.
CaMKII included in the pipette solution was prepared similarly to previous experiments by others (27). 0.5 μl (500,000 units/ml) of CaMKII was diluted into 1× CaMKII reaction buffer, 200 μm ATP, 1.2 μm calmodulin, and 2 mm CaCl2 (P6060, New England Biolabs, Ipswich, MA) and incubated at 37 °C for 10 min. The CaMKII solution was then diluted into 40 μl of pipette solution and kept on ice and used in <3 h. Myristoylated AIP (Enzo Life Sciences) was included directly into the pipette solution at 10 μm. NPPB was bath-perfused at 200 μm to block chloride channels. Paxilline (Santa Cruz Biotechnology) was bath-perfused at 2 μm to block BK channels. All ClC-3 electrophysiology experiments were performed in the presence of 2 μm paxilline.
Western Blot
Protein concentrations were measured using a NanoDrop 1000 spectrophotometer (Thermo Scientific), and 15 μg per sample was diluted to equal volumes using RIPA buffer. 6× sample buffer (60% glycerol, 300 mm Tris, pH 6.8, 12 mm EDTA, 12% SDS, 864 mm 2-mercaptoethanol, 0.05% bromphenol blue) was added to samples and loaded on 10-well 10% gradient pre-cast SDS-polyacrylamide gels (Bio-Rad). Protein separation was performed at 120 mV for 75 min. Gels were then transferred to polyvinylidene difluoride paper (Millipore, Bedford, MA) at 200 mA for 120 min. Blots were blocked with blocking buffer (5% nonfat dried milk in TBS plus 0.1% Tween 20 (TBST)). Primary antibodies, including rabbit anti-ClC-3 (Alpha Diagnostic International; 1:1500), rabbit anti-CaMKII (Abcam; 1:5000), and mouse anti-glyceraldehyde-3-phosphate dehydrogenase (Abcam; 1:5000), were incubated for 1 h at room temperature. After four 10-min washes in TBST, blots were incubated with respective horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology; 1:1500) for 1 h at room temperature. After four 10-min washes in TBST, blots were developed (Supersignal West Femto, Thermo Scientific, or Western Blotting Luminol Reagent, Santa Cruz Biotechnology) and imaged on a 4000MM Image Station (Eastman Kodak Co.). ClC-3 immunoreactivity was normalized to glyceraldehyde-3-phosphate dehydrogenase.
ClC-3 Knockdown
We stably knocked down ClC-3 expression in D54 cells as described previously (9). Briefly, a commercially available pGIPZ-lentiviral small hairpin mir vector containing a hairpin sequence targeting CLCN3 (Open Biosystems) was transfected (nucleofection, Amaxa Biosystems). The hairpin sequence, with the underlined sequence corresponding to the region of the gene targeted, was 5′-TGCTGTTGACAGTGAGCGCCCTACCTCTTTCCAAAGTATATAGTGAAGCCACAGATGTATATACTTTGGAAAGAGGTAGGATGCCTACTGCCTCGGA-3′. To generate stable lines, 10 μg/ml puromycin treatment was begun 96 h after electroporation. These cells are referred to as H8a cells throughout the text.
Migration Assay
Transwell filters with 8-μm pores (BD Biosciences) were placed into 24-well plates. The bottoms of the filters were coated with 3 μg/ml vitronectin (Promega) diluted in phosphate-buffered saline and incubated overnight to serve as a chemoattractant for migration. The following day, the bottoms of the filters were washed with migration assay buffer (Dulbecco's modified Eagle's medium/F-12 media supplemented with 0.1% fatty-acid free bovine serum albumin). 400 μl of migration assay buffer was placed at the bottom of each filter, and 40,000 cells suspended in migration assay buffer were placed on top of the filters. Cells were allowed to adhere for 30 min before drug treatment of water and DMSO loading controls or 100 μm NPPB (8) to block chloride channels or 1 μm myristoylated AIP (Enzo Life Sciences) to block CaMKII. Cells were then allowed to migrate for an additional 5.5 h in the cell incubator. To stop migration, filters were fixed and stained with crystal violet. The tops of the filters were wiped clean before imaging with a 10× objective on a Zeiss Axiovert 200 M microscope. Five fields of view were taken per insert.
Data Analysis
Current responses to voltage steps and current subtractions were quantified using Clampfit (Axon Instruments). Raw data were compiled and graphed in Origin 6.0 (MicroCal, Northampton, MA) or Microsoft Excel. Statistical analyses were run using GraphPad Instat (GraphPad Software). One-way analysis of variance was determined, and all data are reported as mean ± S.E. (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
RESULTS
Glioma Cells Express ClC-3 and CaMKII
Previous studies have demonstrated a role for ClC-3 in the malignant characteristics of glioma cells, but the regulation of ClC-3 in glioma cells has not been examined. We hypothesized that ClC-3 expressed by human glioma cells on the cell membrane is activated by CaMKII leading to an increased chloride conductance. To determine whether ClC-3 and CaMKII are expressed by glioma cells, we immunocytochemically labeled cultured D54 human glioma cells. The cellular location of ClC-3 has been controversial, with some groups finding expression on the cell membrane (16, 28) and others seeing predominantly intracellular expression on endosomes and vesicles (12, 29). We hypothesized that glioma cells express ClC-3 on the cell membrane, providing a resting chloride conductance that plays a role in malignant characteristics (4). To assess cell surface expression, we labeled D54 glioma cells with phalloidin, to visualize cortical actin at the cell membrane, and antibodies targeted against residues 592–661 of ClC-3 and took 0.5-μm confocal optical sections with a spinning disk confocal microscope (Fig. 1). We found expression of ClC-3 (Fig. 1, green) in D54 glioma cells and labeling of cortical actin by phalloidin (Fig. 1, red). ClC-3 localized to the cell surface, indicated by the co-localization of ClC-3 and cortical actin, as seen in a 60× field of view (Fig. 1A) and in digital zooms of individual glioma cells (Fig. 1, B and C; same cells boxed in A). Arrows in Fig. 1, B and C, indicate points at which the cell is depicted in “three-view” (central panel is x,y; top panel is x,z; left panel is z,y) with co-localization of ClC-3 and cortical actin occurring throughout the 10-μm imaging section. A fraction of ClC-3 does not co-localize with cortical actin and is found throughout the cytoplasm. Therefore, ClC-3 is located throughout D54 glioma cells, including the cell surface.
