Abstract
The plasma membrane serotonin (5-HT) transporter (SERT, SLC6A4) clears 5-HT after release at nerve termini and is targeted by both antidepressant medications and psychostimulants (e.g. MDMA, cocaine). Homology modeling of human SERT (hSERT), based on high resolution structures of the microbial SLC6 family member LeuTAa, along with biochemical studies of wild type and mutant transporters, predicts transmembrane (TM) domains 1, 3, 6, and 8 comprise the 5-HT-binding pocket. We utilized the substituted cysteine accessibility method along with surface and site-specific biotinylation to probe TM6 for aqueous accessibility and differential interactions with 5-HT and psychostimulants. Our results are consistent with TM6 being composed of an aqueous-accessible, α-helical extracellular domain (TM6a) that is separated by a central, unwound section from a cytoplasmically localized domain (TM6b) with limited aqueous accessibility. The substitution G338C appears to lock hSERT in an outward-facing conformation that, although accessible to aminoethylmethanethiosulfonate-biotin, 5-HT, and citalopram, is incapable of inward 5-HT transport. Transport of 5-HT by G338C can be partially restored by the TM1 mutation Y95F. With regard to methanethiosulfonate (MTS) inactivation of uptake, TM6a Cys mutants demonstrate Na+-dependent [2-(trimethylammonium)ethyl]-MTS sensitivity. Studies with the centrally located substitution S336C reveal features of a common binding pocket for 5-HT and 3,4-methylenedioxymethamphetamine (MDMA). Interestingly, the substitution I333C reveals an MDMA-induced conformation not observed with 5-HT. In the context of prior studies on TM1, our findings document shared and unique features of TM6 contributing to hSERT aqueous accessibility, ligand recognition, and conformational dynamics.
Keywords: Membrane Proteins, Neurotransmitter Transport, Neurotransmitters, Protein Chemical Modification, Protein Structure, MDMA, MTS, SCAM, Cocaine, Serotonin
Introduction
The serotonin transporter (SERT,2 SLC6A4) is an integral membrane protein responsible for clearing serotonin (5-HT) after vesicular release, thereby constraining the indolamine actions on pre- and postsynaptic 5-HT receptors (1). SERT is closely related to biogenic amine transporters of the solute carrier 6 (SLC6) family that transport the neurotransmitters norepinephrine (SLC6A2) and dopamine (DAT, SLC6A3) and, more distantly, transporters for the neurotransmitters γ-aminobutyric acid (GAT-1, SLC6A1) and glycine (2, 3). SERT is predicted to be a 12-transmembrane (TM) domain protein that is functional as a monomer but likely exists in multimeric complexes, including transporter homomultimers (4, 5). Biochemical studies confirm that SERT possesses intracellular amino and carboxyl termini as well as a heavily glycosylated, extracellular loop 2 (6–10). SERT-supported 5-HT transport is coupled to extracellular sodium and chloride, which is thought to drive inward movement of 5-HT, and intracellular potassium, which is thought to outwardly rectify SERT (11, 12). Because of the clinical efficacy of selective 5-HT reuptake inhibitors, significant effort has been focused on investigating the potentially compromised function of human SERT (hSERT) as a contributor to affective disorders, such as major depressive disorder and generalized anxiety disorder (1). Indeed, genetic studies associate hSERT regulatory and coding polymorphisms with obsessive compulsive disorder, bipolar disorder, and autism (13–15). As many of the functional coding variants identified in hSERT lie within TM domains, we and others have sought a better understanding of the role that these domains play in hSERT function. In addition to its ability to bind and transport 5-HT, SERT is also a major binding target for drugs of abuse, most notably cocaine and 3,4-methylenedioxymethamphetamine (MDMA or “ecstasy”). Because MDMA shares structural similarity to 5-HT, this amphetamine-like compound likely binds to determinants that overlap those used by 5-HT. In contrast, much larger competitive antagonists such as cocaine and the selective 5-HT reuptake inhibitors are believed to gather additional TM binding determinants outside the 5-HT-binding pocket (16–18).
SERT, like other SLC6 family members, is presumed to translocate substrate across the membrane using an alternating access mechanism in which 5-HT binds to an extracellularly facing conformation of the transporter, thereby triggering a conformational change that exposes the intracellular face of the permeation pathway to release 5-HT into the cellular milieu (19). This pathway may also exist briefly in a “two-gate open” conformation to conduct uncoupled substrates at much higher rates (20–24), particularly when coupling conformations are disrupted by mutation or through the actions of associated proteins (25). The recent elucidation of several transporter crystal structures, including the sodium/galactose cotransporter vSGLT, the nucleobase cation symporter Mhp1, the aspartate transporter GltPh, and the bacterial SLC6 family member, the Aquifex aeolicus leucine transporter LeuTAa, offers snapshots of potential conformations involved in the alternating access model and provides significant tools for the prediction of structural contributions to mechanism (26–31). The LeuTAa protein, the closest crystallized homologue of SERT, was initially crystallized with its substrate bound and with passageways to extracellular and intracellular compartments closed (i.e. an “occluded” state). LeuTAa, as well as portions of vSGLT and Mhp1, displays an inverted repeat structure, with (in LeuTAa) TMs 1–5 and 6–10 assembling in pseudo-2-fold symmetry (31). Interestingly, TMs 1 and 6 exist as closely associated, anti-parallel helices localized within the core of the transporter that display significant unwound portions in the middle of the membrane. In LeuTAa, TMs 1, 3, 6, and 8 form the leucine binding pocket. Biochemical analyses and homology modeling of SERT proteins predict a similar binding pocket for 5-HT (32–36). Consistent with these models, pre-structure studies identified residues in hSERT TMs 1 and 3 that confer high affinity interactions and ligand selectivity to substrates and antagonists. Whereas mutational studies of selected TM6 residues in SERT, DAT, and GAT-1 suggest involvement of this TM in substrate and ion interactions (37–39), how these alterations relate to the structural characteristics of TM6 is unknown.
In the current study we present a biochemical characterization of the entirety of hSERT TM6. We probe hSERT TM6 using the substituted cysteine accessibility method (SCAM) to assess the secondary structure and relative residue accessibility, to determine the functional sensitivity of residues to chemical modification, and to elucidate sites of potential ligand interaction through protection experiments (40). Our delineation of the impact of TM6 Cys substitutions on 5-HT transport as well as their relative sensitivity to methanethiosulfonate (MTS) reagents lends support to LeuTAa-based hSERT homology models. Additionally, they provide insight into structural and functional relationships between TM1 and TM6 and offer evidence of TM6 residues bearing unique contributions to transport conformations, including substrate recognition. We discuss opportunities to capitalize on our findings for high resolution structure studies of biogenic amine transporters as well as enhanced computational modeling approaches.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis
Site-directed mutagenesis of hSERT C109A was performed using the QuikChange mutagenesis kit and protocol (Stratagene). Sense and antisense oligonucleotides, purchased from Invitrogen, were designed to generate single Cys mutations at each residue along TM6 from position 323 to 348. Oligonucleotide sequences used for mutagenesis are available upon request. hSERT C109A, cloned into pcDNA3, was used for mutagenesis to permit studies of Cys accessibility in a background deficient in labeling of endogenous Cys residues (41, 42). Sequencing of all mutants was performed at the DNA Sequencing Facility of the Division of Genetic Medicine at Vanderbilt University Medical Center. Successful mutants were transformed into DH5α Escherichia coli cells for amplification and purified using the Qiafilter Maxiprep kit (Qiagen). Results reflect the analysis of multiple, independent plasmid preparations.
Cell Culture and Transient Transfection
hSERT C109A and hSERT C109A bearing TM6 Cys substitutions were transiently transfected into human embryonic kidney 293-T (HEK-293T) cells. HEK-293T cells were grown in complete medium (Dulbecco's modified Eagle's medium, 10% fetal bovine serum, 2 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin) and maintained at 37 °C, 5% CO2. For 5-HT uptake assays, MTS sensitivity and protection assays, and transporter biotinylation assays, HEK-293T cells were plated at a density of 50,000 cells/well in 24-well tissue culture plates coated with 0.1 mg/ml poly-d-lysine and grown in complete medium. Transfection of cells was performed ∼24 h after plating; the TM6 Cys mutants were transfected into cells using 6 μl of Trans-IT (Mirus Bio Corp.) per μg of DNA in Opti-MEM serum-free medium (Invitrogen) at a concentration of 100 ng/well. Cells were incubated in transfection medium for ∼48 h before assay. For competition binding assays, HEK-293T cells were plated at a density of 107 cells/dish in 100-mm tissue culture dishes and incubated for 12–24 h before transfection. Cells were transiently transfected with pcDNA3 hSERT C109A or pcDNA3 hSERT C109A/G338C using 6 μl of Trans-IT per μg DNA as for transport assays at a concentration of 6 μg/dish. Cells were incubated in transfection medium for ∼48 h until binding assays were performed.
