Abstract
The leukocyte common antigen, CD45, is a critical immune regulator whose activity is modulated by cytoskeletal interactions. Components of the spectrin-ankyrin cytoskeleton have been implicated in the trafficking and signaling of CD45. We have examined the lateral mobility of CD45 in resting and activated T lymphocytes using single-particle tracking and found that the receptor has decreased mobility caused by increased cytoskeletal contacts in activated cells. Experiments with cells that have disrupted βΙ spectrin interactions show decreased cytoskeletal contacts in resting cells and attenuation of receptor immobilization in activated cells. Applying two types of population analyses to single-particle tracking trajectories, we find good agreement between the diffusion coefficients obtained using either a mean squared displacement analysis or a hidden Markov model analysis. Hidden Markov model analysis also reveals the rate of association and dissociation of CD45-cytoskeleton contacts, demonstrating the importance of this analysis for measuring cytoskeleton binding events in live cells. Our findings are consistent with a model in which multiple cytoskeletal contacts, including those with spectrin and ankyrin, participate in the regulation of CD45 lateral mobility. These interactions are a major factor in CD45 immobilization in activated cells. Furthermore, cellular activation leads to CD45 immobilization by reduction of the CD45-cytoskeleton dissociation rate. Short peptides that mimic spectrin repeat domains alter the association rate of CD45 to the cytoskeleton and cause an apparent decrease in dissociation rates. We propose a model for CD45-cytoskeleton interactions and conclude that the spectrin-ankyrin-actin network is an essential determinant of immunoreceptor mobility.
Keywords: Biophysics, Cell/Surface, Cytoskeleton/Actin, Immunology, Membrane/Biophysics, Receptors/Leukocyte/Lymphocyte, Ankyrin, Spectrin
Introduction
The lymphocyte receptor-like protein tyrosine phosphatase, CD45,2 is a transmembrane protein that acts as a critical component of T cell activation (1). The phosphatase activity of the CD45 cytoplasmic domain contributes to T cell activation through the removal of an inhibitory phosphate group on a membrane-associated Src family kinase, such as Tyr505 of p56lck (Lck) (2). Other CD45 substrates have been established in addition to Lck, including Fyn, SKAP-55, JAK, ZAP-70, and CK2 (1). The involvement of CD45 as a regulator of critical signaling pathways and autoimmune diseases highlights the importance of understanding its function.
In addition to phosphoprotein substrates, CD45 interacts with a variety of molecular partners that modulate its activity. Its heavily glycosylated extracellular domain interacts with Thy1 (3) and galectins (4), and it self-associates to form dimers. Dimer formation is a process linked to both regulation of its phosphatase activity and sialylation of its ectodomain (1, 5, 6). The cytoplasmic domain of CD45 contains one catalytically active protein-tyrosine phosphatase domain (Domain 1) and one inactive domain (Domain 2) and binds to key cellular regulators of immune function such as CD45AP (7), CD2 (8), CD11a (9), and CSK2 (10). The cytoplasmic domain also interacts with cytoskeletal proteins in the spectrin (11, 12) and ankyrin (13) families. Binding of at least one isoform of spectrin (presumably αΙΙβΙΙ) increases the catalytic activity of CD45 (11), whereas spectrin-ankyrin interactions are required for effective trafficking and signaling of the receptor (13).
The lateral mobility of CD45 was first studied by fluorescence photobleaching recovery (FPR), demonstrating a typical membrane protein diffusion coefficient (4 × 10−10 cm2 s−1) (14, 15). Single-particle tracking (SPT) experiments on CD45 with a cytoplasmic domain deletion showed rapid diffusion that was unaffected by cellular activation (16). SPT measurements on CD45 with an intact cytoplasmic domain revealed a significant decrease in mobility upon cellular activation, suggesting increased cytoskeletal interactions (17). Thus, the lymphocyte cytoskeleton plays an active role in CD45 regulation and lateral mobility.
We have previously used FPR and SPT to study lymphocyte function-associated antigen-1 and CD2 cytoskeletal interactions and have demonstrated the importance of membrane dynamics and cytoskeletal interactions in the biological roles of these molecules (18–21). These experiments revealed heterogeneous immunoreceptor diffusion that was sensitive to cell signaling and molecular conformation. To explore the role of specific cytoskeletal regulators of CD45, we employed lymphocyte cell lines that stably express functional domains derived from βΙ spectrin (13). We report that spectrin-ankyrin interactions are central to the regulation of CD45 lateral mobility, leading to immobilization of the receptor in activated cells. Additionally, we apply a hidden Markov model (HMM) analysis, which provides insight into the apparent association (kon) and dissociation (koff) rates of CD45 to its cytoskeletal contacts. These data reveal that cellular activation induces immobilization of CD45 by a reduction of koff. Finally, short peptides that mimic the βΙ spectrin repeats, actin-binding domain, and ankyrin-binding domain alter CD45 mobility through an apparent increase in koff. Our results provide new insight into the dynamics of CD45 mobility and suggest that HMM analysis is an important tool for understanding cytoskeletal binding events in live cells.
EXPERIMENTAL PROCEDURES
Cells and Antibodies
Antibodies were purchased as mouse IgG and then digested and purified as F(ab)′ fragments using standard procedures. HI30 antibody against CD45 was purchased from BD Biosciences (San Diego, CA). Cytochalasin D (cytoD) and PMA were purchased from EMD Biosciences (San Diego, CA). Isotype control mouse IgG and buffer reagents were purchased from Sigma-Aldrich, Inc.; cell media and buffers were purchased from Invitrogen.
Jurkat cells, clone E6-1, were obtained from ATCC (Manassas, VA) and grown in RPMI medium supplemented with 10% fetal bovine serum and penicillin/streptomycin (1,000 units/ml). Previously characterized stable transfectants (13) were grown under identical conditions with the inclusion of 1.5 mg/ml G418 in the growth medium. All of the cells were harvested from cultures in exponential growth phase and washed three times with Hanks' balanced salt solution supplemented with 1% (mass/volume) BSA (HBSSB). The cells were then resuspended in HBSSB containing either DMSO alone (0.1% volume/volume) or DMSO with PMA (200 ng/ml) or cytoD (5 μg/ml). Aliquots of the cell suspension (0.25 ml) were incubated for 30 min at 37 °C and labeled with polystyrene microspheres for another 15 min at 37 °C. The samples were then diluted to 1 ml with HBSSB and transferred to a 24-well plate containing 12-mm circular coverslips treated with Cell-Tak (BD Pharmingen). The plate was centrifuged at ∼500 rpm for 7 min, and then the wells were carefully washed seven times with fresh HBSSB (1 ml). The coverslip was then transferred to a microslide, seated using a thin circle of vacuum grease, and sealed with Cytoseal 60 (Richard-Allan Scientific, Kalamazoo, MI). The samples were transferred to a heated stage at 37 °C and observed experimentally within 90 min of sealing.
Fluorescence Photobleaching Recovery
Jurkat cells were collected in exponential growth, washed three times with HBSSB, and then incubated for 30 min at 37 °C with HBSSB containing DMSO (0.1% v/v) or DMSO with PMA (200 ng/ml). The cells were then labeled for 15 min with fluorescein isothiocyanate-conjugated F(ab)′ fragments (20 μg/ml) and transferred to a six-well plate containing fresh HBSSB and a BSA-coated coverslip. The plate was centrifuged at ∼500 rpm for 7 min to settle cells onto the glass. The coverslip was then washed twice with fresh HBSSB, transferred to a microslide, and sealed with vacuum grease and Cytoseal. The samples were observed within 90 min of sealing. FPR experiments were conducted on a Meridian Ultima work station (Okemos, MI), using a 40× objective (NA = 1.3). Fluorescence recovery was observed over a 50–200-s period after the photobleach, depending on the rate of recovery.