FIGURE 1.
Human glioma cells express ClC-3 on the plasma membrane. The 1st row contains representative merged images, with examples of ClC-3 and cortical actin co-localization indicating ClC-3 expression on the plasma membrane. The 2nd row demonstrates ClC-3 immunolabeling (green) throughout the cell. The 3rd row shows phalloidin binding of actin (red), including cortical actin on the plasma membrane. The 4th row is the DAPI nuclear stain (blue). A, ×60 view of a field of glioma cells. Boxes demarcate zoomed cells seen in B and C. B and C, digital zooms of individual cells. Arrows point to cross-sections of cells seen in three-view through the entire 10-μm imaging plane.
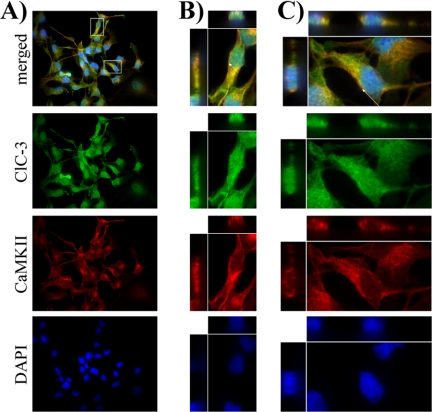
We hypothesized that CaMKII is found in similar cellular domains as ClC-3, allowing it to regulate chloride conductance. To assess this possible co-localization, we fluorescently labeled CaMKII and ClC-3 and took 0.5-μm confocal optical sections. As seen in a 60× field of view (Fig. 2A) and in digital zooms of individual cells (Fig. 2, B and C), ClC-3 (green) and CaMKII (red) co-localize in D54 glioma cells outside of the DAPI-labeled nucleus (blue). Arrows in Fig. 2, B and C, indicate points of co-localization seen in three-view through the entire stacked 10-μm optical section of the cell. Given similar subcellular localization in glioma cells, we then determined the following: 1) if CaMKII binds ClC-3 and 2) if CaMKII can phosphorylate ClC-3.
FIGURE 2.
ClC-3 and CaMKII co-localize in human glioma cells. The 1st row contains representative merged images, with examples of ClC-3 and CaMKII co-localization. The 2nd row demonstrates ClC-3 immunolabeling (green) throughout the cell. The 3rd row shows CaMKII immunolabeling (red). The 4th row is the DAPI nuclear stain (blue). A, ×60 view of a field of glioma cells. Boxes demarcate zoomed cells seen in B and C. B and C, digital zooms of individual cells. Arrows point to cross-sections of cells seen in three-view through the entire 10-μm imaging plane.
CaMKII Phosphorylates and Co-immunoprecipitates with ClC-3
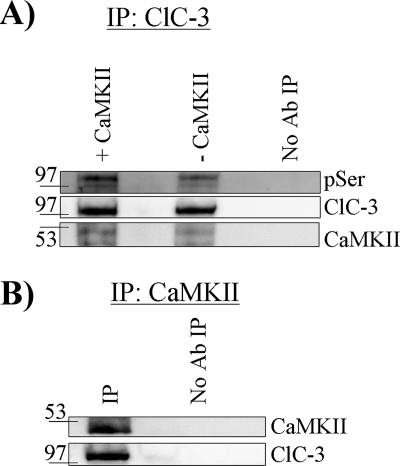
We immunoprecipitated ClC-3 from whole-cell lysates of D54 glioma cells using antibodies conjugated to protein A or protein G magnetic beads and reacted the bound protein with autoactivated CaMKII. ClC-3 was then eluted from the beads, run on an SDS-polyacrylamide gel, and then Western-blotted, probing for phosphoserine residues. CaMKII phosphorylated a protein at the molecular mass of ClC-3 (∼105 kDa) at serine residues (Fig. 3A) compared with untreated control. ClC-3 from D54 glioma cells appears to be endogenously phosphorylated at a fraction of serine residues as seen in the untreated “-CaMKII” condition, and exposure to activated CaMKII enhanced the binding of phosphoserine antibody (Fig. 3A). ClC-3 was also labeled on the same blot to ensure pulldown of ClC-3 and even loading of the lanes.
FIGURE 3.
CaMKII phosphorylates and co-immunoprecipitates with ClC-3. A, after immunoprecipitation (IP) of ClC-3, CaMKII phosphorylated ClC-3 as seen by enhanced binding of phosphoserine (pSer) antibody (Ab). Equal amounts of ClC-3 were pulled down, and CaMKII was detected. B, after immunoprecipitation of CaMKII, ClC-3 was detected only when the precipitating antibody was included. Blots are representative of n ≥3 experiments.
To test the hypothesis that ClC-3 and CaMKII are part of the same protein complex, we performed an affinity pulldown assay whereby antibodies targeting the C terminus of ClC-3 were used to pull down ClC-3. We then probed for CaMKII and found an ∼50-kDa CaMKII band by Western blot (Fig. 3A, bottom row). As a result, we then performed the converse experiment and pulled down CaMKII with CaMKII-specific monoclonal antibodies and probed with anti-ClC-3 antibodies and found an ∼105-kDa ClC-3 band (Fig. 3B). Neither band was seen when the precipitating antibody was excluded from the protein A or protein G magnetic beads (Fig. 3, A and B). These data suggest that CaMKII not only phosphorylates ClC-3 but also is in the same protein complex as ClC-3 in glioma cells, placing it in proximity to modulate ClC-3 function.