5-HT Transport Measurements
Single point and saturation kinetic 5-HT uptake assessments were performed in triplicate within an experiment and repeated in at least three separate experiments. Single point uptake assays were performed as follows. Cells were washed with a modified Krebs-Ringers-HEPES (mKRH) buffer (120 mm NaCl, 4.7 mm KCl, 2.2 mm CaCl2, 1.2 mm MgSO4, 1.2 mm KH2MgSO4, 10 mm HEPES, pH 7.4) containing 1.8 g/liter glucose and then incubated for 10 min at 37 °C with 20 nm [3H]5-HT, 100 μm pargyline, and 100 μm ascorbic acid. Uptake was terminated by two cell washes using 500 μl of mKRH assay buffer, and cells were solubilized in 400 μl of Microscint 20 scintillation fluid and counted on a Packard TopCount scintillation counter. Specific 5-HT uptake was determined by subtracting uptake obtained with mock-transfected cells from uptake of hSERT mutant-transfected cells. Cys mutants exhibiting ≥5% of hSERT C109A uptake activity were further evaluated through saturation kinetic analyses as well as MTS reactivity; mutants displaying <5% uptake activity were considered nonfunctional and were not further examined. For saturation kinetic analyses, serial dilutions of a [3H]5-HT/unlabeled 5-HT mix (1:50) at a constant specific activity were applied to transfected cells in the presence of 100 μm pargyline and 100 μm ascorbic acid and allowed to incubate at 37 °C for 10 min. Uptake was terminated by two cell washes with ice-cold mKRH assay buffer. Cells were solubilized and counted as noted above with nonspecific 5-HT uptake assessed in parallel at each 5-HT concentration using mock-transfected cells. 5-HT transport Km and Vmax values were derived from nonlinear, least squares curve fits using a single site model in Prism 4.0 (Graphpad). Mutant Km and Vmax values were compared with those of hSERT C109A by Student's two-way t test in Prism 4.0 with p < .05 taken as evidence of significant differences.
[3H]Citalopram Competition Binding Assays
hSERT C109A and hSERT C109A mutant-transfected cells were washed once with 4 °C PBS buffer followed by the addition of 4 °C homogenization buffer (50 mm HEPES, 2.5 mm MgCl2, 2 mm EGTA, 100 μm ascorbic acid, pH 7.4). Cells were scraped into 14-ml glass centrifuge tubes, homogenized on ice using a Polytron homogenizer (Brinkmann Instruments), and centrifuged for 10 min at 30,000 × g in a Sorvall RC-5B Plus centrifuge. Supernatants were decanted, fresh 4 °C homogenization buffer was added, and homogenization and centrifugation were repeated. Supernatants were decanted, and membranes were resuspended in 4 °C binding assay buffer (50 mm Tris, pH 7.5, 100 mm NaCl). Protein concentrations were determined via a Coomassie Plus Bradford assay kit (Thermo Fisher Scientific). Competition binding assays were performed on 100 μg of protein/tube in 12 × 75-mm borosilicate glass tubes. [3H]Citalopram (PerkinElmer Life Sciences; 77.0 Ci/mmol; 5 nm final) and cold 5-HT (concentration range from 10−2 to 10−8 m final in 100 μm pargyline and 100 μm ascorbic acid) were added to the membranes, and tubes were gently mixed and allowed to incubate at room temperature for 45 min. Nonspecific binding was determined by co-incubation of samples with 10 μm paroxetine and subtracted from total binding for each sample to determine specific citalopram binding. Samples were processed on a 24-well Brandel cell harvester using Whatman GF/B filters presoaked in 0.3% polyethyleneimine for 60 min. The filters were washed 3 times with ∼1 ml of ice-cold wash buffer (50 mm HEPES, 2.5 mm MgCl2, 2 mm EGTA, pH 7.4), dried, and transferred to EcoScint H scintillant (National Diagnostics). Radioactivity was counted using a Packard Tricarb 2900 Liquid Scintillation Analyzer. 5-HT KI values were estimated using a one-site competition binding fit in Prism 4.0.
Sensitivity of hSERT TM6 Cys Mutants to MTS Reagents
Transiently transfected HEK-293T cells in 24-well tissue culture plates were washed twice with 500 μl of room temperature PBS/CM buffer (PBS, 0.1 mm CaCl2 and 1.0 mm MgCl2, pH 7.4) and incubated in PBS/CM buffer with/without MTS reagents for 10 min at room temperature. Concentrations for the specific MTS reagents are as follows: 1 mm [2-(trimethylammonium)-ethyl]methanethiosulfonate (MTSET), 0.25 mm aminoethylmethanethiosulfonate (MTSEA), and 10 mm (2-sulfonatoethyl)-methanethiosulfonate (MTSES) (Toronto Research Chemicals). The concentrations for MTSET and MTSES are based on previous studies by Karlin and Akabas (40), but due to its high reactivity with hSERT C109A (42), the concentration of MTSEA used by Karlin and Akabas (40) (2.5 mm) was reduced 10-fold. After incubation with MTS reagents, cells were washed with 500 μl of PBS/CM followed by incubation with 500 μl of 37 °C mKRH assay buffer. 5-HT transport activity was then assessed by single point uptake assay, as described above. For ligand protection experiments, cells were preincubated with 50 μm 5-HT, MDMA, or cocaine for 5 min before the addition of MTSET and assayed as described above. For the Na+ substitution assay, cells were washed 3 times with 500 μl of PBS/CM or a PBS/CM buffer in which Na+ was isotonically replaced with NMDG (PBS/CM-NMDG); the MTSET incubation was then performed in the respective sodium or NMDG buffer, and the cells were washed and assayed for 5-HT uptake activity as described above. 5-HT uptake activity of MTS-treated Cys mutants was normalized to its respective untreated cysteine mutant to evaluate the impact of the MTS reagent. MTS-treated and untreated mutants were analyzed using Graphpad Prism software and compared using a one-way ANOVA with Dunnett's post-test.
Immunoblot Analysis of hSERT C109A and TM6 Cys Mutant Protein Levels
Cell surface biotinylation studies were performed to establish the functional impact of Cys substitution on the ability of each mutant to reach the plasma membrane. hSERT C109A or hSERT C109A Cys mutants were transfected in HEK-293T cells as described above. Approximately 48 h after transfection, cells were washed 4 times with ice-cold PBS/CM buffer and incubated with 1.0 mg/ml NHS-SS-biotin in PBS/CM for 30 min at 4 °C. Biotinylation reactions were quenched by washing cells twice with PBS/CM buffer containing 100 mm glycine. Cells were then incubated for 10 min in the PBS/CM/glycine buffer, washed once with PBS/CM buffer, and solubilized with RIPA buffer containing phenylmethylsulfonyl fluoride (PMSF) (10 mm Tris, 150 mm NaCl, 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate, 250 μm PMSF, pH 7.4). Cells were shaken at 100 rpm in RIPA buffer for 30 min at 4 °C followed by centrifugation of lysates at 20,000 × g for 20 min at 4 °C. A fraction of the resulting supernatant was retained for immunoblots to quantify total hSERT protein. Immunopure immobilized streptavidin beads (Pierce) were washed four times with RIPA buffer and resuspended in RIPA buffer to achieve a 50% slurry. The remaining cell supernatant was applied to the streptavidin bead/RIPA slurry at room temperature for 1 h, and beads were washed 4 times with RIPA buffer. Biotinylated proteins were eluted from the streptavidin beads with Laemmli buffer containing 5% β-mercaptoethanol. Biotinylated and total protein samples were resolved via 10% SDS-PAGE, and separated proteins were transferred overnight at 21 V (constant), 4 °C, to Immobilon-P polyvinylidene difluoride membranes (Millipore). Membranes were blocked with 5% nonfat dried milk in PBS/0.5% Tween (PBS/Tween) for 4 h at 4 °C and probed with hSERT monoclonal antibody ST51–2 (mAB Technologies) for 1 h at 4 °C. After multiple washes with PBS/Tween, blots were incubated with goat anti-mouse horseradish peroxidase-conjugated secondary antibody (The Jackson Laboratory) diluted 1:10,000 overnight at 4 °C. After additional washes with PBS/Tween, blots were developed using the Western Lightning chemiluminescence kit and exposed to Eastman Kodak Co. X-Omat Blue XB film. MTSEA biotinylation of cells was performed in the same manner as MTS inactivation studies described above with the following exceptions. Transfected cells were washed twice with room temperature PBS/CM and incubated with 1 mm MTSEA-biotin in PBS/CM for 10 min at room temperature. The cells were then washed twice with PBS/CM and solubilized in 400 μl of RIPA solubilization buffer. Extraction of biotinylated proteins and hSERT immunoblotting was performed as described above. For all blotting studies, multiple exposures were captured in all experiments to ensure collection of data in the film linear range.
