Abstract
The umbilical cord and placenta are extra-embryonic tissues of particular interest for regenerative medicine. They share an early developmental origin and are a source of vast amounts of cells with multilineage differentiation potential that are poorly immunogenic and without controversy. Moreover, these cells are likely exempt from incorporated mutations when compared with juvenile or adult donor cells such as skin fibroblasts or keratinocytes. Here we report the efficient generation of induced pluripotent stem cells (iPSCs) from mesenchymal cells of the umbilical cord matrix (up to 0.4% of the cells became reprogrammed) and the placental amniotic membrane (up to 0.1%) using exogenous factors and a chemical mixture. iPSCs from these 2 tissues homogeneously showed human embryonic stem cell (hESC)-like characteristics including morphology, positive staining for alkaline phosphatase, normal karyotype, and expression of hESC-like markers including Nanog, Rex1, Oct4, TRA-1–60, TRA-1–80, SSEA-3, and SSEA-4. Selected clones also formed embryonic bodies and teratomas containing derivatives of the 3 germ layers, and could as well be readily differentiated into functional motor neurons. Among other things, our cell lines may prove useful for comparisons between iPSCs derived from multiple tissues regarding the extent of the epigenetic reprogramming, differentiation ability, stability of the resulting lineages, and the risk of associated abnormalities.
Keywords: Cell Differentiation, General Transcription Factors, Induced Pluripotent Stem (iPS) Cell, Neurodifferentiation, Stem Cell, Placenta, Reprogramming
Introduction
The recent discovery of induced pluripotency using exogenous factors has meant a tremendous advance for stem cell research (1, 2). By overexpressing defined combinations of transcription factors that are highly expressed in embryonic stem cells (ESCs)3, it is possible to reprogram the nuclei of terminally differentiated cells or stem cells into an ESC-like status (3–7). This technology has been used to create in vitro models for human genetic diseases, and in the future may also allow patient-specific stem cell therapies devoid of ethical concerns (8–11). However, human iPSC generation is not exempt from problems, among which stands out the very low efficiency of colony formation (1). Some cell types including keratinocytes (12), neural stem cells (13), adipose stem cells (14), melanocytes (15), and meningiocytes (16), reprogram more efficiently than the most widely used fibroblasts (<0.01%), and this has prompted laboratories worldwide to systematically search for highly susceptible and easily accessible tissues. However, this has also introduced significant variability and created confusion. In the mouse pluripotency can be exhaustively analyzed: the formation of chimeric mice with germ line transmission is the standard golden test, and entire animals have also been derived after tetraploid complementation (17, 18). The same procedures cannot be applied to human iPSCs, which basically rely on embryonic bodies (EBs) and teratomas as the most stringent tests. Therefore, the advantages and disadvantages of reprogramming human cells from different tissues are currently unclear but at some point will need careful comparison.
The placenta is a mammalian organ that connects the fetus to the maternal uterine wall, while the umbilical cord links the fetus and the placenta. After fertilization, the inner mass of the blastocyst becomes transformed into hypoblast and epiblast. The umbilical cord and vessels of the placenta originate from the hypoblast, while the amniotic membrane or inner layer of the placenta as well as the fetus comes from the epiblast. The amniotic membrane or amnion contains a layer of epithelial cells surrounding the amniotic cavity and internally is composed of mesenchymal-like cells. The outer layer of the blastocyst or trophoectoderm produces the trophoblasts, and these cells penetrate into the uterine wall to progressively form the chorionic membrane (also termed chorion) or external layer of the placenta. Inside the chorion is exchanged gas and nutrients between mother and fetus. Possibly because the formation of the extra-embryonic tissues happens very early after implantation, their cells retain an immature phenotype while the whole embryo develops (19). Therefore, although traditionally discarded upon birth, the placenta and umbilical cord are now regarded as a valuable source of cells with stem cell-like plasticity (20–23). For example, mesenchymal stem cells (MSCs) from the umbilical cord matrix (also termed Wharton jelly) have been differentiated into dopaminergic neurons, and adipogenic, osteogenic, myogenic, and chondrogenic lineages (24, 25). Nowadays, there is also widespread interest in banking umbilical cord blood, as it contains MSCs that in some cases have proven useful for regenerative medicine purposes (26). However, cord blood MSCs can be obtained in very small quantities and are not present in all donors. Interestingly, the umbilical cord matrix has been reported to contain a far larger number of MSCs than the cord blood (25). Because of their inherent characteristics and the abundance of available cells, we aimed to generate iPSCs from umbilical cord matrix and the amniotic mesenchyme. We achieved this using a modification of our recently published protocol based on chemical additions (27). The resulting iPSCs homogeneously displayed hESC-like characteristics and readily differentiated into multiple lineages. These cell lines could represent a useful tool for cross-comparisons between iPSCs generated from multiple tissues, which may assist in the establishment of more rigorous standards.