Activated CaMKII Increases ClC-3 Conductance
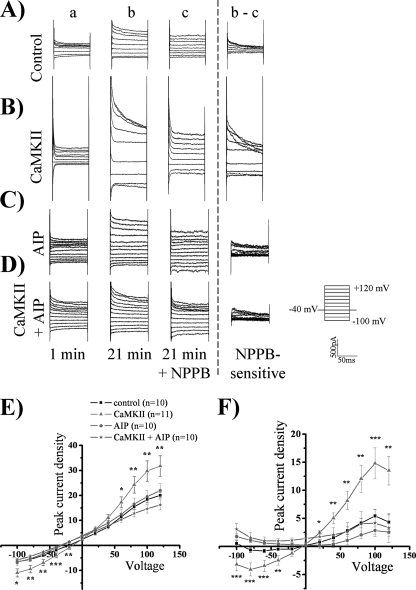
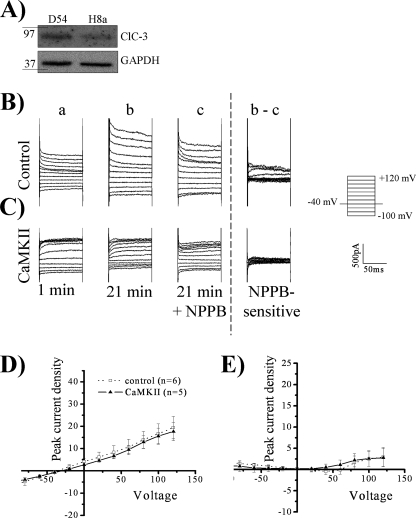
We hypothesized that phosphorylation of ClC-3 would increase whole-cell chloride conductance in normo-osmotic conditions and that this activation could be blocked with NPPB, a chloride channel blocker. To perform these experiments, we autoactivated CaMKII and diluted the activated kinase into pipette solution and whole-cell patch-clamped D54 human glioma cells. After waiting 20 min for the kinase to diffuse into the cytoplasm from the pipette solution, cells were held at −40 mV and stepped from −100 to +120 mV in 20-mV increments. 200 μm NPPB was then bath-applied to block chloride channels, and the NPPB-sensitive current was subtracted (Fig. 4, A and B). We found a significant increase of the peak inward and outward current density in whole-cell recordings upon addition of autoactivated CaMKII (Fig. 4, A, B, and E) as compared with control cells. The whole-cell peak current density at −80 mV was −5.5 pA/pF ± 0.73 in control cells (n = 10) versus −9.4 pA/pF ± 1.5 in cells treated with CaMKII (n = 11; p < 0.05, Tukey-Kramer). Examination of the outward current at +100 mV also revealed a significant enhancement, with control current density measuring 18.4 pA/pF ± 1.1 (n = 10) and CaMKII current density measuring 29.8 pA/pF ± 4.0 (n = 11; p < 0.05, Tukey-Kramer). There also appeared to be a rightward (i.e. positive) shift of the reversal potential upon addition of autoactivated CaMKII toward the chloride equilibrium potential of 0 mV, indicating an increase in chloride permeability (Fig. 4E). Autoactivated CaMKII induced approximately a 3-fold activation of inward and outward NPPB-sensitive current compared with control cells (Fig. 4F). The activated chloride current was outwardly rectifying and voltage- and time-inactivating at depolarized potentials, matching the electrophysiological characteristics of ClC-3 (22, 30). The NPPB-sensitive inward current at −80 mV increased from −0.62 pA/pF ± 0.4 in control cells (n = 10) to −4.1 pA/pF ± 1.2 in CaMKII-treated cells (n = 11; p < 0.05, Tukey-Kramer). Similarly, the outward peak NPPB-sensitive current at +100 mV increased from 5.4 pA/pF ± 1.2 in control cells (n = 10) to 14.8 pA/pF ± 2.8 in CaMKII-treated cells (n = 11; p < 0.01, Tukey-Kramer). The reversal potential of the NPPB-sensitive current was 0 mV, as expected with symmetric chloride solutions. Thus activated CaMKII induces a ClC-3 conductance in human glioma cells.
FIGURE 4.
ClC-3 current is activated by CaMKII. Using whole-cell patch clamp electrophysiology, human glioma cells were held at −40 mV and stepped from −100 mV to +120 mV. For the representative traces, the 1st column (a) is basal current at 1 min, and the 2nd column (b) is current at 21 min. At 21 min, NPPB was added as seen in the 3rd column (c), and the NPPB-sensitive current (b − c) is depicted in the 4th column. A, control condition. B, activated CaMKII included in the pipette solution leading to an enhancement of chloride current. C, AIP, a CaMKII inhibitor, is included in the pipette solution. D, activated CaMKII and AIP are included in the pipette solution leading to a loss of CaMKII-mediated current enhancement. E, peak whole-cell current density corresponding to b. F, peak NPPB-sensitive current density corresponding to b − c. p values indicate significance of CaMKII versus CaMKII + AIP conditions (*, p < 0.05; **, p < 0.01; ***, p < 0.001; Tukey Kramer). n = 10–11.
To determine the specificity of CaMKII-mediated activation of ClC-3, we included AIP, a potent and specific CaMKII inhibitor, in the pipette solution. 10 μm AIP inhibits CaMKII but not protein kinase C, CaMKIV, and protein kinase A (31). Upon 10 μm AIP addition to the pipette solution, CaMKII did not increase chloride conductance (Fig. 4, C and D) as compared with control. AIP inhibited the CaMKII enhancement of peak whole-cell current density and NPPB-sensitive current density (Fig. 4, E and F). At +100 mV CaMKII increased whole-cell current density to 29.8 pA/pF ± 4.0 (n = 11), but upon the addition of AIP, the current density reduced to 14.6 pA/pF ± 1.7 (n = 10; p < 0.01, Tukey-Kramer) and was not significantly different from the control current density of 18.4 pA/pF ± 1.1. AIP inhibited the enhanced NPPB-sensitive current density from 14.8 pA/pF ± 2.8 upon CaMKII addition (n = 11) at +100 mV to 2.9 pA/pF ± 0.7 (n = 10; p < 0.001, Tukey-Kramer), which is not significantly different from control levels of 5.4 pA/pF ± 1.2. The data further suggest that autoactivated CaMKII enhances ClC-3 current density.