RESULTS
Effects of Cys Substitution on hSERT Expression and Function
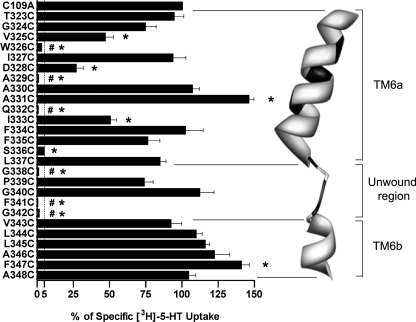
Before characterizing the accessibility of hSERT TM6 Cys substitutions to MTS reagents, the effect of Cys substitution was examined for impact on 5-HT transport activity and transporter surface expression. As noted under “Experimental Procedures,” all TM6 Cys mutants were engineered in the hSERT C109A background, as the C109A substitution in either rat SERT (43) or hSERT (42) essentially eliminates MTS reagent sensitivity while maintaining normal surface expression and transport activity. In single point (20 nm) uptake assays (Fig. 1), 10 Cys mutants displayed uptake equivalent to hSERT C109A when expressed under identical transfection conditions. In contrast, six Cys mutants (W326C, A329C, Q332C, G338C, F341C, and G342C) exhibited less than 5% of hSERT C109A activity and, thus, were considered functionally inactive and inadequate for SCAM analysis. Of the remaining TM6 Cys substitutions, four (V325C, D328C, I333C, and S336C) resulted in significantly reduced uptake relative to hSERT C109A. Interestingly, two Cys mutants, A331C and F347C, exhibited increased 5-HT uptake. Although only the transport enhancement of F347C reached statistical significance, we also observed several mutants in the cytosolic half of the TM to display small, but consistent, elevations in 5-HT uptake in these assays.
FIGURE 1.
[3H]5-HT transport activity of hSERT TM6 Cys mutants. hSERT Cys mutants cDNAs were transiently transfected in HEK-293T cells at equal concentrations and assayed for uptake at a single 3H-5-HT concentration (see “Experimental Procedures”). Results represent the means ± S.E. from three experiments. Means of Cys mutant uptake were compared with C109A using a one-way ANOVA with a post hoc Dunnett's test, with p < 0.05 indicating significance. #, inactive mutant. *, p < 0.05 compared with C109A. The dashed line indicates 5% C109A uptake activity. Homology model of hSERT TM6 is depicted as constructed by Kaufmann et al. (32) based on LeuTAa structures. Residues that are intolerant of Cys substitution are depicted in black.
Full saturation kinetic analyses were performed to assess the impact of Cys substitution on 5-HT transport Km and Vmax (Table 1). A significant increase in Km was observed in the I333C mutant, whereas a significant Km decrease was detected in the P339C, G340C, L344C, F347C, and A348C mutants. Many of the TM6 Cys substitutions exhibited significant changes in 5-HT transport Vmax. Increased Vmax values were observed in the T323C, A330C, A331C, I333C, and F334C substitutions, whereas G324C, V325C, D328C, S336C, P339C, G340C, L344C, L345C, F347C, and A348C displayed decreased Vmax values.
TABLE 1.
Impact of Cys substitution on [3H]5-HT uptake kinetics in functional TM6 Cys mutants
Km and Vmax values were determined from [3H]5-HT transport assays in transiently transfected HEK-293T cells as described under “Experimental Procedures.” All mutants are in the hSERT C109A background. Results are presented as mean values ± S.E. from three or more experiments and were analyzed in reference to C109A using one-way ANOVA with a post hoc Dunnett test.
| hSERT mutant | Km | Vmax |
|---|---|---|
| μm | fmol/cell/min | |
| C109A | 1.93 ± 0.33 | 8.89 ± 0.67 |
| T323C | 2.35 ± 0.82 | 14.3 ± 2.36a |
| G324C | 2.10 ± 0.44 | 6.29 ± 0.60b |
| V325C | 2.96 ± 0.73 | 5.90 ± 0.75b |
| I327C | 1.25 ± 0.53 | 7.99 ± 1.32 |
| D328C | 2.38 ± 1.00 | 2.26 ± 0.45b |
| A330C | 1.61 ± 0.31 | 11.4 ± 0.92a |
| A331C | 2.04 ± 0.43 | 19.2 ± 1.83a |
| I333C | 9.49 ± 3.01a | 20.9 ± 4.88a |
| F334C | 5.38 ± 2.40 | 41.8 ± 11.5a |
| F335C | 2.91 ± 0.72 | 11.5 ± 1.45 |
| S336C | 3.49 ± 1.47 | 0.32 ± 0.07b |
| L337C | 8.62 ± 5.11 | 19.9 ± 8.22 |
| P339C | 0.84 ± 0.18b | 4.36 ± 0.33b |
| G340C | 0.84 ± 0.10b | 6.27 ± 0.27b |
| V343C | 1.40 ± 0.57 | 6.31 ± 1.03 |
| L344C | 0.55 ± 0.24b | 3.20 ± 0.44b |
| L345C | 0.99 ± 0.42 | 5.93 ± 0.90b |
| A346C | 1.34 ± 0.54 | 11.6 ± 1.84 |
| F347C | 0.16 ± 0.13b | 1.66 ± 0.29b |
| A348C | 0.19 ± 0.08b | 1.84 ± 0.17b |
a p < 0.05 above C109A.
b p < 0.05 below C109A.
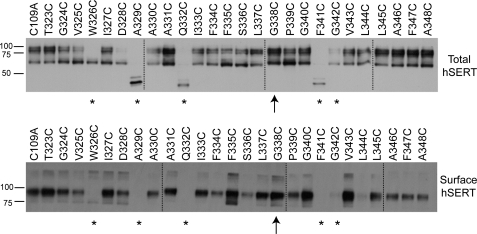
To examine whether the differences in 5-HT uptake observed for the hSERT TM6 Cys mutants reflect an impact on transporter surface expression, total and surface protein levels were examined in biotinylation assays (Fig. 2). In whole cell extracts of transiently transfected HEK-293T cells, hSERT appears in two forms, a 65-kDa form that represents the minimally glycosylated, immature protein, and a 85–90-kDa form that represents more fully glycosylated, mature protein enriched at the cell surface. Consistent with single point uptake data, surface expression of W326C, A329C, Q332C, F341C, and G342C (denoted by asterisks in Fig. 2) exhibited less than 5% of C109A surface expression in biotinylated preparations and were excluded from additional analysis. Interestingly, however, G338C exhibits robust expression of the mature form in both total and surface fractions (denoted by an arrow in Fig. 2), despite exhibiting no uptake activity at 5-HT concentrations up to 20 μm (data not shown). In addition to the surface expression of the C109A/G338C mutant, alanine (Ala) substitutions at Gly-338 and Gly-342, the glycines proposed to define the limits of the unwound portion of TM6, exhibited ∼28 and 10%, respectively, of hSERT C109A 5-HT uptake activity (supplemental Fig. 1). Mutants S336C and L344C exhibited decreased protein expression in comparison to hSERT C109A, likely contributing to their reduced 5-HT transport Vmax.
FIGURE 2.
Total protein and surface expression of hSERT TM6 Cys mutants. To assess the impact of Cys substitution on TM6, whole cell and NHS-SS-biotinylated extracts were examined by Western blot using the monoclonal hSERT antibody ST51–2. Dashed lines indicate separate acrylamide gels run concurrently. *, mutant exhibiting less than 5% 5-HT uptake activity compared with C109A and no detectable surface expression. ↑, mutant exhibiting less than 5% 5-HT uptake activity compared with C109A but significant surface expression.
