Abstract
The UAF1 (Usp1-associated factor 1) protein binds and stimulates three deubiquitinating enzymes: USP1, USP12, and USP46. Although the USP1·UAF1 complex is required for regulation of the Fanconi anemia (FA) DNA repair pathway, less is known about the USP12·UAF1 and the USP46·UAF1 complexes. To understand further the nature of the USP12 and USP46 complexes, we attempted to identify proteins that interact with the USP12 and USP46 deubiquitinating enzyme complexes. We identified WDR20, a WD40-repeat containing protein, as a common binding partner of UAF1, USP12, and USP46. Further analysis showed that WDR20 associates exclusively with USP12 and USP46, not with USP1. Furthermore, we demonstrate the purification of a ternary USP12·UAF1·WDR20 complex. Interestingly, and consistent with the binding assays, WDR20 stimulated the enzymatic activity of USP12·UAF1, but not of USP1·UAF1. Consistent with our previous report that USP12 and USP46 do not regulate the FA pathway, small interference RNA-mediated depletion of WDR20 protein did not affect the FA pathway or DNA damage responses. We provide a model in which WDR20 serves as a stimulatory subunit for preserving and regulating the activity of the subset of the UAF1·USP complexes.
Keywords: Deubiquitination, Enzymes, Protein-Protein Interactions, Ubiquitin, Ubiquitination, WD40-repeat
Introduction
Balanced ubiquitination and deubiquitination regulates numerous cellular processes. Increasing reports suggest that deubiquitination of proteins, the reversal process of ubiquitination, is as an a important step as ubiquitination, for specific regulation of particular cellular pathways (1, 2). For instance, deubiquitinating enzymes (DUBs)6 are critical regulators of the p53/mdm2 (3), NFκB signaling (4, 5), and the Fanconi anemia (FA) DNA repair pathways (6), by directly removing the ubiquitin moieties from their substrates. Deubiquitination either rescues the proteins from proteosome-mediated degradation or alters the activities of the proteins. For instance, USP7·HAUSP regulates the dynamics of p53-dependent pathways by directly deubiquitinating p53 and Mdm2 (3). CYLD suppresses the NFκB pathway by deubiquitinating Lys63-linked polyubiquitination of NEMO, a regulatory subunit for IκB kinase and TRAF2–6 E3 ligases. USP1 deubiquitinates monoubiquitinated FANCD2, which is an important regulator of the FA pathway, a step required for the completion of this DNA repair pathway (6). Inactivation of the DUBs result in abnormal cellular phenotypes in various organism settings (7–10), further suggesting that deubiquitination is a critical step in a variety of biological processes.
There are ∼95 putative DUBs encoded by human cells, and they can be divided into five subfamilies (2, 11). Four families belong to cysteine proteases, including ubiquitin C-terminal hydrolases (Uchs), ubiquitin-specific proteases (USPs), otubain proteases (OTUs), and Josephine domain proteases (MJDs), with the fifth one being JAB1/MPN/Mov34 (JAMM) metalloproteases.
Although the identities of target substrates and biological functions are critical questions for the majority of the DUBs, another important issue is the mechanism of regulation of DUB activity. There are only a few cases so far that provide such mechanistic insight. For instance, the activity of Uch37 is regulated by its association with the 19 S proteasome subunits (12–14). In addition, we have reported that the activity of USP1 is positively regulated by UAF1, a stoichiometric binding partner of USP1, which contains WD40-repeat motifs (15). We subsequently reported that two closely related DUBs, USP12 and USP46, are also stimulated by UAF1, although the level of stimulation was less than for USP1 (16). Based on these observations, we hypothesized that other WD40-repeat proteins might also be associated with DUBs, in general, to regulate their activities in cells.
Here, we report identification and characterization of WDR20, a previously uncharacterized WD40-repeat-containing protein, as a stimulatory subunit of the USP12·UAF1 DUB complex. Our data suggest that the modes of regulation for deubiquitinating enzymes in the cell are more complex and diverse and reinforce our hypothesis that WD40-repeat protein-mediated regulation of DUB activities may be more widespread in the cell.
