Abstract
The transient receptor potential vanilloid 4 (TRPV4) is a non-selective cation channel responsive to various stimuli including cell swelling, warm temperatures (27–35 °C), and chemical compounds such as phorbol ester derivatives. Here we report the three-dimensional structure of full-length rat TRPV4 purified from baculovirus-infected Sf9 cells. Hexahistidine-tagged rat TRPV4 (His-rTRPV4) was solubilized with detergent and purified through affinity chromatography and size-exclusion chromatography. Chemical cross-linking analysis revealed that detergent-solubilized His-rTRPV4 was a tetramer. The 3.5-nm structure of rat TRPV4 was determined by cryoelectron microscopy using single-particle reconstruction from Zernike phase-contrast images. The overall structure comprises two distinct regions; a larger dense component, likely corresponding to the cytoplasmic N- and C-terminal regions, and a smaller component corresponding to the transmembrane region.
Keywords: Electron Microscopy (EM), Ion Channels, Membrane Proteins, Protein Structure, TRP Channels, Single Particle Reconstruction, Zernike Phase Plate, Cryoelectron Microscopy (Cryo-EM)
Introduction
Transient receptor potential (TRP)2 channels form a diverse family of nonselective cation channels, most of which are highly permeable to Ca2+. The TRP superfamily is classified into seven subfamilies according to sequence homology: the TRPC (canonical) family, the TRPV (vanilloid) family, the TRPM (melastatin) family, the TRPP (polycystin) family, the TRPML (mucolipin) family, the TRPA (ankyrin) family, and the TRPN (NOMPC) family (1). Several TRPV and TRPM channels are known to be temperature-sensitive, although each has a different range of temperature sensitivity. All TRP channel subunits have six putative transmembrane segments that are thought to form tetramers similar to Shaker potassium channels (2, 3). The most distinct differences in TRP channels are in the large N- and C-terminal cytoplasmic regions, which contain putative protein interaction and regulatory motifs (1, 3–5).
TRPV4 is a non-selective cation channel that was originally identified as an osmosensor that detects hypotonic stimuli (6–9). More recent studies indicate that TRPV4 is not only activated by osmotic stimuli but also by warm temperatures, the phorbol derivative 4α-phorbol-12,13-didecanoate (4α-PDD), and lipid products of the arachidonic acid cascade (10–13). In addition to these, TRPV4 has been reported to have an association with many proteins (PACSIN3 (14), OS9 (15), microtubule-associated protein 7 (16), inositol 1,4,5-trisphosphate receptor 3 (17), aquaporin 5 (18), TRPP2 (19), caveolin 1 (20), CFTR chloride channel (21), and BK channel (22)). It reminds us that TRPV4 should be flexible to interact with a variety of proteins. TRPV4 has a 40% amino acid sequence homology to TRPV1 and TRPV2 (6, 7). The structure of TRPV4 is predicted to have a transmembrane region homologous to the 6TM tetrameric cation channels and ankyrin repeat domains (ARDs) in the N-terminal sequence (6). Because of the sequence homology among TRPV channels, the effects of swapping of the regions were examined in trafficking to the plasma membrane and electrophysiological properties (23, 24).
In structural biology there has recently been considerable interest in elucidating the molecular mechanisms of TRP channel gating and regulation (25). X-ray crystallography of fragments of the cytoplasmic region has been employed to understand its regulatory mechanism at atomic resolution (26–31), and electron microscopy (EM) of full-length TRP channels has been used to observe the entire structure of channel molecules. TRP channels are thought to be flexible mostly in the cytoplasmic N- and C-terminal regions. This flexibility makes it difficult to crystallize in three dimensions, and then x-ray crystallography of TRP channels has been focused on the fragment of the functionally important domains. Many of TRP channels share sequence homology, and some are homologous to other channel proteins. Even with such highly homologous sequences, homology modeling could be done in independent homologous domains, and the modeling of the entire structure, which consists of structurally homologous domains with flexible linker regions, still needs to be examined with further analysis (32). The EM structural analysis with single particle reconstruction is advantageous in elucidating the entire structure of macromolecular complexes, which is difficult to crystallize. Recently, EM structures have been determined for TRPC3 (33), TRPM2 (34), and TRPV1 (35). Each structure has a unique architecture. The cryo-EM structure of TRPC3 at 1.5-nm of resolution showed a large volume with a mesh-like structure. The negatively stained TRPM2 structure at 2.8-nm resolution showed a featureless region including the transmembrane region, with some features in cytoplasmic region. The cryo-EM structure of TRPV1 at 1.9-nm resolution revealed a “hanging gondola” shape, a term that has been applied to EM reconstructions of voltage-gated channels (36, 37) and cyclic nucleotide-gated channels (38, 39).
In this study we have obtained the complete structure of recombinant rat TRPV4 channel protein at 3.5-nm of resolution by single-particle EM using Zernike phase contrast images. Recombinant expression and purification of rat TRPV4 was obtained using the baculovirus expression system. The purified protein was assayed by chemical cross-linking and N-terminal labeling to be a tetramer; thus, 4-fold symmetry was applied during three-dimensional reconstruction. The resulting three-dimensional model shows two-layered architecture with a small transmembrane region, similar to that of TRPV1. The obvious difference between TRPV1 and TRPV4 is the shape and dimensions of the large component likely corresponding to the cytoplasmic region.
EXPERIMENTAL PROCEDURES
Construction of Baculovirus and Infection of Sf9 Cells
A baculovirus containing His-rTRPV4 was constructed by using the Baculovirus Expression System with Gateway Technology (Invitrogen). The DNA fragment encoding rat TRPV4, a gift from M. Caterina (Johns Hopkins University), was amplified by PCR using primers designed for the BP recombination reaction. This fragment was then inserted into the donor vector pDONR221 (Invitrogen) by recombination. The entire sequence of amplified rat TRPV4 was confirmed by DNA sequencing using a BigDye Terminator cycle sequencing kit and a genetic analyzer (ABI PRISM 3100, Applied Biosystems). The DNA fragment encoding rat TRPV4 was further cloned into the entry vector pDEST10 (Invitrogen), which attached the hexahistidine tag to the N terminus of rat TRPV4. The resulting entry vector was used for transformation of Escherichia coli DH10Bac competent cells (Invitrogen) to obtain the bacmid DNA containing His-rTRPV4. The bacmid DNA was purified and used to produce baculovirus with Sf9 cells according to the procedure from the Bac-to-Bac Baculovirus Expression System (Invitrogen). The resulting baculovirus was amplified five times to obtain a high titer virus stock, which was then used for protein expression. We examined the expression profile of His-rTRPV4 by varying the multiplicity of infection (m.o.i.) and the time of post-infection. The amount of expressed His-rTRPV4 was assessed by Western blotting using an anti-His-tag polyclonal antibody (MBL) and a secondary antibody conjugated with alkaline phosphatase. SDS-PAGE analysis was performed using NuPAGE 4–12% BisTris gel with MOPS running buffer (Invitrogen).
