Abstract
Serotonin is a well known neurotransmitter in mammals and plays an important role in various mental functions in humans. In plants, the serotonin biosynthesis pathway and its function are not well understood. The rice sekiguchi lesion (sl) mutants accumulate tryptamine, a candidate substrate for serotonin biosynthesis. We isolated the SL gene by map-based cloning and found that it encodes CYP71P1 in a cytochrome P450 monooxygenase family. A recombinant SL protein exhibited tryptamine 5-hydroxylase enzyme activity and catalyzed the conversion of tryptamine to serotonin. This pathway is novel and has not been reported in mammals. Expression of SL was induced by the N-acetylchitooligosaccharide (chitin) elicitor and by infection with Magnaporthe grisea, a causal agent for rice blast disease. Exogenously applied serotonin induced defense gene expression and cell death in rice suspension cultures and increased resistance to rice blast infection in plants. We also found that serotonin-induced defense gene expression is mediated by the RacGTPase pathway and by the Gα subunit of the heterotrimeric G protein. These results suggest that serotonin plays an important role in rice innate immunity.
Keywords: Cell Death, Cell Wall, Cytochrome P450, Innate Immunity, Plant, Rac, Serotonin
Introduction
Plants have developed various immune responses to protect themselves from pathogen attack (1). One of them is the hypersensitive response, which is often accompanied by programmed cell death, and is effective to prevent the growth of biotrophic pathogens (2). The hypersensitive response is induced coordinately with production of reactive oxygen species, lignification, production of phytoalexins, and expression of defense-related genes.
To understand the roles of hypersensitive response cell death in immune responses, a large number of lesion mimic mutants, in which cell death is spontaneously induced, have been investigated in many plant species, including rice, Arabidopsis, and maize (3). Most of lesion mimic mutants are known to activate immune responses in the absence of pathogens (2). A rice lesion mimic mutant, Sekiguchi lesion (sl), exhibits unique orange colored lesions that are induced by inoculation of pathogens, including Magnaporthe grisea (4, 5). In the sl mutants, increased activities of tryptophan decarboxylase and monoamine oxidase are correlated with lesion formation (5). However, the identity of the SL gene and its function in cell death and disease resistance remain to be studied.
Plant Rac/Rop GTPases constitute a unique subfamily of the Rho family of small GTPases and are highly conserved in plant species. In rice, OsRac1 regulates a series of immune responses, including cell death, reactive oxygen species production, activation of pathogenesis-related genes, liginification, and phytoalexin production (6). Recently, it was found that OsRac1 forms a complex with RAR1, HSP90, HSP70, OsMAPK6, OsrbohB, Sti1/Hop, and RACK1 (7–10), which play important roles in rice innate immunity. Heterotrimeric G protein is also involved in innate immunity through OsRac1. In fact, rice dwarf1 (d1) mutants lacking a Gα subunit of heterotrimeric G protein reduces elicitor-induced defense gene expression and resistance to M. grisea (11).
Serotonin is known as neurotransmitter in mammals, which regulates mood, sleep, and anxiety in human (12). Although serotonin was found in many plant species (13), the serotonin biosynthesis and its function are unclear. Recently, serotonin has been reported to be accumulated in rice during senescence and defense response (14, 15). In addition, serotonin is incorporated into the cell wall at disease lesions infected with Bipolaris oryzae and M. grisea, suggesting that serotonin plays an important role in control of the strength of the cell wall for the mechanical barrier against pathogens (15).
To understand how the SL gene regulates cell death and disease resistance in rice, we have isolated the SL gene by a map-based approach. We found that the SL gene encodes cytochrome P450 monooxygenase, which catalyzes 5-hydroxylation of tryptamine to synthesize serotonin. In addition, our results suggest that serotonin contributes to innate immunity by activating intracellular signaling of immune responses.