EXPERIMENTAL PROCEDURES
Cell Culture and iPSC Generation
IMR90 cells were purchased from the ATCC and cultured in DMEM (Hyclone) + 10% fetal bovine serum (FBS, Hyclone), penicillin/streptomycin and l-glutamine. Human umbilical cords and placentas were obtained after caesarean section or vaginal delivery of normal pregnancies at Nanfang Hospital of Guangzhou. All tissues were collected following principles approved by the Guangzhou Institutes of Health ethical committee; signed consent forms are available upon request. Mesenchymal-like cells were extracted as described (28, 29). Briefly, collected tissues were washed in phosphate-buffered saline to remove cellular debris and blood, and the amniotic and chorionic membranes separated mechanically. Amniotic membranes were then treated with 0.25% trypsin at 37 °C for 5 min and then with 1.2 units/ml of dispase (GIBCO) for 60 min at 37 °C. After removal and collection of the epithelial layer using forceps, the remaining amniotic membrane was minced and incubated with 2 mg/ml collagenase I (GIBCO) for 60–120 min at 37 °C; cells were then collected after centrifugation. The umbilical cord was minced and incubated with collagenase IV (GIBCO) for 4 h at 37 °C, then DNase I (1 mg/ml, GIBCO) for 15 min, and 0.25% trypsin for 15 min. Cells were then collected by centrifugation after filtration through a 40-μm cell strainer. All extra-embryonic cell types were cultured in DMEM/F12 (Hyclone) + penicillin/streptomycin, l-glutamine, and 10% FBS before infection. At early passage, 40,000 cells of each tissue were transduced in 6-well culture dishes using pMX-based retroviruses (Adgene) as described (16, 27). Transduced cells were maintained in high glucose DMEM (Hyclone) + 20% hESC-defined FBS (DFBS, Hyclone), nonessential amino acids, penicillin/streptomycin, l-glutamine, β-mercaptoethanol, and bFGF (Invitrogen). At day 6 post-infection, 10,000 cells were split in the same medium on 10-cm dishes coated with feeder (mitomycin C-treated mouse embryonic fibroblasts). From day 7, plates were either maintained in DFBS-containing medium or switched to KSR-based medium (DMEM/F12 + 20% knock-out serum replacement (KSR, GIBCO), nonessential amino acids, penicillin/streptomycin, l-glutamine, β-mercaptoethanol, and bFGF). Vitamin C was purchased from Sigma and used at 50 μg/ml, valproic acid was purchased from Merck and added at 1 mm. Picked human iPSC colonies and H9 hESC (purchased from ATCC) were cultured on feeder layers using KSR medium, or on Matrigel (BD Biosciences) using mTeSR1 medium (Stemcell).
iPSC Characterization
Alkaline phosphatase staining, immunofluorescence microscopy, semi-quantitative RT-PCR for transgene integration, karyotyping, and bisulfate sequencing, were performed as described (16, 30). Nanog antibodies were from R&D, SSEA-3, SSEA-4 from Abcam, and TRA-1–60 and TRA-1–81 from Millipore; an Olympus BX51 microscope was used for immunofluorescence staining captures. DNA was extracted using Wizard® Genomic DNA Purification kit (Promega), total RNA was extracted using TRIzol (Invitrogen). Quantitative real time RT-PCR (qPCR) was performed using a Thermal Cycler DiceTM Real Time System and SYBR Premix EX TaqTM (Takara). β-actin was used for qPCR normalization, and all items were measured in triplicate. Primers sequences (for other procedures as well) are available upon request. DNA microarrays were performed using the Affymetrix HuGene 1.0 ST array (Affymetrix) platform in accordance with the manufacturer's instructions and scanned using Affymetrix Scanner 3000 7G. The lists of genes have been submitted to the GEO data base (GEO accession no. GSE 20581). Microsatellite markers were amplified by semi-quantitative PCR using genomic DNA. Primers are as follows: PentaE-forward: ATTACCAACATGAAAGGGTACCAATA, PentaE-reverse: TGGGTTATTAATTGAGAAAACTCCTTACAATTT; CSF1PO-forward: CCGGAGGTAAAGGTGTCTTAAAGT, CSF1PO-reverse: ATTTCCTGTGTCAGACCCTGTT; D16S539-forward: GGGGGTCTAAGAGCTTGTAAAAAG, D16S539-reverse: GTTTGTGTGTGCATCTGTAAGCATGTATC; D18S51-forward: TTCTTGAGCCCAGAAGGTTA, D18S51-reverse: ATTCTACCAGCAACAACACAAATAAAC; D8S1179-forward: ATTGCAACTTATATGTATTTTTGTATTTCATG, D8S1179-reverse: ACCAAATTGTGTTCATGAGTATAGTTTC; vWA-forward: GCCCTAGTGGATGATAAGAATAATCAGTATGTG, vWA-reverse: GGACAGATGATAAATACATAGGATGGATGG; D5S818-forward: GGTGATTTTCCTCTTTGGTATCC, D5S818-reverse: AGCCACAGTTTACAACATTTGTATCT; FGA-forward: GGCTGCAGGGCATAACATTA, FGA-reverse: ATTCTATGACTTTGCGCTTCAGGA.