Ca2+ Imaging
Sf9 cells were grown to the mid-logarithmic phase in a suspension culture at 27 °C in serum-free medium (SF-900II SFM, Invitrogen) and then plated on a cover glass for at least 1 h at room temperature. Cells were washed with fresh medium and then infected with baculovirus containing His-rTRPV4 at an m.o.i. of 0.5. After 48 h at 27 °C in a humidified incubator, cells were washed with fresh medium and then incubated with medium containing 5 μm Fura-2 AM (Invitrogen) at room temperature for 1 h. A cover glass was washed with standard bath solution (10 mm MES, 10 mm NaCl, 60 mm KCl, 17 mm MgCl2, 10 mm CaCl2, 4 mm glucose, 100 mm sucrose, 0.1% bovine serum albumin, pH 6.2), and Fura-2 fluorescence was measured in the same solution. The cover glass was mounted in a chamber (RC-26G, Warner Instruments) connected to a gravity-flow system to deliver various stimuli and heated bath solutions. Heat stimulation was applied by increasing the bath temperature with a preheated solution through an inline heater (SH-27B; Warner Instruments). After heat stimulus, the standard bath solution containing 10 μm 4α-PDD or 5 μm ionomycin was applied to confirm the cellular response to TRPV4 agonist and cell viability, respectively. A xenon lamp was used as an illumination source. Fura-2 loaded in the cells was excited at wavelengths of 340 and 380 nm, and emission was monitored at a wavelength of 510 nm with a CCD camera (CoolSnap ES; Roper Scientific/Photometrics) to obtain fluorescence intensities of Ca2+-bound and Ca2+-free Fura-2, respectively. Data were acquired and analyzed using IPlab software (Scanalytics). The chamber temperature was monitored with an analog-to-digital converter with pClamp software (Molecular Devices).
Protein Purification
Sf9 cells were grown at a density of 2.0 × 106 cells/ml in a suspension culture at 27 °C in SF-900II SFM and were then infected with baculovirus containing His-rTRPV4 at an m.o.i. of 5.0. After 72 h of cultivation, cells were harvested by centrifugation at 4000 × g for 20 min at 4 °C. The cells were washed with Tris-buffered saline (TBS) (25 mm Tris-HCl, 150 mm NaCl, pH 7.4), centrifuged as above, immediately frozen, and stored at −30 °C until use. Frozen cells were suspended into 10 volumes (v/w) of TBS containing a protease inhibitor mixture (Complete EDTA-free protease inhibitor mixture, Roche Applied Science) and disrupted with a Teflon homogenizer. The cells were further processed by French press (Thermo Fisher Scientific) at a pressure of 10,000 p.s.i. The cell lysate was centrifuged at 4000 × g for 20 min at 4 °C to remove cell debris, and then the supernatant was ultracentrifuged at 100,000 × g for 60 min at 4 °C. The precipitate was stored as the membrane fraction at −30 °C.
The membrane fraction was processed with a Teflon homogenizer in 5 ml of TBS containing 20 mm n-dodecyl-β-d-maltoside (DDM) (Dojindo) and protease inhibitor mixture. After incubation for 60 min at 4 °C, the sample was diluted twice with TBS containing 40 mm imidazole to reduce the concentration of DDM to 10 mm. After ultracentrifugation at 10,000 × g for 30 min at 4 °C, the DDM-soluble fraction was applied to a His GraviTrap column (GE Healthcare) equilibrated in advance with TBS containing 20 mm imidazole and 5 mm DDM. The column was washed with 20 ml of TBS containing 5 mm DDM and 20 mm imidazole, and then the bound protein was eluted three times with 5 ml of TBS containing 5 mm DDM and 500 mm imidazole. The eluate was collected and concentrated 10 times with a centrifugal filter device (Microcon YM-100, Millipore). The concentrate was filtered with a 0.22-μm polycarbonate filter (Ultrafree-MC Centrifugal Filter Units, Millipore) and then purified by Superdex 200 (3.2/30) size-exclusion chromatography in a SMART system (GE Healthcare) with TBS containing 1 mm DDM at a flow rate of 40 μl/min. Elution was monitored by absorbance at 280 nm and was fractionated into 20-μl fractions. Protein concentrations were determined spectrophotometrically at 280 nm using the extinction coefficient as previously reported (40).
Chemical Cross-linking Analysis
For chemical cross-linking, the buffer components of purified His-rTRPV4 were exchanged by using a PD-10 desalting column equilibrated with phosphate-buffered saline containing 1 mm DDM. Glutaraldehyde was added to a final concentration of 10 mm, and samples were incubated at room temperature for 30 min. The cross-linking reaction was terminated by the addition of Tris-HCl, pH 8.0, to a final concentration of 50 mm. After 15 min of incubation, samples were subjected to SDS-PAGE using a NuPAGE 4–12% BisTris gel with MOPS running buffer and assessed by Coomassie staining as well as by Western blotting using an anti-His-tag antibody.
N-terminal Labeling and Transmission Electron Microscopy
Purified His-rTRPV4 at a concentration of ∼50 μg/ml was adsorbed onto glow-discharged copper EM grids with thin carbon films. Samples were washed with three drops of MilliQ water, the excess of water was blotted off, and a Ni-NTA-Nanogold (Nanoprobes) solution was applied immediately onto the grids. Excess Ni-NTA-Nanogold was washed out by applying three drops of MilliQ water. The grids were then negatively stained twice for 30 s with NanoVan (Nanoprobes) staining reagent, blotted, and dried in air. Micrographs of stained particles were recorded with a JEM-1010 transmission electron microscope (JEOL) at a nominal magnification of 150,000 and 80-kV acceleration voltage. Images were recorded on a CCD camera (Megaview II, SIS) with a pixel size of 0.5 nm at the specimen.