EXPERIMENTAL PROCEDURES
Plant Materials and Genetic Mapping
The wild type japonica rice cv. Kinmaze and four sl mutants (CM265, CM998, CM1019, and CM2229), induced by N-methyl-N-nitrosourea in the Kinmaze background, were provided by the Japanese National BioResource Project of Rice. The CM265 mutant was used in a cross with a substitution line (SL239) carrying a segment of chromosome 12 from an indica rice (cv. Kasalath) in a japonica rice background (cv. Koshihikari). The F2 and F3 plants were analyzed using a sequence-tagged site and cleaved amplified polymorphic sequence markers to construct a physical map of the region containing the SL gene (supplemental Fig. S1). Details of the mapping are provided in the supplemental “Experimental Procedures”.
Plasmid Constructs and Rice Transformation
The SL cDNA was cloned into the p2K1 vector containing the maize Ubq1 promoter (16). For the RNA interference construct of Sti1a/Hop, a 350-bp 3′-untranslated region of the Sti1a cDNA was PCR-amplified and cloned in the reverse orientation behind the Ubp1 promoter in the pANDA vector (16). The RNA interference construct of OsRac1 was described previously (10). Details of plasmid construction are provided in supplemental “Experimental Procedures”. Transgenic plants were generated from Agrobacterium-transformed rice calli (17).
Reverse Transcription-PCR and Real Time PCR
Total RNAs were isolated from leaves and suspension cultures using TRIzol reagent (Invitrogen) and treated with DNase I (Invitrogen). cDNA was synthesized using SuperScriptII reverse transcriptase (Invitrogen) and used for reverse transcription-PCR and real time PCR with gene-specific primers (see supplemental Table S1).
Rice Blast Infection
Rice seedlings were inoculated at the four- to six-leaf stage by spraying with aqueous spore suspensions containing 105 spores/ml of the compatible rice blast races 003 and 007. Inoculated seedlings were kept in a dark chamber with a moisture-saturated atmosphere at 24 °C for 20 h and then maintained in a greenhouse. Inoculated leaves were harvested at the indicated time points and used for real time PCR. To examine the effect of serotonin on rice blast resistance, rice seedlings were watered with 400 μg/ml serotonin for 22 h and then inoculated with race 007 as described above. Lesion development was scored on rice leaves at 6 days after inoculation by measuring lesion length with a ruler.
Chitin, Serotonin, and Tryptamine Treatments
Rice suspension cultures were treated with 2 μg/ml N-acetylchitooligosaccharide elicitor, 400 μg/ml (1.88 mm) serotonin (5-hydroxytryptamine hydrochloride (Mr 212.68)), or 400 μg/ml (2.03 mm) tryptamine (tryptamine hydrochloride (Mr 196.68)), and then incubated for various lengths of time. As reported previously (15), less than 30% of applied serotonin is considered to be incorporated by the feeding experiment. Total RNAs were extracted from each sample and subjected to real time PCR. For the detection of cell death, suspension-cultured cells were treated with serotonin or tryptamine for 24 h, stained for 10 min with 0.05% (w/v) Evans Blue, and then washed four times with distilled water (18). Dye bound to dead cells was dissolved in a solution of 50% methanol and 1% SDS for 30 min at 50 °C and quantified by absorbance at 595 nm.
Measurement of SL Activities
The SL and SL-G455D (CM265) cDNAs were cloned as SpeI-PstI fragments in the pFastBac1 vector (Invitrogen) and used to transform E. coli strain DH10Bac (Invitrogen). Preparation of the cDNA- containing recombinant baculovirus DNAs and transfection of Spodoptera frugiperda 9 (Sf9) cells were carried out according to the manufacturer's instructions (Invitrogen). Heterologous expression in Sf9 cells and spectrophotometric analysis of P450s were carried out as described previously (19).
Microsomal fractions of insect cells expressing either SL or SL-G455D were obtained as follows. Infected suspension-cultured cells (300 ml) were washed with phosphate-buffered saline buffer and suspended in buffer A, consisting of 20 mm potassium phosphate (pH 7.25), 20% (v/v) glycerol, 1 mm EDTA, and 1 mm dithiothreitol. The cells were sonicated, and cell debris was removed by centrifugation at 10,000 × g for 15 min. The supernatant was further centrifuged at 100,000 × g for 1 h, and the pellet was homogenized with buffer A to provide the microsomal fractions. The microsomal fractions were stored at −80 °C before the enzyme assays were performed.
