Abstract
In the early stages of an infection, pathogenic bacteria use long fibrous structures known as pili as adhesive anchors for attachment to the host cells. These structures also play key roles in colony and biofilm formation. In all those processes, pili must withstand large mechanical forces. The pili of the nasty Gram-positive human pathogen Streptococcus pyogenes are assembled as single, micrometer long tandem modular proteins of covalently linked repeats of pilin proteins. Here we use single molecule force spectroscopy techniques to study the mechanical properties of the major pilin Spy0128. In our studies, we engineer polyproteins containing repeats of Spy0128 flanked by the well characterized I27 protein which provides an unambiguous mechanical fingerprint. We find that Spy0128 is an inextensible protein, even when pulled at forces of up to 800 pN. We also found that this remarkable mechanical resilience, unique among the modular proteins studied to date, results from the strategically located intramolecular isopeptide bonds recently identified in the x-ray structure of Spy0128. Removal of the isopeptide bonds by mutagenesis readily allowed Spy0128 domains to unfold and extend, albeit at relatively high forces of 172 pN (N-terminal domain) or 250 pN (C-terminal domain). Our results show that in contrast to the elastic roles played by large tandem modular proteins such as titin and fibronectin, the giant pili of S. pyogenes evolved to abrogate mechanical extensibility, a property that may be crucial in the pathogenesis of this most virulent bacterium and, therefore, become the target of new therapeutic approaches against its infections.
Keywords: Atomic Force Microscopy, Bacteria, Protein Assembly, Protein Structure, Single Molecule Biophysics, Force Spectroscopy, Gram-positive, Isopeptide, Mechanical Stability, Pilus
Introduction
Streptococcus pyogenes (Group A Streptococcus, GAS) is a human pathogen responsible for acute infections that range from pharyngitis (strep throat) to life-threatening toxic shock syndrome and necrotizing fasciitis (reviewed in Ref. 1). Common to pathogenic bacteria, S. pyogenes uses filamentous micrometer-long pili to adhere mechanically to target cells; for example those of the pharynx (2–6). Apart from cell adhesion, pili are also crucial in other mechanical processes important for colonization and infection, such as biofilm development (2–6). Thus, it is well understood that pili are essential virulence factors (4, 7) regarded as targets for vaccine development (4, 8, 9).
In its essential mechanical roles, a single pilus experiences forces that can reach 100 pN (10), and bundles of pili have been shown to withstand forces in excess of 1000 pN (11). Among the different species of bacteria, distinct protein architectures allow dynamic equilibration of the pili with such high stretching forces, while still retaining their adhesive function (4, 12). Distinctive to Gram-positive bacteria such as S. pyogenes, pili are assembled as a single linear homopolymer of a so-called major pilin, decorated by minor pilins with specific functions, mainly adhesins (4). This architecture results from the covalent polymerization of pilins via isopeptide bonds, in a transpeptidation reaction catalyzed by sortase enzymes (6, 13–15). The assembled pilus is anchored covalently to the cell wall also through a sortase-mediated reaction (16). Such a covalent organization results in pili that are one molecule thick, but over 100 repeats long (17). This molecular architecture bears striking resemblance to a variety of elastic tandem modular proteins that function under a stretching force, such as the giant muscle protein titin (18) and the extracellular protein fibronectin (19). However, at the sustained high forces experienced by pili (10–12), most if not all of the titin and fibronectin modules would be fully unfolded and extended (20). A similar global unfolding and extension in pili would perhaps have deleterious consequences for the adhesivity of S. pyogenes to its host cells. Thus, it is likely that these pili exhibit novel mechanical properties that have not yet been observed in elastic tandem modular proteins.
Despite their crucial role in pathogenesis, the mechanical properties of pili from Gram-positive bacteria have not been studied so far. Recent x-ray crystallography and mass spectrometry studies of Spy0128, the major pilin from pathogenic S. pyogenes (M1 serotype), revealed Spy0128 to be composed of two tandem β-sandwich domains, each containing an intramolecular isopeptide bond (21). Bulk studies showed that the thermal stability and resistance to proteolysis of Spy0128 are drastically decreased by mutations blocking the formation of either or both isopeptide bonds (21, 22). For example, wild-type Spy0128 was found to unfold at 85 °C, whereas mutant forms blocking the formation of either isopeptide bond would lower the unfolding temperature, independently in each domain, by up to 30 °C (22). Because of the large contribution made by the isopeptide bonds to the thermal stability of Spy0128, together with their suggestive location in the structure, the authors and others concluded that the isopeptide bonds were likely to play a key role in conveying mechanical resistance to the pili of S. pyogenes (12, 21, 23). However, no measurement of the mechanical resistance of the isopeptide bonds was provided. In addition, neither x-ray crystallography nor mass spectrometry experiments can accurately determine the proportion of Spy0128 modules forming intramolecular isopeptide bonds in solution. For instance, it is a well known fact that crystallization occasionally induces the formation of covalent bonds in proteins (24). Therefore, the proposed mechanical relevance of the intramolecular isopeptide bonds remains to be tested.
