Abstract
ΔF508 cystic fibrosis transmembrane conductance regulator (CFTR) degradation involves ubiquitin modification and efficient proteasomal targeting of the nascent misfolded protein. We show that a deubiquitinating enzyme, ubiquitin C-terminal hydrolase-L1 (UCH-L1), is highly expressed in cystic fibrosis (CF) airway epithelial cells in vitro and in vivo. We hypothesized that the elevation in UCH-L1 in CF cells represents a cellular adaptation to counterbalance excessive proteasomal degradation. The bronchial epithelial cell lines IB3-1 (CF, high UCH-L1 expression) and S9 (non-CF, low UCH-L1 expression) were transiently transfected with wild type (WT) or ΔF508 CFTR, WT UCH-L1 or small interfering RNA-UCH-L1, and a variety of ubiquitin mutants. We observed a positive correlation between UCH-L1 expression and steady state levels of WT- or ΔF508-CFTR, and this stabilizing effect was confined to the early stages of CFTR synthesis. Immunolocalization of UCH-L1 by confocal microscopy revealed a partial co-localization with a ribosomal subunit and the endoplasmic reticulum. The UCH-L1-associated increase in CFTR levels was correlated with an increase in ubiquitinated CFTR (CFTR-Ub). Co-transfection with mutant ubiquitins and treatment with proteasome inhibitors suggested that UCH-L1 was reducing the proteasomal targeting of CFTR during synthesis by shortening conjugated polyubiquitin chains. Although not sufficient by itself to rescue mutant CFTR therapeutically, the elevation of UCH-L1 and its effect on CFTR processing provides insight into its potential roles in CF and other diseases.
Keywords: Diseases/Cystic Fibrosis, Protease/Inhibitor, Proteases/Ubiquitination, Protein/Intracellular Trafficking, Protein/Targeting, RNA/RNAi
Introduction
Cystic fibrosis (CF)3 is caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR), an epithelial chloride channel that is required to optimally hydrate the apical surface of mammalian secretory tissues. CFTR is a 12-pass transmembrane protein that must undergo a complex sequence of folding events, and many CF disease-causing mutations alter the ability of the CFTR protein to assemble properly and/or prevent its maturation to the cell surface (1, 2). ΔF508, the most common CF mutation, results in a misfolded protein with increased affinity for components of the endoplasmic reticulum-associated degradation (ERAD) machinery, and most (∼99%) of this newly synthesized protein is degraded by the proteasome. Because of the complex nature of folding, wild type CFTR is also processed inefficiently, with only ∼25% of the protein escaping ER quality control and maturing to the plasma membrane (3, 4).
Ubiquitin modification is required for the proteasomal targeting of a large number of proteins including CFTR, which is ubiquitinated at the ER during assembly and also during recycling from the cell surface (5–7). Ubiquitination requires an E1 activating enzyme, an E2 ubiquitin conjugating enzyme, and an E3 ubiquitin ligase (8). The ubiquitination of misfolded CFTR begins as the nascent protein is synthesized, and recent studies have reported that several ERAD molecules, including Rma1, gp78, Derlin-1, and BAP31-Sec61, are involved in this co-translational processing (9–13). CHIP, an Hsc70 associated E3 ligase, also promotes the ubiquitination of CFTR but acts following the completion of synthesis (9, 14–16). Once polyubiquitinated, either co- or post-translationally, CFTR is extracted from the ER by the retrograde translocation protein complex, including VCP/p97, and is directed to the proteasome for degradation (11, 17).
Although the requirement for ubiquitination in CFTR degradation is apparent, little is known about the role of deubiquitinating enzymes (DUBs) in this process. One DUB, Usp19, was recently demonstrated to stabilize several ERAD substrates including ΔF508 CFTR (18). DUBs are responsible for counteracting the action of ubiquitination machinery through the cleavage of conjugated ubiquitin (19). During a proteomics-based screen of newly synthesized protein in CF and non-CF cell lines, we detected a striking elevation in one DUB, ubiquitin C-terminal hydrolase-L1 (UCH-L1) (20).
DUBs belonging to the UCH family are proposed to be involved in the cleavage of short ubiquitin chains and the maintenance of monomeric ubiquitin levels in the cell (21). UCH-L1 was originally described as a neuron-specific DUB making up ∼1% of soluble brain protein (22). More recently, it has been shown to be associated with several cancers including lung, cervical, and prostate (23–25). UCH-L1 possesses Lys48-dependent ubiquitin hydrolase and Lys63-dependent ubiquitin ligase activities in vitro (26), and several studies suggest a role for these activities in the onset of neurological diseases (27–31). However, the substrates of UCH-L1 are unknown, and its precise function in protein processing is not clear. Several studies support a general housekeeping deubiquitination activity as a major function, with UCH-L1 acting to increase the pool of monomeric ubiquitin available for conjugation to target proteins (32, 33). Structural studies, however, suggest that a conformational change is required to access the active site and support the possibility that ubiquitin-conjugated protein substrates of UCH-L1 exist (34).
The hypothesis of this work is that elevations in UCH-L1 in CF represent a cellular compensatory mechanism to rescue misfolded mutant CFTR. We demonstrate that UCH-L1 expression interferes with the proteasomal degradation of wild type and ΔF508 CFTR. UCH-L1-mediated stabilization of CFTR is confined to the early stages of protein synthesis, and UCH-L1 co-localizes with both ER and ribosomal markers. By introducing a series of mutant ubiquitin moieties, we demonstrate that favoring shortened ubiquitin chains similarly stabilizes CFTR during synthesis and enhances the UCH-L1-mediated effect.
EXPERIMENTAL PROCEDURES
Cell Lines and Culture
The CF bronchial epithelial cell line IB3-1 (ΔF508/W1282X; low level expression of ΔF508-CFTR and no W1282X protein) (35), a CF tracheal epithelial cell line CFTE (ΔF508-homozygous) (36), and a CF line with wild type phenotype S9 (IB3-1 corrected by AAV-CFTR) (37) were maintained in LHC-8 medium with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin B at 37 °C in the presence of 5% CO2.
Human Subjects
Pediatric patients with and without cystic fibrosis undergoing fiberoptic bronchoscopy for a clinical indication were invited to donate bronchial mucosal brushings to this research. Informed consent and assent were obtained according to institutional guidelines (Johns Hopkins Medical Institutional Review Board). The samples were deidentified but linked to medical data including gender, age, microbiology of specimen, concomitant medications, and, in some cases, spirometry data. The brushings were immediately immersed in LHC-8 medium, placed on ice, and transported to the laboratory, where they were processed for Western blotting as described for the cell lines.
Plasmids and Transfection
The pEGFP WT- and ΔF508-CFTR expression vectors were described previously (38). A UCH-L1 expression vector, pcDNA3.1-WT UCH-L1 was a kind gift from P. Lansbury (Harvard). The pRK5-Ub-HA (WT, Lys48, and Lys63) mammalian expression vectors were a kind gift from T. Dawson and C. Pickart (Johns Hopkins). A ubiquitin expression vector in which all of the lysines were mutated was created with Lys63 Ub as a template and primers: (+)5′-GACTACAACATCCAGAGAGAGTCCACCCTGCACC-3′ and (−)5′-GGTGCAGGGTGGACTCTCTCTGGATGTTGTAGTC-3′. The site-directed mutagenesis PCR was performed as follows: 95 °C for 45 s, and 18 three-step cycles: 95 °C for 45 s, 57.5 °C for 1 min, and 68 °C for 6 min. Successful mutagenesis was confirmed by sequencing. The mCherry-UCH-L1 expression vector was created by PCR amplification of the mCherry tag from pRSETB-mCherry (a kind gift from R. Tsien, UCSD) with NheI and HindIII terminal sequences. The primers were: (+)5′-GCGCTAGCATGGTGAGCAAGGGCG-3′ and (−)5′-CGAAGCTTCTTGTACAGCTCGTCCATG-3′. The mCherry tag was then inserted into the pcDNA3.1-WT UCH-L1 plasmid (N-terminal tag). Co-transfections were performed with the indicated DNA constructs using Lipofectamine 2000 (Invitrogen) and studied after 48 h unless otherwise indicated.
Reagents and Antibodies
The antibodies used for immunoblotting were monoclonal UCH-L1 (10A1) (Abcam, Cambridge, MA), monoclonal CFTR (M3A7) (Abcam), polyclonal UCH-L3 (Abgent, San Diego, CA), monoclonal HA (Millipore, Billerica, MA), monoclonal ubiquitin (P4D1) (Santa Cruz, Santa Cruz, CA), and polyclonal actin (Sigma). Polyclonal CFTR (CFTR-169, directed against the R domain) (39) was used for immunoprecipitation. For translation inhibition experiments, the cells were incubated with 100 μg/ml cycloheximide (EMD Chemicals Inc., San Diego, CA) for the indicated times. For knockdown experiments, On-TARGETplus duplexes (Dharmacon, Lafayette, CO) targeting UCH-L1 (J-004309–08-0050) or a nonspecific control were used at 100 nm. Proteasome inhibitor experiments were performed with MG132 (Calbiochem) and ALLN (Sigma-Aldrich) at 50 and 200 μm, respectively. N-Ethylmaleimide (Sigma) was added to the lysis buffer at 20 mm to inhibit deubiquitination following protein extraction.