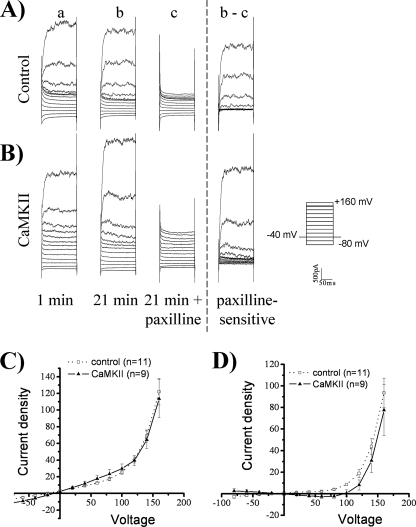
We next examined the potential regulation of the Ca2+-sensitive voltage-gated BK channel by CaMKII. BK channels play a significant role in glioma biology (5, 7, 8, 32) and have consensus sites for CaMKII phosphorylation (33, 34). We whole-cell patch-clamped D54 glioma cells, stepped from −80 to +160 mV, and then washed on paxilline to block BK channels. Paxilline inhibited a strongly outwardly rectifying current, consistent with currents mediated by BK channels (Fig. 5, A and B). However, as compared with control cells, autoactivated CaMKII did not enhance whole-cell current density or paxilline-sensitive current density (Fig. 5, A–D). At +140 mV, control whole-cell current density was 69.1 pA/pF ± 7.2 (n = 11) and was not significantly different upon CaMKII addition, measuring at 64.4 pA/pF ± 10.6 (n = 9). Similarly, paxilline-sensitive current density was not significantly different at +140 mV in control versus CaMKII-treated cells, measuring at 44.01 pA/pF ± 7.11 (n = 11) and 30.8 pA/pF ± 10.9 (n = 9), respectively. We did observe a rightward shift of the reversal potential of whole-cell currents toward the reversal potential of chloride, indicating an increase in chloride permeability. Therefore, CaMKII enhances ClC-3 current density and not BK current density in human glioma cells, despite BK channels having CaMKII phosphorylation sites.
FIGURE 5.
BK channels are not activated by CaMKII. Using whole-cell patch clamp electrophysiology, human glioma cells were held at −40 mV and stepped from −80 mV to +160 mV. For the representative traces, the 1st column (a) is basal current at 1 min, and the 2nd column (b) is current at 21 min. At 21 min, paxilline was added as seen in the 3rd column (c), and the paxilline-sensitive current (b − c) is depicted in the 4th column. A, control condition. B, activated-CaMKII included in the pipette solution does not lead to a change in current. C, whole-cell current density corresponding to b. D, paxilline-sensitive current density corresponding to b − c is not significantly different from control. n = 9–11.
Pharmacological inhibitors of chloride channels such as NPPB are not specific to ClC-3, so to further assess the specificity of CaMKII-mediated enhancement of chloride current density, we stably knocked down ClC-3 in D54 human glioma cells using small hairpin RNA in a pGIPZ-lentiviral vector (Open Biosystems), herein referred to as H8a cells (9). Transfection of ClC-3 small hairpin RNA reduced ClC-3 protein expression by 57–62% and a representative blot corresponding to the knockdown seen in Fig. 6A. ClC-3 knockdown did not affect CaMKII expression as normalized to glyceraldehyde-3-phosphate dehydrogenase loading control. In contrast to wild-type cells (Fig. 4, A and B), autoactivated CaMKII in H8a cells did not significantly increase current density compared with control cells (Fig. 6, B and C). Whole-cell peak current density at +100 mV, 16.7 pA/pF ± 4.4 (n = 6), did not significantly increase in the presence autoactivated CaMKII, which averaged at 15.6 pA/pF ± 3.6 (n = 5; Fig. 6D). There was little NPPB-sensitive current in H8a cells, and this current was not significantly increased by CaMKII (Fig. 6E). NPPB-sensitive current at +100 mV in control cells was 2.5 pA/pF ± 1.7 (n = 6) and 2.6 pA/pF ± 1.8 upon CaMKII treatment (n = 5). These results indicate that the CaMKII enhancement of chloride current density is mediated by ClC-3.
FIGURE 6.
CaMKII does not activate a chloride current after ClC-3 knockdown. A, ClC-3 expression is decreased in H8a cells expressing ClC-3 small hairpin RNA relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) loading control, and CaMKII expression is not altered. B–E, using whole-cell patch clamp electrophysiology, human glioma cells were held at −40 mV and stepped from −100 mV to +120 mV. For the representative traces, the 1st column (a) is basal current at 1 min, and the 2nd column (b) is current at 21 min. At 21 min, NPPB was added as seen in the 3rd column (c), and the NPPB-sensitive current (b − c) is depicted in the 4th column. B, control condition. C, activated CaMKII included in the pipette solution does not lead to an enhancement of chloride current. D, peak whole-cell current density corresponding to b. E, peak NPPB-sensitive current density corresponding to b and c. n = 5–6.