Cryoelectron Microscopy and Image Processing
The purified His-rTRPV4 was embedded in a thin layer of vitreous ice on a holey carbon grid. Commercially available holey carbon grids (R1.2/1.3, Quantifoil MicroTools) were made hydrophilic by glow-discharge in air just before specimen preparation. 2.5 μl of His-rTRPV4 at a concentration of 100 μg/ml was applied, the excess solution was blotted with filter paper, and the grid was quickly plunged into liquid ethane cooled by liquid nitrogen using a Leica CPC cryo-station (Leica Microsystems). The grid was transferred into a JEM-3100FFC (JEOL) cryo-EM equipped with a field emission gun and an in-column (omega-type) energy filter. The microscope was operated at 300 kV acceleration voltage, with in-focus Zernike phase-contrast imaging (41). The specimen temperature was maintained at ∼55 K, as described (41). The Zernike phase plate, inserted at the back focal plane of the objective lens, was made of a vacuum-evaporated amorphous carbon film with a thickness of ∼27 nm, optimized for 300-kV electrons (41). The central hole in the phase plate was 0.7 μm in diameter. The phase plate was continuously maintained above 150 °C to prevent beam-induced contamination (42). Data were collected at a nominal magnification of 40,000 using zero-loss energy filtering with a 20-eV slit width and an incident electron dose of 20 e−/Å2. Images were recorded on a 2k x 2k CCD camera (Megascan 795, Gatan) with a pixel size of 0.524 nm at the specimen.
Image processing and three-dimensional reconstruction were performed by using Version 1.8 of the EMAN software package (43). A total of 3916 particles were selected manually from the CCD images using the BOXER program from the EMAN package. The box size was 48 × 48 pixels. Iterative refinement of the three-dimensional model was performed with the initial model generated using C4 symmetry, as suggested by chemical cross-linking analysis and N-terminal labeling. The particles were classified into 133 classes corresponding to a 6° angular step. The final three-dimensional map was reconstructed after eight iterations from 2808 particles. The resolution of the final three-dimensional map was estimated using Fourier shell correlation between two maps made separately from two halves of the data set. The standard 0.5 criterion was used. The map was surface-rendered using CHIMERA (44).
RESULTS
Expression and Purification of TRPV4
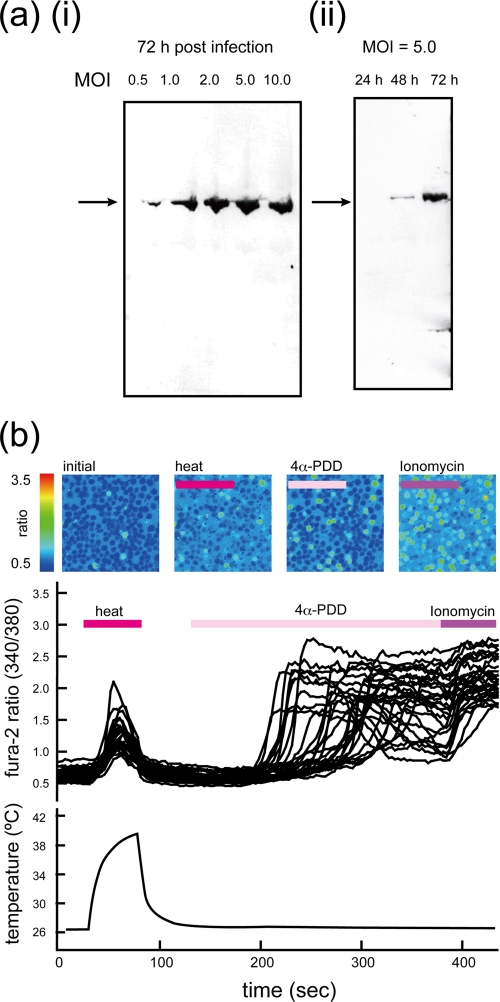
Recombinant expression of rat TRPV4 was successfully accomplished by using the baculovirus expression system with Sf9 cells. A hexahistidine tag at the N terminus was used for both Western blotting and affinity purification. Western blotting analysis (Fig. 1a) revealed that the expression of recombinant His-rTRPV4 increased with post-infection time and with the m.o.i. Protein expression for purification was performed 72 h post-infection at an m.o.i. of 5.0.
FIGURE 1.
Expression profile and [Ca2+]i changes in Sf9 cells infected with baculovirus containing His-rTRPV4. a, Western blot analysis of expression of His-rTRPV4 by using an anti-hexahistidine antibody is shown. Differences in post-infection time and m.o.i. are shown. (i) shows the difference in m.o.i. at 72 h post-infection. (ii) shows the difference in post-infection time at an m.o.i. of 5.0. b, shown is fluorescence imaging of Sf9 cells infected with baculovirus containing His-rTRPV4 observed during heat and agonist stimulation. Pseudocolored images represent the 340/380-nm ratio for Fura-2 fluorescence. The corresponding traces during stimuli are shown below. The period of each stimulus is indicated above the traces, and the measured temperature changes are plotted at the bottom.
The function of expressed His-rTRPV4 was evaluated by Ca2+ imaging. Increased intracellular Ca2+ concentration ([Ca2+]i) was observed as increases in the ratio of Fura-2 fluorescence intensity between 340- and 380-nm excitation wavelengths. Fig. 1b shows pseudocolored images of the fluorescence ratio of Sf9 cells infected with baculovirus containing His-rTRPV4 and the corresponding traces of the ratio during stimulation. The function of His-rTRPV4 was confirmed by the response to heat stimuli and 4α-PDD, which is known to be a robust and potent activator of TRPV4 (11, 45). The heat- or 4α-PDD-evoked [Ca2+]i increase was observed only in Sf9 cells infected with baculovirus containing His-rTRPV4 (data not shown). The responses to 4α-PDD were almost the same as those to ionomycin, which was performed to show the cell viability. These results indicated the His-rTRPV4 was active in Sf9 cells.
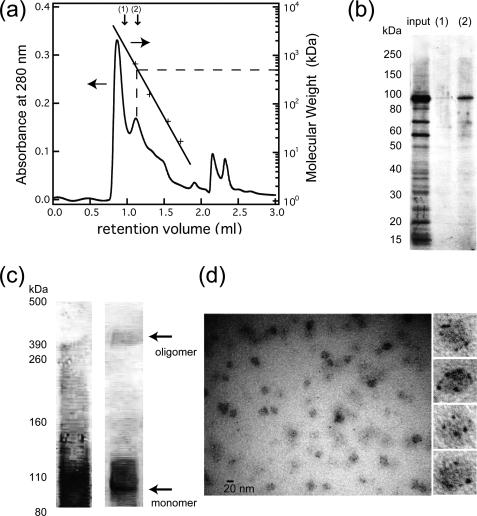
Purification of His-rTRPV4 was performed by affinity chromatography and size-exclusion chromatography. DDM-solubilized His-rTRPV4 appeared as a concentrated major band in Coomassie staining of SDS-PAGE after affinity purification (Fig. 2). The size-exclusion chromatogram (Fig. 2a) indicated that the majority of the proteins appeared in an initial peak corresponding to a retention volume of 0.85 ml, which is equivalent to the void volume of the column. Thus, most of the concentrated His-rTRPV4 was found to be aggregated. However, the second peak, between the molecular mass standards thyroglobulin (670 kDa) and γ-globulin (158 kDa), was revealed by SDS-PAGE analysis to have a molecular mass corresponding to the amino acid composition of His-rTRPV4 (103 kDa) (Fig. 2b). The estimated molecular mass of the second peak, roughly 500 kDa, suggested that it was an oligomeric state of His-rTRPV4.