SL activity was reconstituted by mixing the microsomes with purified Arabidopsis NADPH-P450 reductase (20). The reaction mixture consisted of 20 mm potassium phosphate (pH 7.25), 50 pmol/ml recombinant P450 protein, 0.1 unit/ml NADPH-P450 reductase, 1 mm NADPH, and 100 μm tryptamine. Reactions were initiated by the addition of NADPH, and were carried out at 30 °C for 30 min. The samples were diluted with 20 mm phosphate (pH 2.5) and subjected to HPLC4 as follows: column, COSMOSIL 5C18-PAQ column (150 mm length × 4.6 mm inner diameter; Nacalai Tesque); solvent, 20 mm phosphate (pH 2.5); flow rate, 1.0 ml/min; detection, a fluorescence detector with an excitation wavelength at 285 nm and an emission wavelength at 340 nm. Retention times of authentic samples were 6.5 min for serotonin and 14.5 min for tryptamine. The identity of the reaction product with tryptamine was examined using HPLC equipped with an API-3000 triple stage quadrupole mass spectrometer (Applied Biosystems) as described previously (15). For kinetic analysis, SL activity was assayed using the substrate tryptamine at concentrations ranging from 1 to 40 μm. Reactions were initiated by the addition of NADPH and carried out at 30 °C for 10 min. The kinetic constants were calculated from triplicate data sets. Km and kcat values were calculated from a double-reciprocal plot of the initial velocity (v0) versus substrate concentration.
Immunoblotting
Microsomal fractions of the insect cells, expressing either SL or SL-G455D, were subjected to immunoblotting using a purified anti-SL antibody as described in supplemental “Experimental Procedures.”
RESULTS
Isolation of SL Gene by Map-based Cloning
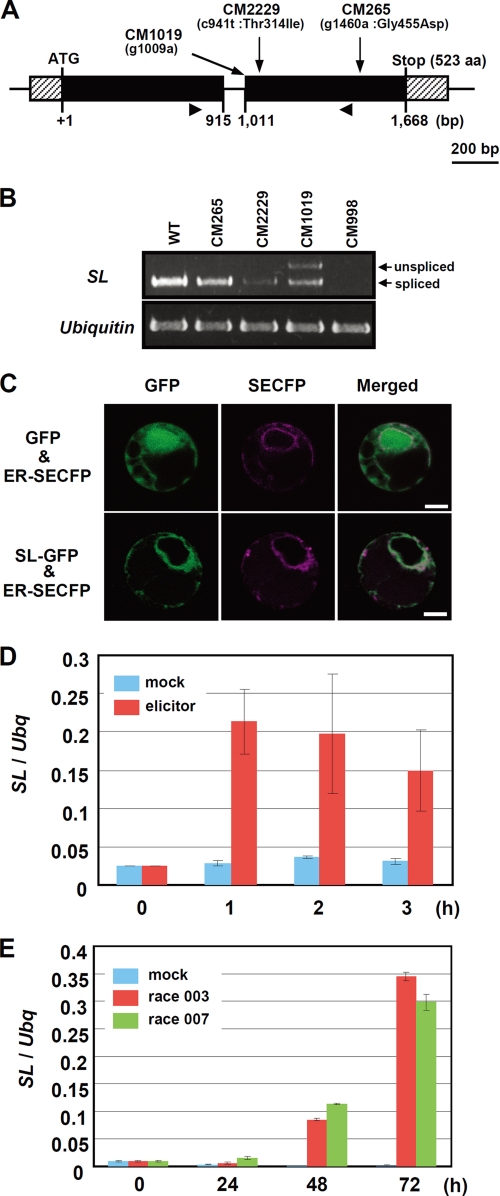
To identify a gene responsible for the sl phenotype, we used four sl mutants (CM265, CM2229, CM998, and CM1019; supplemental Fig. S1), which were generated by mutagenesis using N-methyl-N-nitrosourea (21). The sl locus was mapped to a 350-kb genomic region on chromosome 12, where 7 candidate genes are located (supplemental Fig. S1). PCR experiments indicated that the CM998 mutant has a 30-kb deletion containing genes for a P450 and a zinc finger protein.5 Sequence analyses in the other mutants revealed that SL encodes a P450 monooxygenase (Fig. 1A). SL gene expression levels were significantly reduced in CM2229. CM998 showed no SL expression because of the deletion of the entire P450 gene. CM1019 had two altered transcripts as follows: an unspliced transcript resulting from a point mutation at the 3′ splice site of the single intron, and another generated by cryptic splicing at a site near the 3′ splice site (Fig. 1B). Overexpression of the P450 mRNA complemented the sl phenotype (supplemental Fig. S2).