Teratomas, EB Formation, and Neural Differentiation
For teratoma formation, 3 million cells were injected subcutaneously into SCID mice and the tumors sectioned after 7–9 weeks and stained with hematoxylin/eosin. EB differentiation was performed by detaching (using 5 mg/ml Dispase, Gibco) iPSCs growing on feeders and culturing them in suspension on nonadherent 25 T-flask (Corning) for 8 days with KSR-based medium without bFGF. EBs were then seeded on Matrigel-coated dishes for another 8 days before RNA was extracted. Motor neuron differentiation was induced in differentiating medium as described (31). iPSC-derived EBs at day 10 (without splitting on Matrigel) were treated with retinoic acid (Sigma, 0.1 μm) in DMEM/F12, N2 supplement (GIBCO), heparin (2 μg/ml; Sigma), and cAMP (1 μm, Sigma). One week later, neuroepithelial rosettes were gently blown off and cultured on coverslips in the same medium in the presence of retinoic acid and sonic hedgehog (100 ng/ml; R&D system) for another week. At day 24, brain-derived neurotrophic factor, glial-derived neurotrophic factor, and insulin-like growth factor-1 (10 ng/ml) were added to the culture and the cells maintained like this for another week. Polyclonal antibodies against βIII-tubulin were purchased from Covance PRB-435P; against nestin and choline O-acetyltransferase (ChAT) from Chemicon; and against HB9 from Developmental Studies Hybridoma Bank.
Electophysiology Recording
Electrophysiology was performed at 22–25 °C using standard whole-cell, current-clamp techniques (32). Patch pipettes (resistance 3–5MΩ) were filled with the following: 140 mm potassium methanesulfonate, 10 mm HEPES, 5 mm NaCl, 1 mm CaCl2, 0.2 mm EGTA, 3 mm ATP-Na2, 0.4 mm GTP-Na2, pH 7.3 (adjusted with KOH). The external solution contained 120 mm NaCl, 1.2 mm KH2PO4, 1.9 mm KCl, 26 mm NaHCO3, 2.2 mm CaCl2, 1.4 mm MgSO4, 10 mm d-glucose, 7.5 mm HEPES (pH with NaOH to 7.3). The bath solution was equilibrated with 95% O2 and 5% CO2 before use. Resting potentials were maintained at about −65 mV. Whole-cell patch-clamp techniques were amplified and filtered using an Axopatch 200B amplifier (Molecular Devices, Sunnyvale, CA). Graded current injections used durations of 300 ms (in steps of 5 pA). Signals were sampled at 10 kHz using a Digidata 1440A analog to digital converter and acquired and stored on a computer hard drive using pClamp10 software. Data were analyzed using pClamp10 (Clampfit).