Real Time PCR
Total RNA was extracted with TRIzol (Invitrogen), treated with DNaseI (Qiagen), and purified with RNeasy spin columns (Qiagen). 100 ng of total RNA served as a template using the Superscript III Platinum SYBR Green One-Step quantitative reverse transcription-PCR kit (Invitrogen) and UCH-L1, CFTR, or actin primers (Invitrogen). The primers were: UCH-L1 (+)5′-CCAGCATGAGAACTTCAGGA-3′ and (−)5′-AATTCCCAATGGTCTGCTTC-3′ (product size, 95 bp); actin (+)5′-CACTCTTCCAGCCTTCCTTC-3′ and (−)5′-GGATGTCCACGTCACACTTC-3′ (product size, 90 bp); and CFTR (+)5′-TCTTTCTCTGCAAACTTGG-3′ and (−)5′-TTGCTGGATCCACTGGAG-3′ (product size, 244 bp). The real time reverse transcription-PCR was carried out as follows: 50 °C for 3 min, 95 °C for 10 min, and 40 two-step cycles: 95 °C for 15 s and 60 °C for 1 min. The data were collected using the ABI PRISM 7900HT sequence detection system (Applied Biosystems, Foster City, CA) and analyzed with Sequence Detection Software 2.0 (Applied Biosystems) using the threshold cycle to determine the increase in transcript associated with an exponential growth of PCR product during the log linear phase. The relative expression of CFTR was calculated using the 2−ΔΔCT method where ΔΔCT = (CT,CFTR − CT,Actin)CFTR+WT-UCH-L1 − (CT,CFTR − CT,Actin)CFTR+Vector. For UCH-L1 transcript measurement, ΔΔCT = (CT,UCH-L1 − CT,Actin)IB3-1 − (CT,UCH-L1 − CT,Actin)S9. The standard curves for all primers demonstrated dilution versus calculated ΔΔCT profiles with a slope of 1 ± 0.02 and a correlation coefficient of R2 > 0.99.
Pulse-Chase, Immunoprecipitation, and Immunoblotting
Bronchial epithelial cells were grown on 10-cm2 dishes overnight and transfected with the indicated constructs for 24 h. The cells were washed two times with 2 ml of PBS and 1 ml of Cys/Met-free Dulbecco's modified Eagle's medium was added for 35 min (starvation). Trans-35S label was then added at 200 μCi/ml for 20 min (pulse). The cells were washed twice with 2 ml of PBS and 2 ml of fresh LHC-8 was added for the chase period. The cells were immediately lysed in RIPA buffer (50 mm Tris-HCl, 150 mm NaCl, 1% Nonidet P-40 (Pierce), 0.65% sodium deoxycholate, 0.20% SDS, 1 mm EDTA) containing 20 mm N-ethylmaleimide (DUB inhibitor) and 1× mammalian protease inhibitor mixture (Sigma). Protein lysates were rotated at 4° for 30 min and passed four times through a 26-gauge syringe. The lysates were quantified using the BCA protein assay (Pierce), and 300 μg of the samples were used for immunoprecipitation. The samples were precleared using protein G-Sepharose (GE Healthcare) beads for 3 h, and polyclonal anti-CFTR (CFTR-169) was added at a 1:100 dilution to the cleared lysates for 1 h at 4°. 50 μl of protein G-Sepharose beads were added, and the samples were rotated overnight. The following day, CFTR immunoprecipitates were washed three times with cold PBS, and 2× LDS sample buffer was added to the Sepharose pellet. The samples were then incubated at 44° for 20 min and loaded on 5% polyacrylamide gels (Bio-Rad). After separation, the gels were dried in preparation for autoradiography. Kodak Biomax Films were exposed at −80° for 12 h to 5 days depending on signal intensity. Western blot transfers were performed in a buffer containing 25 mm Tris-base, 200 mm glycine, and 20% methanol.
Confocal Microscopy
IB3-1 cells were plated on 10-cm2 glass bottom dishes (Mattek, Ashland, MA) at 20% (immunolocalization) or 40% (fluorescent tag) confluence and allowed to grow overnight. For the mCherry-UCH-L1 and EmGFP experiments, IB3-1 cells were transiently transfected for 24 h. The cells were fixed at room temperature with 4% paraformaldehyde or at −20 °C with ice-cold methanol for 30 min. For immunolocalization experiments, the cells were fixed in fresh 4% paraformaldehyde for 25 min. The cells were then permeabilized in 0.2% Triton X-100 for 15 min, blocked in 2% bovine serum albumin with PBS for 40 min, and incubated with primary antibody overnight at 4°. The primary antibodies used were polyclonal UCH-L1 at 1:50 (Millipore), monoclonal RPS6 at 1:50 (Cell Signaling, Danvers, MA), and monoclonal KDEL at 1:50 (Abcam). Highly cross-adsorbed Alexa Fluor 488 and 568 (Invitrogen) secondary antibodies were used at 1:400 in 2% bovine serum albumin with PBS + 10% goat serum.
RESULTS
UCH-L1 Expression Is Elevated in CF Cells in Vitro and in Vivo
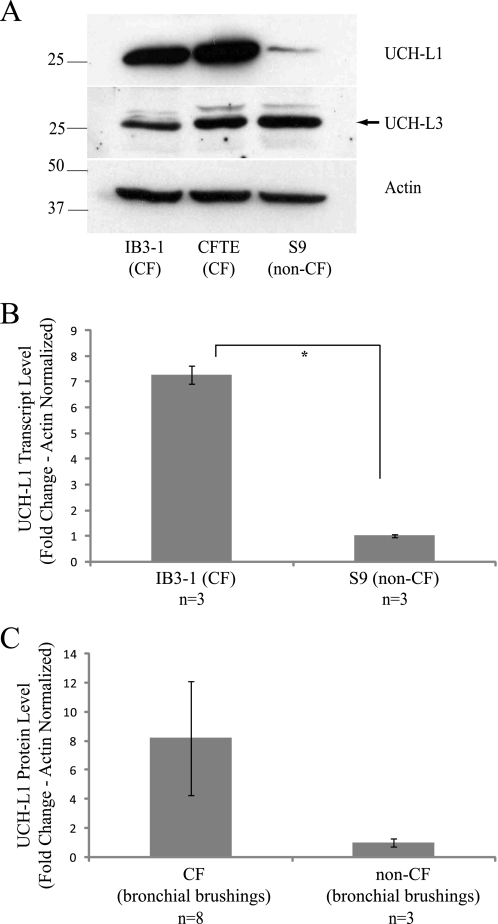
Proteomic analysis identified UCH-L1 protein expression as elevated in the CF IB3-1 cell line as compared with the WT CFTR-corrected S9 (20). To validate these earlier observations, we performed Western blot analysis on cell lysates from IB3-1 (ΔF508/W1282X) and S9 (CF corrected − ΔF508/W1282X + AAV-WT-CFTR) cells and a second CF cell line homozygous for ΔF508 (CFTE). UCH-L1 protein was visibly elevated in the two CF lines when compared with S9 (Fig. 1A, top panel). To determine whether CF is associated with a global increase in DUBs, we examined the closely related UCH-L3, which demonstrated similar expression in all three cell lines (Fig. 1A, middle panel). To determine whether UCH-L1 was elevated at the mRNA transcript level, we performed real time PCR for UCH-L1. We observed a significant increase in UCH-L1 mRNA transcript levels in IB3-1 as compared with S9 (7.3 ± 0.61; Fig. 1B).
FIGURE 1.
UCH-L1 protein and mRNA expression is elevated in CF cells. A, UCH-L1 protein levels are elevated in two CF cell lines, IB3-1 and CFTE. Whole cell protein lysates (30 μg) from IB3-1 (ΔF508/W1282X), CFTE (ΔF508/ΔF508), and S9 (ΔF508/W1282X + AAV-WT CFTR) were resolved by SDS-PAGE and immunoblotted with antibodies to UCH-L1 (upper panel), UCH-L3 (middle panel), and actin (lower panel). B, UCH-L1 transcription is elevated in IB3-1 cells. UCH-L1 mRNA transcripts were measured by real time PCR in the matched CF line IB3-1 and WT CFTR corrected S9 (n = 3 each). The values in the graph are normalized to actin and displayed as ΔΔCT (*, p < 0.001). C, freshly isolated human bronchial epithelial cells from children with and without CF were analyzed for UCH-L1 protein expression. The lysates were resolved by SDS-PAGE and immunoblotted for UCH-L1. The mean UCH-L1 protein expression (± S.E.) of eight CF subjects and three non-CF subjects is quantified by densitometry and normalized to protein loaded (p = 0.06).