FIGURE 1.
SL gene encodes a cytochrome P450 monooxygenase. A, structure of the SL gene. Arrows indicate the locations of mutations found in the sl mutants. Arrowheads indicate locations of primers used in B. aa, amino acids. B, expression of SL in the sl mutants. Reverse transcription-PCR was performed using specific primers for SL and ubiquitin. WT, wild type. C, subcellular localization of SL-GFP. GFP (upper panel) and SL-GFP (lower panel) were transiently expressed in rice protoplasts with ER-localized SECFP. Left panel, GFP image; middle panel, SECFP image; right panel, merged images of GFP and SECFP. Bars, 10 μm. D, expression of SL after elicitor treatment. Total RNAs were purified from rice suspension cultured cells either mock-treated or treated with 2 μg/ml N-acetylchitooligosaccharide elicitor and used for real time PCR. Ubiquitin (Ubq) was used as an internal control. Error bars indicate standard deviations. E, expression of SL in seedlings after mock inoculation or inoculation with compatible rice blast fungus (race 003 and 007). Real time PCR was carried out using total RNAs prepared from the seedlings. Ubq was used as an internal control. Error bars indicate S.D.
SL Belongs to the CYP71 Family
The SL protein possesses motifs conserved in P450 proteins, including the heme-binding motif, the EXXR motif, and the proline-rich motif (supplemental Fig. S3). The G455D mutation in CM265 is in the heme-binding motif and is likely to disturb the heme coordination structure. The threonine residue at position 314, which is mutated in CM2229, is important for the oxygen activation process in the P450 catalytic cycle (supplemental Fig. S3). The SL protein shares 41% identity with CYP71A1 from avocado fruit (GenBankTM accession number AAA32913). In rice there are 90 genes and 28 pseudogenes in the CYP71 family; however, the SL protein shows less than 45% identity with any other members of the family. Hence, the SL protein is a unique member of the CYP71 family and is designated CYP71P1.
The intracellular location of the SL protein was investigated using the transient expression of a GFP-fused protein in rice protoplasts (Fig. 1C). The SL protein, with GFP fused to the C terminus, co-localized with a marker protein that was designed to localize to the endoplasmic reticulum (ER). The marker protein was constructed by adding the N-terminal signal peptide of pro-2S albumin, which localizes to the ER (22), and the C-terminal HDEL ER retention sequence, to SECFP (23). The results indicate that SL localizes to the ER.
SL Gene Is Expressed during Innate Immune Responses
Expression of the SL gene in rice suspension cultures was analyzed after treatment with a chitin elicitor that is known to induce a series of defense responses in rice (Fig. 1D) (24). A clear increase in SL mRNA levels was observed at 1 h after chitin treatment (Fig. 1D). We also analyzed SL expression after infection with M. grisea. Infection by two races of compatible blast fungus caused the activation of SL gene expression in rice plants at 48 h after infection, and expression was further increased at 72 h (Fig. 1E). These results suggest that SL has a role in rice innate immunity.