RESULTS AND DISCUSSION
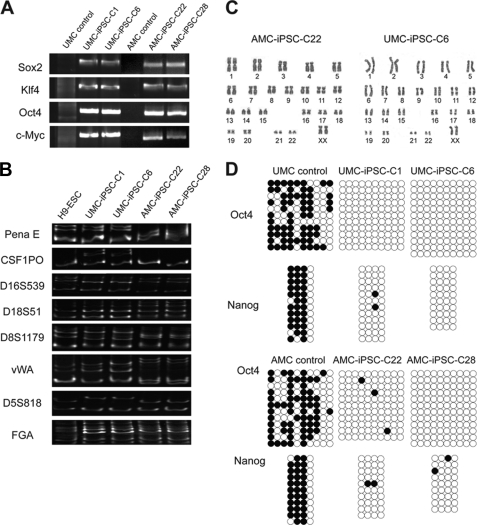
Mesenchymal-like cells from the Wharton jelly of the umbilical cord (UMCs), the amniotic membrane epithelium (AECs), and from the amniotic membrane mesenchyme (AMCs) were isolated and transduced with retroviruses encoding the 4 Yamanaka factors (Sox2, Klf4, Oct4, and c-Myc; SKOM) or GFP as a control (Fig. 1A). GFP-positive cells accounted for close to 100% of the transduced population in UMCs and AMCs (Fig. 1B), but AECs displayed few or no GFP-positive cells (data not shown). The low efficiency of AEC transduction may be related to their slower proliferation, suggesting that infection with lentiviruses may be a more suitable approach, a possibility that we have not tested. At day 5 post-infection, early morphologic changes (reduced cell size and clustering) were noticeable in SKOM-transduced UMCs and AMCs (Fig. 1B). At day 6, infected cells were split on feeders, and the day after, the culture medium was changed to either KSR- or DFBS-based medium with added vitamin C or a combination of vitamin C + valproic acid (Fig. 1A). Valproic acid was added only from days 7 to 14 as otherwise was observed that the iPSC colonies differentiated or died, and vitamin C from days 7 to 19. On day 19, we changed the DFBS-based medium to KSR (Fig. 1A), which contains vitamin C (27), as otherwise we also observed a tendency of the hESC-like colonies to differentiate, possibly in part related to accelerated growth in the presence of serum. We have previously shown that very similar tissue culture conditions drastically increase the efficiency of mouse and human iPSC generation (27). Valproic acid was one of the first chemicals reported to increase the efficiency of nuclear reprogramming, and the underlying mechanism is unclear but possibly relates to its ability to inhibit histone deacetylases (33). Vitamin C is a natural antioxidant that acts at least in part by alleviating cell senescence during reprogramming (34–38). Routinely, colonies with hESC-like characteristics appeared (in the 2 cell types) around days 13–15 post-infection in DFBS + chemicals (and ∼5–7 days later in KSR), and we picked them at day 23 (Fig. 1A). Afterward, all existing hESC-like colonies were marked, and alkaline phosphatase staining was performed to calculate the final reprogramming efficiency, which showed a perfect match (Fig. 1C). For this we quantified three different experiments using UMCs and AMCs from 2 different donors. DFBS + chemicals produced significantly more hESC-like colonies than KSR in both cell types, and under any condition UMCs reprogrammed better than AMCs (Fig. 1C). Picked iPSC colonies were expanded and grown on feeders with KSR or on Matrigel without feeder using mTeSR1 medium. After multiple passages they remained alkaline phosphatase-positive and with stable hESC-like morphologic characteristics (Fig. 2A). They also stained positive for the surface markers TRA-1–60, TRA-1–80, SSEA-3, SSEA-4, and for the transcription factor Nanog, displaying roughly similar intensity and pattern compared with H9 hESC (Fig. 2A). qPCR of endogenous Sox2 and Oct4 as well as Nanog and the transcription factor Rex1 showed high expression levels in all selected iPSCs as well (Fig. 2B), although some variability could be detected between iPSCs and between iPSCs and H9 cells. Importantly, the exogenous transgenes were potently silenced as demonstrated as well by qPCR using primers that amplify an amplicon containing part of the backbone plasmid and the coding sequence of the transgene (Fig. 2C). qPCR analysis also demonstrated high expression of Nanog in the donor UMCs compared with AMCs and human fibroblasts (IMR90 cell line) (Fig. 2D). Expression of ESC surface markers and transcription factors has been reported previously in placenta and umbilical cord cell populations (20, 21, 28, 29, 39). This may explain their susceptibility to reprogramming and could perhaps allow a more extended hESC-like epigenetic remodeling. Expression in UMC and AMC iPSCs of endogenous Sox2 and Oct4 as well as Nanog and Rex1 was verified by DNA arrays, which also demonstrated highly similar transcriptome profile between iPSCs and H9 cells (Fig. 3). In addition, semi-quantitative PCR was used to show genomic DNA integration of the 4 exogenous transgenes in iPSCs (Fig. 4A), and to compare a panel of microsatellite markers (5) between UMC iPSCs, AMC iPSCs, and hESCs (Fig. 4B). The latter is important as it indicates there is no contamination between cells of different origins. Moreover, the karyotypes for all tested iPSC clones displayed normal number of chromosomes (Fig. 4C), and bisulfite treatment of genomic DNA and subsequent sequencing showed high demethylation of the Oct4 and Nanog proximal promoters (Fig. 4D). To further demonstrate the acquisition of pluripotency, selected iPSCs were injected into SCID mice, and this resulted in complex teratomas that contained tissues corresponding to the 3 germ layers (Fig. 5). iPSCs of both origins also readily formed EBs upon culture in suspension (Fig. 6A), and these EBs contained early derivatives of the 3 germ layers as assessed by qPCR analysis of specific markers (Fig. 6B). We also pursued motor neuron differentiation of EBs from UMC iPSCs. Neural rosettes containing neural stem cells were first generated (after 10 days) that stained positively for nestin (Fig. 6C top panel). After an additional 2 weeks with added retinoic acid and sonic hedgehog, neuronal-like cells appeared that stained for βIII-tubulin, ChAT, and HB9 (Fig. 6C, middle and bottom panels) (40, 41). Standard whole-cell patch clamp, current-clamp techniques were then used to study the electrical properties of these motor neurons. Repetitive traces of action potentials were elicited when a 10 pA current injection was applied and increased 5 pA in every new round until 55 pA were reached (Fig. 6D). Action potentials detected after application of a 30 pA current injection is shown individually.