We also examined UCH-L1 expression in freshly isolated bronchial epithelial cells from CF and non-CF pediatric patients undergoing bronchoalveolar lavage for a clinical indication. UCH-L1 protein demonstrated strong expression in five of eight CF samples and none of three non-CF samples by immunoblotting. The mean densitometric value of UCH-L1 protein normalized to protein loaded in each well was 8.2 ± 3.9 times greater for CF brushings than non-CF brushings (Fig. 1C; p = 0.06). The concordance of elevated UCH-L1 with CF in vitro (Fig. 1, A and B) and trend observed in vivo (Fig. 1C) suggests that it may be involved in the processing of mutant CFTR.
CFTR Protein Levels Are Regulated by UCH-L1
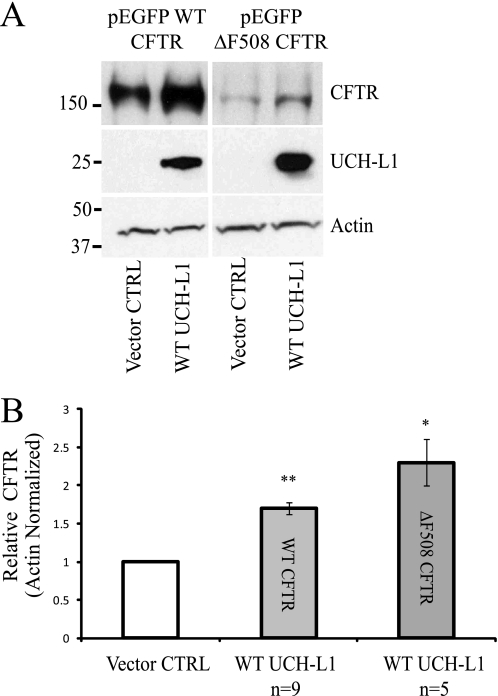
To determine whether UCH-L1 expression modulates steady state CFTR protein levels, we performed Western blot analysis of immunoprecipitated CFTR after transient overexpression of UCH-L1 in the S9 cell line, which expresses low levels of endogenous UCH-L1 (Fig. 2). Co-transfection of UCH-L1 and WT CFTR in S9 cells resulted in a 1.7 ± 0.08-fold increase in bands B/C compared with cells co-transfected with WT CFTR and UCH-L1 vector control (p < 0.001, n = 9). For ΔF508 CFTR, an increase of 2.3 ± 0.3-fold was observed following UCH-L1 overexpression (p < 0.01, n = 5).
FIGURE 2.
CFTR levels are positively correlated with UCH-L1 expression. A, CFTR levels are increased following UCH-L1 overexpression. S9 cells were transfected with WT or ΔF508 CFTR and WT UCH-L1 or its vector control (pcDNA3.1). CFTR was immunoprecipitated, and protein levels were examined by immunoblot. B, densitometric analysis was performed on independent experiments for WT CFTR (n = 9) and ΔF508 CFTR (n = 5) as described for A. *, p < 0.01; **, p < 0.001.
UCH-L1 Increases the Synthesis of CFTR but Has Little Effect on the Rate of Post-translational Degradation
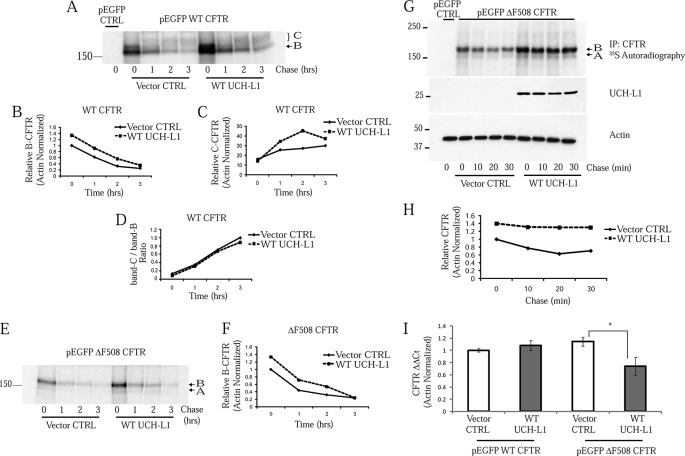
We hypothesized that the CFTR stabilization observed following UCH-L1 expression was due to changes in its protein processing and degradation. To examine the temporal fate of newly synthesized CFTR, we performed a series of pulse-chase experiments in S9 cells. These cells were co-transfected for 24 h, and a pulse-chase was performed over 3 h to follow the fate of CFTR. By densitometry, there was a 1.33-fold increase in immature WT CFTR (band B) at time 0 immediately after the pulse in cells co-transfected with WT UCH-L1 as compared with pcDNA3.1 control vector (Fig. 3, A and B). At each later time point, the UCH-L1 overexpressing cells processed more band B to band C (Fig. 3C). The ratio of C/B CFTR, however, was nearly identical for vector control as compared with WT UCH-L1 (Fig. 3D). This suggested that the increase in mature CFTR was largely due to the increase in early intermediates and did not reflect a role for UCH-L1 in CFTR maturation. The experiment was also performed in S9 cells overexpressing ΔF508 CFTR. This time, band B was not processed to band C, indicating that most of the misfolded CFTR was retained within the ER (Fig. 3E). UCH-L1 co-expression was associated with a 1.34-fold increase in mutant CFTR at time 0 immediately following the pulse (Fig. 3, E and F). Importantly, the rates of degradation for WT or ΔF508-CFTR over the 3-h chase were similar for vector control or WT UCH-L1 co-expression.
FIGURE 3.
UCH-L1 increases CFTR biogenesis but has little effect on the rates of post-translational degradation. Pulse-chase analysis of S9 cells expressing WT CFTR (A–D) or ΔF508 CFTR (E and F) and UCH-L1 or its vector control (pcDNA3.1). The pulse-chase was performed as described under “Experimental Procedures.” The first lane contains the 0-min time point from cells transfected with the empty GFP vector alone and represents endogenous band B mutant CFTR, which is undetectable under these experimental conditions. The results are displayed as CFTR autoradiography (A and E) and densitometry (B–D and F). Note that there is an increase in CFTR at time 0 in the UCH-L1 overexpressing cells but no difference in the rate of disappearance of band B CFTR between the control and WT UCH-L1 overexpression. G, pulse-chase examining CFTR as performed in A and B but with shortened chase time points. H, densitometry of G. I, UCH-L1 expression does not increase CFTR-GFP transcription (n = 3). *, p < 0.05. CFTR mRNA was measured by real time PCR as described under “Experimental Procedures.” Control reactions excluding template or substituting Taq polymerase were negative for CFTR and actin. CFTR levels are normalized to actin and displayed as relative values using the ΔΔCT method.
Our results suggested that UCH-L1 was stabilizing an early biogenic intermediate of CFTR or, alternatively, UCH-L1 was increasing transcription of exogenous CFTR in our model system. To test the early effect hypothesis, we performed a pulse-chase examining 10-, 20-, and 30-min chase points. An increase in CFTR of 1.40-fold was observed at time 0, and UCH-L1 mediated stabilization was maintained throughout the 30-min chase (Fig. 3, G and H). Again, a majority of the stabilization was observed at time 0, with a minimal difference in the rate of post-translational degradation. To evaluate the possibility of a transcriptional effect of UCH-L1 on CFTR expression, we measured WT and ΔF508 CFTR transcription in S9 cells following vector control or WT UCH-L1 co-expression. Real time PCR analysis of CFTR indicated that UCH-L1 expression was not associated with an increase in WT or ΔF508 CFTR transcription (Fig. 3I). Therefore, we conclude that UCH-L1 is increasing the level of CFTR post-transcriptionally.