Single-particle Tracking
One-μm polystyrene microspheres were obtained from Polysciences (2.6% m/v). The beads were diluted to a stock solution of 1.3% (m/v) in deionized water with 0.1% NaN3 and sonicated for 15 min before each use. Beads (10 μl of stock solution) were labeled by incubation with monoclonal F(ab)′ fragments for 1 h in 0.2 ml of borate buffer (100 mm borate, 1 mm EDTA, 0.1% NaN3, pH 8.5) at a final bead concentration of 0.05% (m/v). After adsorption of the protein to the beads, the samples were diluted to 1 ml with blocking buffer (10 mm HEPES, 140 mm NaCl, 1 mm EDTA, 2% dextran, 1% BSA, 0.1% NaN3, 0.1 μg/ml polyethylene glycol compound, pH 7.4) and incubated for 1 h. The samples were then sonicated for 15 min and centrifuged at 5,000 rpm for 7 min. The supernatant was aspirated to 0.1 ml of final volume, resuspended, and sonicated for 15 min immediately before use. All of the labeled bead samples were used within 48 h of preparation. A minimum amount of protein was used to achieve selective binding of beads to cells. Control beads were labeled with polyclonal F(ab)′ or BSA under identical conditions. Selectivity of binding was confirmed at the start of each experiment by manually counting the number of bead-labeled cells in ∼20 random fields. The samples were used for tracking experiments if the selectivity of bead binding was more than 4-fold greater than the isotype control. Bead binding was typically 0.4% of cells for control beads and 3% of cells (7-fold selectivity) for HI30 F(ab)′-labeled beads (1.0 μg).
The cells were observed on a Nikon TE2000-E microscope equipped with differential interference contrast optics, using a 60× oil objective with an oil condenser (NA = 1.4) (22). Images of a single bead on individual cells were captured at 1000 frames per second using a Fastcam Super 10K camera (Photron USA, Inc., San Diego, CA). Video data were processed using Metamorph (Universal Imaging, Downington, PA) and converted to trajectories.
SPT Analysis
The trajectory data were analyzed using a mean squared displacement (MSD) analysis (23) implemented in a custom program written in Matlab (Mathworks, Inc., Natick, MA). For population analysis of Dmicro and DM values, a population density analysis was performed to determine the relative proportion and center of two log normal subpopulations, as described previously (18). To estimate the maximum likelihood parameters of a two-state HMM for a set of tracks, we used a stochastic Markov Chain Monte Carlo optimization scheme as described previously (21). The Markov Chain Monte Carlo optimization scheme yields a distribution for each model parameter whose mean is the maximum likelihood parameter estimate. From these distributions, we also calculated the coefficient of variation and the ratio of the standard deviation to the mean, which is a measure of the variability in the parameter estimates, analogous to parameter confidence intervals.
CD45 Alignment
The amino acid sequences of CD45 (UNIPROT accession P08575, entry name: CD45_HUMAN), AE1 (UNIPROT accession P02730, entry name: B3AT_HUMAN), CD44 (UNIPROT accession P16070, entry name: CD44_HUMAN), and Ankyrin-R (UNIPROT accession P16157, entry name: ANK1_HUMAN) were obtained from the Swiss-Prot data base (24). All of the alignments and aligning scores were obtained by the use of ClustalW (25). Sequence numbering was used throughout this article. The crystal structures of the cytoplasmic domain of CD45 (Protein Data Bank code 1YGU, 1YGR) (26) and ankyrin repeats (Protein Data Bank code 2BKG) (27) were obtained from the RCSB Protein Data Bank. Water molecules were removed, and hydrogens were added with deletion of chain B using PyMOL software (Delano Scientific LLC, Palo Alto, CA).
RESULTS
Lateral Mobility of CD45 Studied Using Photobleaching
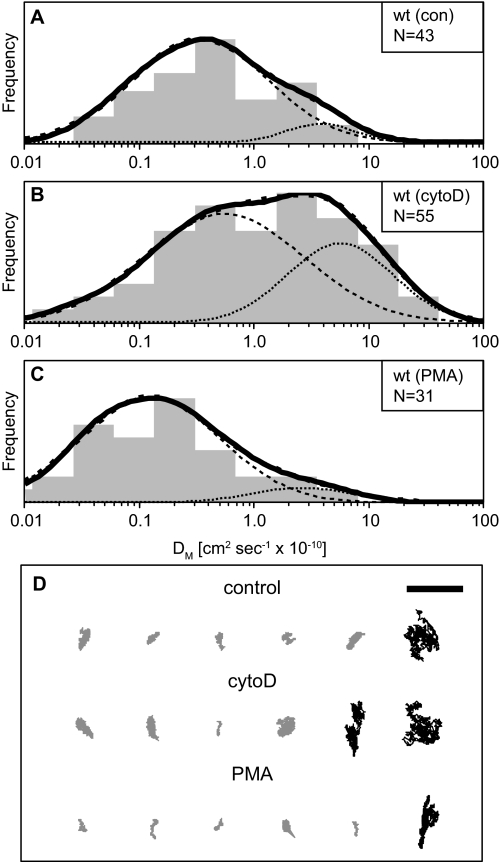
We initiated our studies of CD45 lateral diffusion using FPR experiments (supplemental Table S1). In Jurkat cells, CD45 has an average diffusion coefficient of 3.8 ± 0.7 × 10−10 cm2 s−1 and a mobile fraction of 56 ± 4%. Treatment of these cells with cytoD, an inhibitor of actin polymerization, does not cause a significant change in the diffusion coefficient (p = 0.5), although the mobile fraction increases to 69 ± 3% (p = 0.02). These values are in good agreement with previous reports (15). For comparison, the diffusion coefficient of a lipid reporter (nitrobenzoxadiazole-labeled phosphatidylethanolamine) in the Jurkat membrane was previously measured at 12 × 10−10 cm2 s−1 (18). These results support a role for cytoskeletal interactions with the cytoplasmic domain of CD45.
Lateral Mobility of CD45 Studied Using Single-particle Tracking
Single-particle tracking experiments were performed using 1-μm polystyrene beads labeled with α-CD45 F(ab)′ fragments. We first employed a conventional analysis of the trajectories using MSD, which gave diffusion coefficients for individual trajectories (23). The MSD analysis uses overlapping increments and therefore provides a representation of average diffusion within a single trajectory (28). The coefficient was determined by analyzing diffusion over the course of short or long time steps using the following equation.