EXPERIMENTAL PROCEDURES
Cell Lines, Plasmids, siRNAs, and Antibodies
HeLa and 293T cells were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 15% fetal bovine serum (Invitrogen) and penicillin/streptomycin glutamine (Invitrogen). For the selection of short hairpin RNA cassettes, 1–3 μg/ml puromycin was added in the medium. HeLa cell lines stably expressing N-terminal tandem FLAG-HA USP1, UAF1, USP12, USP46, and WDR20 were generated using pOZ-FH-N system, as described previously (17). siRNAs against UAF1 and WDR20 were synthesized from Invitrogen with the target sequences as following: UAF1, 5′-AAUCAGCACAAGCAAGAUCCAUAUA-3′; WDR20 1, 5′-AUGACAAGUAGGCUGUGUUCCUUUG-3′; and WDR20 2, 5′-UUAAGGAGAGGGUUCCUCGUGGAUU-3′. Antibodies against UAF1, USP1, and USP12 were generated from rabbit as described previously (15, 16). Commercial antibodies were purchased as following: anti-WDR20 and anti-phospho-Chk1 (Ser317) polyclonal antibodies (Bethyl Laboratory), anti-FLAG and anti-tubulin monoclonal antibodies (Sigma), anti-FANCD2 and anti-proliferating cell nuclear antigen monoclonal antibodies (Santa Cruz Biotechnology).
Immunoprecipitation Assay
HeLa cells stably expressing exogenous epitope-tagged proteins were harvested, washed with phosphate-buffered saline, and lysed with a lysis buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 0.5% Nonidet P-40). The lysates were centrifuged at 14,000 rpm for 15 min, and the supernatants were precleared using Sepharose (Sigma) for 30 min. The precleared lysates were subjected to incubation with M2-agarose (Sigma) for 3–4 h, and the beads were washed extensively with the lysis buffer. The bound proteins were eluted by adding 4 × SDS running buffer and analyzed by SDS-PAGE and Western blotting.
Purification of Trimeric Complex from Sf9 Cells
N-terminal His-tagged USP12 was expressed using pFASTBac-HTa vector (Invitrogen), and UAF1 and FLAG-WDR20 were expressed using pFASTBac-1 vector (Invitrogen). Each recombinant virus was produced by transfecting the corresponding bacmid to Sf9 cells. To produce the trimeric complex, all three viruses were used to co-infect fresh Sf9 cells. 72 h after infection, cells were harvested, washed with phosphate-buffered saline, and lysed with a lysis buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 20 mm imidazole, 1% glycerol). The lysate was subjected to incubation with nickel-nitrilotriacetic acid-Sepharose (Qiagen) for 2 h followed by extensive washing of the beads with the lysis buffer. The bound proteins were eluted with an elution buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 250 mm imidazole, 1% glycerol). The eluted proteins were subjected to incubation with M2-agarose (Sigma) for 4 h followed by extensive washing, and the bound proteins were eluted with 3 × FLAG peptides (Sigma). The eluted proteins were run on 4–12% gradient SDS-polyacrylamide gel and analyzed by Coomassie Blue staining.
GST Pulldown Assay
Purifications of USP12 and UAF1 from Sf9 cells were described previously (16). WDR20 was purified from Escherichia coli strain RossettaTM 2(DE3) (Calbiochem). In brief, expression of GST-WDR20 fusion protein from the bacterial cells was induced by 0.9 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h, and the cells were lysed by sonication in a lysis buffer (50 mm Tris, pH 7.5, 1% Nonidet P-40, 150 mm NaCl), before preclearing the lysates by high speed centrifugation (14,000 rpm) for 15 min. The lysates were incubated with glutathione-Sepharose (GE Healthcare) for 2 h followed by extensive washing. The GST-WDR20 and GST proteins on beads were incubated with different combinations of USP12 and UAF1 for 4 h followed by extensive washing. The binding reaction was stopped by adding 4 × SDS loading buffer and analyzed by Western blot analysis.