SL Protein Possesses Tryptamine 5-Hydroxylase Activity
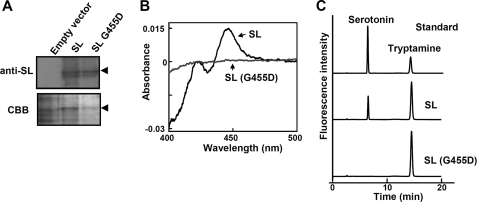
It was shown previously that tryptamine levels were higher in an sl mutant than in wild type plants and that serotonin levels were very low (4, 5, 15). These results suggest that SL converts tryptamine to serotonin. To test this possibility, we generated recombinant SL protein with or without the G455D mutation found in the CM265 allele, using a baculovirus expression system in insect cells. The presence of the recombinant SL and SL-G455D proteins in the microsomal fractions was confirmed by immunoblot analysis (Fig. 2A). Fractions containing the wild type SL protein had a cytochrome P450-specific reduced CO difference spectrum with a clear absorption peak at 450 nm (Fig. 2B). In contrast, the fractions containing the SL-G455D protein exhibited no reduced CO difference spectrum (Fig. 2B). These results indicate that SL protein is indeed a cytochrome P450 enzyme. The microsomal fractions from insect cells expressing the recombinant SL proteins were used in enzyme assays with tryptamine as the potential substrate. HPLC analysis of the reaction products showed a specific peak at the same retention time as that of serotonin when fractions containing the wild type SL protein, but not the SL-G455D mutant protein, were used (Fig. 2C). The SL protein catalyzed the 5-hydroxylation of tryptamine with a high affinity and efficiency (Km = 7.3 ± 0.8 μm and kcat = 45 ± 1.5 min−1), although the SL protein did not metabolize tryptophan, indole 3-acetic acid, or tyramine. Thus, it is likely that the SL protein is tryptamine 5-hydroxylase that catalyzes the conversion of tryptamine to serotonin.
FIGURE 2.
SL is a tryptamine 5-hydroxylase. A, production of recombinant SL proteins in insect cells using a baculovirus expression system. Microsomal fractions from the insect cells expressing the recombinant SL proteins were separated by SDS-PAGE and analyzed on an immunoblot using the anti-SL antibody. CBB, Coomassie Brilliant Blue. B, CO difference spectrum of the microsomal fractions expressing SL proteins. C, HPLC analysis of reaction products after the microsomal SL proteins were assayed for enzyme activity using tryptamine as a substrate.
Serotonin Plays a Role in Innate Immune Responses
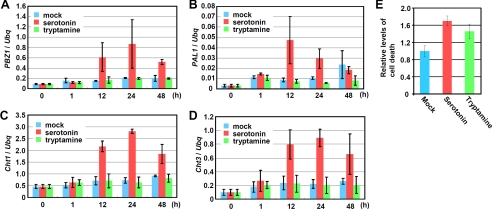
Serotonin has been reported to be accumulated at a high level (1.9 mm) in disease lesions infected with B. oryzae and M. grisea (15), whereas the high accumulation of serotonin is abolished in the sl mutant. In addition, the sl mutant is more susceptible to infection of B. oryzae as compared with wild type, and application of serotonin suppresses the fungal growth in the leaves of the sl mutant (15). Thus, it is possible that serotonin is involved in innate immunity. To elucidate how serotonin regulates immunity responses, we analyzed the expression of four defense genes, probenazol 1 (PBZ1), phenylalanine ammonia-lyase 1 (PAL1), chitinase 1 (Cht1), and chitinase 3 (Cht3) in rice cell suspension cultures. These four genes are induced by chitin elicitor from fungal pathogens (25).6 Expression of these defense genes was clearly induced at 12 h after serotonin treatment, but no induction was found with tryptamine (Fig. 3, A–D). Serotonin also induced cell death in cultured rice cells, as shown by Evans Blue staining (Fig. 3E), although reduced levels of cell death occurred after tryptamine treatment.
FIGURE 3.
Serotonin induces defense gene expression and cell death in rice suspension cell cultures. Total RNAs were prepared from rice suspension cells either mock-treated or treated with 400 μg/ml serotonin or tryptamine. The samples were subjected to real time PCR to analyze expression of PBZ1 (A), PAL1 (B), Cht1 (C), and Cht3 (D). Ubiquitin (Ubq) was used as an internal control. Error bars indicate S.D. E, rice suspension cultured cells were treated with 400 μg/ml serotonin or tryptamine. The levels of cell death were measured using Evans Blue staining.