For the analysis of short time steps (1–4 ms), α is assumed to be 1, and the diffusion coefficient, D, is determined using only the first four increments. This value is referred to as the microscopic diffusion coefficient (Dmicro). For the analysis of long time steps (0.7–1.3 s), a larger segment of the MSD increments is used, and the value of α is determined by fitting. The resulting diffusion coefficient is known as the macroscopic diffusion coefficient (DM). The coefficient α can be interpreted to indicate the mode of diffusion, with confined diffusion giving a small α (< 0.7) and diffusion under flow giving a large α (> 1.3). The results of these two analyses are summarized in Tables 1 and 2. The mean DM values for wild type cells indicate that CD45 is a slowly diffusing receptor, and cytoD treatment causes a significant increase in diffusion coefficient from 0.9 ± 0.2 to 3.4 ± 0.7 × 10−10 cm2 s−1. This observation is consistent with the modest increase in the mobile fraction observed by FPR and highlights the increased sensitivity of SPT. PMA treatment causes a minor reduction in DM to 0.5 ± 0.2 × 10−10 cm2 s−1. Similar trends are found in the Dmicro analysis, with an increase in diffusion coefficient from 5.2 ± 0.5 to 8.6 ± 0.6 × 10−10 cm2 s−1 upon cytoskeletal disruption and a decrease to 1.9 ± 0.5 × 10−10 cm2 s−1 in activated cells. Therefore, both analyses support a role for cytoskeletal interaction of CD45 in resting cells that is enhanced upon cell activation with PMA. The DM analysis shows α values consistently near 1, suggesting that the lower DM values are not due to confined diffusion.
TABLE 1.
DM population analysis
| Set | n | Cells | Conditiona | Mean values |
Subpopulationsb |
||||
|---|---|---|---|---|---|---|---|---|---|
| DMc | α | π1 | D1c | π2 | D2c | ||||
| 1 | 43 | wt | Control | 0.9 ± 0.2 | 0.7 ± 0.1 | 15 | 3.7 ± 1.4 | 85 | 0.3 ± 0.6 |
| 2 | 55 | wt | cytoD | 3.4 ± 0.7 | 0.8 ± 0.1 | 42 | 5.5 ± 1.3 | 58 | 0.5 ± 1.1 |
| 3 | 31 | wt | PMA | 0.5 ± 0.2 | 1.0 ± 0.1 | 12 | 2.6 ± 2.1 | 88 | 0.1 ± 0.8 |
| 4 | 39 | N-5 | Control | 1.7 ± 0.4 | 0.8 ± 0.1 | 36 | 3.7 ± 0.9 | 64 | 0.3 ± 0.7 |
| 5 | 31 | 14–15 | Control | 1.8 ± 0.5 | 0.8 ± 0.1 | 49 | 2.4 ± 0.6 | 51 | 0.2 ± 0.6 |
| 6 | 56 | N5,15 | Control | 2.0 ± 0.4 | 0.8 ± 0.1 | 23 | 5.5 ± 1.6 | 77 | 0.6 ± 0.9 |
| 7 | 33 | 14–15 | PMA | 0.9 ± 0.2 | 0.9 ± 0.1 | 43 | 1.8 ± 1.0 | 57 | 0.2 ± 0.9 |
a The conditions were control (buffer + 0.1% DMSO), cytoD (1 μm in buffer + 0.1% DMSO), and PMA (200 ng/ml in buffer + 0.1% DMSO). The buffer was HBSSB.
b Subpopulations were determined by deconvolution of DM values.
c The units for diffusion coefficients are 10−10 cm2 s−1; error is reported as the S.E.
TABLE 2.
Dmicro population analysis
| Set | n | Cells | Conditiona | Dmicro (mean values)b | Subpopulationsc |
|||
|---|---|---|---|---|---|---|---|---|
| π 1 | D1b | π 2 | D2b | |||||
| 1 | 43 | wt | Control | 5.2 ± 0.5 | 72 | 6.0 ± 0.4 | 28 | 2.0 ± 0.7 |
| 2 | 55 | wt | cytoD | 8.6 ± 0.6 | 86 | 9.6 ± 0.3 | 14 | 1.3 ± 0.8 |
| 3 | 31 | wt | PMA | 1.9 ± 0.5 | 21 | 5.5 ± 0.6 | 79 | 0.5 ± 0.3 |
| 4 | 39 | N-5 | Control | 7.4 ± 0.8 | 50 | 12.3 ± 0.7 | 50 | 3.2 ± 0.7 |
| 5 | 31 | 14–15 | Control | 6.3 ± 0.8 | 58 | 9.2 ± 0.7 | 42 | 2.3 ± 0.8 |
| 6 | 56 | N5,15 | Control | 7.2 ± 0.6 | 81 | 8.5 ± 0.4 | 19 | 1.4 ± 0.8 |
| 7 | 33 | 14–15 | PMA | 3.2 ± 0.6 | 40 | 5.9 ± 0.4 | 60 | 1.2 ± 0.3 |
a The conditions were control (buffer + 0.1% DMSO), cytoD (1 μm in buffer + 0.1% DMSO), and PMA (200 ng/ml in buffer + 0.1% DMSO). The buffer was HBSSB.
b The units for diffusion coefficients are 10−10 cm2 s−1; error is reported as the S.E.
c Subpopulations were determined by deconvolution of Dmicro values.
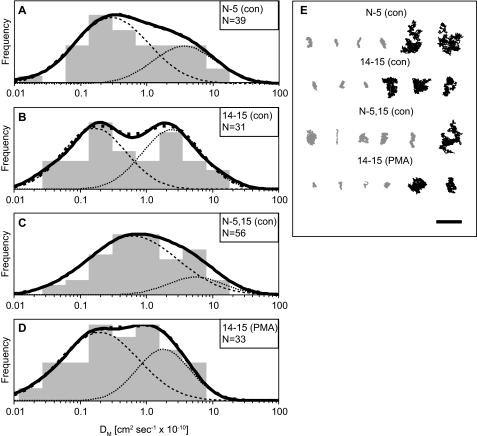
To test the role of specific cytoskeletal regulators, we examined the diffusion of CD45 on cells that stably expressed peptides derived from βΙ spectrin (13). The constructs express sequences of the N terminus to the fifth spectrin repeat (N-5) that includes the actin-binding domain; the fourteenth to fifteenth spectrin repeat (14–15 repeat) that forms the ankyrin-binding domain; and a “mini-spectrin” sequence that contains the N-5 region linked directly to the 14–15 repeats (N-5,15). None of the peptides contain the pleckstrin homology (PH) domain found in native spectrin βIΣ2. The N-5 and 14–15 peptides were previously shown to impair CD45 surface display and signaling and provide a means of observing CD45 diffusion that is decoupled from the spectrin cytoskeleton (13). In contrast, the N-5,15 peptide contains both of the functional domains required to maintain spectrin-ankyrin-mediated coupling of membrane proteins to the actin or microtubule cytoskeleton.
SPT experiments with cells expressing the N-5 peptide show an increased diffusion coefficient for CD45 relative to the wild type cells (DM = 1.7 ± 0.4 × 10−10 cm2 s−1; Dmicro = 7.4 ± 0.8 × 10−10 cm2 s−1). The 14–15-expressing cells show an increase in CD45 diffusion only by macrodiffusion (1.8 ± 0.5 × 10−10 cm2 s−1) and are otherwise unchanged by microdiffusion analysis (6.3 ± 0.8 × 10−10 cm2 s−1). The N-5,15 peptide-expressing cells show an increase in CD45 diffusion by both analyses (DM = 2.0 ± 0.4 × 10−10 cm2 s−1; Dmicro = 7.2 ± 0.6 × 10−10 cm2 s−1). We also tested the effect of PMA activation on CD45 mobility in the presence of the 14–15 peptide. Relative to the untreated 14–15 cells, PMA-treated 14–15 cells show a reduction in DM (0.9 ± 0.2 × 10−10 cm2 s−1) and Dmicro (3.2 ± 0.6 × 10−10 cm2 s−1). Together, the diffusion profiles of CD45 in spectrin peptide-expressing cells support a role for the actin and ankyrin-binding domains of βΙ spectrin in regulating receptor mobility. This role is partly restored by expression of the N-5 and 14–15 domains in a single construct (N-5,15).