Here, we report a study of the mechanical properties of Spy0128 using single molecule force spectroscopy techniques (25). Our results demonstrate that Spy0128 cannot be unfolded by mechanical forces of up to 800 pN, revealing this protein to be completely inextensible. We also found that introducing mutations blocking the formation of isopeptide bonds in either module of Spy0128 allowed the targeted module to readily unfold and extend under force while the other module remained fully folded and inextensible. Although our observations revealed Spy0128 to be a mechanically inextensible protein, in rare cases (∼3%), we observed single module unfolding events in wild-type Spy0128 domains. That small population of proteins could easily be missed using bulk techniques. Therefore, the efficiency of the isopeptide bond formation in wild-type Spy0128 may not be 100%. We speculate that the development of pharmacological agents that enhance the rate of failure of formation of isopeptide bonds may represent a novel strategy to greatly alter the mechanical extensibility of S. pyogenes pili, and block the infectivity of this most prevalent human pathogen.
EXPERIMENTAL PROCEDURES
Protein Engineering and Purification
All the reagents used were molecular biology grade and were obtained from regular commercial sources. The genomic DNA from S. pyogenes, M1 serotype (ATCC 700294) was obtained from the American Type Culture Collection (Manassas, VA). The cDNA coding for Spy0128 (residues 18–308) was obtained by PCR. The primers employed included BamHI, BglII, and KpnI restriction sites to produce polyproteins following the system described before for I27 (25). The original cDNA sequence of Spy0128 has a KpnI site that was removed by introducing silent mutations using the QuikChange site-directed mutagenesis kit (Agilent Technologies, La Jolla, CA). The same mutagenesis method was used to produce Spy0128 E117A and E258A. The cDNAs coding for (I27-Spy0128)2-I27, and its mutant variants were cloned in the expression plasmid pQE-80L (Qiagen, Valencia, CA) using the BamHI and KpnI restriction sites. The final polyproteins have 11 extra amino acids at the N terminus (including six histidines), 4 at the C terminus, and 2 between each module, for a total of 23 extra amino acids. E. coli BLR (DE3) cells were transformed with the expression plasmids. Protein production was induced at A600 ∼0.8 with 1 mm isopropyl-1-thio-β-d-galactopyranoside for 3 h at 37 °C or overnight at 25 °C. Cells were disrupted by sonication, and polyproteins were purified from the soluble fraction by an affinity chromatography using a Talon column (Clontech), followed by an FPLC step using a Superdex 200 column (GE Healthcare). The buffer employed was 10 mm Hepes pH 7.2, 1 mm EDTA, 150 mm NaCl. This procedure rendered homogeneous fractions of proteins, as estimated by SDS-PAGE (supplemental Fig. S1).
Single Molecule Force Spectroscopy
Samples were prepared by depositing 1–5 μl of a ∼0.2 mg/ml polyprotein solution onto an evaporated gold coverslip. Veeco MLCT (Camarillo, CA) or Olympus TR400PSA (Asylum Research, Santa Barbara, CA) cantilevers were used. They were calibrated using the equipartition theorem (26). Typical spring constants were in the range of 20–30 pN/nm. To pick up single molecules, the cantilever was pushed against the surface at contact forces of 0.8–2 nN, and then retracted. In force-extension experiments, the retraction rate was 400 nm/s. Experiments were done at room temperature (22–25 °C). In single molecule atomic force microscopy (AFM)3 determinations, it is frequent to obtain traces reflecting spurious interactions. Therefore, only reproducible traces with three I27 unfolding events were analyzed. Experimental results were fitted to the worm-like chain model of polymer elasticity (27). The experimental histograms were fitted using Gaussian curves. In this work, the error of the mean is given by the standard deviation of the Gaussian fit. To estimate the initial extension of the polyproteins from force-extension traces, the worm-like chain model was fitted to the first unfolding peak. A drawback of this procedure is that the results are dependent on the persistence length that is chosen. It has been shown before that the use of force-clamp provides a model-free estimation of initial extensions (28). The initial extension of (I27-Spy0128)2-I27 measured by force-clamp was equivalent to the one determined by force-extension (supplemental Fig. S5). The retraction of the cantilever from the surface is usually accompanied by the rupture of unspecific interactions that obscure the initial extension of the polyprotein (29). Therefore, only traces showing a clean initial retraction were analyzed. In addition, traces with detachment forces below 250 pN were excluded from the analysis in order to minimize the possibility of selecting polyprotein oligomers. The exact location of the surface is an important issue regarding the determination of initial extensions. In this work, the extension at a pushing force of 100 pN was set to 0. It was estimated that the same result with 5% maximum variation is found when the surface is located by projecting the force-extension trace at pushing forces between 250 and 500 pN to F = 0.
RESULTS
Spy0128 Is a Mechanically Inextensible Protein
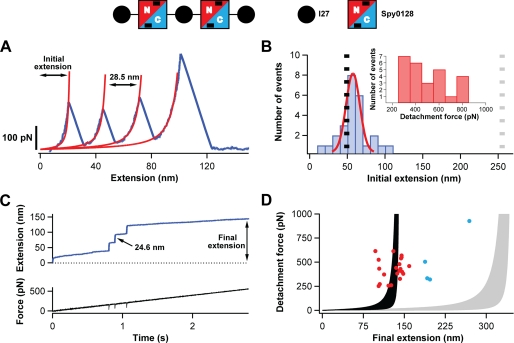
To inspect the mechanical response of Spy0128, a heteropolyprotein composed of two Spy0128 modules flanked by I27 protein domains was produced in Escherichia coli and purified by standard molecular biology techniques (supplemental Fig. S1). This protein was called (I27-Spy0128)2-I27 (Fig. 1). (I27-Spy0128)2-I27 was pulled at constant velocity using an AFM to obtain the corresponding force versus extension curves (18). I27 is an immunoglobin domain present in cardiac titin whose mechanical properties have been thoroughly studied by force spectroscopy (25). In force-extension curves, the unfolding of I27 domains is characterized by peaks around 200 pN with corresponding increments in contour length of ∼28.5 nm. Therefore, the unfolding events of I27 provide a unique fingerprint that serves to identify successful traces (28, 30). As the pilins are flanked between I27 modules, the unfolding of the three I27 domains present in the polyprotein is a signature of events where both Spy0128 have been subject to force. For this reason, only traces showing three I27 unfolding events were analyzed.