FIGURE 1.
Generation of iPSC clones from UMCs and AMCs. A, schematic representation of our iPSC generation protocol including windows of treatment with vitamin C and valproic acid. B, left, immunofluorescence and phase contrast photographs of UMCs and AMCs at day 5 after viral transduction with control GFP retroviruses. Right, phase contrast photographs of UMCs and AMCs at day 5 post-infection with SKOM, notice the early morphology changes compared with the GFP control. Lower and higher magnification captures are shown. C, calculation of iPSC generation efficiency based on the number of AP-positive colonies per 10.000 infected cells seeded on feeder in a 10-cm dish. For this, hESC-like colonies were marked and counted before the staining and proved alkaline phosphatase-positive in all cases. Three experiments using cells from two different donors were considered.
FIGURE 2.
iPSCs from UMCs and AMCs display hESC characteristics. A, phase contrast captures of H9 human ESCs and iPSCs from UMCs (C1) and AMCs (C28) grown on either feeders or Matrigel layers; iPSCs at passage 10 or more are shown. The same cell lines were stained for alkaline phosphatase. Immunofluorescence microscopy also shows activation of the endogenous ESC program. A representative experiment is shown; scale bar corresponds to 150 μm. B, qPCRs of a representative experiment showing high levels of expression of ESC-specific transcription factors in iPSCs (passage 10 or more) from UMCs and AMCs compared with H9 ESCs and the donor cells. C, qPCRs showing silencing of the exogenous transgenes in all iPSCs relative to SKOM infected donor cells extracted at day 6 (at the time of splitting on feeder). D, qPCR for the indicated genes in UMCs, AMCs, and the human fibroblast cell line IMR90.
FIGURE 3.
Comparative DNA microarray analysis between hESCs and UMC/AMC iPSCs. Top, scatter plot comparing DNA microarray analysis data between H9 hESCs and 2 different iPSC clones from UMC and AMC iPSCs. Endogenous Oct4, Sox2, Nanog, and Rex1 are marked. Bottom, hierarchic clustering representation for the same microarrays.
FIGURE 4.
Additional characterization of UMC and AMC iPSCs. A, semi-quantitative RT-PCR shows integration of the exogenous factors into the genomic DNA of selected UMC and AMC iPSC colonies, donor cells were included as the negative control. B, selected microsatellite markers were amplified by semi-quantitative PCR in the indicated cell types, the different banding patterns exclude the possibility of cross-contamination. C, karyotype analysis demonstrates normal number of chromosomes (23 pairs) in selected iPSCs (passage 16). D, DNA methylation profile of the Oct4 and Nanog proximal promoters in the indicated iPSCs (passage 16), donor cells are included as control. White circles indicate unmethylated CpGs, and black circles indicate methylated.
FIGURE 5.
Teratomas produced by UMC and AMC iPSCs. Hematoxylin-eosin-stained sections from teratomas generated with a selected UMC and AMC iPSC clone. Differentiation into multiple derivatives of the 3 germ layers is shown: 1 indicates neural tube-like structures (ectoderm); 2, striated muscle (mesoderm); 3, cartilage (mesoderm); 4, gut epithelium (endoderm); 5, adipose tissue (mesoderm); 6, bone (mesoderm); 7, gland (endoderm). Scale bar corresponds to 200 μm.
FIGURE 6.
EBs and motor neuron differentiation of UMC and AMC iPSCs. A, phase contrast photographs of EBs (at day 4) from selected UMC (C1) and AMC (C28) iPSCs. Lower and higher magnifications are shown. B, qPCR analysis of selected markers using RNA extracted from the same EBs. After 8 days in suspension, EBs were split on Matrigel-coated dishes for another 8 days before RNA extraction and processing. Scale bar corresponds to 150 μm. C, top, phase contrast photographs of neural rosettes from UMC iPSC-derived EBs (C1) and immunofluorescence staining for the neural stem cell marker nestin. Middle, immunofluorescence staining for βIII-tubulin and choline O-acetyltransferase shows efficient generation of motorneurons from the same EBs. Bottom, HB9 staining. DAPI is shown in blue for all immunofluorescences. Scale bar corresponds to 50 μm. D, top, electrophysiological characterization of these motor neurons evaluated by current-clamp recording of membrane voltage in response to serial current injections (300-ms duration current injections with increasing 5 pA every round). Bottom, single current injection (300-ms duration, 30 pA).