Impact of MTS Reagents on 5-HT Transport Activity of Cys Mutants
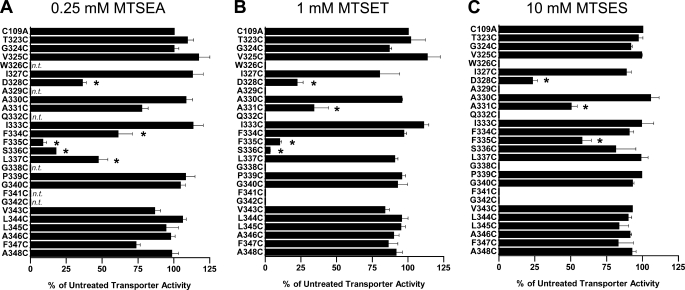
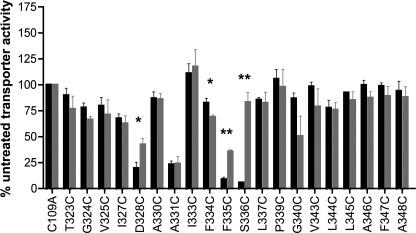
hSERT TM6 Cys mutants that exhibited a minimum of 5% of hSERT C109A activity were characterized for sensitivity to MTS inactivation. To better assess the microenvironment of each Cys substitution, we used MTS reagents that vary in bulk size or charge, with MTSET, MTSES, and MTSEA occupying 173, 140, and 113 Å3, respectively, and with MTSET and MTSEA being positively charged and MTSES negatively charged (44). For those Cys mutants that demonstrated MTS sensitivity, with the exception of hSERT A331C, Ala substitutions were made, expressed, and assayed for MTS sensitivity in the same manner to control for the possibility that mutation alone had exposed endogenous Cys residues. These control Ala mutants failed to demonstrate sensitivity to the MTS reagents (data not shown). The impact of MTSEA, MTSET, and MTSES on 5-HT transport activity (20 nm, Fig. 3) demonstrates that, overall, the greatest degree of inactivation was observed using the smallest of the MTS reagents, MTSEA, which was capable of reducing uptake at multiple positions across the predicted TM6 upper helix (TM6a). Cys mutants at positions 323–327 did not exhibit any impact of MTS modification on subsequent 5-HT uptake. Residues located more cytoplasmically on TM6a exhibited differential MTS sensitivity. D328C was inactivated by all three reagents, A330C was not inactivated by any reagent, and A331C exhibited significant inactivation by MTSET and MTSES but not the smaller MTSEA. A different sensitivity profile emerges as TM6a approaches the unwound region. Interestingly, S336C was inactivated by the positively charged MTSET and MTSEA reagents but not by the negatively charged molecule MTSES. To determine whether MTSES can simply modify, but not functionally perturb, S336C, we pretreated hSERT C109A/S336C-expressing cells with MTSES before treatment with MTSET. Pretreatment with MTSES failed to protect S336C from subsequent inactivation by MTSET (data not shown), demonstrating that the lack of MTSES sensitivity likely arises from an inability to physically access this site. In addition, although F335C was significantly inactivated by all three reagents, the deficit induced by MTSES was significantly less than that induced by MTSET and MTSEA when compared by one-way ANOVA with Bonferroni post-test (p < 0.001), suggesting that MTSES is less effective in modifying F335C. At residues cytoplasmic to Ser-336, only L337C exhibited sensitivity to MTS inactivation. This was a modest impact achieved exclusively by the smallest and only membrane-permeable reagent, MTSEA, consistent with either the significant physical limits on extracellular aqueous accessibility in this region or attack from the interior of the cell.
FIGURE 3.
Impact of MTS application on [3H]5-HT transport activity of hSERT TM6 Cys mutants. Aqueous accessibility of TM6 Cys mutants was assessed by application of 0.25 mm MTSEA (A), 1 mm MTSET (B), or 10 mm MTSES (C). Transiently transfected HEK-293T cells were incubated with an MTS reagent from 10 min at room temperature and then subjected to a [3H]5-HT uptake assay as described under “Experimental Procedures.” Results are expressed as the percent of untreated transporter activity for each mutant and represent the means ± S.E. from at least three uptake experiments. TM6 Cys mutants were compared with C109A using one-way ANOVA with a post hoc Dunnett's test, with * indicating p < 0.05. n.t., not tested.
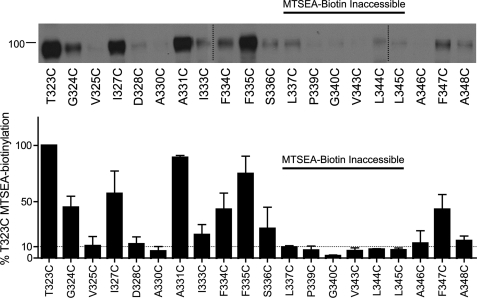
Aqueous Accessibility of Cys Mutants to MTSEA-biotin
It is well understood that aqueous accessibility cannot be inferred entirely from functional inactivation. We, therefore, examined the ability of hSERT TM6 Cys mutants to be labeled by MTSEA-biotin (Fig. 4). TM6 Cys mutants that exhibited less than 10% T323C MTSEA-biotinylation after normalization to total SERT protein were considered inaccessible. These studies revealed four positions in TM6a (T323C, G324C, V325C, and I327C) that are accessible to MTS-biotin but insensitive to inactivation of uptake by the MTS reagents above. Furthermore, the MTS-biotin-inaccessible residues identified in this study are consistent with predictions from the LeuTAa structure and MTS inactivation studies of TM1. Specifically, A330C is likely inaccessible due to its position on the helix oriented away from the aqueous pore. As observed for TM1a (42), the lower TM6 helix (TM6b) forms a barrier to aqueous reagents as functional Cys mutants from position 337 to 345 displayed little to no biotinylation. Within TM6b, A346C, F347C, and A348C exhibited accessibility to MTSEA-biotin, suggesting that aqueous accessibility returns at the limits of the cytoplasmic membrane. This pattern of MTSEA biotinylation does not correlate with surface expression as measured by NHS-SS-biotin (Fig. 2), indicating that this cysteine-directed approach reports differential aqueous accessibility.
FIGURE 4.
Aqueous exposure of TM6 Cys mutants as revealed by MTSEA biotinylation. Accessibility of functional TM6 Cys mutants was assessed by application of 1 mm MTSEA-biotin and a subsequent Western blot using hSERT monoclonal antibody ST51–2. hSERT Cys mutants cDNAs were transiently transfected in HEK-293T cells at equal concentrations (see “Experimental Procedures”). Vertical dashed lines represent separate acrylamide gels run concurrently. Quantification of protein expression (n = 3) performed in Image J and normalized to T323C. The horizontal dashed line indicates 10% T323C MTSEA biotinylation.
G338C Appears to Stabilize an Open Conformation of hSERT
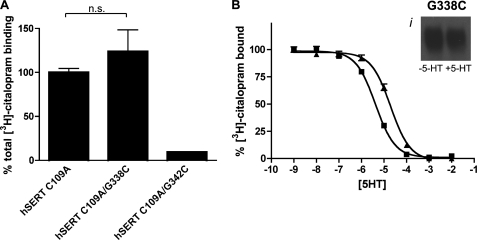
The robust expression of G338C and minimal expression of G342C at the membrane in nonfunctional forms may arise from stabilization of a conformation that is nonpermissive for substrate transport. Alternatively, SERT may simply lose affinity for 5-HT due to disruption of the substrate binding site. To assess these possibilities, membranes from hSERT C109A- hSERT C109A/G338C-, and hSERT C109A/G342C-transfected cells were labeled by [3H]citalopram, with specific binding levels in proportion to the level of surface expression identified with NHS-SS-biotin (Fig. 5A). Labeling of hSERT C109A/G342C indicated detectible expression at the plasma membrane, but the signal was insufficient for additional competition analysis. On the other hand, G338C can be robustly biotinylated by MTSEA-biotin in the presence and absence of 5-HT (Fig. 5B, inset). Competition binding assays also revealed that 5-HT maintained its ability to compete for the primary binding pocket in the G338C mutant, albeit with a small, nonsignificant loss in the apparent Ki (Fig. 5B; hSERT C109A Ki, 0.6 ± 0.2 μm; hSERT C109A/G338C Ki, 2.8 ± 0.8 μm, p = 0.057, Student's two-tailed t test). Because transport of 5-HT is lacking at concentrations up to 20 μm, this evidence of the integrity of substrate and antagonist binding coupled with the efficient labeling of the mutant by MTS-biotin supports the hypothesis that the G338C mutant is locked in an “outward-facing” conformation.