FIGURE 1.
Study of the extensibility of Spy0128 domains. A schematic of the protein construct used to probe the mechanical response of Spy0128 and (I27-Spy0128)2-I27 is shown. Two Spy0128 molecules (squares) were inserted between I27 domains (black circles). Spy0128 is composed of two domains (N terminus, red; C terminus, blue). Intramolecular isopeptide bonds are represented by black bars. A, typical force-extension trace (blue) showing three peaks corresponding to the unfolding of the three I27 domains in the protein construct. In red, fits to the worm-like chain model are shown. Even though the Spy0128 domains must have been subject to force, no unfolding peaks corresponding to Spy0128 are identified. B, histogram of the initial extension for traces with three I27 domains unfolding events (n = 26). The expected initial extensions if the two Spy0128 behaved as random coils (gray dotted line) or as mechanically stable domains (black dotted line) are shown. A Gaussian fit to the histogram is shown in red. Inset, histogram showing the detachment force in the force-extension experiments. C, top, typical force-ramp trace characterized by three steps marking the unfolding of the I27 modules. No steps corresponding to the Spy0128 domains were observed. Bottom, time course of the force experienced by the polyprotein. The unfolding events appear as negative spikes, reflecting the time response of the feedback system (33). D, scatter plot showing the final extension in the force-ramp experiments versus the detachment force (n = 25). The shaded regions represent the worm-like chain model of polymer elasticity (persistence lengths between 0.2 and 2.3 nm) if the Spy0128 domains remain folded (black) or, in contrast, unfold during the force-ramp protocol (gray) (28). The experimental points not following the general trend are colored in blue.
A typical force-extension curve for (I27-Spy0128)2-I27 is shown in Fig. 1A. The unfolding events corresponding to the three I27 modules are clearly detected as peaks at ∼200 pN. Fits of these peaks to the worm-like chain of polymer elasticity (27) render peak-to-peak contour length increments, ΔLc, of 28.5 nm. The last peak in the force-extension curve corresponds to the detachment of the polyprotein from the surface and/or the cantilever. No unfolding peaks assignable to the Spy0128 domains were generally observed in these force-extension experiments, even when the polyprotein was pulled at forces exceeding 800 pN (Fig. 1B, inset). Two scenarios are compatible with this result. Either Spy0128 behaves as a random coil that extends before the first I27 unfolding, or it remains folded throughout the experiment. To determine which description is accurate, the initial extension of the protein before the first I27 unfolding event was estimated by fitting the worm-like chain model to the first unfolding peak (Fig. 1, A and B). A similar approach has been found useful to study the mechanical properties of homopolypeptide chains (28). If the Spy0128 modules were extending before the first I27 unfolding event, the expected initial extension would be given by the sum of the length of three folded I27 modules (3 × 4.4 nm) (31), the handles and linkers routinely included in polyproteins used for force spectroscopy (23 amino acids × 0.4 nm/amino acid) (25, 32) and the length of the extended Spy0128 (2 × 291 amino acids × 0.4 nm/amino acid) for a total length of 255.2 nm (gray dashed line in Fig. 1B). In contrast, if Spy0128 remained folded, the initial extension would correspond to the extension of the folded I27 domains, the handles and linkers, the folded Spy0128 (2 × 8.2 nm, measured between the α-carbons of residues 30 and 307) and the unstructured regions in Spy0128 (residues 18 to 29 and 308; 2 × 13 amino acids × 0.4 nm/amino acid), i.e. 49.2 nm (black dashed line in Fig. 1B) (21). An initial extension of 52 ± 9 nm was obtained from a Gaussian fit to the experimental histogram in Fig. 1B, strongly suggesting that the Spy0128 modules do not unfold upon application of force. Only a small proportion of the traces showed initial extensions of 80–100 nm, probably corresponding to the unraveling of one Spy0128 domain. Those few traces may arise from rare misfolded Spy0128 domains produced in the E. coli expression system used to obtain the polyprotein.
In general, the force-extension approach allows the application of high forces during short periods of time. For instance, in Fig. 1A, the polyprotein is held at forces above 200 pN for only ∼15 ms. If the unfolding of Spy0128 was a slow process, its unfolding events may not be detected in the force-extension mode of the AFM. To further check the inextensibility of (I27-Spy0128)2-I27, the polyprotein was pulled in force-ramp mode (Fig. 1C). In this mode, the force is finely tuned by an electronic feedback system that controls the displacement of the piezoelectric positioner to attain the desired force. The unfolding events are now registered as steps in an extension versus time curve (33). The force was linearly varied at a rate of 200 pN/s for up to 5 s. A typical experimental trace is shown in Fig. 1C. In this particular experiment, the polyprotein was subject to forces above 200 pN during almost 2 s. As in the force-extension experiments, only traces with three steps of ∼24.5 nm marking the unfolding of the three I27 domains (34) were analyzed. No steps assignable to the unfolding of the Spy0128 modules were found (Fig. 1C). Additionally, in traces showing the I27 fingerprint, most final extensions are scattered around the values predicted by the worm-like chain model if the Spy0128 modules remained folded (Fig. 1D, red dots). As in force-extension experiments, only few traces showed longer final extensions that would be compatible with the extension of some misfolded Spy0128 domains (Fig. 1D, blue dots). Taking the force-extension and force-ramp results together, it is concluded that Spy0128 cannot be extended even when pulled at forces of 800 pN.