ClC-3 and CaMKII Play a Role in Glioma Migration
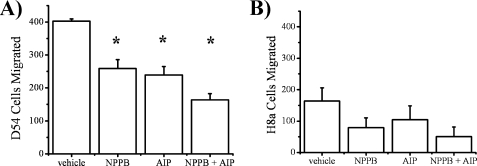
ClC-3 is implicated in the migration of several malignant cells, including gliomas (8), so we hypothesized that inhibition of ClC-3 or inhibition of CaMKII, via its action on ClC-3, would decrease glioma migration. To examine the role of ClC-3 and CaMKII in glioma migration, we plated cells on 8 μm Transwell filters in Boyden chambers and counted the number of cells that migrated through pores toward a chemoattractant. In control conditions, 402 ± 7 cells migrated through the pores but significantly dropped to 259 ± 27, 239 ± 26, and 164 ± 18 cells upon application of 100 μm NPPB, 1 μm AIP, or 100 μm NPPB + 1 μm AIP, respectively (n = 3; p < 0.01 for each condition compared with control, Tukey-Kramer; Fig. 7A). There was no significant difference in migration between 100 μm NPPB, 1 μm AIP, and 100 μm NPPB + 1 μm AIP conditions, indicating that the ClC-3 and CaMKII may be blocking similar pathways, precluding an additive effect. To better assess ClC-3 specificity, we also examined migration in ClC-3 knockdown H8a cells and found lower levels of migration (Fig. 7B) compared with control cells (n = 3). This is consistent with previous reports showing that blockage of ClC-3 currents causes reduced glioma migration (8). Importantly, in H8a cells we found no significant difference in migration between control (164 cells ± 42), 100 μm NPPB (79 cells ± 31), 1 μm AIP (104 cells ± 44), and 100 μm NPPB + 1 μm AIP (50 cells ± 31) conditions. The lack of an NPPB effect can be attributed to the knockdown of ClC-3 (Fig. 6, A and B), and the lack of an AIP effect suggests the role of CaMKII in migration may be ClC-3-dependent. These data suggest that ClC-3 and CaMKII play an important role in glioma migration.
FIGURE 7.
ClC-3 or CaMKII inhibition leads to an attenuation in Transwell migration. A, representative images of Transwell migration of D54 and H8a (ClC-3 knock-down) cells after drug treatment. B, NPPB, AIP, and NPPB + AIP decrease D54 cell migration. C, ClC-3 knockdown leads to a decrease in migration that is not altered after drug treatment. (*, p < 0.05; Tukey Kramer). n = 3.
ClC-3 Expression Is Elevated in Human GBM Tissue
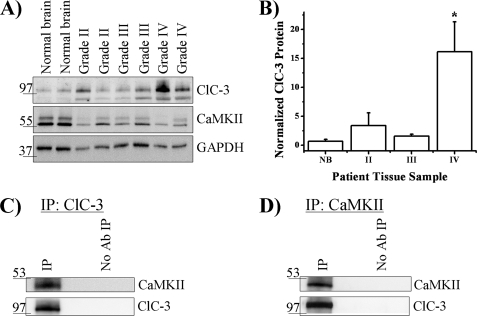
To determine whether a relationship between ClC-3 and CaMKII in gliomas occurs beyond our in vitro system, we examined human glioblastoma multiforme tissue from patient biopsies. We compared ClC-3 expression in normal human brain specimens to human biopsy tissue samples from World Health Organization grade II, grade III, and grade IV tumors and found that grade IV tumors, the most aggressive and lethal glioma, had significantly more expression of ClC-3 compared with normal brain (Fig. 8, A and B). After normalizing to normal human brain tissue, we found that grade IV tumors (n = 4) express 16.1 ± 5.2 times more ClC-3 (n = 3; p < 0.05, Tukey-Kramer). However, grade II tumors (n = 4) and grade III tumors (n = 3) did not express ClC-3 levels significantly different from normal brain tissue (p > 0.05). This increased expression of ClC-3 in grade IV tumors may correlate with the enhanced ability of these cells to diffusely migrate in the human brain. Normal brain, grade II, grade III, and grade IV tumor samples also expressed CaMKII (Fig. 8A). Finally, given the association of ClC-3 and CaMKII in D54 glioma cells, we hypothesized that ClC-3 and CaMKII co-immunoprecipitate in patient-derived glioma tissue. We immunoprecipitated ClC-3, ran the protein on SDS-polyacrylamide gel, and then Western-blotted for ClC-3 and CaMKII. CaMKII immunoprecipitated with ClC-3 but not when anti-ClC-3 antibody was excluded (Fig. 8C). We then immunoprecipitated CaMKII and found associated ClC-3 on a Western blot (Fig. 8D). These data suggest higher grade gliomas express elevated levels of ClC-3 that are associated with CaMKII, which positively regulates ClC-3 conductance and may play a role in glioma migration.
FIGURE 8.
ClC-3 is overexpressed in grade IV human glioma tissue and associated with CaMKII. A, representative blot demonstrating ClC-3 and CaMKII immunoreactivity in normal brain, grade II, grade III, and grade IV human biopsy samples. B, quantification of ClC-3 expression normalized to normal brain reveals overexpression in grade IV samples. (*, p < 0.05; Tukey Kramer). (Normal brain, n = 3; grade II, n = 4; grade III, n = 3; and grade IV, n = 4.) C, after immunoprecipitation of ClC-3 from a grade IV glioblastoma patient biopsy, CaMKII was detected only when the precipitating antibody was included. D, after immunoprecipitation of CaMKII from a grade IV glioblastoma patient biopsy, ClC-3 was detected only when the precipitating antibody was included. Blots are representative of n ≥ 3 experiments.