FIGURE 5.
hSERT C109A/G338C is accessible to MTSEA-biotin and capable of binding 5-HT. A, total [3H]citalopram binding of hSERT C109A/G338C and C109A/G342C, expressed as a percentage of hSERT C109A is shown. n.s., not significant. B, the ability of the hSERT C109A/G338C mutant to bind 5-HT was assessed by 5-HT competition of [3H]citalopram binding. ■, hSERT C109A; ▴, hSERT C109A/G338C. Bi, accessibility of hSERT C109A/G338C was assessed by application of 1 mm MTSEA-biotin in the presence and absence of 50 μm 5-HT and a subsequent Western blot using hSERT monoclonal antibody ST51–2 (see “Experimental Procedures”).
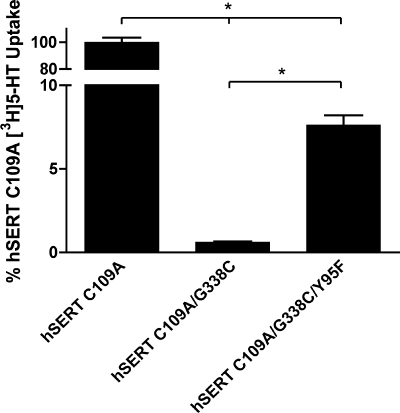
If the G338C mutant indeed stabilizes an outward-facing conformation, we hypothesized that a second-site mutation could be introduced that could relieve the loss of 5-HT transport activity. After inspection of TM1 and TM6 in homology models of hSERT (32, 34), we hypothesized that the thiolate of G338C might establish an obstructive hydrogen bond with the hydroxyl group of Y95 in TM1. The hSERT Y95F mutation has been extensively characterized and exhibits ∼85% of wild type hSERT uptake activity (17, 45, 46). Indeed, the triple mutant hSERT C109A/G338C/Y95F restored significant, although modest, 5-HT uptake activity (7.6% of C109A uptake activity) (Fig. 6). These data provide additional evidence of TM1/TM6 interactions that are critical in the transition from outward-facing to inward-facing conformations during the 5-HT translocation process.
FIGURE 6.
hSERT C109A/G338C/Y95F exhibits partial restoration of 5-HT uptake. hSERT C109A, C109A/G338C, and C109A/G338C/Y95F mutant cDNAs were transiently transfected in HEK-293T cells at equal concentrations and assayed for uptake at a single [3H]5-HT concentration (see “Experimental Procedures”). Results represent the means ± S.E. from three experiments. Means of mutant uptake were compared with C109A using two-way Student's t test, with the asterisks indicating p < 0.001.
Impact of Na+ on MTSET Inactivation
The LeuTAa crystal structure and hSERT homology models derived from it predict that TM6 is involved in the coordination of a sodium ion (27, 32–34, 36). Interestingly, replacement of sodium with NMDG in the MTS incubation buffer resulted in protection of three TM6 Cys mutants and exposure of a single mutant (Fig. 7). Specifically, D328C, F335C, and S336C were significantly protected from MTSET inactivation in the presence of NMDG. In fact, S336C was fully protected from MTSET inactivation by NMDG substitution, supporting predictions from homology models that Ser-336 is involved in sodium interaction. The absence of the sodium ion also induces a change in accessibility at positions 334 and 335, exposing and protecting respectively, which could result from a larger movement of this segment of the helix. Distal to the binding site, D328C was also partially protected from MTSET inactivation in the absence of sodium, suggesting the influence of ion binding throughout TM6a.
FIGURE 7.
Impact of Na+ substitution on MTSET inactivation of [3H]5-HT transport in TM6 Cys mutants. Accessibility of Cys mutants was assessed by alteration of [3H]5-HT transport after exposure to MTSET in the presence of NMDG (gray) or presence of sodium (black). Results for each mutant are expressed as the means ± S.E., n = 3, and normalized to C109A. Means of Cys mutants in the presence and absence of sodium were compared with each other using a two-tailed Student's t test, with the single asterisks indicating p < 0.05 and the double asterisks indicating p < 0.005.
Impact of Ligands on MTSET Inactivation
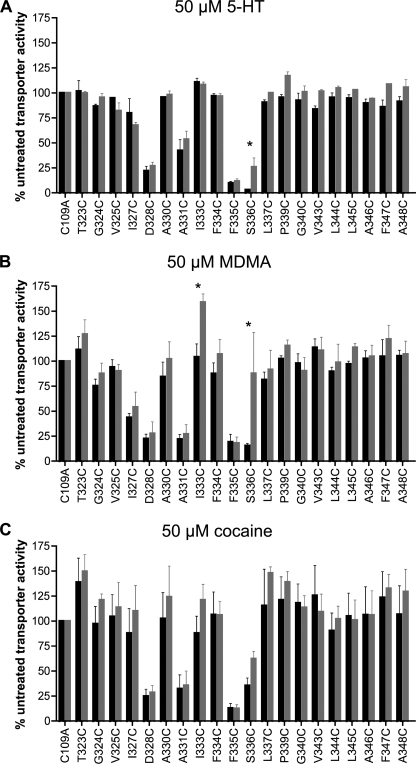
Residues in TM1 for multiple SLC6 family members have been identified as critical determinants of substrate binding as well as recognition sites for competitive antagonists (42, 43, 45, 46). Because hSERT TM6 is predicted to lie adjacent to and share topology with TM1 (27, 32–36), we explored potential contributions of TM6 to the interactions of 5-HT, MDMA, and cocaine via an assessment of their ability to protect hSERT C109A Cys mutants from inactivation by MTSET and biotinylation with MTSEA-biotin. Before initiation of protection experiments, we examined the sensitivity of each Cys mutant to MTSET at 0.1, 1, and 10 mm to ensure we were using a concentration of MTSET appropriate for detecting either protection or exposure. Because of the high reactivity observed to 1 mm MTSET, the mutants D328C, F335C, and S336C were assayed for protection in the presence of 0.1 mm MTSET, paralleling these studies with hSERT C109A at the same concentration. All other experiments utilized 10 mm MTSET. Preincubation with 50 μm 5-HT protected only the S336C mutant from MTSET inactivation, and although this effect was significant, the protection was small in magnitude (Fig. 8A). 5-HT preincubation experiments utilizing MTSEA-biotin did not reveal significant changes in TM6 Cys accessibility (data not shown), although in our experience this assay yields higher variability than functional MTSET inactivation.
FIGURE 8.
Impact of ligand on MTSET inactivation of [3H]5-HT transport in TM6 Cys mutants. Accessibility of Cys mutants was assessed by alteration of [3H]5-HT transport after exposure to MTSET in the absence (black) or presence (gray) of 50 μm 5-HT (A), 50 μm MDMA (B), or 50 μm cocaine (C). Results for each mutant are expressed as the means ± S.E., n = 3, and normalized to C109A. Means of Cys mutants in the presence and absence of substrate were compared with each other using a two-tailed Student's t test, with the asterisks indicating p < 0.05.
Similar to 5-HT studies, but to a much greater degree, S336C was protected from MTSET inactivation after pretreatment with 50 μm MDMA (Fig. 8B). Interestingly, although hSERT C109A/I333C showed no sensitivity to MTSET, this substitution demonstrated a robust stimulation of uptake when preincubated with MDMA. This stimulation required the combined presence of MDMA and MTSET, as pretreatment of cells with MDMA does not enhance SERT activity (supplemental Fig. 2). It must be noted, however, that further investigation of this phenomenon revealed this stimulation to be quite variable, suggesting that other factors, such as the substrate-dependent phosphorylation state of the transporter (47, 48), may contribute to the effect. Once again, MTSEA-biotinylation experiments were less conclusive in these analyses, with MDMA pretreatment demonstrating nonsignificant reductions in biotinylation at S336C and nonsignificant enhancements in biotinylation at I333C (data not shown). Finally, cocaine has been found to protect against inactivation of hSERT TM1 residues that lie within the 5-HT binding pocket (G100C, N101C), whereas cocaine enhances inactivation of two outer TM1 residues, Y107C and I108C (42). In marked contrast to these findings, cocaine pretreatment did not significantly impact MTSET inactivation at any TM6 position (Fig. 8C).