Population Analysis of SPT Data
We and others have used SPT on lymphocytes (18, 19) and erythrocytes (29, 30) and have found heterogeneous distributions of diffusion coefficients for membrane proteins. Accordingly, data from these experiments should be analyzed for the appearance of subpopulations that may reveal additional information about receptor mobility. We performed additional analysis of the CD45 diffusion coefficients measured by SPT using a population density analysis (18). We first deconvoluted the distribution of macrodiffusion coefficients as two overlapping populations centered at D1 and D2 (Table 1). This analysis finds a large immobile population of CD45 in untreated wild type cells (85%, D2 = 0.3 ± 0.6 × 10−10 cm2 s−1) that is reduced by cytoD treatment (58%, D2 = 0.5 ± 1.1 × 10−10 cm2 s−1) but maintained upon PMA treatment (88%, D2 = 0.1 ± 0.8 × 10−10 cm2 s−1) (Fig. 1). CD45 macrodiffusion on spectrin peptide-expressing cells shows relatively small changes in the proportions of D1 and D2 (Fig. 2). Cells expressing the N-5 (64%, D2 = 0.3 ± 0.7 × 10−10 cm2 s−1) and 14–15 (51%, D2 = 0.2 ± 0.6 × 10−10 cm2 s−1) peptides have a reduced D2 population, whereas cells expressing the N-5,15 (77%, D2 = 0.6 ± 0.9 × 10−10 cm2 s−1) peptide have a population intermediate between wild type and N-5- or 14–15-expressing cells. Treatment of the 14–15-expressing cells with PMA gives an increased D2 population when compared with untreated 14–15 cells (57%, D2 = 0.2 ± 0.9 × 10−10 cm2 s−1), similar to the response of wild type cells.
FIGURE 1.
Single-particle tracking of CD45: population analysis of macrodiffusion (DM) values. The diffusion of CD45 was observed using F(ab)′-coated beads and high speed microscopy. A–C, the distribution of calculated diffusion coefficients (DM) is plotted as a histogram (gray) for each treatment: A, control (0.1% DMSO in buffer); B, cytoD (1 μm, in buffer containing 0.1% DMSO); and C, PMA (200 ng/ml, in buffer containing 0.1% DMSO). The population density is shown in black (bold solid line); the best fit of the population density is shown in black (bold dashed line) with the best fitted subpopulations of D1 (dotted) or D2 (dashed). D, representative trajectories are shown for each condition. Trajectories are oriented with the first time point at the bottom and the last time point at the top. Each trajectory was recorded for 2–4 s at 1000 frames per second. Representative trajectories from each subpopulation are shown in proportion to the fits (D1, black; D2, gray). The scale bar represents 1 μm. Each condition represents the results of two to four independent experiments. wt, wild type.
FIGURE 2.
Single-particle tracking of CD45 on stable transfectants: population analysis of macrodiffusion (DM) values. The diffusion of CD45 on stable transfectants expressing fragments of βΙ spectrin was observed by SPT. Macrodiffusion (A–D) and trajectory data (E) are plotted as described in the legend to Fig. 1.
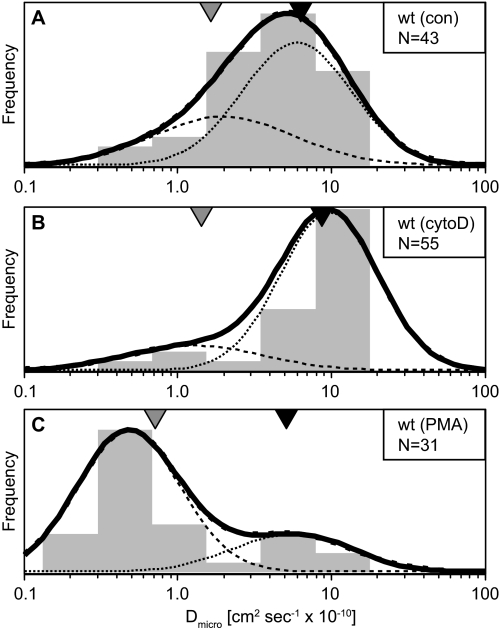
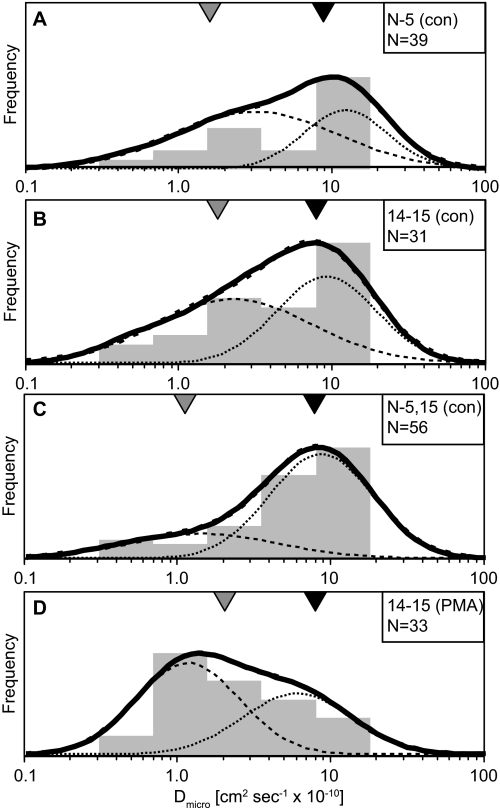
We also analyzed CD45 trajectories using microdiffusion analysis. Microdiffusion values are more sensitive to changes in diffusion at a shorter time scale (1–4 ms) than values from the macrodiffusion analysis. The diffusion coefficients obtained by microdiffusion analysis tend to be larger than those from macrodiffusion analysis, suggesting that factors that influence receptor mobility, such as membrane heterogeneity or anomalous subdiffusion, manifest at longer time scales (Table 2) (23). The distribution of microdiffusion values in wild type cells finds the majority of trajectories in the faster moving D1 population, and the size of this population increases from untreated (72%, D1 = 6.0 ± 0.4 × 10−10 cm2 s−1) to cytoD treated cells (86%, D1 = 9.6 ± 0.3 × 10−10 cm2 s−1) (Fig. 3). Treatment with PMA causes a reversal of the populations, with the majority of trajectories found in D2 (79%, D2 = 0.5 ± 0.3 × 10−10 cm2 s−1). Interestingly, the N-5-expressing (50%, D2 = 3.2 ± 0.7 × 10−10 cm2 s−1) and 14–15-expressing (42%, D2 = 2.3 ± 0.8 × 10−10 cm2 s−1) cells have an increased population in D2. However, the N-5,15-expressing cells have a much larger D1 (81%, D1 = 8.5 ± 0.4 × 10−10 cm2 s−1), similar to that of the wild type. Finally, 14–15-expressing cells treated with PMA show an apparent attenuation of CD45 immobilization, maintaining a larger D1 relative to PMA-treated wild type cells (40%, D1 = 5.9 ± 0.4 10−10 cm2 s−1) (Fig. 4).
FIGURE 3.
Single-particle tracking of CD45: population analysis of microdiffusion (Dmicro) values. SPT trajectories of CD45 on Jurkat cells are plotted using a microdiffusion analysis and labeled as described in the legend to Fig. 1. The center of the D1 and D2 subpopulations as determined by a HMM are indicated by black and gray triangles, respectively. wt, wild type.
FIGURE 4.