In Vitro Deubiquitination Assay
The in vitro enzymatic assays were performed using purified proteins from Sf9 cells as described previously (15). WDR20 proteins were purified from bacteria as described above, except that the GST-WDR20 proteins on beads were cut off the GST tag using Precision protease (GE Healthcare) for overnight at 4 °C. The assays used ubiquitin-AMC (Ub-7-amido-4-methylcoumarin; Boston Biochem) as substrates, and the reaction was done in a reaction buffer (20 mm HEPES-KOH, pH 7.8, 20 mm NaCl, 0.1 mg/ml ovalbumin, 0.5 mm EDTA, 10 mm dithiothreitol). The fluorescence was measured by FluoStar Galaxy Fluorometer (BMG Labtech). For the Ub-vinylsulfone (VS) assays, the immunoprecipitated proteins were eluted using 3×xFLAG peptides (Sigma), and the eluted fractions were incubated with final 0.5 μm Ub-VS for 1 h at 37 °C.
RESULTS
WDR20 Selectively Interacts with USP12 and USP46, but Not with USP1
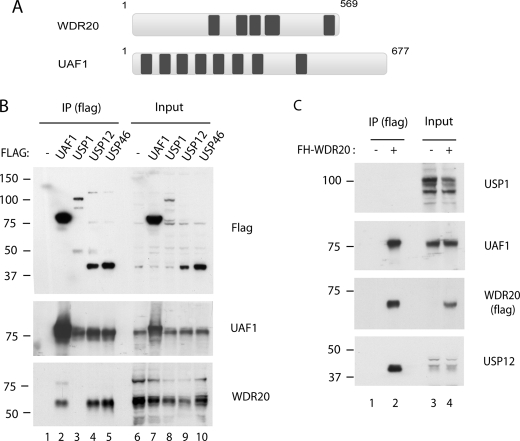
To understand further the nature of UAF1-associated DUBs, we performed the immunoprecipitation of UAF1, USP12, and USP46 proteins and identified the associated proteins. Mass spectrometry analysis revealed a WD40-repeat-containing protein, WDR20, as a common binding partner of UAF1, USP12, and USP46 (data not shown). This observation independently validated a recently published work in which WDR20 was identified as binding partner of the respective DUBs (18). WDR20 is a 569-amino acid protein harboring five recognizable WD40-repeat motifs (Fig. 1A). Sequence alignment among different species showed that it is well conserved from human to yeast Schizosaccharomyces pombe, with Saccharomyces cerevisiae lacking an apparent homolog. To validate this analysis, we performed immunoprecipitation and Western blot analysis using HeLa cell lines stably expressing epitope-tagged proteins. Interestingly, WDR20 specifically interacted with UAF1, USP12, and USP46 (Fig. 1B, lanes 2, 4, and 5), but not with USP1 (lane 3). In an inverse experiment, UAF1 and USP12 (and possibly USP46) proteins were co-purified, but not USP1, in the WDR20-immunoprecipitated complexes (Fig. 1C). Together, these results demonstrate that WDR20 selectively interacts with the USP12·UAF1 and the USP46·UAF1 subcomplexes, but not with the USP1·UAF1 subcomplex.
FIGURE 1.
WDR20 interacts with UAF1, USP12, and USP46, but not with USP1. A, schematic of the domain distribution of WDR20 and UAF1 is shown. Dark boxes indicate the WD40-repeat motifs. B, anti-FLAG immunoprecipitations (IP) were performed using the lysates from HeLa cells stably expressing each FLAG-HA tandem-tagged proteins, followed by Western blots using anti-FLAG, anti-UAF1, and anti-WDR20 antibodies. C, anti-FLAG immunoprecipitations were performed from HeLa cells stably expressing FLAG-HA-tagged WDR20 followed by Western blotting using anti-UAF1, anti-USP1, anti-USP12, and anti-FLAG antibodies. Note that there are two forms of USP1, with the full-length (long form) and C-terminally cleaved product (short form) of USP1, which is also detected by our anti-USP1 antibody, as described previously (21).