In summary, herein we report the efficient generation of iPSCs from 2 human extra-embryonic tissues: the umbilical cord matrix and the amniotic membrane mesenchyme. It will be interesting to study whether these tissues are equally or more susceptible to non integrating reprogramming approaches (adenoviruses, recombinant proteins, or episomal vectors) than standard donor cells (skin fibroblasts) or can be reprogrammed using fewer factors (42–45), possibilities that we have not tested. Other human cells have also being reprogrammed to iPSCs (keratinocytes, melanocytes, adipocytes, peripheral blood) with varied efficiency (<1% using juvenile cells as a source, ∼0.05, ∼0.2, and ∼0.01%, respectively) (12, 14, 15, 46) but in terms of their quality there is no indication that one cell type has advantage over others, and in fact it may be the opposite. Therefore, it is an interesting possibility that because extra-embryonic tissues are less differentiated than adult somatic cells, iPSCs from these sources are more faithfully reprogrammed than others. Moreover, human cells can incorporate mutations over the course of a life time, and these mutations may be further selected during in vitro donor cell expansion and iPSC generation. It is thus an attractive idea that neonatal tissues are routinely stored in the near future for later reprogramming and allogenic transplantation. Storage of neonatal tissues would require biopsy and except for more simple procedures such as male foreskin excision there could be risk of associated infections. On the other hand, the preservation of cord blood is a frequent procedure in many countries but the number of MSCs in cord blood is low, and therefore successful reprogramming with all samples is not guaranteed (24). While our work was in process, iPSCs were generated from amniocytes (floating cells in the amniotic cavity which detach from both the amnion and the fetus; with up to 0.2% efficiency) and from umbilical cord blood (up to 0.03%) (47–49). In addition, we recently reported the generation of iPSCs from chorionic mesenchymal cells, using as well a combination of vitamin C + valproic acid (27). The reprogramming efficiency of chorionic cells is higher compared with UMCs and AMCs but it may be possible to increase the efficiency in UMCs and AMCs by sorting specific populations expressing specific cell surface markers. Comparisons between all these different iPSCs including fibroblasts should prove invaluable to answer relevant questions in the field such as the existence of tissue-specific epigenetic memory in iPSCs and to evaluate safety issues. For this it will be important on one side to perform whole genome analyses, for example deep sequencing mRNA analysis (including noncoding small RNAs), DNA methylation profile, and ChIP-on-Chip sequencing for histone modification marks, and on the other to evaluate multilineage differentiation ability. In case the extra-embryonic tissues prove to be a better cell source for reprogramming, storage of MSCs from umbilical cord matrix and placenta would perhaps facilitate safer future personalized stem cell-based therapies.
Acknowledgments
We thank members of our laboratories for their support.
This work was supported by the Knowledge Innovation Program of the Chinese Academy of Sciences, Chinese Academy of Sciences/SAFEA International Partnership Program for Creative Research Teams, Chinese Academy of Sciences (KSCX2-YW-R-48, KSCX2-YW-R-244), National Natural Science Foundation of China (30725012, 230725012, 90813033, 30630039), Ministry of Science and Technology 973 Program (2006CB701504, 2006CB943600, 2007CB948002, 2007CB947804, 2007CB947900, 2009CB941102, 2009CB940902, 2010CB944800), National High Technology Research and Development Program of China (2006AA02A103, 2007G-P034, 2007Z3-C7031), Bureau of Science and Technology of Guangzhou Municipality (2006A50104002, 2008A1-E4011), and an EFBIC (European Federation of Biotechnology) RED travel grant.
- ESC
- embryonic stem cell
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- GFP
- green fluorescent protein
- MSC
- mesenchymal stem cells
- iPSC
- induced pluripotent stem cells
- ChAT
- choline O-acetyltransferase
- UMC
- umbilical cord
- AMC
- amniotic membrane mesenchyme
- SKOM
- Sox2, Klf4, Oct4, and c-Myc
- EB
- embryonic bodies.