Single-particle tracking of CD45 on stable transfectants: population analysis of microdiffusion (Dmicro) values. SPT trajectories of CD45 on stable transfectants expressing fragments of βΙ spectrin are plotted using a microdiffusion analysis and labeled as described in Fig. 1. The plots are shown for untreated N-5, N-5,15, and 14–15 cells (A–C) and 14–15 cells treated with PMA (D). The centers of the D1 and D2 subpopulations as determined by HMM are indicated by black and gray triangles, respectively.
HMM Analysis of SPT Data
The population analysis described above relies on MSD calculations over each trajectory and classifies the trajectories into two subpopulations. However, this analysis ignores transient changes in mobility that could occur on a smaller time scale within individual trajectories. To capture this transient behavior, we have previously used a HMM to analyze the diffusion of lymphocyte function-associated antigen-1 (21). This analysis provides an improved resolution of the two underlying diffusion coefficients compared with the population analysis described above (supplemental Fig. S1). In addition, it detects transitions between the two states within a single trajectory and thus extracts detailed state switching kinetics that are inaccessible by the population analysis. The HMM analysis is based on a bimolecular reaction between a membrane protein (P) and a cytoskeletal substrate (S), forming a complex (C). This is a typical model used for membrane protein diffusion where interactions with the cytoskeletal- or cytoskeletal associated proteins play a role in lateral mobility. The reaction is characterized by an intrinsic bimolecular forward rate constant kon.intr, and a first order dissociation constant koff.
 |
We assume that S is spatially homogeneous, such that, at equilibrium, the forward reaction is pseudo-first order with a rate constant kon = kon.intr·[S]eq. When the particle is imaged with a fixed sampling interval τ (frame rate = 1/τ), it can be shown that transitions between the two states occur with the following probabilities.
 |
 |
We further assume that P and C both undergo Brownian diffusion with diffusion coefficients D1 and D2, respectively, and that D2 < D1. We estimated the most likely values of the four model parameters (D1, D2, p12, and p21) for each data set (see “Experimental Procedures”) (21). Using estimates of the transition probabilities, the relative populations of the two states are calculated as follows.
 |
 |
We also report an effective diffusion coefficient for each trajectory, defined as
 |
that weights the diffusion coefficient of each state by the relative fraction of time spent in that state. Deff is comparable with DM or Dmicro values from the MSD analysis, whereas the individual diffusion coefficients, D1 and D2, correspond to the centers of the subpopulations in the MSD analysis.
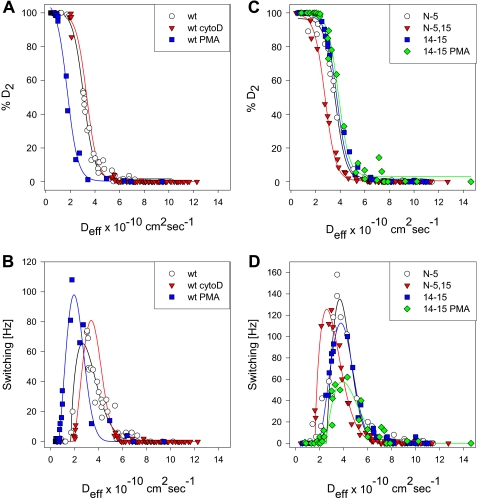
Parameter estimates from the HMM analysis of CD45 trajectories are summarized in Table 3. The results are strikingly similar to the values obtained from the population density analysis of Dmicro values (Table 2). The diffusion coefficients found by HMM are indicated on the Dmicro plots shown in Figs. 3 and 4. Comparison of the parameter estimates from Dmicro and HMM analyses reveals that both the diffusion coefficients and relative populations are highly correlated (r2 = 0.90 and 0.95). In contrast, comparison of the same parameters between Dmicro and DM shows lower correlation for the diffusion coefficients (r2 = 0.73), and no correlation for relative population values (r2 = 0.03) (supplemental Figs. S2 and S3). The HMM analysis also exposes the detailed state switching behavior within single trajectories. This allowed us to compute the fraction of time that a particle spends in the slowly diffusing state, and the total number of state switches within a trajectory. These quantities are shown in Fig. 5 for each trajectory as a function of the calculated Deff. Most of the data sets used here have very similar profiles, with a large number of trajectories found primarily in D1, and a smaller proportion found in D2. The number of intermediate trajectories varies slightly, with cytoD treatment abolishing all intermediate states. Tabulation of the frequency of state switches for each trajectory was also performed. These plots show that there is a characteristic range of diffusion coefficients where state switches are common for each condition; this range tends to fall within 1–5 × 10−10 cm2 s−1. In contrast, at the extremes the receptor remains in either D2 or D1 almost exclusively. State switches can be extremely frequent, with values as high as 160 Hz in a single trajectory. We interpret an increased rate of switching as a reduced energy of activation separating the two states, leading to a rapid exchange of binding partners. Trajectories that fall within the range of high frequency switching are those most likely to be inaccurately represented by a standard MSD analysis.
TABLE 3.
HMM Analysis
| Set | n | Cells | Conditiona | Deffb | Subpopulationsc |
p12 | p21 | kon | koff | K | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| π 1 | D1b,d | π 2 | D2b,d | ||||||||||
| s−1 | s−1 | ||||||||||||
| 1 | 43 | wt | Control | 5.03 | 75 | 6.2 | 25 | 1.6 | 0.008 | 0.024 | 7.81 | 23.85 | 0.33 |
| 2 | 55 | wt | cytoD | 7.48 | 88 | 8.5 | 13 | 1.4 | 0.001 | 0.002 | 0.35 | 2.11 | 0.17 |
| 3 | 31 | wt | PMA | 1.78 | 26 | 5.0 | 74 | 0.7 | 0.009 | 0.003 | 9.29 | 3.22 | 2.89 |
| 4 | 39 | N-5 | Control | 6.48 | 67 | 8.8 | 33 | 1.6 | 0.006 | 0.012 | 5.83 | 12.09 | 0.48 |
| 5 | 31 | 14–15 | Control | 5.63 | 63 | 7.9 | 37 | 1.8 | 0.006 | 0.011 | 6.27 | 10.69 | 0.59 |
| 6 | 56 | N5,15 | Control | 6.57 | 83 | 7.7 | 17 | 1.1 | 0.003 | 0.015 | 3.09 | 15.17 | 0.20 |
| 7 | 33 | 14–15 | PMA | 4.67 | 47 | 7.8 | 54 | 2.0 | 0.005 | 0.004 | 4.61 | 3.99 | 1.16 |
a The conditions were control (buffer + 0.1% DMSO), cytoD (1 μm in buffer + 0.1% DMSO), and PMA (200 ng/ml in buffer + 0.1% DMSO). The buffer was HBSSB.
b The units for diffusion coefficients are 10−10 cm2 s−1.
c Subpopulations were determined by HMM analysis of trajectories.
d The coefficient of variation for these values was between 0.3% and 1.8%.
FIGURE 5.
Fraction of bound CD45 as determined by HMM. SPT trajectories were analyzed using a two-state HMM (21). Individual trajectories were then classified according to the fraction of total steps spent in D2 (% D2 = π2/(π1 + π2)) (A and C) or the total number of transitions between states (switching frequency [Hz]) (B and D). These data are plotted versus the effective diffusion coefficient calculated from the HMM analysis for each individual trajectory (Deff). Plots are shown for control and PMA- and cytoD-treated wild type cells (A and B) and for untreated N-5, N5,15, and 14–15 cells and 14–15 cells treated with PMA (C and D). Trend lines were added for clarity. wt, wild type.