The USP12·UAF1 Complex Interacts Directly with WDR20
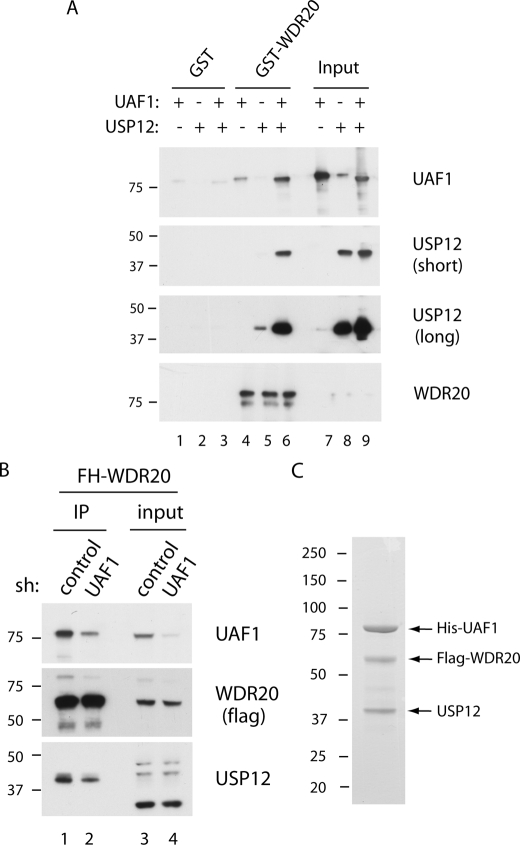
To understand the nature of interactions among WDR20, UAF1, and USP12, we performed in vitro pulldown assays using purified proteins (Fig. 2A). For this study, GST-fused WDR20 was purified from bacteria and UAF1, or the USP12·UAF1 complex was purified from Sf9 cells, as described previously (16). In the pulldown assay using GST-WDR20, minor interactions of UAF1 and USP12 with the WDR20, respectively, were detected (lanes 4 and 5). However, when the preformed USP12·UAF1 complex was incubated with GST-WDR20, the interaction was enhanced significantly (lane 6), suggesting that WDR20 binds more efficiently to the DUB complex than to the individual subunits.
FIGURE 2.
WDR20 directly interacts with the USP12·UAF1 complex. A, GST pulldown assays were performed using purified GST, GST-WDR20, USP12, and UAF1, as described under “Experimental Procedures.” In brief, individual USP12, UAF1, or preformed USP12·UAF1 complex was incubated with either GST or GST-WDR20, which are prebound on glutathione-agarose. After 2 h, the beads were washed, and the bound proteins were analyzed by Western blotting using anti-UAF1, anti-USP12, and anti-WDR20 antibodies. B, FLAG-HA-tagged WDR20 (FH-WDR20) proteins were immunoprecipitated (IP) using anti-FLAG-agarose from the HeLa cells in which either control or UAF1 was stably knocked down by short hairpin (sh) RNA. C, Coomassie stain for a trimeric complex of USP12, His-UAF1, and FLAG-WDR20 is shown. Individual viruses were used to co-infect SF9 cells, and cells were harvested 48 h after infection. Tandem-affinity purifications using nickel-nitrilotriacetic acid beads followed by anti-FLAG beads were performed, and the bound proteins were eluted off the beads using 3×FLAG peptides, as described under “Experimental Procedures.”
To investigate further whether UAF1 is required for the interaction between USP12 and WDR20, we generated a HeLa cell line in which epitope-tagged WDR20 is stably expressed and UAF1 is knocked down by short hairpin RNA (Fig. 2B). Under the condition in which approximately half of the endogenous UAF1 was co-purified as WDR20 complex from the UAF1-knockdown cells (compare lanes 1 and 2), a similar reduction in the amount of USP12 was observed, suggesting that UAF1 is required for the integrity of the USP12 and WDR20 interaction.
To test whether the stable trimeric complex can be isolated, we co-expressed USP12, His-UAF1, and FLAG-WDR20 in Sf9 cells and performed sequential His and Flag affinity purification (Fig. 2C). As predicted, a near stoichiometric complex of UAF1·USP12·WDR20 proteins was purified, suggesting the existence of this ternary complex in human cells. Taken together, these results demonstrated that WDR20 directly interacts with the USP12·UAF1 complex, and a stable USP12·UAF1·WDR20 ternary complex can be isolated.