DISCUSSION
Expression of ClC-3 on the Plasma Membrane
For ClC-3 to play a role in cell migration (8, 11, 35), proliferation (9, 17, 36), and volume regulation (37, 38), we hypothesize that it is at least partially expressed on the cell membrane. Using confocal imaging, we found that ClC-3 co-localizes with cortical actin at the cell membrane (Fig. 1). We did not assess interactions between ClC-3 and actin, but McCloskey et al. (39) found that actin directly binds the C terminus of ClC-3 and that actin interaction with ClC-3 is necessary for maximal response of volume-sensitive current in NIH-3T3 cells. Association with actin can regulate the electrophysiological properties of membrane-associated channels, e.g. as observed in the sodium channel ENaC (40, 41). Membrane-associated ClC-3 has been quantified in COS-7 cells transfected with ClC3-GFP, where 25% of synthesized ClC-3 trafficked through the membrane, but only 6% of ClC-3 was at the membrane at any given time (42). The membrane localization of ClC-3 has been confirmed in glioma cells (9, 10), immature neurons (16), and nonpigmented ciliary epithelial cells (28) and in heterologous expression systems, such as HEK293-tsA201 cells (20), NIH-3T3 cells (43), and Xenopus oocytes (19). However, other groups found predominantly intracellular expression on vesicles and endosomes in neuroendocrine cells (29) and hippocampal neurons (12, 44) and in heterologous expression systems such as CHO-K1 and Huh-7 cells (14). Therefore, diversity in ClC-3 localization may vary between cell types and expression systems, yielding biophysical intricacies and functional consequences ranging from migration and proliferation to vesicular acidification.
Electrophysiological Properties of ClC-3
We found that membrane-associated ClC-3 was phosphorylated and activated by CaMKII, producing a slightly outwardly rectifying chloride current that was time- and voltage-inactivating, matching the electrophysiological signature of the long isoform of ClC-3 (9, 10, 22). Shimada et al. (30) found short and long isoforms of ClC-3 in rat hepatocytes, with the short isoform characterized by a strongly outwardly rectifying current without voltage inactivation resembling ClC-4 and ClC-5 and the long isoform, found in human tissue, characterized by a slightly outwardly rectifying current that is voltage-inactivating. Characterization of the exact electrophysiological signature of ClC-3 has been controversial (45) because cells from ClC-3−/− animals maintain an outwardly rectifying time- and voltage-inactivating current attributed to the swelling-induced current Iswell (12, 46, 47). However, Iswell regulation in ClC-3−/− mice by protein kinase C, anti-ClC-3 antibodies, [ATP]i, and [Mg2+]i is altered (48), raising the possibility of compensatory changes, such as up-regulation of other members of the ClC family in response to ClC-3 ablation. ClC-3−/− mice also lack CaMKII-mediated ClC-3 activation (16), which is supported by our ClC-3 knockdown data (Fig. 6).
ClC-3 activity is modulated by a variety of kinases, including CaMKII (21, 22), protein kinase C (49), serum- and glucocorticoid-inducible kinase (50), and inositol 3,4,5,6-tetrakisphosphate (20). Here, we find that CaMKII not only activates chloride conductance similar to that found in mouse aorta smooth muscle (22) but also co-immunoprecipitates with ClC-3. We did not investigate if this co-immunoprecipitation was due to a direct or indirect protein interaction, but CaMKII is known to bind a variety of target substrate channels, including N-methyl-d-aspartic acid receptors in the context of synaptic plasticity (51, 52), the Eag potassium channel (53), and voltage-gated calcium channels (54). We also did not investigate if binding of CaMKII to ClC-3 enhances CaMKII activity or ClC-3 conductance, but our data suggest that changes in intracellular calcium concentrations in glioma cells may manifest in enhanced chloride conductance via a phosphorylation-dependent mechanism.
CaMKII May Translate Ca2+ Signals into Changes in Chloride Conductance
Oscillation of intracellular calcium levels in glioma cells plays a role in migration (24, 56), reminiscent of immature neurons (57). One potential source of Ca2+ influx enabling glioma migration is from Ca2+-permeable, GluR2-lacking AMPA-R (24, 25). Because glioma cells release high concentrations of glutamate (58, 59), activation of Ca2+-permeable AMPA-R may occur in an autocrine manner, enhancing migration and pro-survival pathways (60). If the calcium influx from Ca2+-permeable AMPA-R is sufficient to activate CaMKII, then ClC-3 conductance may be enhanced (Fig. 4) leading to malignant glioma behaviors associated with ClC-3, such as migration and proliferation. Our data regarding ClC-3 and CaMKII inhibition suggest that both proteins play a role in glioma migration (Fig. 7). CaMKII may function as an interface translating changes in intracellular Ca2+ into motility via ClC-3 activation. Ca2+ signals may simultaneously activate parallel pathways, including pro-survival signaling cascades and CaMKII-induced ClC-3-mediated cell volume and shape changes. Our examination of World Health Organization grade IV glioblastoma multiforme tissue suggests that ClC-3 is up-regulated more than 10-fold relative to normal brain tissue and co-immunoprecipitates with CaMKII (Fig. 8). Therefore, understanding the role ClC-3 plays in glioma disease progression may lead to better clinical outcomes. Our data suggest that drugs interfering with the function of ClC-3 or CaMKII in glioma cells are potential therapies to decrease the invasiveness of gliomas. Indeed, an inhibitor of chloride channels for the treatment of malignant gliomas is currently in phase II clinical trials (55), and other therapies interfering with CaMKII phosphorylation of ClC-3 may be novel alternatives for more effective clinical management of gliomas.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1-NS-31234 and RO1-NS-36692 (to H. S.).
- NPPB
- 5-nitro-2-(3-phenylpropylamino)benzoic acid
- AIP
- autocamtide-2 related inhibitory peptide
- CaMKII
- Ca2+/calmodulin-dependent protein kinase II
- DAPI
- 4′,6-diamidino-2-phenylindole
- TRITC
- tetramethylrhodamine isothiocyanate
- AMPA-R
- α-amino-3-hydroxy-5-methyl-4-isoxazole propionate receptor.