DISCUSSION
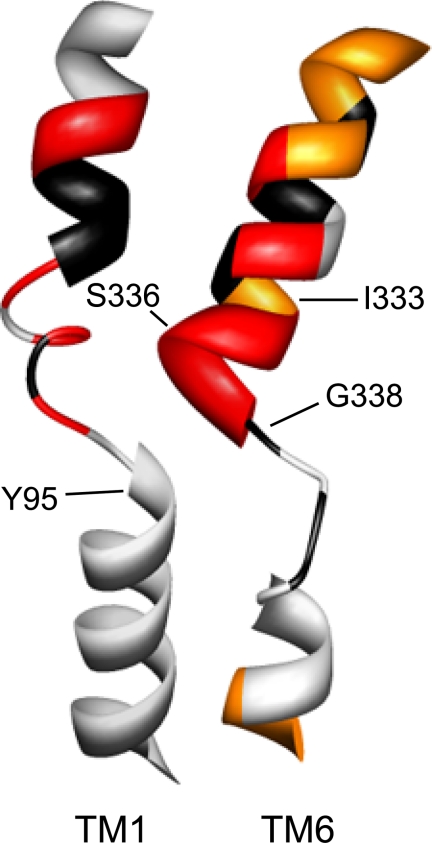
The availability of high resolution structures for the SLC6 family member LeuTAa provided new tools for an increased understanding of the structure of neurotransmitter transporters, including hSERT. Whereas the contributions of TMs 1 and 3 to substrate and antagonist binding sites had been foreshadowed by pre-structure biochemical studies (42, 43, 45, 46, 49–52), the involvement of TMs 6 and 8 in the LeuTAa binding pocket was unexpected. Here, we utilized homology modeling and well-validated biochemical methods to probe TM6 in hSERT, revealing features of TM6 both shared with, and distinct from, its predicted structural homolog and physical neighbor, TM1 (Fig. 9). Our studies document a contribution of TM6 to an aqueously accessible binding pocket for substrates as well as a critical contribution to conformations supportive of 5-HT transport.
FIGURE 9.
Homology model of hSERT TM6. Homology models of hSERT TM1 and TM6 are depicted as constructed by Kaufmann et al. (32), based on LeuTAa structures. TMs 1 and 6 are viewed perpendicular to the plasma membrane with the remaining TMs removed for clarity. Depiction of TM1 and TM6 SCAM results are from this study and from Henry et al. (42). Residues that are intolerant of Cys substitution are depicted in black; residues that are sensitive to MTS inactivation of uptake are depicted in red, and additional residues that are detectable by MTSEA-biotin but not functionally sensitive to MTS reagents are represented in orange.
Similar to the effects of Cys substitution observed in TM1 (42), Cys substitution produced a greater impact on uptake activity in TM6a than the relatively unperturbed TM6b. Based on the complete elimination of transport engendered by the Cys substitutions W326C, A329C, and Q332C, we hypothesized that these mutants would not be expressed at the plasma membrane, and indeed, they were not. In the Rosenberg and Kanner (39) SCAM analysis of GAT-1 TM6a, the analogous Cys mutants W285C, A288C, and Q291C also demonstrated significant reductions in γ-[3H]aminobutyric acid uptake. Similarly, Cys substitution of Gln-317 in an otherwise Cys-less hDAT, analogous to hSERT Q332C, resulted in loss of surface expression (38). Together, these results reveal that this group of residues, whose side chains (based on LeuTAa-based homology models) orient away from the binding pocket, may be critical for inter-TM interactions that support transporter folding or protein stability.
Saturation kinetic analyses of 5-HT transport for the functional Cys mutants offer insight into the contributions of TM6 residues to substrate affinity and transport function. The elevation observed in single point 5-HT transport for the A331C substitution was found to derive solely from an elevated transport Vmax. Although we detected an average elevation in surface expression, this difference did not reach significance, and thus, this substitution may relieve an energy barrier to 5-HT translocation. In contrast, the significantly elevated 5-HT transport activity of F347C in single point assays, despite a decrease in Vmax, appears to derive from a reduction in 5-HT Km. Because this Cys substitution does not exhibit MTS inactivation that can be protected by 5-HT, this Cys mutation likely does not increase 5-HT affinity through a direct interaction with substrate but, rather, stabilizes a conformation that reduces the 5-HT Km at the expense of maximal translocation rates.
Alternately, the opposite basal effect is seen for I333C, where an elevation in the 5-HT Km is accompanied by an increase in transport Vmax. Here, the reduced 5-HT transport capacity of I333C, especially at the low 5-HT concentrations initially tested, reflects an increase in 5-HT Km despite its having an elevated Vmax and surface expression equivalent to C109A. In addition, the low level of 5-HT transport activity observed for V325C, D328C, and S336C is attributable to a decrease in Vmax that is supported by comparatively lower surface expression. The reduced surface expression and 5-HT transport activity of S336C we observed replicates the impact of Cys substitution at GAT-1 S295 on γ-aminobutyric acid transport as well as the loss of surface expression in the hDAT substitution S321C. Interestingly, for the G340C mutant, a decrease in 5-HT transport Vmax is counterbalanced by elevated protein expression, resulting in a single point uptake indistinguishable from C109A. Together, these findings underscore a need to obtain full kinetic profiles for Cys substitutions as well as surface expression information to better understand their overall functional perturbations.
In addition to the impact of Cys substitution, the differences in MTS inactivation of 5-HT uptake along TM6 closely parallel the SCAM profile of TM1 and are supported by recent hSERT homology models. It is important to recognize, however, that our data derive from hSERT in substrate-free, outward-facing conformations, whereas models based on current LeuTAa structures derive from either a completely closed conformation or a conformation with both substrate and a noncompetitive antagonist bound (27, 53). Nonetheless, the limits of aqueous accessibility for TM1 and TM6, assessed in independent SCAM studies, closely correspond, as seen in Fig. 9. Our data indicate that all but one of the functional Cys mutants in TM6a (A330C) is accessible to an MTS reagent, whereas TM6b is largely inaccessible, mirroring the TM1 SCAM data and contributing evidence to an occluded pore. Interestingly, our MTS accessibility data suggest a greater aqueous accessibility of positions 334–337 in TM6 than would be predicted from the LeuTAa homology models, suggesting that the unwound region of TM6 in hSERT may initiate earlier as TM6a spirals through the core of the transporter.
In addition to refining our homology models of hSERT, our SCAM analysis of TM6 identified several residues with interesting characteristics, including Gly-338 and Gly-342. The C109A/G338C mutant was expressed at the plasma membrane at levels equivalent to hSERT C109A but was not competent for 5-HT uptake, whereas the G342C mutant demonstrated a substantial deficit in reaching the plasma membrane. Valine substitutions at analogous positions in hDAT TM6 yielded very similar results; hDAT G323V is minimally expressed at the cell surface but does not transport tyramine, whereas hDAT G327V does not reach the plasma membrane (38). It has been previously suggested from hDAT and hSERT studies that TM6 Gly residues contribute to a dimerization motif that is required for surface expression and function (38, 54). However, the realization that these residues lie within the central core of the transporter, surrounded by outer helices, makes contribution to transporter dimerization highly unlikely. Given the robust expression of hSERT C109A/G338C and the 5-HT uptake exhibited by hSERT C109A/G338A and C109A/G342A, we can conclude that these Gly residues are important but not absolutely required for hSERT expression or function, particularly if limited bulk is introduced at these positions. Additional modeling and experimental studies will be necessary to define the functional limits of residues that are tolerated at positions 338 and 342. Here, we suggest that these Gly residues (and their inherent flexibility) are critical for domain movement during 5-HT translocation.
Importantly, in addition to being expressed at the plasma membrane, G338C remains competent for 5-HT and citalopram binding as well as accessibility to MTSEA-biotin but cannot initiate substrate transport. We propose that hSERT C109A/G338C exists at the surface in a stable conformation with the proposed ligand binding site intact and accessible. Additional experiments, such as a more detailed analysis of citalopram binding affinity, electrophysiological characterization of hSERT C109A/G338C, or electron paramagnetic resonance (EPR) spectroscopy on a bacterial homologue mutant will be necessary to validate an outward-facing conformation. These results could enhance efforts to achieve a structural determination of a ligand-free, outward-facing conformation of SLC6 family members that can supplement current ligand-bound structures (27, 30, 53). Furthermore, our ability to achieve functional recovery of 5-HT uptake with the G338C/Y95F mutant reinforces the assertion that TM1 and TM6 are physically proximal and collaborate in substrate transport. Additional substitutions at Tyr-95 were not tested as other replacements of the tyrosine have proven to be nonfunctional (17); it is noted, however, that these substitutions were not tested in the G338C background.