Intramolecular Isopeptide Bonds Are Responsible for the Mechanical Inextensibility of Spy0128
The presence of one intramolecular isopeptide bond in both domains of Spy0128 has been proposed by x-ray crystallography and mass spectrometry (21). Mutations abolishing the formation of these isopeptide bonds have been shown to decrease both the resistance to proteases and the thermal stability of Spy0128 (21, 22). From a mechanical point of view, their location within the structure of the Spy0128 domains is striking, for they would connect the first and last β-strands in both β-sandwiches (12). Taking into account the mechanical strength of covalent bonds, in such a configuration, an axial force would be transmitted exclusively through the isopeptide bonds and both β-strands, leaving the rest of the domain unaffected at forces presumably as high as 2000 pN (35). Using the same design described above for wild-type Spy0128, polyproteins for Spy0128 E117A and E258A were produced (supplemental Fig. S1). These two mutations have been shown to abrogate the formation of the intramolecular isopeptide bonds (21, 22). As above, only traces with three I27 unfolding events were selected. The results showed that, contrary to wild-type, both Spy0128 mutants unfolded upon the application of force, as indicated by the appearance of two additional peaks in the force-extension traces (Figs. 2A and 3A). These results agree with the mutants retaining the overall fold of wild-type Spy0128, which has also been shown by x-ray crystallography (21, 22). More importantly, they show that the Spy0128 fold is mechanically stable per se, with unfolding forces of 172 ± 33 pN for the N-terminal domain and 250 ± 96 pN for the C-terminal domain (Figs. 2B and 3B, Table 1). For both mutant polyproteins, the initial extension before the first unfolding event shows that the domain still containing the isopeptide bond remains inextensible (supplemental Fig. S2).
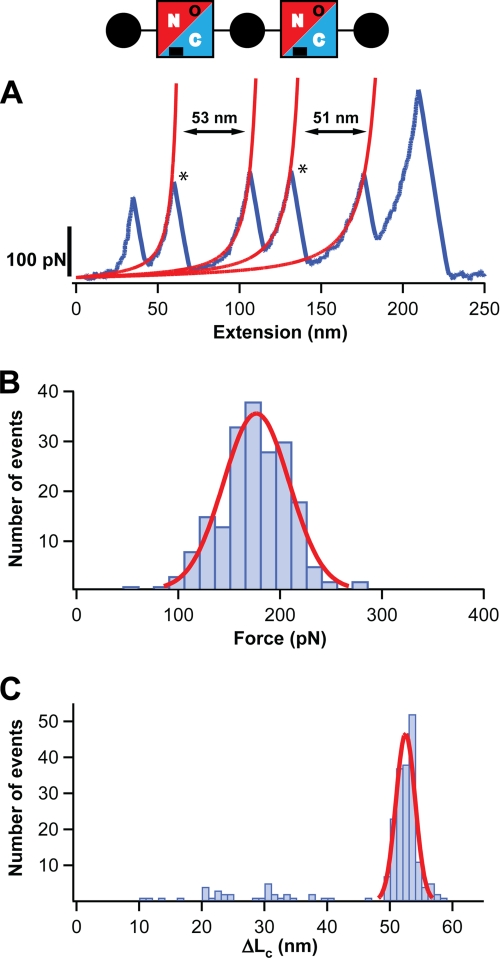
FIGURE 2.
The N-terminal domain of Spy0128 E117A extends upon the application of force. A diagram of the protein construct employed, (I27-Spy0128-E117A)2-I27, is shown (see also legend to Fig. 1). The mutation E117A abolishes the formation of the isopeptide bond at the N-terminal domain of Spy0128 (in the diagram, this is denoted by an open circle). A, typical force-extension trace for (I27-Spy0128-E117A)2-I27 is shown in blue. The peaks marking the unfolding of the Spy0128 N-terminal domains in the polyprotein are labeled with an asterisk. The increment in contour length after the Spy0128 unfolding events is calculated from the worm-like chain fits shown in red. B, histogram of the unfolding forces for the N-terminal domain in Spy0128 E117A (n = 197). The solid line corresponds to a Gaussian fit of the experimental data. C, histogram of the ΔLc associated with the unfolding of the N-terminal domain in Spy0128 E117A (n = 214). A Gaussian fit for the data above 45 nm is shown (solid line).
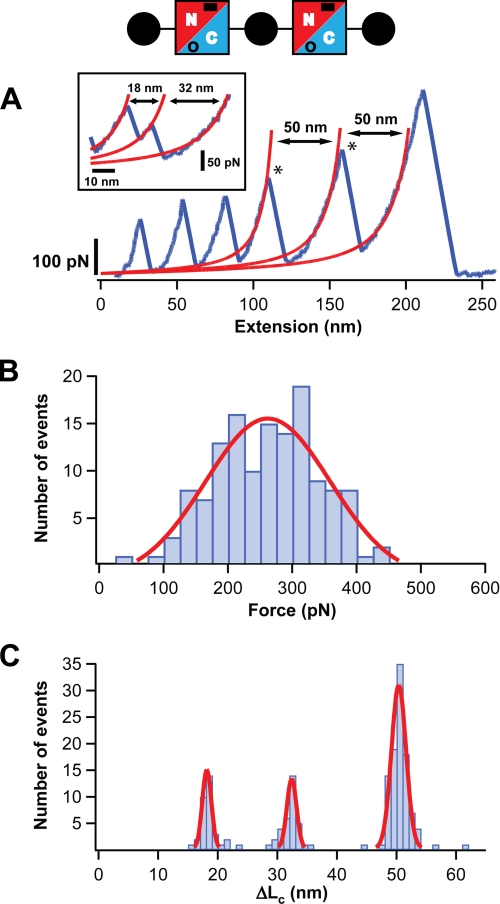
FIGURE 3.