REFERENCES
- 1.Wen P. Y., Kesari S. (2008) N. Engl. J. Med. 359, 492–507 [DOI] [PubMed] [Google Scholar]
- 2.Farin A., Suzuki S. O., Weiker M., Goldman J. E., Bruce J. N., Canoll P. (2006) Glia 53, 799–808 [DOI] [PubMed] [Google Scholar]
- 3.Bernstein J. J., Goldberg W. J., Laws E. R., Jr. (1989) J. Neurosci. Res. 22, 134–143 [DOI] [PubMed] [Google Scholar]
- 4.Sontheimer H. (2008) Exp. Biol. Med. 233, 779–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X., Chang Y., Reinhart P. H., Sontheimer H., Chang Y. (2002) J. Neurosci. 22, 1840–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ransom C. B., Liu X., Sontheimer H. (2002) Glia 38, 281–291 [DOI] [PubMed] [Google Scholar]
- 7.Weaver A. K., Bomben V. C., Sontheimer H. (2006) Glia 54, 223–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ransom C. B., O'Neal J. T., Sontheimer H. (2001) J. Neurosci. 21, 7674–7683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Habela C. W., Olsen M. L., Sontheimer H. (2008) J. Neurosci. 28, 9205–9217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olsen M. L., Schade S., Lyons S. A., Amaral M. D., Sontheimer H. (2003) J. Neurosci. 23, 5572–5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao J., Chen L., Xu B., Wang L., Li H., Guo J., Li W., Nie S., Jacob T. J., Wang L. (2008) Biochem. Pharmacol. 75, 1706–1716 [DOI] [PubMed] [Google Scholar]
- 12.Stobrawa S. M., Breiderhoff T., Takamori S., Engel D., Schweizer M., Zdebik A. A., Bösl M. R., Ruether K., Jahn H., Draguhn A., Jahn R., Jentsch T. J. (2001) Neuron 29, 185–196 [DOI] [PubMed] [Google Scholar]
- 13.Hara-Chikuma M., Yang B., Sonawane N. D., Sasaki S., Uchida S., Verkman A. S. (2005) J. Biol. Chem. 280, 1241–1247 [DOI] [PubMed] [Google Scholar]
- 14.Li X., Wang T., Zhao Z., Weinman S. A. (2002) Am. J. Physiol. Cell Physiol. 282, C1483–C1491 [DOI] [PubMed] [Google Scholar]
- 15.Deriy L. V., Gomez E. A., Jacobson D. A., Wang X., Hopson J. A., Liu X. Y., Zhang G., Bindokas V. P., Philipson L. H., Nelson D. J. (2009) Cell Metab. 10, 316–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X. Q., Deriy L. V., Foss S., Huang P., Lamb F. S., Kaetzel M. A., Bindokas V., Marks J. D., Nelson D. J. (2006) Neuron 52, 321–333 [DOI] [PubMed] [Google Scholar]
- 17.Qian J. S., Pang R. P., Zhu K. S., Liu D. Y., Li Z. R., Deng C. Y., Wang S. M. (2009) Cell. Physiol. Biochem. 24, 461–470 [DOI] [PubMed] [Google Scholar]
- 18.Mao J., Chen L., Xu B., Wang L., Wang W., Li M., Zheng M., Li H., Guo J., Li W., Jacob T. J., Wang L. (2009) Biochem. Pharmacol. 77, 159–168 [DOI] [PubMed] [Google Scholar]
- 19.Kawasaki M., Uchida S., Monkawa T., Miyawaki A., Mikoshiba K., Marumo F., Sasaki S. (1994) Neuron 12, 597–604 [DOI] [PubMed] [Google Scholar]
- 20.Mitchell J., Wang X., Zhang G., Gentzsch M., Nelson D. J., Shears S. B. (2008) Curr. Biol. 18, 1600–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang P., Liu J., Di A., Robinson N. C., Musch M. W., Kaetzel M. A., Nelson D. J. (2001) J. Biol. Chem. 276, 20093–20100 [DOI] [PubMed] [Google Scholar]
- 22.Robinson N. C., Huang P., Kaetzel M. A., Lamb F. S., Nelson D. J. (2004) J. Physiol. 556, 353–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bordey A., Sontheimer H., Trouslard J. (2000) J. Membr. Biol. 176, 31–40 [DOI] [PubMed] [Google Scholar]
- 24.Lyons S. A., Chung W. J., Weaver A. K., Ogunrinu T., Sontheimer H. (2007) Cancer Res. 67, 9463–9471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishiuchi S., Tsuzuki K., Yoshida Y., Yamada N., Hagimura N., Okado H., Miwa A., Kurihara H., Nakazato Y., Tamura M., Sasaki T., Ozawa S. (2002) Nat. Med. 8, 971–978 [DOI] [PubMed] [Google Scholar]
- 26.Hamill O. P., Marty A., Neher E., Sakmann B., Sigworth F. J. (1981) Pflügers Arch. 391, 85–100 [DOI] [PubMed] [Google Scholar]
- 27.Chan H. C., Kaetzel M. A., Gotter A. L., Dedman J. R., Nelson D. J. (1994) J. Biol. Chem. 269, 32464–32468 [PubMed] [Google Scholar]
- 28.Vessey J. P., Shi C., Jollimore C. A., Stevens K. T., Coca-Prados M., Barnes S., Kelly M. E. (2004) Biochem. Cell Biol. 82, 708–718 [DOI] [PubMed] [Google Scholar]
- 29.Maritzen T., Keating D. J., Neagoe I., Zdebik A. A., Jentsch T. J. (2008) J. Neurosci. 28, 10587–10598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimada K., Li X., Xu G., Nowak D. E., Showalter L. A., Weinman S. A. (2000) Am. J. Physiol. Gastrointest Liver Physiol. 279, G268–G276 [DOI] [PubMed] [Google Scholar]
- 31.Ishida A., Kameshita I., Okuno S., Kitani T., Fujisawa H. (1995) Biochem. Biophys. Res. Commun. 212, 806–812 [DOI] [PubMed] [Google Scholar]