To map the region of WDR20 that mediates interaction with UAF1 and USP12, a series of FLAG-tagged WDR20 truncation mutants (Fig. 3A) was transiently expressed in 293T cells, and immunoprecipitation was performed to test the interactions with endogenous UAF1 and USP12 (Fig. 3B). Although all of the deletion mutants we generated showed reduced binding to both UAF1 and USP12 compared with the full-length WDR20, the mutant deleted of second WD40-repeat (WDR20-ΔWD2) was significantly compromised in interacting with both UAF1 and USP12, suggesting that this region is critical for interacting with the DUB complex.
FIGURE 3.
Mapping of WDR20 required for interaction with the USP12·UAF1 complex. A, wild type and the deletion mutants of WDR20 were used for the mapping study. B, individual FLAG-WDR20 plasmids were transiently transfected in 293T cells, and anti-FLAG immunoprecipitations (IP) were performed from the lysates prepared 48 h after the transfections. Western blotting was performed using anti-UAF1, anti-USP12, and anti-FLAG antibodies.
WDR20 Stimulates Activity of the USP12·UAF1, but Not of the USP1·UAF1 DUB Complex
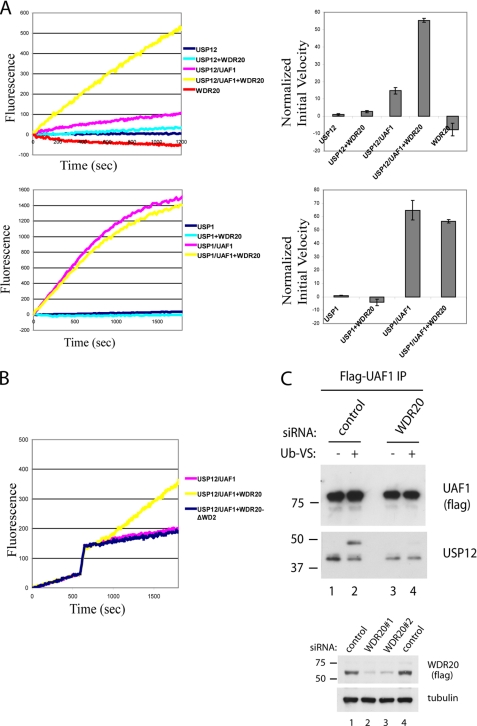
We previously demonstrated that UAF1 stimulates deubiquitinating activity of USP1, USP12, and USP46 enzymes in vitro (16, 16). Because WDR20 also contains multiple WD40-repeats similar to UAF1, we hypothesized that WDR20 may contribute to the catalytic activity of the complexes. To test this, we used purified WDR20 along with previously described USP12 and UAF1 proteins (15) and measured the deubiquitinating activity using Ub-AMC as a substrate (Fig. 4). As shown previously, USP12 alone did not show efficient deubiquitinating activity, and addition of UAF1 stimulated the activity of USP12, albeit to a low degree (Fig. 4A). Interestingly, addition of WDR20 in the reaction further stimulated the catalytic activity of the USP12·UAF1 complex 4–5-fold. WDR20 only minimally stimulated the activity of USP12 alone, in agreement with the observation that WDR20 and USP12 interact weakly in the absence of UAF1 (Fig. 2A). Also, consistent with the interaction studies, WDR20 did not stimulate the activity of either USP1 alone or the USP1·UAF1 complex (Fig. 4B). These results suggest that WDR20 serves as a cofactor that is required for the full enzymatic activity of the USP12·UAF1 DUB complex.
FIGURE 4.