The C-terminal domain of Spy0128 E258A extends upon the application of force. A diagram of the protein construct employed, (I27-Spy0128-E258A)2-I27, is shown (see also legend to Fig. 1). The mutation E258A abolishes the formation of the isopeptide bond at the C-terminal domain of Spy0128 (in the diagram, this is denoted by an open circle). A, typical force-extension trace for (I27-Spy0128-E258A)2-I27 is shown in blue. The peaks marking the unfolding of the Spy0128 C-terminal domains in the polyprotein are labeled with an asterisk. The increment in contour length after the Spy0128 unfolding events is calculated from the worm-like chain fits shown in red. Inset: the unfolding of the C-terminal domain through an intermediate is shown. B, histogram of the unfolding forces for the C-terminal domain in Spy0128 E258A (n = 135). The solid line corresponds to a Gaussian fit of the experimental data. C, histogram of the ΔLc associated with the unfolding of the C-terminal domain in Spy0128 E258A (n = 169). Solid lines are Gaussian fits to the data.
TABLE 1.
Mechanical properties of mutant Spy0128 domains lacking isopeptide bonds
The experimental histograms were fitted to Gaussian curves (Figs. 2 and 3). The mean value is presented in the table. Errors are given by the standard deviation of the Gaussian fit.
| Construct | ΔLc,a I27 | Fu,b I27 | ΔLc, Spy0128, Nt | Fu, Spy0128, Nt | ΔLc, Spy0128, Ct | Fu, Spy0128, Ct |
|---|---|---|---|---|---|---|
| nm | pN | nm | pN | nm | pN | |
| I278 (from Ref. 25) | 28.4 ± 0.3 | 204 ± 26 | ||||
| (I27-Spy0128-E117A)2-I27 | 28.7 ± 0.7 | 201 ± 29 | 52 ± 2 | 172 ± 33 | ||
| (I27-Spy0128-E258A)2-I27 | 29.2 ± 0.8 | 204 ± 30 | 50 ± 2 | 250 ± 96 |
a ΔLc, increment in contour length.
b Fu, unfolding force.
The ΔLc associated with the unfolding of Spy0128 domains was calculated from worm-like chain fits to the force-extension traces of mutant Spy0128 polyproteins (Figs. 2A and 3A). Histograms were built and ΔLc values were estimated using Gaussian fits (Figs. 2C and 3C, Table 1). For the N-terminal domain (E117A mutant), it was found that ΔLc = 52 ± 2 nm. In the case of the C-terminal domain (E258A mutant), a value of 50 ± 2 nm was determined. The N-terminal domain of Spy0128 comprises residues from 30 to 171, and the distance between them is 3.6 nm. Therefore, the expected ΔLc is given by (142 amino acids × 0.4 nm/amino acid, minus 3.6 nm), i.e. 53.2 nm, in close agreement with the experimental value. A similar situation is found for the C-terminal domain, which spans residues 173–307 and has a folded size of 4 nm. In this case, the expected ΔLc is 50 nm, which matches the experimental value. These calculations suggest that, when pulled, mutant Spy0128 domains do not have to extend appreciably to reach the transition state. On the contrary, the initial structure is almost coincident with the mechanical unfolding transition state. An inspection of the three-dimensional structure of Spy0128 corroborates this interpretation (Fig. 4). It has been described before that the high mechanical resistance of β-sheet proteins usually derives from the simultaneous rupture of several hydrogen bonds between neighboring parallel β-strands (36, 37). In Fig. 4A, it can be observed that six hydrogen bonds connect the first and last β-strands in the N-terminal domain. These strands are in parallel configuration. Up to nine hydrogen bonds are found at the equivalent location in the C-terminal domain. Presumably, the simultaneous rupture of those hydrogen bonds leads to the unfolding of the mutant domains, which forms the basis of the theoretical ΔLc calculated above. It is interesting to note that the C-terminal domain, with 9 hydrogen bonds between the parallel β-strands, is significantly more stable than the N-terminal domain, with only 6 hydrogen bonds (Fig. 4A and Table 1). In summary, the results presented here strongly suggest that the location of the intramolecular isopeptide bonds specifically locks the unfolding mechanical transition state of both domains in Spy0128, preventing any extension of the domains to happen and providing the Spy0128 molecules with covalent-like mechanical resilience.
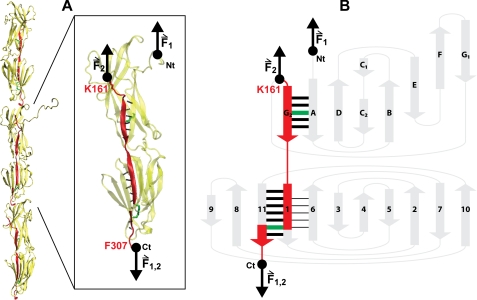
FIGURE 4.