FIGURE 2.
Oligomerization analysis of purified His-rTRPV4. a, shown is a size-exclusion chromatogram of proteins in the detergent-solubilized state after affinity purification. The retention volume of the molecular mass standards is shown by crosses for thyroglobulin (670 kDa), γ-globulin (158 kDa), ovalbumin (44 kDa), and myoglobin (17 kDa). The molecular mass of His-rTRPV4 (including detergent molecules) was estimated as 500 kDa. b, SDS-PAGE analysis of the input, fraction 1, and fraction 2 indicated in the size-exclusion chromatogram was assessed by Coomassie staining. c, chemical cross-linking analysis of purified His-rTRPV4 was assessed by Coomassie staining (left) and a Western blot using anti-His-tag antibody (right). The molecular weight of a cross-linked band above 390 kDa is consistent with a tetramer of His-rTRPV4 subunits (103 kDa). d, shown is a general view of a Nanogold-labeled and NanoVan-stained His-rTRPV4 and an enlarged view of particles having multiple Nanogolds.
Subunit Stoichiometry of Purified TRPV4
TRPV4 is thought to be a tetramer similar to Shaker potassium channels (2, 3). To evaluate the subunit stoichiometry of the purified His-rTRPV4 in the DDM-solubilized state, we applied both chemical cross-linking analysis and N-terminal labeling. Chemical cross-linking followed by SDS-PAGE analysis reveals the molecular weight of the complex. Specific labeling of a genetically encoded hexahistidine at the N terminus of the protein indicates the subunit stoichiometry by the number of gold particles attached to the protein particle as observed by EM.
Chemical cross-linking analyses of purified His-rTRPV4 are shown in Fig. 2c. Chemical cross-linking using glutaraldehyde showed a smeared band by Coomassie staining, but Western blotting of the same sample showed a clear band just above the 390-kDa molecular mass marker.
The combination of Ni-NTA-Nanogold labeling and NanoVan staining resulted in clear images of the small Nanogold label (1.8-nm diameter) in negatively stained particles (Fig. 2d). The images were taken as in-focus bright field images to clarify the shape of the Nanogold label and to evaluate its size. Most of the small, high contrast particles had diameters corresponding to the Nanogold label. The number of Nanogolds associated with protein particles varied, most likely due to the orientation of proteins and the labeling efficiency. However, proteins with four Nanogolds often showed a clear 4-fold symmetry so that we concluded that His-rTRPV4 remains a tetramer in the DDM-solubilized state.
Cryoelectron Microscopy and Three-dimensional Reconstruction
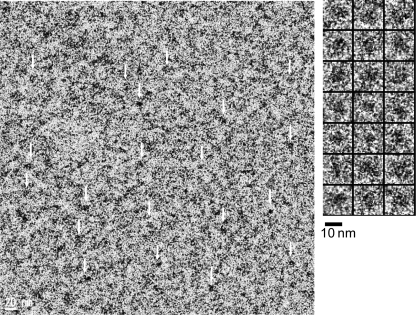
A Zernike phase-contrast cryo-EM image of purified His-rTRPV4 is shown in Fig. 3. Enlarged views of the particles indicated with white arrows are shown at the right. Cryo-EM imaging of membrane proteins in the detergent-solubilized state is known to be difficult due to structural heterogeneity and low contrast. The cryo-specimens sometimes have an excessive thickness of vitreous ice, resulting in low contrast. One approach to overcome this issue is to take images at high defocus to increase the contrast at lower spatial frequencies. Here, we obtain higher contrast by using a Zernike phase plate. Compared with conventional under-focus imaging, Zernike phase-contrast imaging has a much higher contrast at lower spatial frequencies, making it easier to identify and align the particles and obviating the need for contrast transfer function correction and the acquisition of images at various defocus values (46).
FIGURE 3.
Cryo-EM observation of His-rTRPV4 in the detergent-solubilized state. A typical raw image of purified His-rTRPV4 (left) is shown. Some of the individual proteins (white arrows) were picked from the raw image, as shown on the right.
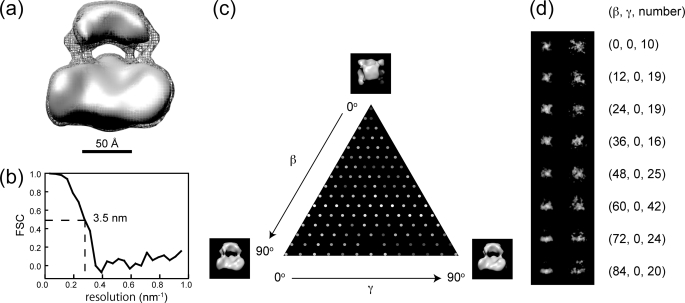
For three-dimensional reconstruction, we applied C4 symmetry. Surface representations of His-rTRPV4 are shown in Fig. 4a at two different contour levels enclosing a volume corresponding to 500 and 630 kDa for gray solid surface and gray mesh, respectively. Because the resolution of the three-dimensional model was estimated to be 3.5 nm, as shown by Fourier shell correlation (Fig. 4b), the model was low pass-filtered to 3.0 nm for additional clarity. The three-dimensional model corresponding to a molecular mass of 500 kDa, which was estimated from size-exclusion chromatography, was segregated in two components without connections; even it was bigger than its mass of tetrameric protein moieties. The molecular mass should contain residual lipids and additional detergent molecules surrounding the transmembrane region of the channel but was estimated as a globular shape. Therefore, the model is also presented to have a connection between the two components with gray mesh enclosing a volume corresponding to 630 kDa.
FIGURE 4.