REFERENCES
- 1.Yamanaka S. (2009) Cell 137, 13–17 [DOI] [PubMed] [Google Scholar]
- 2.Belmonte J. C., Ellis J., Hochedlinger K., Yamanaka S. (2009) Nat. Rev. Genet. 10, 878–883 [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 4.Okita K., Ichisaka T., Yamanaka S. (2007) Nature 448, 313–317 [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- 6.Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin I. I., Thomson J. A. (2007) Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- 7.Wernig M., Meissner A., Foreman R., Brambrink T., Ku M., Hochedlinger K., Bernstein B. E., Jaenisch R. (2007) Nature 448, 318–324 [DOI] [PubMed] [Google Scholar]
- 8.Chamberlain S. J., Li X. J., Lalande M. (2008) Neurogenetics 9, 227–235 [DOI] [PubMed] [Google Scholar]
- 9.Dimos J. T., Rodolfa K. T., Niakan K. K., Weisenthal L. M., Mitsumoto H., Chung W., Croft G. F., Saphier G., Leibel R., Goland R., Wichterle H., Henderson C. E., Eggan K. (2008) Science 321, 1218–1221 [DOI] [PubMed] [Google Scholar]
- 10.Park I. H., Arora N., Huo H., Maherali N., Ahfeldt T., Shimamura A., Lensch M. W., Cowan C., Hochedlinger K., Daley G. Q. (2008) Cell 134, 877–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye L., Chang J. C., Lin C., Sun X., Yu J., Kan Y. W. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9826–9830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aasen T., Raya A., Barrero M. J., Garreta E., Consiglio A., Gonzalez F., Vassena R., Bilić J., Pekarik V., Tiscornia G., Edel M., Boué S., Izpisúa, Belmonte J. C. (2008) Nat. Biotechnol. 26, 1276–1284 [DOI] [PubMed] [Google Scholar]
- 13.Kim J. B., Zaehres H., Wu G., Gentile L., Ko K., Sebastiano V., Araúzo-Bravo M. J., Ruau D., Han D. W., Zenke M., Schöler H. R. (2008) Nature 454, 646–650 [DOI] [PubMed] [Google Scholar]
- 14.Sun N., Panetta N. J., Gupta D. M., Wilson K. D., Lee A., Jia F., Hu S., Cherry A. M., Robbins R. C., Longaker M. T., Wu J. C. (2009) Proc. Natl. Acad. Sci. 106, 15720–15725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Utikal J., Maherali N., Kulalert W., Hochedlinger K. (2009) J. Cell Sci. 122, 3502–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin D., Gan Y., Shao K., Wang H., Li W., Wang T., He W., Xu J., Zhang Y., Kou Z., Zeng L., Sheng G., Esteban M. A., Gao S., Pei D. (2008) J. Biol. Chem. 283, 33730–33735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang L., Wang J., Zhang Y., Kou Z., Gao S. (2009) Cell Stem Cell 5, 135–138 [DOI] [PubMed] [Google Scholar]
- 18.Zhao X. Y., Li W., Lv Z., Liu L., Tong M., Hai T., Hao J., Guo C. L., Ma Q. W., Wang L., Zeng F., Zhou Q. (2009) Nature 461, 86–90 [DOI] [PubMed] [Google Scholar]
- 19.Red-Horse K., Zhou Y., Genbacev O., Prakobphol A., Foulk R., McMaster M., Fisher S. J. (2004) J. Clin. Invest. 114, 744–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Can A., Karahuseyinoglu S. (2007) Stem Cells 25, 2886–2895 [DOI] [PubMed] [Google Scholar]
- 21.Yen B. L., Huang H. I., Chien C. C., Jui H. Y., Ko B. S., Yao M., Shun C. T., Yen M. L., Lee M. C., Chen Y. C. (2005) Stem Cells 23, 3–9 [DOI] [PubMed] [Google Scholar]
- 22.Fukuchi Y., Nakajima H., Sugiyama D., Hirose I., Kitamura T., Tsuji K. (2004) Stem Cells 22, 649–658 [DOI] [PubMed] [Google Scholar]
- 23.Miki T., Strom S. C. (2006) Stem Cell Rev. 2, 133–142 [DOI] [PubMed] [Google Scholar]
- 24.Secco M., Zucconi E., Vieira N. M., Fogaça L. L., Cerqueira A., Carvalho M. D., Jazedje T., Okamoto O. K., Muotri A. R., Zatz M. (2008) Stem Cells 26, 146–150 [DOI] [PubMed] [Google Scholar]
- 25.Fu Y. S., Cheng Y. C., Lin M. Y., Cheng H., Chu P. M., Chou S. C., Shih Y. H., Ko M. H., Sung M. S. (2006) Stem Cells 24, 115–124 [DOI] [PubMed] [Google Scholar]
- 26.Hollands P., McCauley C. (2009) Stem Cell Rev. 5, 195–203 [DOI] [PubMed] [Google Scholar]
- 27.Esteban M. A., Wang T., Qin B., Yang J., Qin D., Cai J., Li W., Weng Z., Chen J., Ni S., Chen K., Li Y., Liu X., Xu J., Zhang S., Li F., He W., Labuda K., Song Y., Peterbauer A., Wolbank S., Redl H., Zhong M., Cai D., Zeng L., Pei D. (2010) Cell Stem Cell 6, 71–79 [DOI] [PubMed] [Google Scholar]