Endogenous UCH-L1 Expression Is Positively Correlated with CFTR Biogenesis
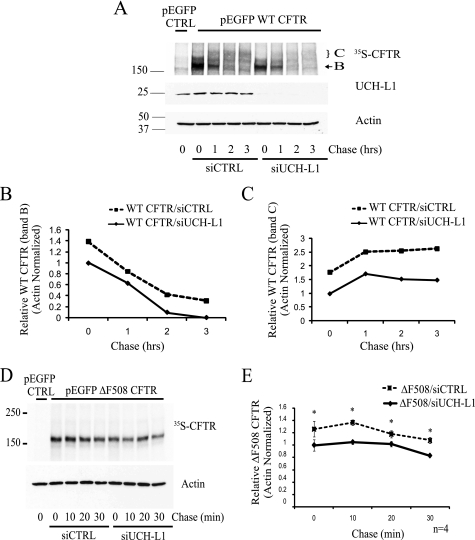
To examine the role of endogenous UCH-L1 in CFTR biogenesis, we utilized siRNA targeting UCH-L1 to knock down expression in IB3-1. With pulse-chase analysis, a decrease in immature band B and mature band C WT CFTR expression was observed following UCH-L1 knockdown (Fig. 4, A–C). Analysis of ΔF508 CFTR by pulse-chase following siRNA knockdown of UCH-L1 also revealed a reduction in ΔF508 CFTR expression following UCH-L1 knockdown (Fig. 4, D and E; n = 4; *, p < 0.05). The effect of UCH-L1 knockdown on CFTR was observed predominately during the pulse-period (time 0) for both WT and ΔF508 CFTR. Thus, the steady state protein level of endogenous UCH-L1 is positively correlated with the amount of WT- or ΔF508-CFTR protein synthesized.
FIGURE 4.
Endogenous UCH-L1 levels are positively correlated with CFTR biogenesis. IB3-1 cells were transfected with WT or ΔF508 CFTR and a nontargeting siRNA (siCTRL) or siRNA targeting UCH-L1 for 48 h. Following transfection, CFTR biogenesis was analyzed by pulse-chase. UCH-L1 levels were examined by immunoblot, and actin served as a loading control. A, knockdown of UCH-L1 reduces WT CFTR protein expression. A pulse-chase examining WT CFTR expression following UCH-L1 knockdown was performed as described under “Experimental Procedures.” B, densitometric analysis of immature band B WT CFTR from A. C, densitometric analysis of mature band C CFTR in A. D, knockdown of UCH-L1 with siRNA reduces ΔF508 CFTR protein expression. ΔF508 CFTR biogenesis was analyzed by pulse-chase. E, the mean ΔF508 CFTR expression ± S.E. from four independent experiments was calculated by densitometry. *, p < 0.05.
UCH-L1 Is Membrane-associated and Co-localizes with the Ribosomal Subunit RPS6 and the ER Retention Signal KDEL
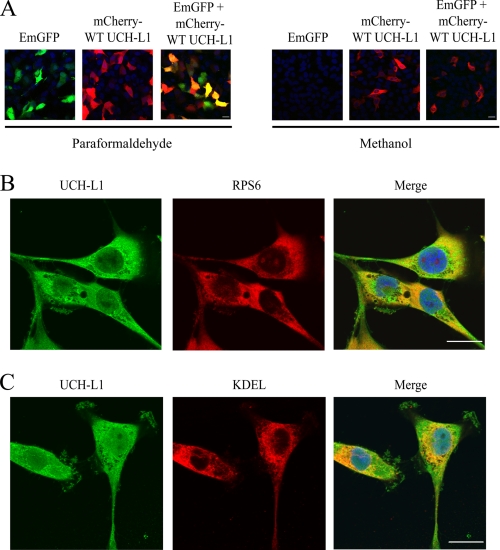
Our results suggested that the stabilizing effect of UCH-L1 was occurring during early CFTR biogenesis. We therefore hypothesized that UCH-L1 would be localized to the site of CFTR synthesis: the ER-associated ribosome. We analyzed the ability of UCH-L1 to remain membrane-associated in bronchial epithelial cells following methanol fixation, which results in leakage of free cytosolic proteins (40–42). IB3-1 cells were transiently transfected with mCherry tagged UCH-L1, EmGFP, or both expression vectors and fixed with paraformaldehyde or methanol. Following paraformaldehyde fixation, confocal microscopy demonstrated expression of both proteins (Fig. 5A, Paraformaldehyde). Following methanol treatment, mCherry-UCH-L1 was retained, whereas EmGFP was completely removed from the cells (Fig. 5A, Methanol). This suggests that a fraction of UCH-L1 is membrane-localized or associated with a membrane-bound protein. To further explore UCH-L1 localization, we performed immunostaining of UCH-L1 and a small ribosomal subunit (RPS6) or ER retention signal (KDEL). Immunofluorescence of endogenous UCH-L1 in paraformaldehyde fixed IB3-1 cells demonstrated that UCH-L1 partially co-localized with both RPS6 (Fig. 5B) and KDEL (Fig. 5C). We conclude that a portion of UCH-L1 is membrane-associated and partially co-localizes with the ribosome and ER in bronchial epithelial cells.
FIGURE 5.
UCH-L1 is partially co-localized to the site of CFTR protein synthesis. A, UCH-L1 remains associated with cellular membranes following methanol fixation, whereas free cytosolic GFP leaks from the cells. IB3-1 cells were transiently transfected with EmGFP, mCherry-WT UCH-L1, or both. The cells were fixed in paraformaldehyde (cytosolic proteins retained) or methanol (cytosolic proteins leak). Blue indicates nuclear 4′,6′-diamino-2-phenylindole staining. The cells were analyzed using a Zeiss 510 Meta confocal microscope, and the images were processed using Zeiss LSM image examiner software. All of the images were obtained using identical confocal laser and scan settings. B, confocal microscopy demonstrates UCH-L1 partially co-localizes with the ribosome. IB3-1 cells were plated on 10-cm2 Mattek dishes. The cells were fixed in fresh 4% paraformaldehyde for 25 min, permeabilized in 0.2% Triton X-100 for 15 min, and blocked in 2% bovine serum albumin with PBS for 40 min. UCH-L1 and RPS6 (small ribosomal subunit) primary antibodies were added followed by Alexa 488 and 568 secondary antibodies. The nucleus was stained with 4′,6′-diamino-2-phenylindole. C, co-localization of UCH-L1 and the ER were examined by confocal microscopy as described in B. An antibody specific for the ER retention signal, KDEL, was used to immunostain the ER. The scale bar is equal to 20 μm.
Proteasome Inhibition Stabilizes CFTR during Synthesis and Abrogates the UCH-L1-mediated Effect
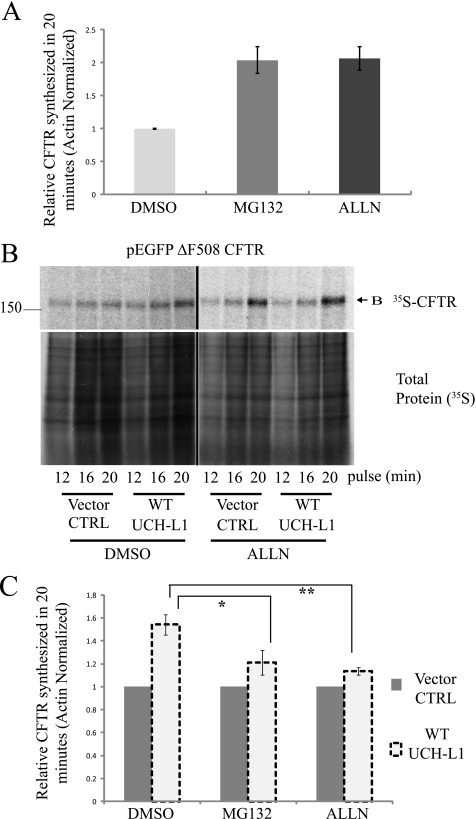
Given the observation that UCH-L1 stabilized newly synthesized CFTR and was localized to the site of protein synthesis, we explored the possibility that UCH-L1 was protecting an early CFTR intermediate from proteasome-mediated degradation. Previous reports have suggested that a fraction of CFTR becomes targeted to the proteasome before translation is fully completed (2, 3, 9, 43, 44). Similarly, we observed that a 20-min pretreatment with the proteasome inhibitors MG132 (50 μm) or ALLN (200 μm) significantly increased the yield of CFTR during a 20-min radiolabeling period by 2.0- and 2.1-fold, respectively, supporting the notion of co-translational proteasome targeting (Fig. 6A; p < 0.01). We next attempted to determine whether UCH-L1 was protecting CFTR from proteasome-mediated degradation. To remove the proteasome from this equation, we added ALLN or MG132 20 min before pulse-labeling the newly synthesized CFTR. Importantly, the addition of either 50 μm MG132 or 200 μm ALLN prior to the pulse period abrogated the stabilizing effect of UCH-L1 on CFTR (Fig. 6, B and C; p < 0.05). The reduced effect after proteasome inhibition suggests a role for UCH-L1 in the protection of CFTR from proteasome-mediated degradation.
FIGURE 6.