Mechanical architecture of Spy0128. A, model of the pili in S. pyogenes showing the spatial arrangement of three Spy0128 units (21). The structure of one Spy0128 monomer (PDB code: 3B2M) is enlarged. The hydrogen bonds involved in the mechanical stabilization of the domains are shown as dotted black lines. The intramolecular isopeptide bonds (Lys-36—Asn-168; Lys-179—Asn-303) are depicted in green. Pilin monomers homopolymerize to produce the pilus shaft through intermolecular isopeptide bonds between Lys-161 and the carboxyl group of Thr-311 (21). As a consequence of both the inter- and intramolecular isopeptide bonds, an axial force applied to the pilus is transmitted along the chain colored in red (instead of Thr-311, the last residue modeled in the crystal structure of Spy0128, Phe-307, is shown). The pulling directions in the polyproteins used in this work (F1) and in native pili (F2) are indicated by black arrows. VMD was used to prepare the figure (49). B, topology diagram of Spy0128. The main mechanical transitions states are represented by thick black bars. The mechanical transition state giving rise to the intermediate captured in the mechanical unfolding of Spy0128 E258A is represented by black lines between strands 1 and 6. Force pathways and intramolecular isopeptide bonds are colored as in A. The N and C termini are indicated in both panels.
Intermediates in the Mechanical Unfolding of Spy0128 Mutant Variants
During the mechanical unfolding of Spy0128 E117A and E258A, the presence of intermediates was observed (Figs. 2C and 3, A and C). These intermediates were identified as two consecutive peaks in the force-extension traces with total ΔLc around 50 nm (Fig. 3A, inset). For the N-terminal domain, the frequency of appearance of unfolding intermediates was low (8%), and no clear pattern for the distribution of ΔLc was found (Fig. 2C). In contrast, a better-defined unfolding intermediate was identified for the C-terminal domain in 25% of the events, showing ΔLc1 = 18 ± 1 nm and ΔLc2 = 32 ± 1 nm (Fig. 3C). Its unfolding force is moderate, 105 ± 16 pN (supplemental Fig. S3). The analysis of the ΔLc values suggests that this intermediate may arise from the limited unfolding of the C-terminal domain from residues ∼249 to 307. If that is the case, the hydrogen bonds between strands 1 and 6 would be responsible for the mechanical stability of the intermediate (Fig. 4B). Those β-strands are antiparallel, which would explain the lower force needed to break this second set of hydrogen bonds (29).
The Formation of the Intramolecular Isopeptide Bonds Is Not 100% Efficient
Single molecule experiments offer the opportunity to study rare events that are extremely hard to detect in bulk determinations. Previous bulk reports assumed that all wild-type Spy0128 domains have intramolecular isopeptide bonds (21, 22). If that was the case, and considering the high mechanical strength of a covalent bond (35), no mechanical unfolding of wild-type Spy0128 domains should ever be detected. However, such unfolding events were found occasionally (supplemental Fig. S4). For wild-type Spy0128, the appearance of a peak showing unfolding forces around 200 pN and ΔL ∼50 nm (the same values found for the unfolding of mutant Spy0128 domains) marked the unfolding of a wild-type domain (supplemental Fig. S4A). In the case of both E117A and E258A mutants, the presence of three peaks with similar unfolding parameters suggested that one of them was due to one of the two remaining wild-type domains (supplemental Fig. S4, B and C). Overall, the proportion of these rare wild-type unfolding events was low (around 3%).
DISCUSSION
In the last few years, there have been a growing number of reports on the structural organization of pili from Gram-positive bacteria. The cornerstone was laid by the work of Ton-That and Schneewind in 2003, which demonstrated that pili in Corynebacterium diphtheriae were assembled by the covalent association of three different pilins via isopeptide bonds (13). It was found that one of the pilins formed the pilus shaft, whereas two minor pilins were located at regular intervals along the shaft, or at the tip of the pilus. Similar organizations have been found in a number of other pathogenic Gram-positive bacteria, including S. pyogenes (8, 38–40). Another breakthrough in the field was the unexpected discovery of intramolecular isopeptide bonds in pilins using x-ray crystallography and mass spectrometry (21, 23, 41, 42). Their strategic location led to the hypothesis that their primary role may be mechanical (Fig. 4) (12, 22, 23). However, those previous reports were unable to test the extent of intramolecular isopeptide bond formation, and suffered from the drawback that isopeptide bonds may well be formed as an artifact during crystallization, as reported for other covalent bonds in proteins (24). In this work, the mechanical characterization of Spy0128, the major pilin from S. pyogenes (M1 serotype), univocally demonstrates that intramolecular isopeptide bonds are present in the vast majority of soluble Spy0128 proteins. Remarkably, those isopeptide bonds confer on the pilin modules the mechanical resilience characteristic of covalent bonds.
Using single molecule force spectroscopy, it is here reported that Spy0128 is mechanically inextensible (Fig. 1). Therefore, Spy0128 is the most mechanically stable protein characterized so far (43). However, when the formation of the isopeptide bonds is prevented, both domains in Spy0128 show the typical behavior of other force-bearing proteins, with unfolding forces around 200 pN (Figs. 2 and 3). This suggests that pilins would not need intramolecular isopeptide bonds if they only had to resist the same range of mechanical stresses as other proteins such as immunoglobin domains in titin or fibronectin. In other words, the presence of isopeptide bonds within otherwise mechanically stable domains points to the idea that functional pili must possess a stringent resistance to mechanical perturbations, and that these perturbations might be higher than those experienced by other mechanically stable proteins.