Three-dimensional reconstruction using 4-fold symmetry. a, shown is a surface representation of rTRPV4 with the contour level enclosing a volume corresponding to 500 and 630 kDa for the gray solid surface and gray mesh, respectively. b, Fourier shell correlation (FSC) is shown between reconstructions from even and odd halves of the data set, plotted against spatial frequency. The value falls to the criterion level of 0.5 at a resolution of 3.5 nm. c, shown is Euler angular distribution over the asymmetric unit used in the reconstruction. The brightness of each dot corresponds to the number of particles belonging to the class average Black represents 7 particles, and white represents 43 particles. d, shown is a comparison of class averages (left) with projections of the final three-dimensional model (right). Corresponding Euler angles and numbers of particles were indicated at the right.
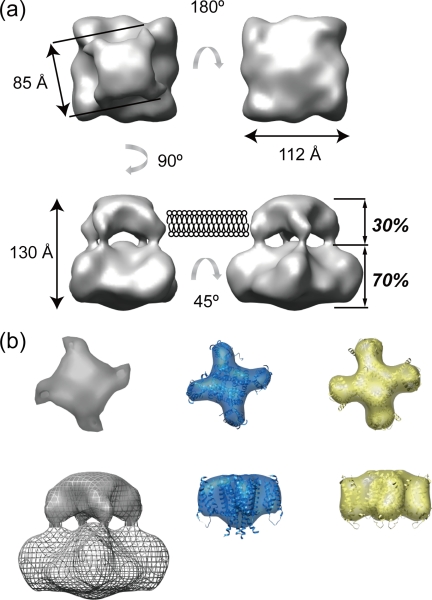
The model shows two components, both with square cross-sections twisted 15° from each other (Fig. 5a). The smaller, upper component has a dimension of ∼85 Å, and the larger component has a dimension of ∼112 Å. The overall height of the model is ∼130 Å. Volume analysis of the three-dimensional model performed using CHIMERA revealed that the small component accounted for 30% of the total volume. A primary sequence analysis performed using the program SOSUI (47) concluded that 27% of the amino acids comprise the transmembrane region, and the remaining residues are located in the cytoplasmic region of the molecule. The assignment of the transmembrane region is also confirmed by comparison of the shape and dimensions between transmembrane regions of structurally homologous 6TM tetrameric cation channels. Fig. 5b shows calculated volume representations at 3-nm of resolution from the atomic coordinates of both MlotiK1 (blue, PDB code 3BEH) and Kv1.2-chimera (yellow, PDB code 2R9R) superposed with each ribbon representation of the atomic coordinates. The shape and dimensions of both channels are almost identical in this resolution, and some of the helices stick away from volume representations. This means that the 3-nm resolution could not provide such a slight difference in arrangement of the TM segments. Although the comparison shows that our 3.5-nm map of TRPV4 depicts the general architecture of 6TM tetrameric cation channel, the tetrameric pore-forming domain with four domains at the corner. These results suggest that the small component likely corresponds to the transmembrane region.
FIGURE 5.
The shape and dimensions of surface representation of His-rTRPV4. a, shown is a surface representation of rTRPV4 viewed from four different angles. Dimensions and estimated of volume analysis (italics) are shown. b, shown are surface representations of the transmembrane region of 6TM tetrameric cation channels, TRPV4 (gray), MlotiK1 (blue, PDB code 3BEH), and Kv1.2-chimera (yellow, PDB code 2R9R). Surface representations calculated from atomic coordinates are shown at 3 nm of resolution and superimposed with its ribbon diagram. The upper three models show extracellular views of transmembrane regions, and the lower three show side views. The surface representation of transmembrane region of TRPV4 is superimposed with gray-meshed entire structure.
Fitting the Atomic Structure of Structurally Homologous Proteins
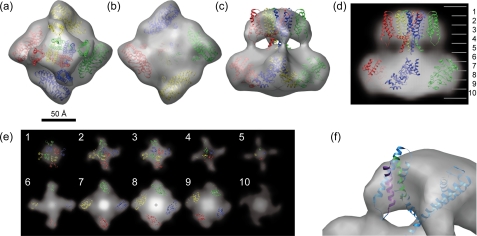
Based on biochemical and mutagenesis data, the topology of TRP channels has been proposed to resemble that of voltage-gated potassium channels (1, 5). TRPV4 shares the tetrameric stoichiometry in the detergent-solubilized state, as confirmed by chemical cross-linking (Fig. 2c) and N-terminal labeling (Fig. 2d). Two high resolution structures of the 6TM tetrameric cation channel are now available. The x-ray crystallography of purified eukaryotic potassium channel Kv1.2 reveals its open conformation and the arrangement of TM segments 1–4, the voltage sensor (48). Compared with the Kv1.2, the recently solved prokaryotic potassium channel MlotiK1 shows its closed conformation. The MlotiK1 is a ligand-gated channel that has a C-terminal cytoplasmic cyclic nucleotide binding site and a non-voltage-dependent channel in which TM segments 1–4 do not act as voltage sensors. Because TRPV1 has been shown to have weak voltage dependence (49), the superposition with Kv1.2 was performed (35). On the other hand, the voltage dependence of TRPV4 has not been clearly shown so far. Furthermore, the protein was purified without 4α-PDD in the present study; it means that the structure of the TM segment 1–4 should be similar to that of MlotiK1 at a closed conformation. Here we showed the superposition of the atomic structure of the transmembrane region of the MoltiK1 on our 3.5-nm map of TRPV4 (Fig. 6a–c). Using the “fit in map” command of CHIMERA, the shape and dimensions of the tetramer of TM segments of MlotiK1 fitted well onto our model's small component. The most distinct fitting error is the size of central pore-forming domain (Fig. 6a). Fig. 6f shows a close-up view of the superposition of one of the MlotiK1 monomer on our map to show the fitting of TM segment 3 and 4, which corresponds to a putative binding site of 4α-PDD. These helices are fitted well on our map, but the site between TM3 and 4 could not be resolvable in the current resolution.
FIGURE 6.
Superposition of high resolution structures onto the three-dimensional model of TRPV4. Shown is a superposition of a tetramer of the transmembrane segments of MlotiK1 (PDB code 3BEH) onto the small component and a tetramer of the ARD of TRPV1 (PDB code 2PNN) onto the large component of our 3.5-nm map of TRPV4. Superposition onto surface representation is shown as a top view (a), a bottom view (b), and a side view (c). Superposition onto the density map is shown as a side-view projection (d), and horizontal cross-sections are parallel to the membrane plane (e). Each section is 1.3-nm thick. The number in each section corresponds to that on the left side of d. A bird's-eye view of the superposition of MlotiK1 monomer (blue) onto our 3.5-nm map of TRPV4 is shown. 4α-PDD binding pockets are colored with purple and green for TM segments 3 and 4, respectively.