WDR20 stimulates the deubiquitinating activity of the USP12·UAF1 complex. A, in vitro deubiquitination assays were performed using WDR20 protein purified from bacteria, and USP12, USP1, USP12·UAF1, and USP1·UAF1 purified from SF9 cells as described previously (16). Ub-AMC was used as a substrate at the final concentration of 1 μm, and the fluorescence was measured using a fluorometer. The final concentrations of USP12, USP12·UAF1, USP1, USP1·UAF1, and WDR20 in all reactions are 80 nm, 80 nm, 10 nm, 10 nm, and 80 nm, respectively. The reactions were performed in triplicate. B, similar deubiquitination assays were performed as in A, except that WDR20 proteins were added 10 min after the DUB complexes were preincubated with Ub-AMC. C: top panels, FLAG-HA-tagged UAF1 complexes were immunopurified using anti-FLAG-agarose from HeLa cells that were treated with either control or WDR20 siRNAs. The bound proteins were eluted off the beads using 3×FLAG peptides, followed by incubation of the eluates with Ub-VS, as described under “Experimental Procedures.” Bottom panels, efficiency of each WDR20 siRNA was tested in FLAG-HA WDR20-expressing cell line. C indicates control siRNA. In upper panel, the mixture of siRNAs 1 and 2 was used to deplete WDR20. Western blotting was performed using anti-FLAG, anti-USP12, and anti-tubulin antibodies.
To confirm the specificity and gain insight into the dynamics of the USP12·UAF1·WDR20 complex formation, we set up a similar assay in which either full-length wild type WDR20 or mutant WDR20ΔWD2 proteins were added 10 min after the USP12·UAF1 complexes were preformed (Fig. 4B). Consistent with the mapping results, the mutant failed to stimulate the enzymatic activity of the USP12·UAF1 complex, whereas the wild type WDR20 instantaneously stimulated the activity, suggesting that the affinity between WDR20 and the preformed USP12·UAF1 complex is very high. To test further the role of WDR20 in the deubiquitinating activity in cell lysates, we generated a HeLa cell line in which epitope-tagged UAF1 is stably expressed and WDR20 is knocked down by siRNA (Fig. 4C). We used Ub-VS, a chemical derivative of ubiquitin, which can be used for measuring the activity of DUBs (19). Consistent with the Ub-AMC assays, knockdown of WDR20 nearly abolished the shift of endogenous USP12, which is co-purified as UAF1 complex (compare lanes 3 and 4). Together, these results demonstrate that WDR20 stimulates enzymatic activity of the USP12·UAF1 complex.
WDR20 Does Not Regulate the FA Pathway or DNA Damage Responses
We reported previously that the USP12·UAF1 and the USP46·UAF1 complexes do not regulate the FA pathway (16), suggesting that the subset of UAF1 complexes participates in different cellular pathways. Because WDR20 does not associate with the USP1·UAF1 subcomplex, we postulated that WDR20 does not regulate the FA pathway. Indeed, it appears to be the case because siRNA-mediated depletion of WDR20 did not affect the level of monoubiquitinated FANCD2 in the absence or presence of the DNA damage-inducing agent hydroxyurea (Fig. 5A). The level of monoubiquitinated proliferating cell nuclear antigen, another suggested substrate of the UAF1·USP1 complex, or the level of phospho-Chk1 was not affected, suggesting that WDR20 depletion did not affect the overall cellular DNA damage response. As reported previously (15), depletion of UAF1 caused elevation of both FANCD2 and proliferating cell nuclear antigen monoubiquitination, shown as positive controls for this experiment. Taken together, this result reinforces the notion that WDR20 selectively affects the activity of the USP12·UAF1 and/or the USP46·UAF1 subcomplexes, but not the USP1·UAF1 complex. The results are summarized in Fig. 5B, which provides our current model for the role of WDR20.
FIGURE 5.
WDR20 does not regulate the FA pathway. A, HeLa cells were treated with the indicated siRNAs followed by treatment with hydroxyurea (HU) of final 2 mm concentration for the indicated times. The lysates were analyzed by Western blotting using the indicated antibodies. PCNA, proliferating cell nuclear antigen. B, model for the existence of distinctive DUB complexes containing WDR20. The biological function of the WDR20-containing DUB complexes is unknown, indicated as a question mark.