The results presented in this work highlight the significance of the covalent connectivity in pilins to achieve a high mechanical stability in pili from Gram-positive bacteria. In the polyproteins used, force is applied from the N and C termini of the protein (Fig. 4). However, this is not the actual pulling direction experienced by Spy0128 as the major constituent of the pilus. In pili, Spy0128 is polymerized by a sortase-catalyzed reaction that binds the C-terminal carboxyl group of one monomer to the Lys-161 Nζ of the next monomer (13, 21). Thus, the force is transmitted through residues 161–179, then through the C-terminal isopeptide bond to residue 303 and finally to the next monomer (red in Fig. 4). Remarkably, the N-terminal intramolecular isopeptide bond does not participate in this pathway (Fig. 4). It has recently been described that only two out of the three domains that constitute the major pilin in C. diphtheriae, SpaA, have intramolecular isopeptide bonds equivalent to those in Spy0128. It is to note that, given the covalent architecture of C. diphtheriae pili, the domain devoid of isopeptide bonds in SpaA is also the only one that does not sense any axial force in the pilus (23). This suggests that intramolecular isopeptide bonds may only appear when their mechanical role is needed. Considering how force is transmitted through S. pyogenes pili, the mechanical role of the N-terminal isopeptide bond in Spy0128 remains unclear.
Taking into account the absence of quaternary structure in the pili from Gram-positive bacteria (4, 12, 17, 21, 23) and the inextensibility of the constituent pilins (Fig. 1), it can be hypothesized that pili in Gram-positive bacteria are the first reported example of an inextensible modular protein. Then, pili in Gram-positive bacteria would be much stiffer than their Gram-negative counterparts, which have been shown to be elastic and able to reversibly extend several times their resting length (44–47). The different mechanical response of pili from Gram-positive and Gram-negative bacteria suggests that they may use different strategies to achieve a sustained attachment to their target cells (46, 48).
The establishment of the intramolecular isopeptide bonds seems to be autocatalytic (21). However, in a small proportion of cases (∼3%), Spy0128 domains were found to unfold under force (supplemental Fig. S4). Such a small proportion would probably have gone unnoticed for bulk techniques such as x-ray crystallography. Most probably, in those rare events, the isopeptide bonds failed to form, allowing the affected module to readily extend under force. Then, it is possible to envisage a pharmacological intervention aimed to increase the number of such failures. This therapeutic approach may render pili in S. pyogenes mechanically inefficient and help fight against infections by this dangerous human pathogen. These observations could be extended to other pathogenic Gram-positive bacteria showing similar pili organizations. As demonstrated in this work, single molecule force spectroscopy emerges as an optimal tool to evaluate the extent of intramolecular isopeptide bond formation in pili.
Supplementary Material
Acknowledgments
We thank Pallav Kosuri, Sergi Garcia-Manyes, and the rest of the members of the Fernandez laboratory for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants HL66030 and HL61228 (to J. M. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- AFM
- atomic force microscopy.
REFERENCES
- 1.Cunningham M. W. (2008) Adv. Exp. Med. Biol. 609, 29–42 [DOI] [PubMed] [Google Scholar]
- 2.Manetti A. G., Zingaretti C., Falugi F., Capo S., Bombaci M., Bagnoli F., Gambellini G., Bensi G., Mora M., Edwards A. M., Musser J. M., Graviss E. A., Telford J. L., Grandi G., Margarit I. (2007) Mol. Microbiol. 64, 968–983 [DOI] [PubMed] [Google Scholar]
- 3.Abbot E. L., Smith W. D., Siou G. P., Chiriboga C., Smith R. J., Wilson J. A., Hirst B. H., Kehoe M. A. (2007) Cell Microbiol. 9, 1822–1833 [DOI] [PubMed] [Google Scholar]
- 4.Proft T., Baker E. N. (2009) Cell Mol. Life Sci. 66, 613–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craig L., Pique M. E., Tainer J. A. (2004) Nat. Rev. Microbiol. 2, 363–378 [DOI] [PubMed] [Google Scholar]
- 6.Mandlik A., Swierczynski A., Das A., Ton-That H. (2008) Trends Microbiol. 16, 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nallapareddy S. R., Singh K. V., Sillanpää J., Garsin D. A., Höök M., Erlandsen S. L., Murray B. E. (2006) J. Clin. Invest. 116, 2799–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mora M., Bensi G., Capo S., Falugi F., Zingaretti C., Manetti A. G., Maggi T., Taddei A. R., Grandi G., Telford J. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15641–15646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Budzik J. M., Schneewind O. (2006) J. Clin. Invest. 116, 2582–2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maier B., Potter L., So M., Long C. D., Seifert H. S., Sheetz M. P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16012–16017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biais N., Ladoux B., Higashi D., So M., Sheetz M. (2008) PLoS Biol. 6, e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeates T. O., Clubb R. T. (2007) Science 318, 1558–1559 [DOI] [PubMed] [Google Scholar]
- 13.Ton-That H., Schneewind O. (2003) Mol. Microbiol. 50, 1429–1438 [DOI] [PubMed] [Google Scholar]