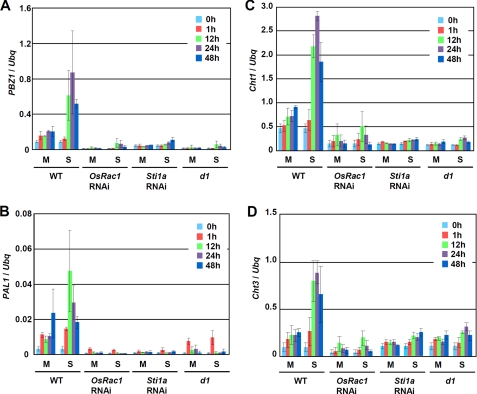
Because serotonin was able to induce immune responses in rice cell cultures, we examined the possibility that known components of rice innate immunity are involved in serotonin-induced defense gene induction. It has been shown that the small GTPase OsRac1 and its signaling partners, including Sti1/Hop and a heterotrimeric G protein, are involved in rice innate immunity (8, 10, 26). For these experiments, we used rice cell cultures expressing RNA interference knockdown constructs for OsRac1 and Sti1/Hop, and a cell culture of the dwarf1 (d1) mutant, which lacks a functional Gα subunit of the heterotrimeric G protein (11). The results clearly showed that these proteins are involved in serotonin-induced defense gene induction (Fig. 4, A–D).
FIGURE 4.
Involvement of OsRac1, Sti1/Hop, and the Gα subunit in serotonin-induced defense gene expression. Expression of PBZ1 (A), PAL1 (B), Cht1 (C), and Cht3 (D) in wild type (WT), OsRac1, and Sti1a RNA interference and d1 mutant cells treated with serotonin (S) or water (mock; M). Total RNA samples were analyzed by real time PCR. Ubiquitin (Ubq) was used as an internal control. Error bars indicate S.D.
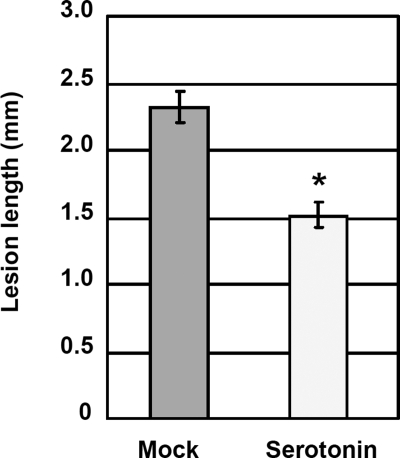
Because our results suggest that serotonin plays a role in rice innate immunity, we examined whether serotonin treatment of rice seedlings (∼1 month old) had any effect on infection with a compatible race of M. grisea. The results indicate that seedlings treated with serotonin are significantly more resistant to fungal infection than mock-treated seedlings (Fig. 5), suggesting that serotonin has the ability to induce resistance against rice blast infection.
FIGURE 5.
Induction of rice blast resistance by serotonin treatment. Rice seedlings were inoculated with the compatible blast fungus race 007. Disease lesion development was scored at 6 days after inoculation by measuring lesion length. Error bars indicate standard error (n = 78). An asterisk indicates a significant difference by the t test at p < 0.01.
DISCUSSION
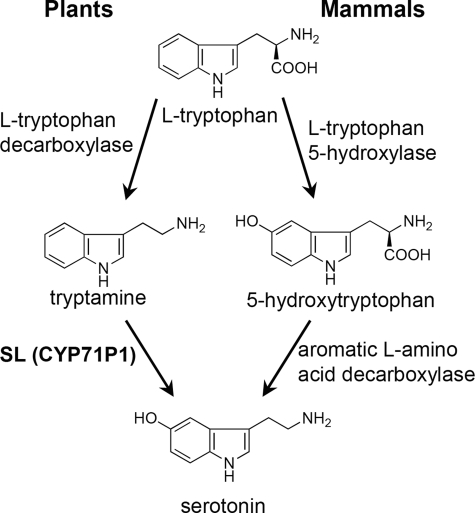
Serotonin is a well known neurotransmitter in mammals and plays an important role in various mental functions in humans (27). In mammals, serotonin is synthesized from tryptophan in two steps involving l-tryptophan 5-hydroxylase and aromatic l-amino acid decarboxylase (Fig. 6) (28). Tryptophan hydroxylase belongs to a family of aromatic amino acid hydroxylases and requires iron and the coenzyme tetrahydropterin for catalytic activity (29). It is interesting that the SL gene encodes CYP71P1 in the P450 superfamily, which is different from the tetrahydropterin-dependent hydroxylase family in mammals (Fig. 6). Serotonin has been found in more than 42 plant species (13), suggesting that enzymes for serotonin biosynthesis are conserved in plants. However, the SL orthologs have not been found in other plants, including Arabidopsis. Among the Arabidopsis P450s, CYP83B1/SUR2 shows the highest identity (39%) with the SL protein. CYP83B1/SUR2 catalyzes the N-hydroxylation of indole-3-acetaldoxime to synthesize indole glucosinolate (30).