DISCUSSION
Here, we demonstrate that WDR20, a protein containing WD40-repeat motifs and with previously unknown function, serves as a stimulatory subunit for the USP12·UAF1 DUB complex. Our data are consistent with a recently published report in which WDR20 was shown to interact with USP12 and USP46, but not with USP1, in a large scale interaction analysis (18). We further characterize the functional nature of the interactions and demonstrated that WDR20 is specifically required for the hyperenzymatic activity of the USP12·UAF1 DUB complex in vitro. We previously reported that UAF1 stimulates the catalytic activities of USP1, USP12, and USP46 in vitro (15, 16). Although all three DUBs require UAF1 as a stoichiometric binding partner, it appeared that the degree of activation of USP12 and USP46 by UAF1 was significantly lower than the activation of USP1 (16). This observation had suggested the possibility that the USP12·UAF1 and the USP46·UAF1 had additional stimulatory subunits or different regulatory mechanisms. We now have demonstrated that the WDR20 subunit has a stimulatory activity to, at least, the USP12·UAF1 complex. Consistent with the results indicating that WDR20 selectively interacts with UAF1, USP12, and USP46, but not with USP1 (Fig. 1), WDR20 did not further stimulate the catalytic activity of the USP1·UAF1 DUB complex (Fig. 4B), suggesting that the activity of WDR20 is specific. Furthermore, USP12 and USP46 were the only DUNs recognized in the mass spectrometry analysis of WDR20-interacting proteins (data not shown), suggesting that WDR20 does not promiscuously associate with and activate all in cells.
Unlike UAF1, which appears to be the stoichiometric binding partner of USP1, WDR20 does not associate with the USP12·UAF1 complex as a stoichiometric subunit in cells, based on our silver stain analysis of the purified USP12 complex from HeLa cells (16). However, we were able to isolate a stable ternary complex of USP12, UAF1, and WDR20, when the three proteins were co-expressed in Sf9 cells (Fig. 2C). This suggests that the ternary complex exists in human cells, but formation of the complex may be regulated so that the complex only exists under certain conditions. Alternatively, only a fraction of the USP12·UAF1 complex might associate with WDR20 that has distinct functions.
In the study done by Sowa et al. (18), USP12 and USP46 were found to be associated with other proteins in addition to WDR20, such as two phosphatases PHLPP and PHLPPL. The phosphatases were also found in our mass spectrometry analysis, but the functional relationships of the phosphatases with the DUBs are currently unknown. Our preliminary analysis indicated that they may not be involved in regulating the DUB activities (data not shown). We postulate that the other interactors are involved in regulating substrate recognition, rather than regulating the inherent catalytic activities of the DUBs.
Although WDR20 seems to be highly conserved throughout evolutions, its cellular function remains elusive. Its function may be suggested from the studies in Aspergillus nidulans (20), where CreC protein of A. nidulans, an apparent orthologue of WDR20, is required for the stabilization of CreB, an orthologue of USP12. The Cre proteins are involved in transcriptional regulation under catabolite repression, where CreA protein, a transcription factor, was hypothesized to be a substrate of the DUB CreB. Although such transcriptional regulation does not exist in human cells, it is possible that the USP12·UAF1·WDR20 complex may be involved in other types of transcriptional regulation.
Finally, our siRNA-mediated knockdown studies showed that WDR20 is not involved in regulating the FA pathway, consistent with our previous report that USP12 and USP46 do not regulate the FA pathway (16). We speculate that WDR20 not only stimulates the catalytic activity of the DUB complexes, but may also serve as a specific adaptor subunit for recruiting specific substrates. Alternatively, selective association of WDR20 may allow the UAF1-containing DUB complexes to have distinct subunit compositions, therefore directing different cellular functions.
Acknowledgments
We thank Alfred Goldberg and David Smith for helpful discussions and assistance with the fluorometric assays. We thank the members of D'Andrea laboratory for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants R01DK43889, R01HL52725, P01CA092584, and U19A1067751.
- DUB
- deubiquitinating enzyme
- FA
- Fanconi anemia
- USP
- ubiquitin-specific protease
- UAF
- USP-associated factor
- HA
- hemagglutinin
- siRNA
- small interfering RNA
- GST
- glutathione S-transferase
- Ub
- ubiquitin
- AMC
- 7-amido-4-methylcoumarin
- VS
- vinylsulfone.