Enhanced expression of UCH-L1 protects CFTR from proteasome-mediated degradation. The contribution of the proteasome to early CFTR synthesis was examined by the addition of the proteasome inhibitors ALLN or MG132. The pulse-chase was performed as described previously, but with the addition of proteasome inhibitors during the Cys/Met starvation period (20 min prior to the pulse). A, proteasome inhibition results in a 2-fold increase in ΔF508-CFTR observed following a 20-min pulse. Densitometric analysis of ΔF508 biosynthesis (means ± S.E.) from seven independent CFTR radiolabeling experiments. *, p < 0.01. B, proteasome inhibition diminishes the UCH-L1-mediated stabilization of CFTR. Representative autoradiography from the experiment outlined in A, but with UCH-L1 overexpression. Proteasome inhibitor (200 μm ALLN) was added 20 min prior to the pulse to assess whether blockade of degradation contributes to UCH-L1-mediated preservation of CFTR during biogenesis. The lower blot shows total protein biosynthesis. The effect of UCH-L1 on CFTR in the presence of ALLN was examined (right panel) compared with in the presence of dimethyl sulfoxide (DMSO) vehicle control (CTRL, left panel). C, quantification of the effect of proteasome inhibition on UCH-L1-mediated CFTR biosynthesis/stabilization (pulse = 20 min). The data are displayed as the means ± S.E. of band B CFTR. For each experimental condition, the CFTR level was normalized to the empty vector control to assess the UCH-L1 dependent effect. The mean CFTR expression levels were calculated for 50 μm MG132 (n = 7; *, p < 0.05) and 200 μm ALLN (n = 7; **, p < 0.01) treatments.
UCH-L1 Overexpression Increases the Levels of Ubiquitinated CFTR
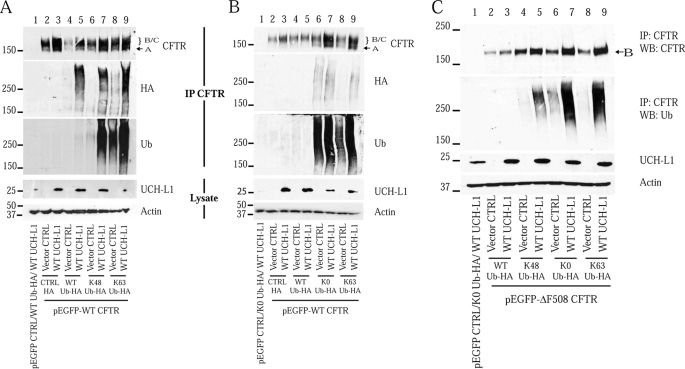
UCH-L1 contains ubiquitin hydrolase and ligase enzymatic domains (21, 26). We considered the possibility that UCH-L1 was modifying the ubiquitination of CFTR during translation, thereby preventing its extraction and/or degradation and resulting in the observed net increase in synthesis. To test the effects of modified ubiquitin chains on UCH-L1-mediated CFTR stabilization, S9 cells were co-transfected for 48 h with CFTR (WT or ΔF508), WT or mutant HA-tagged ubiquitin constructs, and WT UCH-L1 or their vector controls. Transfected cells were lysed in a denaturing RIPA buffer, and the lysates were then immunoprecipitated and immunoblotted for CFTR to analyze the steady state CFTR protein levels. The ubiquitination status of CFTR was analyzed in two ways: anti-HA tag and anti-ubiquitin immunoblots of immunoprecipitated CFTR. To confirm the identity of the immunoreactive species as CFTR, we transfected the cells with an empty CFTR vector and observed no immunoreactivity (Fig. 7A, lane 1). Importantly, by analyzing immunoprecipitated CFTR, we observed that the UCH-L1-mediated increase in CFTR levels was associated with an increase in its ubiquitinated form (Fig. 7A, HA and Ub blots; compare lanes 4 and 5, lanes 6 and 7, and lanes 8 and 9). We also utilized a set of ubiquitin mutants that contain only a single lysine residue available for polyubiquitin chain formation. Because Lys48-linked chains are often a signal for protein degradation, we hypothesized that UCH-L1 was promoting the conjugation of a degradation-incompetent (i.e. Lys63-ubiquitin) linkage. We reasoned that a UCH-L1-mediated stabilization that required Lys63-linked chains would be diminished in the case of Lys48-Ub overexpression, because this mutant is unable to form Lys63-linked chains. Overexpression of Lys48-Ub-HA or Lys63-Ub-HA led to a dramatic increase in ubiquitinated CFTR compared with WT-Ub-HA overexpression (Fig. 7A, Ub blot, lanes 6–9), and UCH-L1 co-expression retained a stabilizing effect on CFTR for both mutants (Fig. 7A, CFTR blot, compare lanes 6 and 7 and lanes 8 and 9).
FIGURE 7.
Elevated UCH-L1 expression is associated with an increase in ubiquitinated CFTR. S9 cells were co-transfected for 48 h with WT (or ΔF508) CFTR, one of four HA tagged ubiquitin constructs, and WT UCH-L1 or their vector controls (pEGFP-C1, pRK5, and pcDNA3.1, respectively). The cells were lysed in denaturing RIPA buffer, immunoprecipitated for CFTR, and analyzed by Western blot (WB) for CFTR, HA tag, and ubiquitin. Whole cell lysates (5% input) were immunoblotted for UCH-L1 and actin. As a control for nonspecific immunoprecipitation (IP) of ubiquitinated proteins, the cells were transfected with the CFTR vector control (lane 1). A, UCH-L1 overexpression increases CFTR and CFTR-Ub protein expression, and stabilization is not abrogated by overexpression of WT-Ub-HA or mutants only capable of making Lys48 or Lys63-linked chains (Lys48 Ub-HA and Lys63 Ub-HA). B, K0-Ub and UCH-L1 co-expression maximize CFTR and CFTR-Ub protein expression. S9 cells were transfected with a lysine-less ubiquitin construct (K0-Ub) to favor monoubiquitination and shortened polyubiquitin chains. In this series of experiments, conducted as described for A, K0-Ub is compared with expression of Lys63-Ub. CFTR immunoprecipitates or whole cell lysates (5% input) were separated by SDS-PAGE and immunoblotted for CFTR, ubiquitin, HA tag, UCH-L1, and actin. C, the effect of UCH-L1 on ΔF508 CFTR in the presence of K0 Ub-HA was analyzed as described for B.
Lysine-less Ubiquitin (K0-Ub) Co-expression Enhances the UCH-L1-mediated Stabilization of CFTR and CFTR-Ub
The observation that UCH-L1 retained a stabilizing effect on CFTR and CFTR-Ub when promoting the formation of Lys48-linked chains (Fig. 7A, lanes 6 and 7) was not consistent with a role for UCH-L1 in enhancing protective (i.e. Lys63-linked) chains. We hypothesized that the increase in CFTR-Ub was due to an accumulation of short chains on CFTR (e.g. one to three ubiquitin molecules) that are unable to be extracted from the ER and targeted to the proteasome. To test this hypothesis, we created a ubiquitin mutant in which all seven lysine residues were mutated to arginine (K0-Ub). Attachment of this dead-end molecule results in forced monoubiquitination or chain termination, and we reasoned that a protective effect of UCH-L1 that acts through shortening of chains or monoubiquitination would increase in the presence of K0-Ub overexpression. Indeed, for both WT- and ΔF508-CFTR, we observed an increase in both CFTR and its ubiquitinated form with K0-Ub expression alone, supporting a role for short chains in CFTR stabilization (Fig. 7, B, compare lanes 4 and 6, and C, compare lanes 2 and 6). Importantly, co-expression of K0-Ub and UCH-L1 resulted in an increase in CFTR and CFTR-Ub that was greater than that observed for the other ubiquitin mutants (Fig. 7, B, lane 7, and C, lane 7). This result suggested that the UCH-L1-mediated CFTR stabilization was not due to enhanced formation of protective polyubiquitin chains using one of the seven lysines of ubiquitin. Instead, the results are consistent with an increase in ubiquitination because of monoubiquitination or the maintenance of short ubiquitin chains. We conclude that the stabilizing effect of UCH-L1 on CFTR and CFTR-Ub is greatest with K0-Ub co-expression.
K0-Ub and UCH-L1 Expression Synergistically Protects Early Biosynthetic Intermediates of CFTR
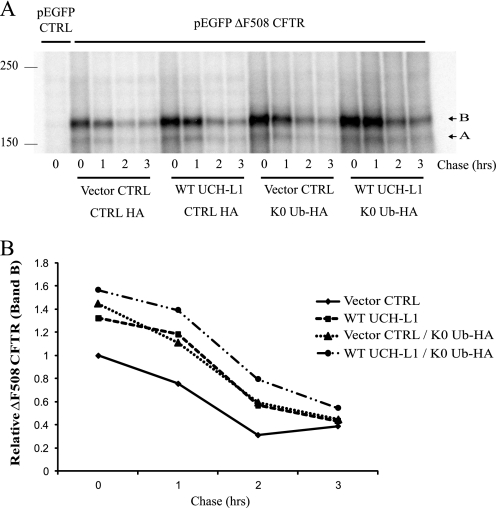
It remained possible that UCH-L1 and K0-Ub overexpression stabilized CFTR during different phases of CFTR processing. Using a model where ubiquitin chain length during CFTR synthesis is important for determining its fate, we hypothesized that K0-Ub overexpression would stabilize CFTR during the synthetic phase, similarly to UCH-L1. To analyze the temporal effects of K0-Ub overexpression on CFTR, we performed a pulse-chase analysis after co-expressing ΔF508 CFTR, UCH-L1, K0-Ub, or their vector controls (Fig. 8). Again, following a 20-min pulse, newly synthesized ΔF508 CFTR band B is increased when WT UCH-L1 is present. As hypothesized, K0-Ub expression also resulted in CFTR stabilization during synthesis as demonstrated by an increase in ΔF508 CFTR following the pulse. This stabilization looked very similar to UCH-L1 overexpression, with the minimal effect on degradation 1 h beyond the completion of translation. Furthermore, the co-expression of UCH-L1 and K0-Ub demonstrated a synergistic stabilizing effect on CFTR. We conclude that promoting the shortening of ubiquitin chains via K0-Ub expression stabilizes an early CFTR intermediate similar to UCH-L1.