The ARD is another important domain thought to be structurally homologous in TRP channels. Several structures of ARDs of TRPV family proteins have been published (26–29). TRPV4 is thought to have at least three ARDs but possibly six in its N-terminal cytoplasmic region (29). Here we showed the superposition of the ARDs of TRPV1 (PDB code 2PNN) onto the large component of our 3.5-nm map of TRPV4. The superposition was performed manually to fit the x-ray structure onto the periphery of the large component, placing the C terminus of ARD close to the connection between the two components and the hydrophobic surfaces of the ARDs facing the inside of the large component. The ARDs are fitted well onto the large component with its surface morphology (Fig. 6, a–c) and also density (Fig. 6, d and e).
DISCUSSION
Here we report the structure of the tetrameric TRPV4 channel determined by single-particle cryo-EM. The three-dimensional model consists of two components; a larger dense part, likely corresponding to the cytoplasmic N- and C-terminal regions, and a smaller component corresponding to transmembrane region. Assignment of these regions was supported by volume analysis of the three-dimensional map and also by comparing the transmembrane region of x-ray structure of the structurally homologous 6TM tetrameric cation channels with that of TRPV4 cryo-EM map.
Recently, several EM studies of TRP channels have portrayed the architecture of the channel complex. The structures of three TRP channels have been determined by single-particle EM (TRPC3 (33), TRPM2 (34), and TRPV1 (35)), each with a unique architecture. The TRPC3 structure, having the highest resolution (1.5 nm), has a surprising mesh-like density with an apparent disparity between the molecular weight and the total volume of the particle. This disparity had already been suggested by previous work with negative staining (50). Immunolabeling of the C-terminal region of TRPC3 suggested a larger putative cytoplasmic region and a smaller, dense transmembrane region. The TRPC3 shares sequence features of 6TM tetrameric cation channels, but the superposition of the atomic coordinate of Kv1.2 suggests a similar arrangement of the pore-forming domain but different arrangement of TM segments 1–4. The sections corresponding to the transmembrane region was much bigger than that of Kv1.2 and suggested that the pore-forming domain was covered with a wireframe outer shell. Surprisingly, not only the transmembrane region but also the cytoplasmic region has a core-like small domain and wireframe outer shell. The structure of TRPM2 was obtained by the same group using negative staining at a reported resolution of 2.8 nm (3.7 nm using the 0.5 criterion). The overall shape is similar to that of TRPC3 as observed in negatively stained specimens and shows a relatively featureless, dense transmembrane region. The transmembrane region and upper part of the cytoplasmic region were covered with continuous smooth surface. That might be due to the negative staining because the image taken with negative staining shows a shape of the cast surrounding the protein particles.
Compared with these two structures, our 3.5-nm map of TRPV4 is quite similar to that of TRPV1 recently obtained by cryo-EM, revealing a two-layered architecture with a four-pillar connection. The superposition of Kv1.2 on the cryo-EM map of TRPV1 revealed structural homology of transmembrane regions (35). The small component, likely corresponding to the transmembrane region of our 3.5-nm map shows an almost identical shape and dimensions to other 6TM tetrameric cation channels, Kv1.2 and MlotiK1. The comparison reveals structural homology of transmembrane region to the 6TM tetrameric cation channels and also to TRPV1. X-ray crystallography of the 6TM tetrameric cation channels, Kv1.2 and MlotiK1, reveals the mechanism of channel gating in the arrangement of TM segments. In the case of TRPV4, the 4α-PDD binding site between TM segments 3 and 4 is important to see in a three-dimensional structure in a similar sense. The fitting of MlotiK1 onto our 3.5-nm map showed a good fit but revealed the difficulty in resolving TM helices (Fig. 6f). In seeing the hydrophobic surface of pore-forming region, the fitting error is obvious (Fig. 6a). This might be due to the attached residual lipids and detergent molecules. Such a fitting error is not obvious at the surface of TM segments 1–4 but residual attachments might affect visibility. To visualize such an important structure-function relationship, it would be advantageous to purify the 4α-PDD-bound form of TRPV4.
The transmembrane region shows an almost identical shape and dimensions in the current resolution, but the large component, likely corresponding to the cytoplasmic N- and C-terminal regions, shows distinct differences in the shape and density distributions. In the case of TRPV1, the ARDs were placed into the map in a vertical orientation (23). This produced a good fit because the cytoplasmic region of TRPV1 is tall but shows a central cavity. In our map of TRPV4, the ARDs could not be fitted in a vertical orientation but were tilted toward the connection between the cytoplasmic and transmembrane regions (Fig. 6, a–c). The density distribution of the cytoplasmic region (Fig. 6e) suggests a tight assembly in central upper part of cytoplasmic region with a hint of a connection between the transmembrane and cytoplasmic regions. Because the C-terminal region of TRPV1 could be swapped with that of TRPV4 (24), that region should have structural similarity to that of TRPV1. Homology modeling of the C-terminal region of TRPV1 shows a tight assembly of the tetramer to make a coiled-coil domain just downstream of the TM segment 6 (51). Analogous to TRPV1, the C-terminal region of TRPV4 should be located at the upper central part of cytoplasmic region. These results allow us to place the ARDs on the periphery of cytoplasmic region, leaving the vacant space in the upper central part of the cytoplasmic region for the C-terminal region (Fig. 6e, 6–9). The C-terminal region of TRPV4 has been revealed to have an interaction with the microtubule-associated protein 7 (MAP7) (Suzuki et al. (16)). Furthermore, the last four amino acid residues of the C terminus of TRPV4 show a PDZ binding-like motif that might contribute to the interaction of TRPV4 with a huge variety of PDZ-domain proteins (52). To have such accessibility, the C-terminal region should be located not to the inside but to the surface of the cytoplasmic region. The vacant spaces at the upper cytoplasmic region between the ARDs are in good agreement with this issue (Fig. 6, c and d). In addition to the C-terminal region, the rest of N-terminal region should be located to the surface of cytoplasmic region. The proline-rich region just upstream of the first ARD interacts with the SH3 domain of PACSIN3, which is a cytoskeleton protein involved in synaptic vesicular membrane trafficking and endocytosis (14). This also leads us to leave the vacant spaces at the bottom surface of the cytoplasmic region for the rest of the N-terminal region (Fig. 6, b and e, 9 and 10).