Inspection of the rates (kon and koff) given in Table 3 reveals the mechanism of CD45 regulation. In wild type cells, CD45 lateral mobility is maintained by high kon (7.8 s−1) and a fast koff (24 s−1). Treatment with cytoD disrupts this equilibrium by reducing the number of available cytoskeletal binding sites and thus reducing the apparent kon (0.4 s−1). Activation of the cell by PMA treatment increases the kon (9.3 s−1) concurrent with a large reduction in koff (3.2 s−1). The combined effect is the large decrease in observed lateral mobility. The spectrin peptides manifest their effects primarily through a large decrease in koff (11–15 s−1) and a small decrease in kon (3.1–6.3 s−1). This result suggests that the unnatural peptides have a reduced affinity for their target sites relative to native spectrin. Finally, PMA-treated cells expressing the 14–15 peptide show a koff similar to that of the wild type PMA-treated cells (4.0 s−1). However, there is a large decrease in the kon in the 14–15 cells (4.6 s−1), consistent with the interpretation that the peptide directly disrupts CD45 binding to spectrin.
Modeling of CD45-Cytoskeletal Interactions
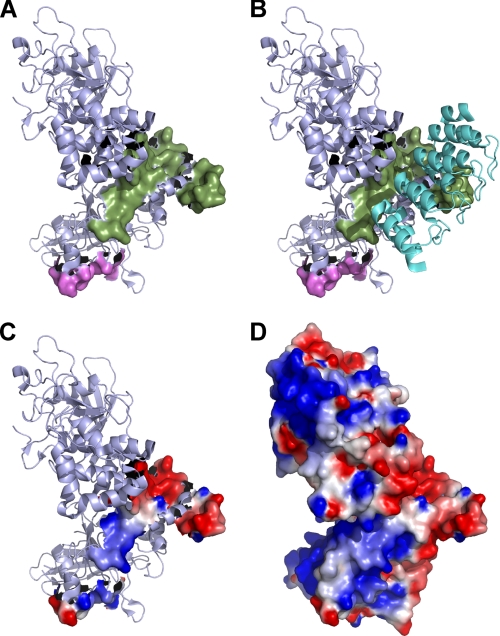
Our SPT results support a critical role for the spectrin-ankyrin cytoskeleton in regulating the lateral mobility of CD45, and previous work has shown that both spectrin and ankyrin bind directly to the cytoplasmic domain of CD45 (11–13). Although the binding site of spectrin has been mapped on CD45, the ankyrin-binding site is not known. Because the structure of the cytoplasmic domain of CD45 has been determined (26), we considered using sequence alignment to identify a potential binding site. Many membrane proteins bind ankyrin, but no single canonical ankyrin-binding motif has emerged (31). However, the erythroid anion exchanger (AE1, band 3) and CD44 have known ankyrin-binding sites (32, 33), so we considered their homology to CD45 using CLUSTALW (supplemental Fig. S4) (25). We observed homology to ankyrin-binding sites within the CD45 cytoplasmic domain (residues 904–955) and mapped these residues onto the reported protein structure (34). The majority of these residues form a solvent-accessible patch on the cytoplasmic domain, whereas residues 940–947 are buried as part of an α-helix (Fig. 6). The surface of the domain forms a deep right-handed groove that is contiguous with the known spectrin-binding domain.
FIGURE 6.
Proposed CD45-ankyrin binding interaction. We propose an ankyrin-binding site on CD45 based on an alignment of AE1 (B3AT) and CD44 to the CD45 cytoplasmic domain (32, 33) using the ClustalW algorithm (42) (supplemental Fig. S4). A, the known fodrin/spectrin-binding site is shown in magenta (33), and the putative ankyrin-binding domain is shown in green. Both sites are mapped on the structure of CD45 Domain 1 and Domain 2 (26). The residues that form the putative site map to a contiguous groove on the back of the catalytic domain. B, a hypothetical model was constructed by docking four ankyrin repeat domains to the proposed binding domain. Ankyrin is shown in cyan (27). The model suggests that the groove has a complementary topography to the ankyrin repeats. (Protein Data Bank codes 1YGR for CD45 and 2BKG for ankyrin). C and D, an electrostatic map of the proposed binding groove is shown (C), as well as a complete electrostatic map of the CD45 cytoplasmic domain (D).
DISCUSSION
We observed the lateral mobility of CD45 on lymphocytes using FPR and SPT. Our results indicate that CD45 mobility is tightly regulated by cytoskeletal contacts and that the receptor becomes immobilized upon cell stimulation with PMA. Importantly, we show that disruption of spectrin-ankyrin interactions in these cells by expression of specific domains from βΙ spectrin alters the diffusion profile of CD45 and attenuates the activation-dependent immobilization of the receptor. This effect can be partly relieved by peptides that combine the actin-binding (N-5) and ankyrin-binding (14–15 region) regions of spectrin into a single peptide, presumably restoring contacts between the actin cytoskeleton and the membrane protein.
Previous work has examined the diffusion of CD45 by FPR (15), and experiments with activated cells are in agreement with our observations of reduced lateral mobility (17). CD45 receptors lacking a cytoplasmic domain have been reported to have much higher diffusion coefficients than observed in our experiments, implicating the cytoplasmic domain in the regulation of mobility (16). Earlier work has also revealed a key role for the spectrin-ankyrin skeleton in facilitating the trafficking of newly synthesized CD45 to the cell membrane (13). Our findings now support an additional and specific role for the spectrin-ankyrin skeleton in regulating CD45 surface mobility. Disruption of the actin-spectrin interaction (N-5 peptide) or the spectrin-ankyrin interaction (14–15 peptide) leads to an increase in CD45 lateral mobility. This effect is greatly reduced in the presence of a peptide that contains both the actin- and ankyrin-binding domains of spectrin (N-5,15 peptide). Treatment of cells with PMA induces a large increase in cytoskeletal contacts for CD45, and this effect is disrupted by the presence of the spectrin 14–15 peptide that competes for the spectrin-ankyrin interaction.
The diffusion profile of cells that have the spectrin-ankyrin and spectrin-actin interactions disrupted reveals an increase in the population of D1 (Fig. 2, A and B). However, a microdiffusion analysis (Fig. 4, A and B) shows an increase in the population of D2, presumably because of an increase in cytoskeletal interactions at short time scales. This observation seems paradoxical. We propose that an explanation of this result rests with the presence of competing cytoskeleton-CD45 and spectrin-membrane linkages. Relative expression levels of CD45 at the membrane are significantly lower in the N-5 and 14–15 transfectants (13); therefore, CD45 observed by SPT in these cells is likely exposed to a much larger pool of binding sites within the cortical actin cytoskeleton (supplemental Table S3). Candidate interactions include different β spectrin isoforms (e.g. βΙ versus βΙΙ spectrin (also known as fodrin) versus βΙΙΙ spectrin) (12, 13), as well as direct linkages of spectrin to the membrane that do not involve ankyrin, such as the interaction of the β-spectrin PH domain with phosphatidylinositol 4,5-bisphosphate (PIP2) (35) and PKCβ (36, 37) or the interaction of its associated αΙΙ spectrin subunit with G-protein-coupled receptors (38). Actin-membrane linkages mediated by the ERM family of proteins (39) or variable partitioning of the spectrin membrane complex into lipid raft domains (13) could also contribute to the observed heterogeneity. Interestingly, all of the latter interactions (PIP2 binding, ERM-mediated attachment, and lipid raft partitioning) are modulated during lymphocyte activation (39). Disruption of the actin-spectrin-ankyrin complex by the N-5 and 14–15 peptides would thus either destabilize cytoskeletal interactions with CD45 directly or leave vacant a binding site on CD45 for alternative cytoskeletal linkages. Moreover, all of the spectrin peptides, to the extent they compete for native spectrin interactions with sites on CD45, would also impair the PH domain-mediated interactions of spectrin with membrane phospholipids or PKCβ, interactions critical to lymphocyte activation (13).