FIGURE 8.
The shortening of polyubiquitin chains via lysine-less ubiquitin (K0-Ub) expression stabilizes an early CFTR intermediate, similar to UCH-L1. S9 cells were co-transfected with ΔF508 CFTR, UCH-L1, and K0-Ub or their vector controls (pEGFP-C1, pcDNA3.1, and pRK5, respectively) as indicated for 24 h. A pulse-chase examining ΔF508 CFTR biogenesis was performed as described under “Experimental Procedures.” The cells were lysed in RIPA and immunoprecipitated for CFTR. A, UCH-L1 and K0-Ub overexpression similarly increase the amount of ΔF508 CFTR synthesized during a 20-min pulse. Radiolabeled ΔF508 CFTR was analyzed by autoradiography. B, densitometric analysis of ΔF508 CFTR levels displayed in A.
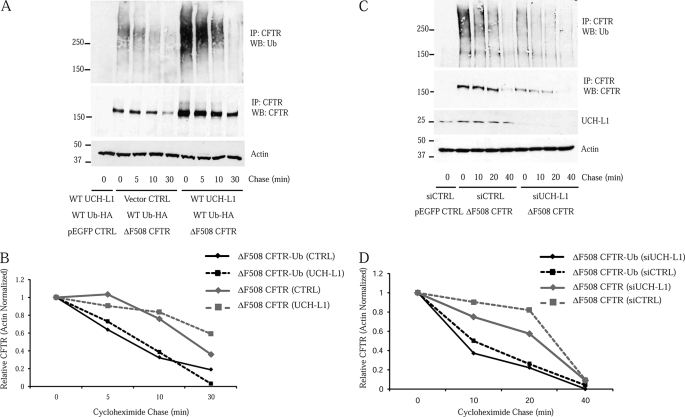
A Subpool of Ubiquitinated CFTR Is Rapidly Processed
UCH-L1 expression was associated with a large increase in ubiquitinated CFTR (Fig. 7); however, the composition of this pool was unknown. Based on our results, we observed a protective function of UCH-L1 that was confined to early stages of CFTR biogenesis (Figs. 3 and 4). Therefore, we hypothesized that a constituent of the CFTR-Ub observed with UCH-L1 expression was a co-translational species unable to target to the proteasome. To test this hypothesis, we employed cycloheximide treatment to inhibit translation and analyzed the fate of the ubiquitinated ΔF508 CFTR. We anticipated two possible scenarios: 1) a homogenous pool of ΔF508 CFTR-Ub destined to the proteasome would exhibit kinetics similar to ΔF508 CFTR or 2) a pool of ΔF508 CFTR-Ub enriched in a rapidly processed and potentially co-translational species would exhibit a shortened half-life compared with ΔF508 CFTR. A cycloheximide chase of ΔF508 CFTR-Ub (Fig. 9A, top panel) revealed that its half-life was shorter than for ΔF508 CFTR (Fig. 9A, middle panel) with and without UCH-L1 overexpression (Fig. 9B). We then repeated this experiment utilizing siRNA-UCH-L1 to analyze the fates in the presence of endogenous UCH-L1 (Fig. 9, C and D). Again, the half-life of the ΔF508 CFTR-Ub (Fig. 9C, top panel) was shorter than observed for ΔF508 CFTR (Fig. 9C, second panel). These observations support the notion of a nonproteasomal fate for a subset of the ΔF508 CFTR-Ub. It is important to note that although readily visible by Western blot with HA or ubiquitin antibodies, we estimate the ubiquitinated form to be a minor fraction of the total CFTR (supplemental Fig. S1). We conclude that the pool of CFTR-Ub contains a rapidly processed species, supporting the possibility that UCH-L1 stabilizes a ubiquitinated intermediate associated with translation.
FIGURE 9.
The increased pool of ubiquitinated CFTR associated with UCH-L1 expression is enriched in a short-lived and potentially co-translational species. The pool of ubiquitinated ΔF508 CFTR associated with UCH-L1 expression was examined following cycloheximide inhibition of protein synthesis. S9 cells were transfected for 40 h and treated with 100 μg/ml cycloheximide for the indicated chase periods. Following the short chase, the cells were immediately placed on ice, and proteasome inhibitor (MG132) was added at 50 μm to prevent any further degradation. The cells were washed in ice-cold PBS containing 50 μm MG132 and lysed in RIPA. ΔF508 CFTR was immunoprecipitated (IP), and ubiquitinated CFTR was analyzed by blotting for ubiquitin. The first lane represents S9 cells that were transfected with WT Ub-HA, WT UCH-L1, and CFTR control vector. The lack of immunoreactivity confirms the identity of the immunoreactive protein as CFTR-Ub. A, the total cellular pool of ΔF508 CFTR-Ub disappears faster than ΔF508 CFTR, suggesting a nonproteasomal fate for a subpool of ΔF508 CFTR-Ub. ΔF508 CFTR-Ub (top panel) and ΔF508 CFTR (middle panel) were examined by Western blot (WB) following cycloheximide treatment in S9 cells overexpressing WT-Ub-HA ± UCH-L1. B, densitometric analysis of ΔF508 CFTR and ΔF508 CFTR-Ub in A. C, the fate of ΔF508 CFTR-Ub (top panel) and ΔF508 CFTR (second panel) were analyzed following UCH-L1 knockdown in IB3-1 cells. D, densitometric analysis of ΔF508 CFTR and ΔF508 CFTR-Ub levels displayed in C.
DISCUSSION
UCH-L1 Alters the Proteasomal Degradation of CFTR during Early Stages of Synthesis
CFTR folding is thought to occur entirely during translation. Recognition of misfolded CFTR by ERAD components has been demonstrated to begin as early as co-translational insertion into the ER (43, 44). Previous studies indicate that CFTR is co-translationally ubiquitinated, supporting the possibility for proteasomal targeting during the process (6). Early and late sensors of CFTR folding during biogenesis have been described, including a study that demonstrates Rma1 and CHIP function in distinct phases of CFTR degradation (9). Similarly, we observed a role for UCH-L1 in a very early phase of CFTR biogenesis. Our observation that the post-translational rates of CFTR degradation were similar in the presence or absence of excess UCH-L1 may reflect an additional UCH-L1-independent phase of processing.
Proteasome-mediated degradation has been demonstrated to play a critical role in CFTR turnover (45). As expected, proteasome inhibition during the pulse-labeling phase resulted in an increased yield of CFTR; however, the magnitude of the increase in CFTR supports a biphasic degradative mechanism including a co-translational phase (Fig. 6A). Because the half-life of post-synthetic ΔF508 CFTR-GFP was 1–1.5 h (see pulse-chase; Figs. 3 and 8), we would expect that ∼21 and 14% of the CFTR, respectively, would be degraded during a 20-min pulse labeling. Therefore, the 2-fold increase in CFTR observed with the inclusion of proteasome inhibitors during the 20 min of synthesis cannot be directly attributed to single-phase degradation model, and it appeared that ∼30–35% of the CFTR was degraded in an independent co-translational phase. Previous reports also support the notion that a large percentage of CFTR is rapidly degraded during synthesis and suggest that this process is coupled to translational activity (2, 3, 9). An alternative explanation for these results would be an increase in transcription or protein synthesis following proteasome inhibition; however, analysis of total protein labeling during the pulse labeling period revealed that proteasome inhibition decreased total protein biogenesis (Fig. 6B, Total Protein panel).
Evidence supporting a stabilizing role for UCH-L1 during early CFTR synthesis was obtained in pulse-chase experiments where increases in CFTR were observed following the pulse with minimal differences in the rates of degradation during the chase periods (Figs. 3 and 4). The membrane localization of UCH-L1 and its partial co-localization with a ribosomal subunit (RPS6) and the ER (KDEL) are consistent with the possibility that UCH-L1 is modifying CFTR during translation (Fig. 5). A recent report described the importance of farnesylation in the membrane localization of UCH-L1 (42), and we hypothesize that this modification may play a role in CFTR stabilization.