Our hSERT homology model predicts a significant involvement of TM6 Ser-336 in the coordination of 5-HT (32, 33) as well as sodium and chloride.3 It is interesting to note, therefore, that S336C demonstrates the same differential sensitivity to MTS reagents as previously documented for the TM1 substitution N101C; specifically, that S336C exhibits transport inactivation with positively charged MTSET and MTSEA, whereas the negatively charged MTSES fails to inactivate transport. These findings are consistent with hSERT homology models that depict hSERT where Ser-336 in TM6 is apposed to Asn-101 in TM1 and where access to the two residues is impacted by local negative charge. In this regard, Yamashita et al. (27) have suggested that the unwound regions of TMs 1 and 6 (positioned just below Asn-101 and Ser-336 in hSERT) generate intramembrane dipoles, and thus, such an effect could contribute to what appears to be repulsion of MTSES (55). Consistent with this idea and similar analysis of hSERT N101C (42), prior treatment of hSERT C109A/S336C with MTSES did not protect against subsequent MTSET inactivation, indicating that the negatively charged MTSES cannot access the Cys thiolate that the positively charged MTS reagents readily attack.
As predicted by the proximity of 5-HT in our and others' homology models (32, 33), hSERT S336C could be protected by the substrate from MTSET inactivation, although this protection was incomplete. It is interesting to note, however, that S336C is fully protected in the absence of sodium and only partially protected in the presence of 5-HT; this suggests that the binding of the sodium ion may significantly reorient the side chain of S336C, exposing it to the binding site for attack by MTSET or interaction with 5-HT. It is not surprising that reorientation at S336C would also impact the exposure of F334C and F335C, but the protection by NMDG at D328C suggests that ion binding induces a conformational change experienced across TM6a. The psychostimulant MDMA has an indolamine-like structure and is an hSERT substrate, but it is distinct in that it can trigger transporter-mediated efflux of 5-HT from the cytoplasm (56). A greater understanding of how 5-HT and MDMA differentially interact with hSERT is needed to clarify the structural conformations that support 5-HT efflux. Interestingly, the significantly greater protection of S336C afforded by MDMA co-incubations suggests both an overlap of the 5-HT/MDMA binding sites and a greater extent of physical proximity of MDMA to Ser-336. A further distinction between 5-HT and MDMA arose at I333C, where 5-HT produced no change in hSERT activity in the presence or absence of MTSET. In contrast, MDMA enhanced hSERT I333C activity when the psychostimulant was co-incubated with MTSET. Because neither MDMA nor MTSET on their own can trigger an increase in hSERT I333C activity (supplemental Fig. 2), we propose that Ile-333 may contribute to conformational changes that distinguish MDMA and 5-HT interactions with hSERT, although these conformations may be transient and require covalent modification to promote a detectible enhancement of hSERT activity. Further studies are needed to explore this possibility, including whether it generalizes to other amphetamines (e.g. fenfluramine).
Remarkably, the hSERT competitive antagonist cocaine fails to protect or expose TM6 Cys residues as assessed by MTSET inactivation. Modeling and mutagenesis studies of cocaine and cocaine analog binding in hDAT predict that cocaine would interact significantly with hSERT TM6 residues (16, 57, 58). Specifically, hDAT residues Phe-320, Ser-321, and Leu-322, which correspond by homology to hSERT Phe-335, Ser-336, and Leu-337, are predicted to interact with the N-methylamine of (−)-2β-carbomethoxy-3β-(4-fluorophenyl)tropane (CFT), a cocaine analog, and thus, we predicted that the homologous hSERT residues would be protected by cocaine. Although the data suggest protection at S336C, this effect was small and did not reach significance. In addition, hDAT Phe-326 and Val-328, which correspond by homology to hSERT Phe-341 and Val-343, are predicted to have side chains that interact with the tropane ring of the cocaine-like molecule CFT (16). These findings suggest that, in hSERT, cocaine may orient in a conformation distinct from that adopted in hDAT. However, it is important to recognize that F341C was unable to be assayed due to a lack of surface expression. Secondly, Val-343 lies in a region below our deepest site of MTSET accessibility, limiting our ability to infer interactions via protection. Nonetheless, our studies point to Ser-336 as a site that exhibits preferential physical and/or functional interactions with hSERT substrates versus cocaine. Furthermore, they indicate more limited helix movements in TM6 in response to antagonist binding, especially when compared with TM1, which contained sites of both protection and exposure in the presence of cocaine.
In conclusion, our data contribute critical biochemical information needed to refine the use of LeuTAa structures for the modeling of human SLC6 homologues and point to important similarities and differences regarding the relative contributions TMs 1 and 6 to the 5-HT binding pocket. We identify a single residue, Ser-336, that appears to be in proximity to the 5-HT binding site and is influenced to a greater degree by the psychostimulant MDMA. Finally, the effects of substitutions at Gly-338 highlight the importance of the flexibility of central portions of TM6 for 5-HT transport. The G338C substitution appears to represent a stable, ligand accessible and plasma membrane-expressed form of hSERT that can aid in structural analysis of externally open conformations of this transporter and possibly other SLC6 family members.
Supplementary Material
Acknowledgments
We thank Tammy Jessen, Jane Wright, Qiao Han, Angela Steele, Tracy Moore-Jarrett, Chris Svitek, Sarah Whitaker, and Kathryn Lindler for general laboratory support and Jens Meiler, Kristian Kaufmann, Steven Combs, and Eric Dawson for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants DA07390 (to R. D. B.), DA022378 (to L. K. H.), and DA23337 (to J. R. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
L. K. Henry, unpublished data.
- SERT
- serotonin transporter
- hSERT
- human SERT
- 5-HT
- 5-hydroxytryptamine (serotonin)
- DAT
- dopamine transporter
- hDAT
- human DAT
- GAT-1
- γ-aminobutyric acid transporter type 1
- TM
- transmembrane domain
- MDMA
- 3,4-methylenedioxymethamphetamine
- SLC6
- solute carrier 6
- SCAM
- substituted cysteine accessibility method
- MTS
- methanethiosulfonate
- MTSET
- [2-(trimethylammonium)ethyl]methanethiosulfonate
- MTSEA
- aminoethylmethanethiosulfonate
- MTSES
- (2-sulfonatoethyl)methanethiosulfonate
- PBS
- phosphate-buffered saline
- mKRH
- modified Krebs-Ringers-HEPES buffer
- ANOVA
- analysis of variance
- RIPA
- radioimmune precipitation assay
- NMDG
- N-methyl-d-glucamine.