The detailed state switching behavior revealed by the HMM analysis further supports this hypothesis (Fig. 5, B and D). Comparison of the wild type cells and the N-5 or 14–15 cells with PMA treatment show that there are more state switches in the wild type cells (supplemental Table S2). This may be evidence of a reduced number of binding sites for D2, consistent with the competition by βΙ spectrin peptides and the abrogation of the PH domain-mediated interactions that follow the displacement of native spectrin by the spectrin peptides.
The spectrin-ankyrin interaction sites have recently been defined (40, 41), and although the structure of the CD45 cytoplasmic domain is known (26), no structures of a CD45 complex with spectrin or ankyrin have been reported. The spectrin-binding site of CD45 maps to the base of Domain 2, and although ankyrin is known to bind directly to CD45 (13), its binding site remains undefined. We considered that our observations of the N-5,15- and 14–15-expressing cells could be consistent with a competitive binding of spectrin and ankyrin to CD45. An alignment of the CD45 cytoplasmic domain to the ankyrin-binding sites of AE1 and CD44 was performed, and we observed partial homology between these known ankyrin-binding sites and CD45 (32, 33). This region maps to surface residues on CD45 proximal to the linker region between Domain 1 and Domain 2. Importantly, these residues form a contiguous groove along the protein surface that is adjacent to the spectrin-binding site (Fig. 6). Truncation of the CD45 cytoplasmic domain close to this region (residue 895) reduced binding to ankyrin (data not shown). If the linker region is indeed the binding site for ankyrin, then its proximity to the putative spectrin-binding site suggests the potential for interplay between these cytoskeleton linkage sites. Recent work suggesting that the ankyrin-binding site in spectrin may also function as a mechano-sensitive switch (41) that responds to cytoskeletal deformation heightens further the potential explanations for complexity in the dynamics of the CD45-cytoskeletal linkage observed here.
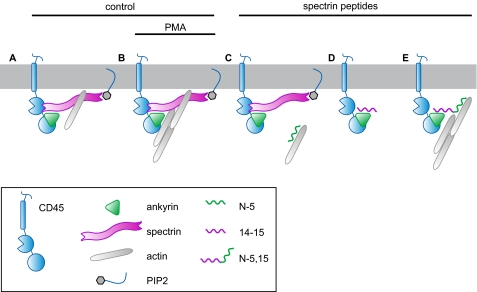
Based on our lateral mobility data, structural analysis, and the considerations discussed above, we propose a model of CD45 cytoskeletal regulation (Fig. 7). In the resting cell, the plasma membrane pool of CD45 may be associated with at least two isoforms of spectrin (βΙ and βΙΙ) and ankyrin. Actin linkages are present, but direct interactions of the PH domain of spectrin with PIP2 are modest. Cell activation leads to ERM-mediated release of competing cortical actin interactions and activates novel attachments via spectrin to PIP2 binding. Collectively, these processes cause enhanced CD45 immobilization via its linkage to spectrin-ankyrin. The N-5 peptide disrupts the actin-spectrin interaction. The 14–15 peptide attenuates the linkage between ankyrin (which is bound to CD45) and spectrin. Some of these interactions can be reconstituted with the N-5,15 peptide; however, because this peptide still lacks the C-terminal PH domain that binds PIP2, it cannot participate fully by binding either PIP2 or activated PKCβ. The observation that the spectrin peptides primarily alter CD45 diffusion through a reduced dissociation rate (koff) may suggest that an active process is responsible for maintaining CD45 mobility.
FIGURE 7.
Model of CD45 lateral diffusion. We propose a model of CD45 cytoskeletal interactions, based on the work of Pradhan and Morrow (13) and the data presented here. A, in resting cells CD45 has only transient attachment to the cytoskeleton, likely through spectrin, and modest membrane interactions through the PH domain of spectrin isoforms. B, upon cellular activation with PMA, CD45 becomes immobilized by multiple cytoskeletal and membrane contacts, which include interactions with ankyrin, spectrin, PIP2, or PKCβ. C, inhibition of the ankyrin-spectrin interaction by the N-5 peptide leads to an increase in D1 by macrodiffusion, although microdiffusion and HMM analysis suggest that transient cytoskeletal contacts may occur at the now vacant CD45 sites. D, disruption of the spectrin-ankyrin interaction by the 14–15 peptide shows an effect similar to that of the N-5 peptide, implicating ankyrin binding as a regulatory step for CD45 contacts. E, cells that express the N-5,15 peptide are found to be similar to wild type cells by all analyses employed and thus reconstitute native CD45 regulation of lateral mobility. A potential molecular mechanism for these effects is the overlap in the spectrin and ankyrin-binding sites on CD45 (see Fig. 6). Activation of cells expressing spectrin peptides prevents the almost complete immobilization of CD45 seen in wild type cells, although an increase in the immobile fraction is seen. This interaction likely represents a direct contact between spectrin and the CD45 cytoplasmic domain (11).
Our primary motivation for this study was the analysis of CD45 interactions; however, the analysis of our SPT results also provides an interesting example of the complexity of membrane protein diffusion. The traditional MSD analysis has been in use for some time and still provides an intuitive and computationally simple means of data analysis (23, 28). We have recognized in previous studies of membrane protein diffusion that treatment of SPT results as a single population can obscure significant differences in diffusion profiles. Our approach has been to use MSD analysis on individual trajectories and then attempt to sort those trajectories into separate populations (18, 19, 29). A drawback to this approach is that heterogeneity within individual trajectories will be masked. The HMM analysis introduced by Das et al. (21) exposes these transient heterogeneities within single trajectories. We found that the parameter estimates from a population analysis of Dmicro values were highly correlated with those from the HMM analysis, but not with those from a DM population analysis. This should be expected, considering that DM calculations rely on a much longer time frame of diffusion, whereas Dmicro and HMM rely on much shorter intervals. It is gratifying that the Dmicro and HMM analyses agree on the location and proportion of populations. However, the HMM analysis provides a better resolution of the underlying populations and reveals the kinetics of state switching behavior that are inaccessible through a population analysis of Dmicro. We expect that HMM will be of critical importance for systems where multiple subpopulations are relatively close in diffusion coefficient and especially when the dynamics of switching between these states are of interest.
The lateral mobility of CD45 is regulated by the spectrin-ankyrin cytoskeleton in lymphocytes. Stable expression of peptides containing the N-5 or 14–15 βΙ spectrin domain disrupts CD45 immobilization in resting and activated cells. In contrast, cells expressing peptides that include both of these domains more closely resemble wild type cells but presumably remain deficient in the PH domain-mediated interactions with PIP2 and PKCβ. We propose a model for understanding the regulation of CD45 lateral mobility as a competitive process involving multiple cytoskeletal proteins and multiple dynamic cytoskeletal-membrane linkages. Finally, we contrast several methods of trajectory analysis for receptor diffusion. Using a newly developed HMM analysis, we are able to estimate the kinetics of CD45 binding interactions. This analysis confirms that the spectrin peptides disrupt CD45 lateral mobility through a reduced rate of association of the receptor with its cytoskeletal contacts.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants DEG R01-HL32854, QJB F31-GM78720, JSM R01-HL28560, and R01-DK43812. This work was also supported by funds from the Natural Science and Engineering Research Council of Canada (to C. W. C. and D. C.), the Alberta Ingenuity Centre for Carbohydrate Science (to C. W. C.), and Mathematics of Information Technology and Complex Systems (to D. C. and R. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S4.