Favoring Shortened Ubiquitin Chains by Overexpressing K0-Ub Stabilizes Early CFTR Intermediates and Enhances the UCH-L1-mediated Effect
UCH-L1-mediated stabilization of steady state CFTR was associated with an increase in ubiquitinated CFTR (Fig. 7). Typically, increased ubiquitination of a protein is associated with enhanced degradation via proteasomal targeting. Polyubiquitin chain type and length, however, are important for proteasomal targeting of proteins, and previous studies demonstrate that a minimal chain length of four ubiquitin molecules in a Lys48 linkage is required for efficient targeting (46, 47). In the case of Lys48-Ub co-expression in our experiments, we observed less CFTR-Ub than for the other ubiquitin mutants tested, and we propose that this is due to efficient proteasomal targeting of CFTR under these conditions (Fig. 7, K48-Ub). UCH-L1 expression stabilized CFTR and increased the amount of CFTR-Ub when co-expressed with all of the ubiquitin mutants tested, including Lys48-Ub (Fig. 7). Our data reveal that the magnitude of the UCH-L1-mediated increase in CFTR and CFTR-Ub was the greatest when conditions were imposed that favored short ubiquitin chains (Fig. 7; K0-Ub). Additionally, when expressed independently, K0-Ub and UCH-L1 similarly stabilized an early CFTR biogenic intermediate (Fig. 8). By inhibiting the proteasome during CFTR biogenesis, we observed a reduction in the stabilizing effect of UCH-L1 on CFTR (Fig. 6). Therefore, we propose that the increase in CFTR-Ub observed following UCH-L1 expression reflects a protein that is inaccessible to the proteasome because of insufficient polyubiquitin chain length.
We also considered the possibility that the results were consistent with the formation of linear ubiquitin chains, a recently described polyubiquitin linkage important in NFκB signaling (48–50). Linear ubiquitin chains would be unimpeded by untagged K0-Ub expression; however, the presence of the N-terminal HA tag in our experiments would prevent the formation of this type of chain (51). Unfortunately, the large number of possible ubiquitin conjugation sites in CFTR (87 lysine residues accessible to the cytosolic ERAD machinery) makes a direct measurement of ubiquitin chain linkage and length difficult.
Based on a model where UCH-L1 modifies CFTR ubiquitination during synthesis, we would predict that this ubiquitinated form is not degraded by the proteasome and is instead rapidly processed. Indeed, the CFTR-Ub demonstrated a half-life that was shorter than the total cellular CFTR (Fig. 9). An alternative explanation for this result is that free ubiquitin is no longer replaced during the cycloheximide chase. Previous experiments, however, estimate the half-life of ubiquitin at 2 h or greater in yeast and ∼10 h in mouse embryonic fibroblasts, which reduces this likelihood given the short duration of cycloheximide inhibition in our experiments (32, 52, 53).
UCH-L1- and K0-stabilized CFTR Is Nonaggregated
We observed that UCH-L1- and K0-mediated stabilization occurred exclusively during the early phases of CFTR synthesis and did not have an effect on CFTR maturation. It was unknown whether the significant increase in the B-form was ER arrested or moved to the aggresome or whether it could be rescued to the cell surface. Preliminary experiments examining the solubility of CFTR following UCH-L1 or K0-Ub expression demonstrated that the majority of the CFTR was not aggregated as compared with MG132 treatment (positive control), which induced aggregates (supplemental Fig. S2). Further experimentation is required to determine whether modulating the trafficking, by chemical chaperones or temperature rescue, would result in the maturation of the stabilized product to the cell surface.
UCH-L1 May Directly or Indirectly Stabilize CFTR
Previous studies demonstrate that UCH-L1 increases the cellular levels of monomeric ubiquitin (32), and it was possible that UCH-L1 indirectly stabilized CFTR by regulating ubiquitin levels. We tested the hypothesis that UCH-L1 was modifying ubiquitin levels in our cell lines; however, we found no differences following UCH-L1 overexpression (supplemental Fig. S3).
A direct interaction between UCH-L1 and CFTR was undetectable by standard co-immunoprecipitation techniques (data not shown). A previous study utilizing a yeast two-hybrid screen for UCH-L1 binding partners demonstrated very few interactions, presumably because of the transient nature of many enzymatic interactions (54). Recent evidence demonstrates that UCH-L1 interacts with Hsp90 and Hsc70, and these two proteins are known to be key mediators of CFTR processing (55–61). We are unable to exclude the possibility of a direct interaction with CFTR, and additional experimentation is necessary to determine whether the UCH-L1-mediated effect is direct or indirect. Further examination of the role of UCH-L1 in CFTR synthesis may provide insight into its cellular interactions and the mechanism of up-regulation in CF cells.
Supplementary Material
Acknowledgments
We thank Peter Lansbury, Ted Dawson, Cecile Pickart, and Roger Tsien for providing reagents (to P. L. Z. and N. V.). We also thank Kelvin MacDonald and Om Singh for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL59410, BAA-HL-0204, and CTSA UL1 RR 025005 (to P. L. Z.). This work was also supported by a Cystic Fibrosis Foundation grant (to N. V.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- CF
- cystic fibrosis
- CFTR
- CF transmembrane conductance regulator
- UCH-L1
- ubiquitin C-terminal hydrolase-L1
- WT
- wild type
- siRNA
- small interfering RNA
- ER
- endoplasmic reticulum
- ERAD
- endoplasmic reticulum-associated degradation
- E1
- ubiquitin-activating enzyme
- E2
- ubiquitin-conjugating enzyme
- E3
- ubiquitin ligase
- DUB
- deubiquitinating enzyme
- HA
- hemagglutinin
- PBS
- phosphate-buffered saline
- RIPA
- radioimmunoprecipitation assay
- Ub
- ubiquitin
- ALLN
- N-acetyl-l-leucyl-l-leucyl-l-norleucinal.
REFERENCES
- 1.Qu B. H., Strickland E., Thomas P. J. (1997) J. Bioenerg. Biomembr. 29, 483–490 [DOI] [PubMed] [Google Scholar]
- 2.Farinha C. M., Amaral M. D. (2005) Mol. Cell. Biol. 25, 5242–5252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward C. L., Kopito R. R. (1994) J. Biol. Chem. 269, 25710–25718 [PubMed] [Google Scholar]
- 4.Ward C. L., Omura S., Kopito R. R. (1995) Cell 83, 121–127 [DOI] [PubMed] [Google Scholar]
- 5.Ravid T., Hochstrasser M. (2008) Nat. Rev. Mol. Cell Biol. 9, 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sato S., Ward C. L., Kopito R. R. (1998) J. Biol. Chem. 273, 7189–7192 [DOI] [PubMed] [Google Scholar]
- 7.Sharma M., Pampinella F., Nemes C., Benharouga M., So J., Du K., Bache K. G., Papsin B., Zerangue N., Stenmark H., Lukacs G. L. (2004) J. Cell Biol. 164, 923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pickart C. M., Eddins M. J. (2004) Biochim. Biophys. Acta 1695, 55–72 [DOI] [PubMed] [Google Scholar]
- 9.Younger J. M., Chen L., Ren H. Y., Rosser M. F., Turnbull E. L., Fan C. Y., Patterson C., Cyr D. M. (2006) Cell 126, 571–582 [DOI] [PubMed] [Google Scholar]
- 10.Morito D., Hirao K., Oda Y., Hosokawa N., Tokunaga F., Cyr D. M., Tanaka K., Iwai K., Nagata K. (2008) Mol. Biol. Cell 19, 1328–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vij N., Fang S., Zeitlin P. L. (2006) J. Biol. Chem. 281, 17369–17378 [DOI] [PubMed] [Google Scholar]
- 12.Sun F., Zhang R., Gong X., Geng X., Drain P. F., Frizzell R. A. (2006) J. Biol. Chem. 281, 36856–36863 [DOI] [PubMed] [Google Scholar]