Recently, the similarity of ARD between TRPV1 and TRPV4 was revealed (53). The ARD of TRPV4 has a binding affinity to ATP and Ca2+-calmodulin. TRPV4 is sensitized by intracellular ATP and desensitized by repeated or prolonged stimulations. This similarity reminds us of the common regulatory mechanism. The distinct difference in the shape and dimensions of the cytoplasmic region between TRPV1 and TRPV4 shown here could be explained by a different arrangement of N- and C-terminal regions in the cytoplasmic region. A similar difference in arrangement was proposed by molecular modeling of TRPV1 (32). The resulting model of a closed conformation of TRPV1 shows a shape of the cytoplasmic region similar to that of our 3.5-nm map of TRPV4. The arrangement of the N-terminal, transmembrane, and C-terminal region was modeled initially having a cavity inside the cytoplasmic region, but the resulting model shows the interaction between the N- and C-terminal regions to eliminate the cavity inside. This result suggests that our 3.5-nm map of TRPV4 shows closed conformation. Clearly, this hypothetical arrangement needs to be supported by further structural analysis.
Zernike phase-contrast imaging is a novel approach in cryo-EM, which yields a much higher contrast for identifying and aligning protein particles. We applied this approach to overcome the difficulties in single particle EM of this membrane protein. Our 3.5-nm map does not have enough resolution to show the structural features for TRPV4 functions, but it does show the distinct difference in the shape of cytoplasmic region to the structurally homologous TRPV1. TRPV4 is a polymodal channel and is reported to have an interaction with many proteins at the cytoplasmic region. Because of such a variety of interaction, TRPV4 should have flexibility in the cytoplasmic region. This flexibility may limit the resolution in reconstruction. The recombinant expression and purification of His-rTRPV4 achieved here results in sufficient quantities for EM analysis, but the issue of structural heterogeneity needs still to be examined.
The TRPV4 structure combined with the atomic structures of structurally homologous proteins reveals a similarity and a distinct difference to that of TRPV1. Our 3.5-nm map of TRPV4 shows an identical shape and dimensions in its transmembrane region to the other 6TM tetrameric cation channels and proposes a different arrangement of the N- and C-terminal regions in the cytoplasmic region. Further effort to achieve high resolution three-dimensional maps will be required to clarify the differences in detail such as different arrangement of helices with ligand binding, physical stimulus, or protein interactions. The required resolution to clarify each helix in the EM three-dimensional map is thought to be less than 1 nm. This resolution is still challenging to the cryo-EM structural analysis, but the combination with the lower resolution EM map and x-ray crystallography or homology modeling with other structurally homologous domains helps a lot to elucidate the substantial conformational changes in different channel states or ligand binding.
Acknowledgments
We thank Hiroshi Okawara for preparing the phase plate and Michael Marko and Fred Sigworth for helpful discussions and editing.
This work was supported in part by a grant-in-aid for creative scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (2001–2005) and by Core Research for Evolutional Science and Technology (CREST) of Japan Science and Technology Corporation (2006–2009).
- TRP
- transient receptor potential
- TRPV4
- TRP vanilloid 4
- His-rTRPV4
- hexahistidine-tagged rat TRPV4
- 4α-PDD
- 4α-phorbol-12,13-didecanoate
- ARD
- ankyrin repeat domain
- EM
- electron microscopy
- m.o.i.
- multiplicity of infection
- TBS
- Tris-buffered saline
- DDM
- n-dodecyl-β-d-maltoside
- Ni-NTA
- nickel-nitrilotriacetic acid
- TM
- transmembrane
- Bistris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- MES
- 4-morpholineethanesulfonic acid
- MOPS
- 4-morpholinepropanesulfonic acid
- PDB
- Protein Data Bank.
REFERENCES
- 1.Montell C. (2005) Sci. STKE 2005, re3. [DOI] [PubMed] [Google Scholar]
- 2.Kedei N., Szabo T., Lile J. D., Treanor J. J., Olah Z., Iadarola M. J., Blumberg P. M. (2001) J. Biol. Chem. 276, 28613–28619 [DOI] [PubMed] [Google Scholar]
- 3.Phelps C. B., Gaudet R. (2007) J. Biol. Chem. 282, 36474–36480 [DOI] [PubMed] [Google Scholar]
- 4.Cahalan M. D. (2001) Nature 411, 542–543 [DOI] [PubMed] [Google Scholar]
- 5.Clapham D. E. (2003) Nature 426, 517–524 [DOI] [PubMed] [Google Scholar]
- 6.Liedtke W., Choe Y., Martí-Renom M. A., Bell A. M., Denis C. S., Sali A., Hudspeth A. J., Friedman J. M., Heller S. (2000) Cell 103, 525–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strotmann R., Harteneck C., Nunnenmacher K., Schultz G., Plant T. D. (2000) Nat. Cell Biol. 2, 695–702 [DOI] [PubMed] [Google Scholar]
- 8.Wissenbach U., Bödding M., Freichel M., Flockerzi V. (2000) FEBS Letters 485, 127–134 [DOI] [PubMed] [Google Scholar]
- 9.Nilius B., Prenen J., Wissenbach U., Bödding M., Droogmans G. (2001) Pflugers Arch. 443, 227–233 [DOI] [PubMed] [Google Scholar]
- 10.Güler A. D., Lee H., Iida T., Shimizu I., Tominaga M., Caterina M. (2002) J. Neurosci. 22, 6408–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe H., Davis J. B., Smart D., Jerman J. C., Smith G. D., Hayes P., Vriens J., Cairns W., Wissenbach U., Prenen J., Flockerzi V., Droogmans G., Benham C. D., Nilius B. (2002) J. Biol. Chem. 277, 13569–13577 [DOI] [PubMed] [Google Scholar]
- 12.Watanabe H., Vriens J., Suh S. H., Benham C. D., Droogmans G., Nilius B. (2002) J. Biol. Chem. 277, 47044–47051 [DOI] [PubMed] [Google Scholar]
- 13.Watanabe H., Vriens J., Prenen J., Droogmans G., Voets T., Nilius B. (2003) Nature 424, 434–438 [DOI] [PubMed] [Google Scholar]