- CD
- cluster of differentiation
- BSA
- bovine serum albumin
- cytoD
- cytochalasin D
- DMSO
- dimethyl sulfoxide
- AE1
- erythroid anion exchanger
- FPR
- fluorescence photobleaching recovery
- HMM
- hidden Markov model
- MSD
- mean squared displacement
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- PMA
- phorbol-12-myristate-13-acetate
- SPT
- single-particle tracking
- PH
- pleckstrin homology
- PKC
- protein kinase C
- wt
- wild type.
REFERENCES
- 1.Hermiston M. L., Xu Z., Weiss A. (2003) Annu. Rev. Immunol. 21, 107–137 [DOI] [PubMed] [Google Scholar]
- 2.Rodgers W., Rose J. K. (1996) J. Cell Biol. 135, 1515–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynes M. A., Tibbetts D. J., Swenson L. M., Sidman C. L. (1993) Cell Immunol. 151, 65–79 [DOI] [PubMed] [Google Scholar]
- 4.Earl L. A., Baum L. G. (2008) Immunol. Cell Biol. 86, 608–615 [DOI] [PubMed] [Google Scholar]
- 5.Irles C., Symons A., Michel F., Bakker T. R., van der Merwe P. A., Acuto O. (2003) Nat. Immunol. 4, 189–197 [DOI] [PubMed] [Google Scholar]
- 6.Xu Z., Weiss A. (2002) Nat. Immunol. 3, 764–771 [DOI] [PubMed] [Google Scholar]
- 7.Bruyns E., Hendricks-Taylor L. R., Meuer S., Koretzky G. A., Schraven B. (1995) J. Biol. Chem. 270, 31372–31376 [DOI] [PubMed] [Google Scholar]
- 8.Schraven B., Samstag Y., Altevogt P., Meuer S. C. (1990) Nature 345, 71–74 [DOI] [PubMed] [Google Scholar]
- 9.Geng X., Tang R. H., Law S. K., Tan S. M. (2005) Immunology 115, 347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greer S. F., Wang Y., Raman C., Justement L. B. (2001) J. Immunol. 166, 7208–7218 [DOI] [PubMed] [Google Scholar]
- 11.Lokeshwar V. B., Bourguignon L. Y. (1992) J. Biol. Chem. 267, 21551–21557 [PubMed] [Google Scholar]
- 12.Iida N., Lokeshwar V. B., Bourguignon L. Y. (1994) J. Biol. Chem. 269, 28576–28583 [PubMed] [Google Scholar]
- 13.Pradhan D., Morrow J. S. (2002) Immunity 17, 303–315 [DOI] [PubMed] [Google Scholar]
- 14.Cairo C. W., Golan D. E. (2008) Biopolymers 89, 409–419 [DOI] [PubMed] [Google Scholar]
- 15.Goldman S. J., Uniyal S., Ferguson L. M., Golan D. E., Burakoff S. J., Kiener P. A. (1992) J. Biol. Chem. 267, 6197–6204 [PubMed] [Google Scholar]
- 16.Douglass A. D., Vale R. D. (2005) Cell 121, 937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drbal K., Moertelmaier M., Holzhauser C., Muhammad A., Fuertbauer E., Howorka S., Hinterberger M., Stockinger H., Schütz G. J. (2007) Int. Immunol. 19, 675–684 [DOI] [PubMed] [Google Scholar]
- 18.Cairo C. W., Mirchev R., Golan D. E. (2006) Immunity 25, 297–308 [DOI] [PubMed] [Google Scholar]
- 19.Zhu D. M., Dustin M. L., Cairo C. W., Thatte H. S., Golan D. E. (2006) ACS Chem. Biol. 1, 649–658 [DOI] [PubMed] [Google Scholar]
- 20.Zhu D. M., Dustin M. L., Cairo C. W., Golan D. E. (2007) Biophys. J. 92, 1022–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das R., Cairo C. W., Coombs D. (2009) PLoS Comput. Biol. 5, e1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirchev R., Golan D. E. (2001) Blood Cells Mol. Dis. 27, 143–147 [DOI] [PubMed] [Google Scholar]
- 23.Saxton M. J., Jacobson K. (1997) Annu. Rev. Biophys. Biomol. Struct. 26, 373–399 [DOI] [PubMed] [Google Scholar]
- 24.Bairoch A., Apweiler R. (1997) Nucleic Acids Res. 25, 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higgins D. G. (1994) Methods Mol. Biol. 25, 307–318 [DOI] [PubMed] [Google Scholar]
- 26.Nam H. J., Poy F., Saito H., Frederick C. A. (2005) J. Exp. Med. 201, 441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Binz H. K., Kohl A., Plückthun A., Grütter M. G. (2006) Proteins Struct. Funct. Genet. 65, 280–284 [DOI] [PubMed] [Google Scholar]
- 28.Qian H., Sheetz M. P., Elson E. L. (1991) Biophys. J. 60, 910–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karnchanaphanurach P., Mirchev R., Ghiran I., Asara J. M., Papahadjopoulos-Sternberg B., Nicholson-Weller A., Golan D. E. (2009) J. Clin. Investig. 119, 788–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kodippili G. C., Spector J., Sullivan C., Kuypers F. A., Labotka R., Gallagher P. G., Ritchie K., Low P. S. (2009) Blood 113, 6237–6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Matteis M. A., Morrow J. S. (2000) J. Cell Sci. 113, 2331–2343 [DOI] [PubMed] [Google Scholar]
- 32.Ding Y., Kobayashi S., Kopito R. (1996) J. Biol. Chem. 271, 22494–22498 [DOI] [PubMed] [Google Scholar]
- 33.Lokeshwar V. B., Fregien N., Bourguignon L. Y. (1994) J. Cell Biol. 126, 1099–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Notredame C., Higgins D. G., Heringa J. (2000) J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]
- 35.Zhang P., Talluri S., Deng H., Branton D., Wagner G. (1995) Structure 3, 1185–1195 [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez M. M., Ron D., Touhara K., Chen C. H., Mochly-Rosen D. (1999) Biochemistry 38, 13787–13794 [DOI] [PubMed] [Google Scholar]
- 37.Leshchyns'ka I., Sytnyk V., Morrow J. S., Schachner M. (2003) J. Cell Biol. 161, 625–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Street M., Marsh S. J., Stabach P. R., Morrow J. S., Brown D. A., Buckley N. J. (2006) J. Cell Sci. 119, 1528–1536 [DOI] [PubMed] [Google Scholar]
- 39.Hao J. J., Liu Y., Kruhlak M., Debell K. E., Rellahan B. L., Shaw S. (2009) J. Cell Biol. 184, 451–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ipsaro J. J., Huang L., Mondragón A. (2009) Blood 113, 5385–5393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stabach P. R., Simonović I., Ranieri M. A., Aboodi M. S., Steitz T. A., Simonović M., Morrow J. S. (2009) Blood 113, 5377–5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.