- 32.Weaver A. K., Olsen M. L., McFerrin M. B., Sontheimer H. (2007) J. Biol. Chem. 282, 31558–31568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Q., Chen B., Ge Q., Wang Z. W. (2007) J. Neurosci. 27, 10404–10413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J., Asuncion-Chin M., Liu P., Dopico A. M. (2006) Nat. Neurosci. 9, 41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Volk A. P., Heise C. K., Hougen J. L., Artman C. M., Volk K. A., Wessels D., Soll D. R., Nauseef W. M., Lamb F. S., Moreland J. G. (2008) J. Biol. Chem. 283, 34315–34326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang Y. B., Liu Y. J., Zhou J. G., Wang G. L., Qiu Q. Y., Guan Y. Y. (2008) Cell Prolif. 41, 775–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossow C. F., Duan D., Hatton W. J., Britton F., Hume J. R., Horowitz B. (2006) Acta Physiol. 187, 5–19 [DOI] [PubMed] [Google Scholar]
- 38.Xiong D., Wang G. X., Burkin D. J., Yamboliev I. A., Singer C. A., Rawat S., Scowen P., Evans R., Ye L., Hatton W. J., Tian H., Keller P. S., McCloskey D. T., Duan D., Hume J. R. (2009) Clin. Exp. Pharmacol. Physiol. 36, 386–393 [DOI] [PubMed] [Google Scholar]
- 39.McCloskey D. T., Doherty L., Dai Y. P., Miller L., Hume J. R., Yamboliev I. A. (2007) J. Biol. Chem. 282, 16871–16877 [DOI] [PubMed] [Google Scholar]
- 40.Cantiello H. F., Stow J. L., Prat A. G., Ausiello D. A. (1991) Am. J. Physiol. 261, C882–C888 [DOI] [PubMed] [Google Scholar]
- 41.Berdiev B. K., Prat A. G., Cantiello H. F., Ausiello D. A., Fuller C. M., Jovov B., Benos D. J., Ismailov I. I. (1996) J. Biol. Chem. 271, 17704–17710 [DOI] [PubMed] [Google Scholar]
- 42.Zhao Z., Li X., Hao J., Winston J. H., Weinman S. A. (2007) J. Biol. Chem. 282, 29022–29031 [DOI] [PubMed] [Google Scholar]
- 43.Duan D., Winter C., Cowley S., Hume J. R., Horowitz B. (1997) Nature 390, 417–421 [DOI] [PubMed] [Google Scholar]
- 44.Salazar G., Love R., Styers M. L., Werner E., Peden A., Rodriguez S., Gearing M., Wainer B. H., Faundez V. (2004) J. Biol. Chem. 279, 25430–25439 [DOI] [PubMed] [Google Scholar]
- 45.Jentsch T. J. (2008) Crit. Rev. Biochem. Mol. Biol. 43, 3–36 [DOI] [PubMed] [Google Scholar]
- 46.Gong W., Xu H., Shimizu T., Morishima S., Tanabe S., Tachibe T., Uchida S., Sasaki S., Okada Y. (2004) Cell. Physiol. Biochem. 14, 213–224 [DOI] [PubMed] [Google Scholar]
- 47.Arreola J., Begenisich T., Nehrke K., Nguyen H. V., Park K., Richardson L., Yang B., Schutte B. C., Lamb F. S., Melvin J. E. (2002) J. Physiol. 545, 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamamoto-Mizuma S., Wang G. X., Liu L. L., Schegg K., Hatton W. J., Duan D., Horowitz T. L., Lamb F. S., Hume J. R. (2004) J. Physiol. 557, 439–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duan D., Cowley S., Horowitz B., Hume J. R. (1999) J. Gen. Physiol. 113, 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang G. X., McCrudden C., Dai Y. P., Horowitz B., Hume J. R., Yamboliev I. A. (2004) Am. J. Physiol. Heart Circ. Physiol. 287, H533–H544 [DOI] [PubMed] [Google Scholar]
- 51.Gardoni F., Caputi A., Cimino M., Pastorino L., Cattabeni F., Di Luca M. (1998) J. Neurochem. 71, 1733–1741 [DOI] [PubMed] [Google Scholar]
- 52.Leonard A. S., Lim I. A., Hemsworth D. E., Horne M. C., Hell J. W. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3239–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun X. X., Hodge J. J., Zhou Y., Nguyen M., Griffith L. C. (2004) J. Biol. Chem. 279, 10206–10214 [DOI] [PubMed] [Google Scholar]
- 54.Grueter C. E., Abiria S. A., Wu Y., Anderson M. E., Colbran R. J. (2008) Biochemistry 47, 1760–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mamelak A. N., Rosenfeld S., Bucholz R., Raubitschek A., Nabors L. B., Fiveash J. B., Shen S., Khazaeli M. B., Colcher D., Liu A., Osman M., Guthrie B., Schade-Bijur S., Hablitz D. M., Alvarez V. L., Gonda M. A. (2006) J. Clin. Oncol. 24, 3644–3650 [DOI] [PubMed] [Google Scholar]
- 56.Rondé P., Giannone G., Gerasymova I., Stoeckel H., Takeda K., Haiech J. (2000) Biochim. Biophys. Acta 1498, 273–280 [DOI] [PubMed] [Google Scholar]
- 57.Komuro H., Rakic P. (1996) Neuron 17, 275–285 [DOI] [PubMed] [Google Scholar]
- 58.Ye Z. C., Sontheimer H. (1999) Cancer Res. 59, 4383–4391 [PubMed] [Google Scholar]
- 59.Takano T., Lin J. H., Arcuino G., Gao Q., Yang J., Nedergaard M. (2001) Nat. Med. 7, 1010–1015 [DOI] [PubMed] [Google Scholar]
- 60.Ishiuchi S., Yoshida Y., Sugawara K., Aihara M., Ohtani T., Watanabe T., Saito N., Tsuzuki K., Okado H., Miwa A., Nakazato Y., Ozawa S. (2007) J. Neurosci. 27, 7987–8001 [DOI] [PMC free article] [PubMed] [Google Scholar]