- 14.Cuajungco M. P., Grimm C., Oshima K., D'hoedt D., Nilius B., Mensenkamp A. R., Bindels R. J., Plomann M., Heller S. (2006) J. Biol. Chem. 281, 18753–18762 [DOI] [PubMed] [Google Scholar]
- 15.Wang Y., Fu X., Gaiser S., Köttgen M., Kramer-Zucker A., Walz G., Wegierski T. (2007) J. Biol. Chem. 282, 36561–36570 [DOI] [PubMed] [Google Scholar]
- 16.Suzuki M., Hirao A., Mizuno A. (2003) J. Biol. Chem. 278, 51448–51453 [DOI] [PubMed] [Google Scholar]
- 17.Fernandes J., Lorenzo I. M., Andrade Y. N., Garcia-Elias A., Serra S. A., Fernández-Fernández J. M., Valverde M. A. (2008) J. Cell Biol. 181, 143–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X., Bandyopadhyay B. C., Bandyopadhyay B., Nakamoto T., Singh B., Liedtke W., Melvin J. E., Ambudkar I. (2006) J. Biol. Chem. 281, 15485–15495 [DOI] [PubMed] [Google Scholar]
- 19.Köttgen M., Buchholz B., Garcia-Gonzalez M. A., Kotsis F., Fu X., Doerken M., Boehlke C., Steffl D., Tauber R., Wegierski T., Nitschke R., Suzuki M., Kramer-Zucker A., Germino G. G., Watnick T., Prenen J., Nilius B., Kuehn E. W., Walz G. (2008) J. Cell Biol. 182, 437–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saliez J., Bouzin C., Rath G., Ghisdal P., Desjardins F., Rezzani R., Rodella L. F., Vriens J., Nilius B., Feron O., Balligand J. L., Dessy C. (2008) Circulation 117, 1065–1074 [DOI] [PubMed] [Google Scholar]
- 21.Arniges M., Vázquez E., Fernández-Fernández J. M., Valverde M. A. (2004) J. Biol. Chem. 279, 54062–54068 [DOI] [PubMed] [Google Scholar]
- 22.Fernández-Fernández J. M., Andrade Y. N., Arniges M., Fernandes J., Plata C., Rubio-Moscardo F., Vázquez E., Valverde M. A. (2008) Pflugers Arch. 457, 149–159 [DOI] [PubMed] [Google Scholar]
- 23.Hellwig N., Albrecht N., Harteneck C., Schultz G., Schaefer M. (2005) J. Cell Sci. 118, 917–928 [DOI] [PubMed] [Google Scholar]
- 24.García-Sanz N., Valente P., Gomis A., Fernández-Carvajal A., Fernández-Ballester G., Viana F., Belmonte C., Ferrer-Montiel A. (2007) J. Neurosci. 27, 11641–11650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaudet R. (2008) J. Physiol. 586, 3565–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lishko P. V., Procko E., Jin X., Phelps C. B., Gaudet R. (2007) Neuron 54, 905–918 [DOI] [PubMed] [Google Scholar]
- 27.Jin X., Touhey J., Gaudet R. (2006) J. Biol. Chem. 281, 25006–25010 [DOI] [PubMed] [Google Scholar]
- 28.McCleverty C. J., Koesema E., Patapoutian A., Lesley S. A., Kreusch A. (2006) Protein Sci. 15, 2201–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phelps C. B., Huang R. J., Lishko P. V., Wang R. R., Gaudet R. (2008) Biochemistry 47, 2476–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamaguchi H., Matsushita M., Nairn A. C., Kuriyan J. (2001) Mol. Cell 7, 1047–1057 [DOI] [PubMed] [Google Scholar]
- 31.Fujiwara Y., Minor D. L., Jr. (2008) J. Mol. Biol. 383, 854–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernández-Ballester G., Ferrer-Montiel A. (2008) J. Membr. Biol. 223, 161–172 [DOI] [PubMed] [Google Scholar]
- 33.Mio K., Ogura T., Kiyonaka S., Hiroaki Y., Tanimura Y., Fujiyoshi Y., Mori Y., Sato C. (2007) J. Mol. Biol. 367, 373–383 [DOI] [PubMed] [Google Scholar]
- 34.Maruyama Y., Ogura T., Mio K., Kiyonaka S., Kato K., Mori Y., Sato C. (2007) J. Biol. Chem. 282, 36961–36970 [DOI] [PubMed] [Google Scholar]
- 35.Moiseenkova-Bell V. Y., Stanciu L. A., Serysheva I. I., Tobe B. J., Wensel T. G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7451–7455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobertz W. R., Williams C., Miller C. (2000) Biochemistry 39, 10347–10352 [DOI] [PubMed] [Google Scholar]
- 37.Kim L. A., Furst J., Gutierrez D., Butler M. H., Xu S., Goldstein S. A., Grigorieff N. (2004) Neuron 41, 513–519 [DOI] [PubMed] [Google Scholar]
- 38.Higgins M. K., Weitz D., Warne T., Schertler G. F., Kaupp U. B. (2002) EMBO J. 21, 2087–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiu P. L., Pagel M. D., Evans J., Chou H. T., Zeng X., Gipson B., Stahlberg H., Nimigean C. M. (2007) Structure 15, 1053–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. (1995) Protein Sci. 4, 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Danev R., Glaeser R. M., Nagayama K. (2009) Ultramicroscopy 109, 312–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hosokawa F., Danev R., Arai Y., Nagayama K. (2005) J. Electron Microsc. (Tokyo) 54, 317–324 [DOI] [PubMed] [Google Scholar]
- 43.Ludtke S. J., Baldwin P. R., Chiu W. (1999) J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 44.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 45.Nilius B., Watanabe H., Vriens J. (2003) Pflugers Arch 446, 298–303 [DOI] [PubMed] [Google Scholar]
- 46.Danev R., Nagayama K. (2008) J. Struct. Biol. 161, 211–218 [DOI] [PubMed] [Google Scholar]
- 47.Hirokawa T., Boon-Chieng S., Mitaku S. (1998) Bioinformatics 14, 378–379 [DOI] [PubMed] [Google Scholar]
- 48.Long S. B., Campbell E. B., Mackinnon R. (2005) Science 309, 897–903 [DOI] [PubMed] [Google Scholar]
- 49.Voets T., Owsianik G., Janssens A., Talavera K., Nilius B. (2007) Nat. Chem. Biol. 3, 174–182 [DOI] [PubMed] [Google Scholar]
- 50.Mio K., Ogura T., Hara Y., Mori Y., Sato C. (2005) Biochem. Biophys. Res. Commun. 333, 768–777 [DOI] [PubMed] [Google Scholar]
- 51.Valente P., García-Sanz N., Gomis A., Fernández-Carvajal A., Fernández-Ballester G., Viana F., Belmonte C., Ferrer-Montiel A. (2008) FASEB J. 22, 3298–3309 [DOI] [PubMed] [Google Scholar]
- 52.Garcia-Elias A., Lorenzo I. M., Vicente R., Valverde M. A. (2008) J. Biol. Chem. 283, 31284–31288 [DOI] [PubMed] [Google Scholar]
- 53.Phelps C. B, Wang R. R., Choo S. S., Gaudet R. (2009) J. Biol. Chem. 285, 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]