FIGURE 6.
Model for serotonin biosynthetic pathways in plants and mammals.
It has been reported that some serotonin derivatives such as coumaroylserotonin and feruloylserotonin, which are hydroxycinnamic acid amides, are involved in plant defense responses against pathogen infection (31). The accumulation of hydroxycinnamic acid amides in the cell wall could provide an undegradable mechanical barrier for pathogens (31, 32). In fact, it was shown that serotonin itself is polymerized and incorporated into cell walls in rice, leading to the suppression of growth of the fungus B. oryzae (15). It was also shown that hydroxycinnamic acid amides possess anti-fungal activity (33). These studies suggest that serotonin may act as a phytoalexin, an antimicrobial compound that is produced by plant cells in response to pathogen infection. However, we found no effect of serotonin on the germination of M. grisea spores (supplemental Table S2). In this study, we found that serotonin induces immune responses including defense gene expression and cell death through the pathway involved in Gα, OsRac1, and Sti1. Taken together, it is suggested that serotonin may have dual functions that regulate the strength of the cell wall for the physical barrier against pathogens as well as activate intracellular signaling for immune responses.
We have identified SL as tryptamine 5-hydroxylase enzyme for serotonin biosynthesis, and we showed that in rice, serotonin is synthesized in different pathways found in animals. Recently, it was reported that serotonin is accumulated during senescence in rice (14). However, whether SL is involved in senescence response remains to be analyzed. Two genes, tryptophan decarboxylase and SL, required for serotonin synthesis from tryptophan have now been identified in plants. It is therefore possible to control the serotonin levels in plants by manipulating the expression of these genes.
Supplementary Material
Acknowledgments
We thank Yuko Tamaki for technical assistance and Nagao Hayashi for help with the rice blast infections. We thank the Rice Genome Resource Center for providing a substitution line and a full-length cDNA clone, and the National BioResource Project of Rice for providing the sl mutant lines.
This work was supported by grants-in-aid from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Rice Genome Project IP4001), the Japan Society for Promotion of Science Grant 13G0023 (to K. S.), and the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) (to T. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Tables S1 and S2, and Figs. S1–S3.
M. Isshiki, unpublished results.
S. H. Kim, unpublished results.
- HPLC
- high pressure liquid chromatography
- TDC
- tryptophan decarboxylase
- ER
- endoplasmic reticulum
- GFP
- green fluorescent protein
- SECFP
- super enhanced cyan fluorescent protein.