- 13.Wang B., Heath-Engel H., Zhang D., Nguyen N., Thomas D. Y., Hanrahan J. W., Shore G. C. (2008) Cell 133, 1080–1092 [DOI] [PubMed] [Google Scholar]
- 14.Younger J. M., Ren H. Y., Chen L., Fan C. Y., Fields A., Patterson C., Cyr D. M. (2004) J. Cell Biol. 167, 1075–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meacham G. C., Patterson C., Zhang W., Younger J. M., Cyr D. M. (2001) Nat. Cell Biol. 3, 100–105 [DOI] [PubMed] [Google Scholar]
- 16.Arndt V., Daniel C., Nastainczyk W., Alberti S., Höhfeld J. (2005) Mol. Biol. Cell 16, 5891–5900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlson E. J., Pitonzo D., Skach W. R. (2006) EMBO J. 25, 4557–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hassink G. C., Zhao B., Sompallae R., Altun M., Gastaldello S., Zinin N. V., Masucci M. G., Lindsten K. (2009) EMBO Rep. 10, 755–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amerik A. Y., Hochstrasser M. (2004) Biochim. Biophys. Acta 1695, 189–207 [DOI] [PubMed] [Google Scholar]
- 20.Pollard H. B., Eidelman O., Jozwik C., Huang W., Srivastava M., Ji X. D., McGowan B., Norris C. F., Todo T., Darling T., Mogayzel P. J., Zeitlin P. L., Wright J., Guggino W. B., Metcalf E., Driscoll W. J., Mueller G., Paweletz C., Jacobowitz D. M. (2006) Mol. Cell. Proteomics 5, 1628–1637 [DOI] [PubMed] [Google Scholar]
- 21.Larsen C. N., Krantz B. A., Wilkinson K. D. (1998) Biochemistry 37, 3358–3368 [DOI] [PubMed] [Google Scholar]
- 22.Wilkinson K. D., Lee K. M., Deshpande S., Duerksen-Hughes P., Boss J. M., Pohl J. (1989) Science 246, 670–673 [DOI] [PubMed] [Google Scholar]
- 23.Miyoshi Y., Nakayama S., Torikoshi Y., Tanaka S., Ishihara H., Taguchi T., Tamaki Y., Noguchi S. (2006) Cancer Sci. 97, 523–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rolén U., Kobzeva V., Gasparjan N., Ovaa H., Winberg G., Kisseljov F., Masucci M. G. (2006) Mol. Carcinog. 45, 260–269 [DOI] [PubMed] [Google Scholar]
- 25.Tokumaru Y., Yamashita K., Kim M. S., Park H. L., Osada M., Mori M., Sidransky D. (2008) Int. J. Cancer 123, 753–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y., Fallon L., Lashuel H. A., Liu Z., Lansbury P. T., Jr. (2002) Cell 111, 209–218 [DOI] [PubMed] [Google Scholar]
- 27.Choi J., Levey A. I., Weintraub S. T., Rees H. D., Gearing M., Chin L. S., Li L. (2004) J. Biol. Chem. 279, 13256–13264 [DOI] [PubMed] [Google Scholar]
- 28.Ardley H. C., Scott G. B., Rose S. A., Tan N. G., Robinson P. A. (2004) J. Neurochem. 90, 379–391 [DOI] [PubMed] [Google Scholar]
- 29.Maraganore D. M., Farrer M. J., Hardy J. A., Lincoln S. J., McDonnell S. K., Rocca W. A. (1999) Neurology 53, 1858–1860 [DOI] [PubMed] [Google Scholar]
- 30.Nazé P., Vuillaume I., Destée A., Pasquier F., Sablonnière B. (2002) Neurosci. Lett. 328, 1–4 [DOI] [PubMed] [Google Scholar]
- 31.Xue S., Jia J. (2006) Brain Res. 1087, 28–32 [DOI] [PubMed] [Google Scholar]
- 32.Osaka H., Wang Y. L., Takada K., Takizawa S., Setsuie R., Li H., Sato Y., Nishikawa K., Sun Y. J., Sakurai M., Harada T., Hara Y., Kimura I., Chiba S., Namikawa K., Kiyama H., Noda M., Aoki S., Wada K. (2003) Hum. Mol. Genet. 12, 1945–1958 [DOI] [PubMed] [Google Scholar]
- 33.Meray R. K., Lansbury P. T., Jr. (2007) J. Biol. Chem. 282, 10567–10575 [DOI] [PubMed] [Google Scholar]
- 34.Das C., Hoang Q. Q., Kreinbring C. A., Luchansky S. J., Meray R. K., Ray S. S., Lansbury P. T., Ringe D., Petsko G. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4675–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeitlin P. L., Lu L., Rhim J., Cutting G., Stetten G., Kieffer K. A., Craig R., Guggino W. B. (1991) Am. J. Respir. Cell Mol. Biol. 4, 313–319 [DOI] [PubMed] [Google Scholar]
- 36.Gruenert D. C., Basbaum C. B., Welsh M. J., Li M., Finkbeiner W. E., Nadel J. A. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 5951–5955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flotte T. R., Afione S. A., Solow R., Drumm M. L., Markakis D., Guggino W. B., Zeitlin P. L., Carter B. J. (1993) J. Biol. Chem. 268, 3781–3790 [PubMed] [Google Scholar]
- 38.Moyer B. D., Loffing J., Schwiebert E. M., Loffing-Cueni D., Halpin P. A., Karlson K. H., Ismailov I. I., Guggino W. B., Langford G. M., Stanton B. A. (1998) J. Biol. Chem. 273, 21759–21768 [DOI] [PubMed] [Google Scholar]
- 39.Zeitlin P. L., Crawford I., Lu L., Woel S., Cohen M. E., Donowitz M., Montrose M. H., Hamosh A., Cutting G. R., Gruenert D. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 344–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalejta R. F., Shenk T., Beavis A. J. (1997) Cytometry 29, 286–291 [DOI] [PubMed] [Google Scholar]
- 41.Kalejta R. F., Brideau A. D., Banfield B. W., Beavis A. J. (1999) Exp. Cell Res. 248, 322–328 [DOI] [PubMed] [Google Scholar]
- 42.Liu Z., Meray R. K., Grammatopoulos T. N., Fredenburg R. A., Cookson M. R., Liu Y., Logan T., Lansbury P. T., Jr. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4635–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Oene M., Lukacs G. L., Rommens J. M. (2000) J. Biol. Chem. 275, 19577–19584 [DOI] [PubMed] [Google Scholar]
- 44.Kleizen B., van Vlijmen T., de Jonge H. R., Braakman I. (2005) Mol. Cell 20, 277–287 [DOI] [PubMed] [Google Scholar]
- 45.Gelman M. S., Kannegaard E. S., Kopito R. R. (2002) J. Biol. Chem. 277, 11709–11714 [DOI] [PubMed] [Google Scholar]
- 46.Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. (2000) EMBO J. 19, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deveraux Q., Ustrell V., Pickart C., Rechsteiner M. (1994) J. Biol. Chem. 269, 7059–7061 [PubMed] [Google Scholar]
- 48.Iwai K., Tokunaga F. (2009) EMBO Rep. 10, 706–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rahighi S., Ikeda F., Kawasaki M., Akutsu M., Suzuki N., Kato R., Kensche T., Uejima T., Bloor S., Komander D., Randow F., Wakatsuki S., Dikic I. (2009) Cell 136, 1098–1109 [DOI] [PubMed] [Google Scholar]
- 50.Tokunaga F., Iwai K. (2009) Tanpakushitsu Kakusan Koso 54, 635–642 [PubMed] [Google Scholar]
- 51.Kirisako T., Kamei K., Murata S., Kato M., Fukumoto H., Kanie M., Sano S., Tokunaga F., Tanaka K., Iwai K. (2006) EMBO J. 25, 4877–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hanna J., Leggett D. S., Finley D. (2003) Mol. Cell. Biol. 23, 9251–9261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swaminathan S., Amerik A. Y., Hochstrasser M. (1999) Mol. Biol. Cell 10, 2583–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caballero O. L., Resto V., Patturajan M., Meerzaman D., Guo M. Z., Engles J., Yochem R., Ratovitski E., Sidransky D., Jen J. (2002) Oncogene 21, 3003–3010 [DOI] [PubMed] [Google Scholar]
- 55.Kabuta T., Furuta A., Aoki S., Furuta K., Wada K. (2008) J. Biol. Chem. 283, 23731–23738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loo M. A., Jensen T. J., Cui L., Hou Y., Chang X. B., Riordan J. R. (1998) EMBO J. 17, 6879–6887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X., Venable J., LaPointe P., Hutt D. M., Koulov A. V., Coppinger J., Gurkan C., Kellner W., Matteson J., Plutner H., Riordan J. R., Kelly J. W., Yates J. R., 3rd, Balch W. E. (2006) Cell 127, 803–815 [DOI] [PubMed] [Google Scholar]
- 58.Meacham G. C., Lu Z., King S., Sorscher E., Tousson A., Cyr D. M. (1999) EMBO J. 18, 1492–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rubenstein R. C., Zeitlin P. L. (2000) Am. J. Physiol. Cell Physiol. 278, C259–C267 [DOI] [PubMed] [Google Scholar]
- 60.Singh O. V., Pollard H. B., Zeitlin P. L. (2008) Mol. Cell. Proteomics 7, 1099–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Youker R. T., Walsh P., Beilharz T., Lithgow T., Brodsky J. L. (2004) Mol. Biol. Cell 15, 4787–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.