REFERENCES
- 1.Cooper J., Bloom F., Roth R. (2003) The Biochemical Basis of Neuropharmacology, 7th Ed., pp. 271–304, Oxford University Press, New York [Google Scholar]
- 2.Lill H., Nelson N. (1998) Methods Enzymol. 296, 425–436 [DOI] [PubMed] [Google Scholar]
- 3.Saier M. H. (2002) in Transmembrane Transporters pp. 1–17, (Quick M. ed) pp. 1–17, Wiley-Liss, Hoboken, NJ [Google Scholar]
- 4.Kilic F., Rudnick G. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 3106–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seidel S., Singer E. A., Just H., Farhan H., Scholze P., Kudlacek O., Holy M., Koppatz K., Krivanek P., Freissmuth M., Sitte H. H. (2005) Mol. Pharmacol. 67, 140–151 [DOI] [PubMed] [Google Scholar]
- 6.Chen J. G., Liu-Chen S., Rudnick G. (1998) J. Biol. Chem. 273, 12675–12681 [DOI] [PubMed] [Google Scholar]
- 7.Smicun Y., Campbell S. D., Chen M. A., Gu H., Rudnick G. (1999) J. Biol. Chem. 274, 36058–36064 [DOI] [PubMed] [Google Scholar]
- 8.Androutsellis-Theotokis A., Rudnick G. (2002) J. Neurosci. 22, 8370–8378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tate C. G., Blakely R. D. (1994) J. Biol. Chem. 269, 26303–26310 [PubMed] [Google Scholar]
- 10.Melikian H. E., Ramamoorthy S., Tate C. G., Blakely R. D. (1996) Mol. Pharmacol. 50, 266–276 [PubMed] [Google Scholar]
- 11.Gu H. H., Wall S., Rudnick G. (1996) J. Biol. Chem. 271, 6911–6916 [DOI] [PubMed] [Google Scholar]
- 12.Rudnick G. (1998) Methods Enzymol. 296, 233–247 [DOI] [PubMed] [Google Scholar]
- 13.Sutcliffe J. S., Delahanty R. J., Prasad H. C., McCauley J. L., Han Q., Jiang L., Li C., Folstein S. E., Blakely R. D. (2005) Am. J. Hum. Genet. 77, 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCauley J. L., Olson L. M., Dowd M., Amin T., Steele A., Blakely R. D., Folstein S. E., Haines J. L., Sutcliffe J. S. (2004) Am. J. Med. Genet. B Neuropsychiatr. Genet. 127B, 104–112 [DOI] [PubMed] [Google Scholar]
- 15.Prasad H. C., Steiner J. A., Sutcliffe J. S., Blakely R. D. (2009) Philos. Trans. R. Soc. Lond. B. Biol. Sci. 364, 163–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beuming T., Kniazeff J., Bergmann M. L., Shi L., Gracia L., Raniszewska K., Newman A. H., Javitch J. A., Weinstein H., Gether U., Loland C. J. (2008) Nat. Neurosci. 11, 780–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen J., Olsen L., Hansen K. B., Taboureau O., Jørgensen F. S., Jørgensen A. M., Bang-Andersen B., Egebjerg J., Strømgaard K., Kristensen A. S. (2010) J. Biol. Chem. 285, 2051–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersen J., Taboureau O., Hansen K. B., Olsen L., Egebjerg J., Strømgaard K., Kristensen A. S. (2009) J. Biol. Chem. 284, 10276–10284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jardetzky O. (1966) Nature 211, 969–970 [DOI] [PubMed] [Google Scholar]
- 20.Sitte H. H., Hiptmair B., Zwach J., Pifl C., Singer E. A., Scholze P. (2001) Mol. Pharmacol. 59, 1129–1137 [DOI] [PubMed] [Google Scholar]
- 21.Sitte H. H., Scholze P., Schloss P., Pifl C., Singer E. A. (2000) J. Neurochem. 74, 1317–1324 [DOI] [PubMed] [Google Scholar]
- 22.Adams S. V., DeFelice L. J. (2003) Biophys. J. 85, 1548–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams S. V., DeFelice L. J. (2002) Biophys. J. 83, 3268–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kahlig K. M., Binda F., Khoshbouei H., Blakely R. D., McMahon D. G., Javitch J. A., Galli A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3495–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quick M. W. (2003) Neuron 40, 537–549 [DOI] [PubMed] [Google Scholar]
- 26.Yernool D., Boudker O., Jin Y., Gouaux E. (2004) Nature 431, 811–818 [DOI] [PubMed] [Google Scholar]
- 27.Yamashita A., Singh S. K., Kawate T., Jin Y., Gouaux E. (2005) Nature 437, 215–223 [DOI] [PubMed] [Google Scholar]
- 28.Weyand S., Shimamura T., Yajima S., Suzuki S., Mirza O., Krusong K., Carpenter E. P., Rutherford N. G., Hadden J. M., O'Reilly J., Ma P., Saidijam M., Patching S. G., Hope R. J., Norbertczak H. T., Roach P. C., Iwata S., Henderson P. J., Cameron A. D. (2008) Science 322, 709–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faham S., Watanabe A., Besserer G. M., Cascio D., Specht A., Hirayama B. A., Wright E. M., Abramson J. (2008) Science 321, 810–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh S. K., Piscitelli C. L., Yamashita A., Gouaux E. (2008) Science 322, 1655–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krishnamurthy H., Piscitelli C. L., Gouaux E. (2009) Nature 459, 347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaufmann K. W., Dawson E. S., Henry L. K., Field J. R., Blakely R. D., Meiler J. (2009) Proteins 74, 630–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Celik L., Sinning S., Severinsen K., Hansen C. G., Møller M. S., Bols M., Wiborg O., Schiøtt B. (2008) J. Am Chem. Soc. 130, 3853–3865 [DOI] [PubMed] [Google Scholar]
- 34.Jørgensen A. M., Tagmose L., Jørgensen A. M., Bøgesø K. P., Peters G. H. (2007) Chem. Med. Chem. 2, 827–84017436258 [Google Scholar]
- 35.Jørgensen A. M., Tagmose L., Jørgensen A. M., Topiol S., Sabio M., Gundertofte K., Bøgesø K. P., Peters G. H. (2007) Chem. Med. Chem. 2, 815–826 [DOI] [PubMed] [Google Scholar]
- 36.Beuming T., Shi L., Javitch J. A., Weinstein H. (2006) Mol. Pharmacol. 70, 1630–1642 [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y. W., Rudnick G. (2006) J. Biol. Chem. 281, 36213–36220 [DOI] [PubMed] [Google Scholar]
- 38.Hastrup H., Karlin A., Javitch J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10055–10060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenberg A., Kanner B. I. (2008) J. Biol. Chem. 283, 14376–14383 [DOI] [PubMed] [Google Scholar]
- 40.Karlin A., Akabas M. H. (1998) Methods Enzymol. 293, 123–145 [DOI] [PubMed] [Google Scholar]
- 41.Chen J. G., Liu-Chen S., Rudnick G. (1997) Biochemistry 36, 1479–1486 [DOI] [PubMed] [Google Scholar]
- 42.Henry L. K., Adkins E. M., Han Q., Blakely R. D. (2003) J. Biol. Chem. 278, 37052–37063 [DOI] [PubMed] [Google Scholar]
- 43.Chen J. G., Sachpatzidis A., Rudnick G. (1997) J. Biol. Chem. 272, 28321–28327 [DOI] [PubMed] [Google Scholar]
- 44.Kaplan R. S., Mayor J. A., Brauer D., Kotaria R., Walters D. E., Dean A. M. (2000) J. Biol. Chem. 275, 12009–12016 [DOI] [PubMed] [Google Scholar]
- 45.Barker E. L., Perlman M. A., Adkins E. M., Houlihan W. J., Pristupa Z. B., Niznik H. B., Blakely R. D. (1998) J. Biol. Chem. 273, 19459–19468 [DOI] [PubMed] [Google Scholar]
- 46.Adkins E. M., Barker E. L., Blakely R. D. (2001) Mol. Pharmacol. 59, 514–523 [DOI] [PubMed] [Google Scholar]
- 47.Ramamoorthy S., Blakely R. D. (1999) Science 285, 763–766 [DOI] [PubMed] [Google Scholar]
- 48.Jayanthi L. D., Ramamoorthy S. (2005) Aaps. J. 7, E728–E738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen J. G., Rudnick G. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1044–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanner B. I. (2003) J. Biol. Chem. 278, 3705–3712 [DOI] [PubMed] [Google Scholar]
- 51.Dodd J. R., Christie D. L. (2005) J. Biol. Chem. 280, 32649–32654 [DOI] [PubMed] [Google Scholar]
- 52.Henry L. K., Field J. R., Adkins E. M., Parnas M. L., Vaughan R. A., Zou M. F., Newman A. H., Blakely R. D. (2006) J. Biol. Chem. 281, 2012–2023 [DOI] [PubMed] [Google Scholar]
- 53.Singh S. K., Yamashita A., Gouaux E. (2007) Nature 448, 952–956 [DOI] [PubMed] [Google Scholar]
- 54.Horschitz S., Lau T., Schloss P. (2008) Neurochem Int. 52, 770–775 [DOI] [PubMed] [Google Scholar]
- 55.Henry L. K., Meiler J., Blakely R. D. (2007) Mol. Interv. 7, 306–309 [DOI] [PubMed] [Google Scholar]
- 56.Rudnick G., Wall S. C. (1992) Proc. Natl. Acad. Sci. 89, 1817–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parnas M. L., Gaffaney J. D., Zou M. F., Lever J. R., Newman A. H., Vaughan R. A. (2008) Mol. Pharmacol. 73, 1141–1150 [DOI] [PubMed] [Google Scholar]
- 58.Vaughan R. A., Sakrikar D. S., Parnas M. L., Adkins S., Foster J. D., Duval R. A., Lever J. R., Kulkarni S. S., Hauck-Newman A. (2007) J. Biol. Chem. 282, 8915–8925 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.