REFERENCES
- 1.Hammond-Kosack K. E., Parker J. E. (2003) Curr. Opin. Biotechnol. 14, 177–193 [DOI] [PubMed] [Google Scholar]
- 2.Hofius D., Tsitsigiannis D. I., Jones J. D., Mundy J. (2007) Semin. Cancer Biol. 17, 166–187 [DOI] [PubMed] [Google Scholar]
- 3.Lorrain S., Vailleau F., Balagué C., Roby D. (2003) Trends Plant Sci. 8, 263–271 [DOI] [PubMed] [Google Scholar]
- 4.Arase S., Ueno M., Toko M., Honda Y., Itho K., Ozoe Y. (2001) J. Phytopathol. 149, 409–413 [Google Scholar]
- 5.Ueno M., Shibata H., Kihara J., Honda Y., Arase S. (2003) Plant J. 36, 215–228 [DOI] [PubMed] [Google Scholar]
- 6.Kawasaki T., Henmi K., Ono E., Hatakeyama S., Iwano M., Satoh H., Shimamoto K. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 10922–10926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lieberherr D., Thao N. P., Nakashima A., Umemura K., Kawasaki T., Shimamoto K. (2005) Plant Physiol. 138, 1644–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thao N. P., Chen L., Nakashima A., Hara S., Umemura K., Takahashi A., Shirasu K., Kawasaki T., Shimamoto K. (2007) Plant Cell 19, 4035–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong H. L., Pinontoan R., Hayashi K., Tabata R., Yaeno T., Hasegawa K., Kojima C., Yoshioka H., Iba K., Kawasaki T., Shimamoto K. (2007) Plant Cell 19, 4022–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakashima A., Chen L., Thao N. P., Fujiwara M., Wong H. L., Kuwano M., Umemura K., Shirasu K., Kawasaki T., Shimamoto K. (2008) Plant Cell 20, 2265–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suharsono U., Fujisawa Y., Kawasaki T., Iwasaki Y., Satoh H., Shimamoto K. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13307–13312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gingrich J. A., Hen R. (2001) Psychopharmacology 155, 1–10 [DOI] [PubMed] [Google Scholar]
- 13.Roshchina V. V. (2001) Neurotransmitters in Plant Life ( Roshchina V. V. ed) pp.4– 81, Science Publisher, Enfield, NH [Google Scholar]
- 14.Kang K., Kim Y. S., Park S., Back K. (2009) Plant Physiol. 150, 1380–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishihara A., Hashimoto Y., Tanaka C., Dubouzet J. G., Nakao T., Matsuda F., Nishioka T., Miyagawa H., Wakasa K. (2008) Plant J. 54, 481–495 [DOI] [PubMed] [Google Scholar]
- 16.Miki D., Shimamoto K. (2004) Plant Cell Physiol. 45, 490–495 [DOI] [PubMed] [Google Scholar]
- 17.Hiei Y., Ohta S., Komari T., Kumashiro T. (1994) Plant J. 6, 271–282 [DOI] [PubMed] [Google Scholar]
- 18.Hayashi M., Takahashi H., Tamura K., Huang J., Yu L. H., Kawai-Yamada M., Tezuka T., Uchimiya H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 7020–7025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saito S., Hirai N., Matsumoto C., Ohigashi H., Ohta D., Sakata K., Mizutani M. (2004) Plant Physiol. 134, 1439–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizutani M., Ohta D. (1998) Plant Physiol. 116, 357–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satoh H., Omura T. (1979) J. Fac. Agr. Kyushu Univ. 24, 165–174 [Google Scholar]
- 22.Hara-Hishimura I., Takeuchi Y., Inoue K., Nishimura M. (1993) Plant J. 4, 793–800 [DOI] [PubMed] [Google Scholar]
- 23.Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K., Miyawaki A. (2002) Nat. Biotechnol. 20, 87–90 [DOI] [PubMed] [Google Scholar]
- 24.Yamaguchi T., Yamada A., Hong N., Ogawa T., Ishii T., Shibuya N. (2000) Plant Cell 12, 817–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Midoh N., Iwata M. (1996) Plant Cell Physiol. 37, 9–18 [DOI] [PubMed] [Google Scholar]
- 26.Ono E., Wong H. L., Kawasaki T., Hasegawa M., Kodama O., Shimamoto K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 759–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geyer M. A., Vollenweider F. X. (2008) Trends Pharmacol. Sci. 29, 445–453 [DOI] [PubMed] [Google Scholar]
- 28.Mockus S. M., Vrana K. E. (1998) J. Mol. Neurosci. 10, 163–179 [DOI] [PubMed] [Google Scholar]
- 29.Fitzpatrick P. F. (2003) Biochemistry 42, 14083–14091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bak S., Tax F. E., Feldmann K. A., Galbraith D. W., Feyereisen R. (2001) Plant Cell 13, 101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hahlbroch K., Scheel D. (1989) Annu. Rev. Plant Physiol. Plant Mol. Biol. 40, 347–369 [Google Scholar]
- 32.Keller H., Hohlfeld H., Wray V., Hahlbrock K., Scheel D., Strack D. (1996) Phytochemistry 42, 389–396 [Google Scholar]
- 33.Miyagawa H., Ishihara A., Nishimoto T., Ueno T., Mayama S. (1995) Biosci. Biotechnol. Biochem. 59, 2305–2306 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.