- 28.Bilic G., Zeisberger S. M., Mallik A. S., Zimmermann R., Zisch A. H. (2008) Cell Transplant. 17, 955–968 [DOI] [PubMed] [Google Scholar]
- 29.Bailo M., Soncini M., Vertua E., Signoroni P. B., Sanzone S., Lombardi G., Arienti D., Calamani F., Zatti D., Paul P., Albertini A., Zorzi F., Cavagnini A., Candotti F., Wengler G. S., Parolini O. (2004) Transplantation 78, 1439–1448 [DOI] [PubMed] [Google Scholar]
- 30.Esteban M. A., Xu J., Yang J., Peng M., Qin D., Li W., Jiang Z., Chen J., Deng K., Zhong M., Cai J., Lai L., Pei D. (2009) J. Biol. Chem. 284, 17634–17640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X. J., Hu B. Y., Jones S. A., Zhang Y. S., Lavaute T., Du Z. W., Zhang S. C. (2008) Stem Cells 26, 886–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perrier A. L., Tabar V., Barberi T., Rubio M. E., Bruses J., Topf N., Harrison N. L., Studer L. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12543–12548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huangfu D., Maehr R., Guo W., Eijkelenboom A., Snitow M., Chen A. E., Melton D. A. (2008) Nat. Biotechnol. 26, 795–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong H., Takahashi K., Ichisaka T., Aoi T., Kanagawa O., Nakagawa M., Okita K., Yamanaka S. (2009) Nature 460, 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawamura T., Suzuki J., Wang Y. V., Menendez S., Morera L. B., Raya A., Wahl G. M., Belmonte J. C. (2009) Nature 460, 1140–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H., Collado M., Villasante A., Strati K., Ortega S., Cañamero M., Blasco M. A., Serrano M. (2009) Nature 460, 1136–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marión R. M., Strati K., Li H., Murga M., Blanco R., Ortega S., Fernandez-Capetillo O., Serrano M., Blasco M. A. (2009) Nature 460, 1149–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Utikal J., Polo J. M., Stadtfeld M., Maherali N., Kulalert W., Walsh R. M., Khalil A., Rheinwald J. G., Hochedlinger K. (2009) Nature 460, 1145–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carlin R., Davis D., Weiss M., Schultz B., Troyer D. (2006) Reprod. Biol. Endocrinol. 4, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee H., Shamy G. A., Elkabetz Y., Schofield C. M., Harrsion N. L., Panagiotakos G., Socci N. D., Tabar V., Studer L. (2007) Stem Cells 25, 1931–1939 [DOI] [PubMed] [Google Scholar]
- 41.Li X. J., Du Z. W., Zarnowska E. D., Pankratz M., Hansen L. O., Pearce R. A., Zhang S. C. (2005) Nat. Biotechnol. 23, 215–221 [DOI] [PubMed] [Google Scholar]
- 42.Stadtfeld M., Nagaya M., Utikal J., Weir G., Hochedlinger K. (2008) Science 322, 945–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou H., Wu S., Joo J. Y., Zhu S., Han D. W., Lin T., Trauger S., Bien G., Yao S., Zhu Y., Siuzdak G., Schöler H. R., Duan L., Ding S. (2009) Cell Stem Cell 4, 381–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim D., Kim C. H., Moon J. I., Chung Y. G., Chang M. Y., Han B. S., Ko S., Yang E., Cha K. Y., Lanza R., Kim K. S. (2009) Cell Stem Cell 4, 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin, Thomson J. A. (2009) Science 324, 797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loh Y. H., Agarwal S., Park I. H., Urbach A., Huo H., Heffner G. C., Kim K., Miller J. D., Ng K., Daley G. Q. (2009) Blood 113, 5476–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li C., Zhou J., Shi G., Ma Y., Yang Y., Gu J., Yu H., Jin S., Wei Z., Chen F., Jin Y. (2009) Hum. Mol. Genet. 18, 4340–4349 [DOI] [PubMed] [Google Scholar]
- 48.Giorgetti A., Montserrat N., Aasen T., Gonzalez F., Rodríguez-Pizà I., Vassena R., Raya A., Boué S., Barrero M. J., Corbella B. A., Torrabadella M., Veiga A., Izpisua Belmonte J. C. (2009) Cell Stem Cell 5, 353–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haase A., Olmer R., Schwanke K., Wunderlich S., Merkert S., Hess C., Zweigerdt R., Gruh I., Meyer J., Wagner S., Maier L. S., Han D. W., Glage S., Miller K., Fischer P., Schöler H. R., Martin U. (2009) Cell Stem Cell 5, 434–441 [DOI] [PubMed] [Google Scholar]