REFERENCES
- 1.D'Andrea A., Pellman D. (1998) Crit. Rev. Biochem. Mol. Biol. 33, 337–352 [DOI] [PubMed] [Google Scholar]
- 2.Reyes-Turcu F. E., Ventii K. H., Wilkinson K. D. (2009) Annu. Rev. Biochem. 78, 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li M., Brooks C. L., Kon N., Gu W. (2004) Mol. Cell 13, 879–886 [DOI] [PubMed] [Google Scholar]
- 4.Brummelkamp T. R., Nijman S. M., Dirac A. M., Bernards R. (2003) Nature 424, 797–801 [DOI] [PubMed] [Google Scholar]
- 5.Kovalenko A., Chable-Bessia C., Cantarella G., Israël A., Wallach D., Courtois G. (2003) Nature 424, 801–805 [DOI] [PubMed] [Google Scholar]
- 6.Nijman S. M., Huang T. T., Dirac A. M., Brummelkamp T. R., Kerkhoven R. M., D'Andrea A. D., Bernards R. (2005) Mol. Cell 17, 331–339 [DOI] [PubMed] [Google Scholar]
- 7.Kon N., Kobayashi Y., Li M., Brooks C. L., Ludwig T., Gu W. (November30, 2009) Oncogene 10.1038/onc.2009.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Massoumi R., Chmielarska K., Hennecke K., Pfeifer A., Fässler R. (2006) Cell 125, 665–677 [DOI] [PubMed] [Google Scholar]
- 9.Kim J. M., Parmar K., Huang M., Weinstock D. M., Ruit C. A., Kutok J. L., D'Andrea A. D. (2009) Dev. Cell 16, 314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oestergaard V. H., Langevin F, Kuiken H. J., Pace P., Niedzwiedz W., Simpson L. J., Ohzeki M., Takata M., Sale J. E., Patel K. J. (2007) Mol. Cell 28, 798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nijman S. M., Luna-Vargas M. P., Velds A., Brummelkamp T. R., Dirac A. M., Sixma T. K., Bernards R. (2005) Cell 123, 773–786 [DOI] [PubMed] [Google Scholar]
- 12.Yao T., Song L., Xu W., DeMartino G. N., Florens L., Swanson S. K., Washburn M. P., Conaway R. C., Conaway J. W., Cohen R. E. (2006) Nat. Cell Biol. 8, 994–1002 [DOI] [PubMed] [Google Scholar]
- 13.Yao T., Song L., Jin J., Cai Y., Takahashi H., Swanson S. K., Washburn M. P., Florens L., Conaway R. C., Cohen R. E., Conaway J. W. (2008) Mol. Cell 31, 909–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu X. B., Ouyang S. Y., Li C. J., Miao S., Wang L., Goldberg A. L. (2006) EMBO J. 25, 5742–5753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohn M. A., Kowal P., Yang K., Haas W., Huang T. T., Gygi S. P., D'Andrea A. D. (2007) Mol. Cell 28, 786–797 [DOI] [PubMed] [Google Scholar]
- 16.Cohn M. A., Kee Y., Haas W., Gygi S. P., D'Andrea A. D. (2009) J. Biol. Chem. 284, 5343–5351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakatani Y., Ogryzko V. (2003) Methods Enzymol. 370, 430–444 [DOI] [PubMed] [Google Scholar]
- 18.Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009) Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borodovsky A., Kessler B. M., Casagrande R., Overkleeft H. S., Wilkinson K. D., Ploegh H. L. (2001) EMBO J. 20, 5187–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lockington R. A., Kelly J. M. (2002) Mol. Microbiol. 43, 1173–1182 [DOI] [PubMed] [Google Scholar]
- 21.Huang T. T., Nijman S. M., Mirchandani K. D., Galardy P. J., Cohn M. A., Haas W., Gygi S. P., Ploegh H. L., Bernards R., D'Andrea A. D. (2006) Nat. Cell Biol. 8, 339–347 [DOI] [PubMed] [Google Scholar]