- 14.Scott J. R., Zähner D. (2006) Mol. Microbiol. 62, 320–330 [DOI] [PubMed] [Google Scholar]
- 15.Telford J. L., Barocchi M. A., Margarit I., Rappuoli R., Grandi G. (2006) Nat. Rev. Microbiol. 4, 509–519 [DOI] [PubMed] [Google Scholar]
- 16.Swaminathan A., Mandlik A., Swierczynski A., Gaspar A., Das A., Ton-That H. (2007) Mol. Microbiol. 66, 961–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hilleringmann M., Ringler P., Müller S. A., De Angelis G., Rappuoli R., Ferlenghi I., Engel A. (2009) EMBO J. 28, 3921–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rief M., Gautel M., Oesterhelt F., Fernandez J. M., Gaub H. E. (1997) Science 276, 1109–1112 [DOI] [PubMed] [Google Scholar]
- 19.Oberhauser A. F., Badilla-Fernandez C., Carrion-Vazquez M., Fernandez J. M. (2002) J. Mol. Biol. 319, 433–447 [DOI] [PubMed] [Google Scholar]
- 20.Li H., Linke W. A., Oberhauser A. F., Carrion-Vazquez M., Kerkvliet J. G., Lu H., Marszalek P. E., Fernandez J. M. (2002) Nature 418, 998–1002 [DOI] [PubMed] [Google Scholar]
- 21.Kang H. J., Coulibaly F., Clow F., Proft T., Baker E. N. (2007) Science 318, 1625–1628 [DOI] [PubMed] [Google Scholar]
- 22.Kang H. J., Baker E. N. (2009) J. Biol. Chem. 284, 20729–20737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang H. J., Paterson N. G., Gaspar A. H., Ton-That H., Baker E. N. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16967–16971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H., Fernandez J. M. (2003) J. Mol. Biol. 334, 75–86 [DOI] [PubMed] [Google Scholar]
- 25.Carrion-Vazquez M., Oberhauser A. F., Fowler S. B., Marszalek P. E., Broedel S. E., Clarke J., Fernandez J. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3694–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Florin E. L., Rief M., Lehmann H., Ludwig M., Dornmair C., Moy V. T., Gaub H. E. (1995) Biosens. Bioelectron. 10, 895–901 [Google Scholar]
- 27.Bustamante C., Marko J. F., Siggia E. D., Smith S. (1994) Science 265, 1599–1600 [DOI] [PubMed] [Google Scholar]
- 28.Dougan L., Li J., Badilla C. L., Berne B. J., Fernandez J. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12605–12610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carrion-Vazquez M., Oberhauser A. F., Fisher T. E., Marszalek P. E., Li H., Fernandez J. M. (2000) Prog. Biophys. Mol. Biol. 74, 63–91 [DOI] [PubMed] [Google Scholar]
- 30.Perez-Jimenez R., Garcia-Manyes S., Ainavarapu S. R., Fernandez J. M. (2006) J. Biol. Chem. 281, 40010–40014 [DOI] [PubMed] [Google Scholar]
- 31.Improta S., Krueger J. K., Gautel M., Atkinson R. A., Lefèvre J. F., Moulton S., Trewhella J., Pastore A. (1998) J. Mol. Biol. 284, 761–777 [DOI] [PubMed] [Google Scholar]
- 32.Ainavarapu S. R., Brujic J., Huang H. H., Wiita A. P., Lu H., Li L., Walther K. A., Carrion-Vazquez M., Li H., Fernandez J. M. (2007) Biophys. J. 92, 225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oberhauser A. F., Hansma P. K., Carrion-Vazquez M., Fernandez J. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 468–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Manyes S., Brujić J., Badilla C. L., Fernández J. M. (2007) Biophys. J. 93, 2436–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grandbois M., Beyer M., Rief M., Clausen-Schaumann H., Gaub H. E. (1999) Science 283, 1727–1730 [DOI] [PubMed] [Google Scholar]
- 36.Marszalek P. E., Lu H., Li H., Carrion-Vazquez M., Oberhauser A. F., Schulten K., Fernandez J. M. (1999) Nature 402, 100–103 [DOI] [PubMed] [Google Scholar]
- 37.Forman J. R., Clarke J. (2007) Curr. Opin. Struct. Biol. 17, 58–66 [DOI] [PubMed] [Google Scholar]
- 38.Lauer P., Rinaudo C. D., Soriani M., Margarit I., Maione D., Rosini R., Taddei A. R., Mora M., Rappuoli R., Grandi G., Telford J. L. (2005) Science 309, 105. [DOI] [PubMed] [Google Scholar]
- 39.LeMieux J., Hava D. L., Basset A., Camilli A. (2006) Infect Immun. 74, 2453–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mishra A., Das A., Cisar J. O., Ton-That H. (2007) J. Bacteriol. 189, 3156–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Budzik J. M., Marraffini L. A., Souda P., Whitelegge J. P., Faull K. F., Schneewind O. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10215–10220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Budzik J. M., Poor C. B., Faull K. F., Whitelegge J. P., He C., Schneewind O. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 19992–19997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sikora M., Sulkowska J. I., Cieplak M. (2009) PLoS Comput. Biol. 5, e1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jass J., Schedin S., Fällman E., Ohlsson J., Nilsson U. J., Uhlin B. E., Axner O. (2004) Biophys. J. 87, 4271–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fällman E., Schedin S., Jass J., Uhlin B. E., Axner O. (2005) EMBO Rep. 6, 52–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller E., Garcia T., Hultgren S., Oberhauser A. F. (2006) Biophys. J. 91, 3848–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Touhami A., Jericho M. H., Boyd J. M., Beveridge T. J. (2006) J. Bacteriol. 188, 370–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oberhauser A. F., Marszalek P. E., Erickson H. P., Fernandez J. M. (1998) Nature 393, 181–185 [DOI] [PubMed] [Google Scholar]
- 49.Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.