Abstract
The coordinated and dynamic regulation of adhesions is required for cell migration. We demonstrated previously that limited proteolysis of talin1 by the calcium-dependent protease calpain 2 plays a critical role in adhesion disassembly in fibroblasts (Franco, S. J., Rodgers, M. A., Perrin, B. J., Han, J., Bennin, D. A., Critchley, D. R., and Huttenlocher, A. (2004) Nat. Cell Biol. 6, 977–983). However, little is known about the contribution of other calpain substrates to the regulation of adhesion dynamics. We now provide evidence that calpain 2-mediated proteolysis of focal adhesion kinase (FAK) regulates adhesion dynamics in motile cells. We mapped the preferred calpain cleavage site between the two C-terminal proline-rich regions after Ser-745, resulting in a C-terminal fragment similar in size to the FAK-related non-kinase (FRNK). We generated mutant FAK with a point mutation (V744G) that renders FAK resistant to calpain proteolysis but retains other biochemical properties of FAK. Using time-lapse microscopy, we show that the dynamics of green fluorescent protein-talin1 are impaired in FAK-deficient cells. Expression of wild-type but not calpain-resistant FAK rescues talin dynamics in FAK-deficient cells. Taken together, our findings suggest a novel role for calpain proteolysis of FAK in regulating adhesion dynamics in motile cells.
Keywords: Adhesion, Calpain, Cell Migration, Microscopic Imaging, Post-translational Modification, FAK, Focal Adhesion, Migration, Proteolysis, Talin
Introduction
The coordinated and dynamic regulation of adhesions is central for cell migration in both normal and pathological processes (1). Cell migration is initiated by forming protrusions, which are stabilized by integrin-mediated adhesions that establish structural and signaling linkages between the extracellular matrix and the actin cytoskeleton. Subsequently, actomyosin contractility drives forward propulsion, and finally, adhesions are disassembled to allow for directional movement. Thus, it is not surprising that efficient cell migration necessitates precise spatial and temporal control of these events. However, the mechanisms governing the formation and disassembly of adhesions are still not well understood.
A prominent component involved in this regulation is focal adhesion kinase (FAK).2 FAK promotes cell migration by its capacity to orchestrate signals between integrin and growth factor receptors (2). Downstream of integrin or growth factor stimulation, FAK is phosphorylated at Tyr-397, which is an important binding site for Src family kinases (3). Previous studies have demonstrated a critical role for FAK as a regulator of adhesion dynamics (4–7). In addition, FAK has been shown to mediate the tyrosine phosphorylation of p190RhoGAP as a negative regulator of Rho activity in focal adhesion turnover and polarity (5, 8). Furthermore, microtubule-induced focal adhesion disassembly requires FAK and dynamin (9). Nevertheless, the mechanisms by which FAK regulates the disassembly of focal adhesions remain to be elucidated.
Evidence has emerged supporting the role of the calpain family of intracellular calcium-dependent proteases in regulating cell migration (10–14). Calpains have been proposed to regulate migration, at least in part, through their ability to modulate the dynamics of adhesions (15). Numerous calpain targets have been identified, some of which are proteins that are present in focal adhesions, including talin, paxillin, and FAK (16). Thus, it is likely that proteolysis of these substrates contributes to the regulation of adhesion dynamics and cell migration.
The involvement of both FAK and calpain in regulating the turnover of adhesions prompted us to investigate the cleavage of FAK by calpain as a possible mechanism by which FAK affects adhesion dynamics. We demonstrated previously that calpain-mediated proteolysis of talin regulates adhesion dynamics (17). Here, we show that FAK also regulates talin dynamics. We have identified the calpain cleavage site of FAK and have generated a mutant form of FAK that is resistant to calpain proteolysis. Expression of wild-type but not calpain-resistant FAK restores the adhesion dynamics of talin in FAK-deficient cells. Taken together, our findings suggest a novel role for calpain-mediated cleavage of FAK in regulating adhesion dynamics.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Fibronectin was purified from human plasma by affinity chromatography as described previously (18). Mouse anti-FAK (clone 77), anti-Pyk2 (clone 11), anti-p130Cas (clone 21), and anti-paxillin (clone 349) monoclonal antibodies were purchased from BD Transduction Laboratories. Mouse anti-actin (clone AC-15), anti-talin (clone 8d4), anti-hemagglutinin (HA; clone HA-7), and anti-FLAG (clone M2) monoclonal antibodies were purchased from Sigma-Aldrich. Rabbit anti-FAK polyclonal antibody (clone C-20) was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit anti-calpain 2 polyclonal antibody was purchased from Triple Point Biologics (Forest Grove, OR). Rabbit anti-phospho-Tyr-397 FAK polyclonal antibody was obtained from Invitrogen. Rhodamine-phalloidin, rabbit anti-green fluorescent protein (GFP) antibody, Rhodamine Red-X goat anti-mouse IgG and Alexa Fluor 680 goat anti-mouse IgG were purchased from Invitrogen. IRDye 800CW goat anti-rabbit IgG was obtained from Rockland Immunochemicals (Gilbertsville, PA). Calpain inhibitor (N-acetyl-Leu-Leu-Met) and recombinant rat calpain 2 were purchased from Calbiochem. Ionomycin was purchased from Sigma-Aldrich.
DNA Constructs
Murine GFP-FAK (pEGFP-C1-FAK-HA) was provided by D. Schlaepfer (University of California San Diego). GFP-FAK-V744G was generated from GFP-FAK using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) with the following primers: forward, 5′-CAATCACTACCAGGGCTCTGGCTACCCG-3′; and reverse, 5′-CGGGTAGCCAGAGCCCTGGTAGTGATTG-3′.
GFP-FAKΔ724–750 was also generated by site-directed mutagenesis using the following primers: forward, 5′-CAGACCTGGTTATCCTAGCCCGCATGGAATCCCAGCCATG-3′; and reverse, 5′-CATGGCTGGGATTCCATGCGGGCTAGGATAACCAGGTCTG-3′. FAK-FLAG was generated by PCR amplification of FAK with the following primers: forward, 5′-GTAGATCTGCCACCATGGCAGCTGCTTATC-3′; and reverse, 5′-CAGTCGACGTGTGGCCGTGTCTGCCCTAGC-3′. The PCR product was subsequently digested with BglII and SalI and ligated into the BamHI and XhoI sites of pcDNA3.1(+) (Invitrogen), which contained a C-terminal FLAG tag. FAK-FLAG was subcloned into the BamHI and KpnI sites of pFastBac (Invitrogen) with the following primers: forward, 5′-GACGGATCCGCCACCATGGCAGCTGCTTATC-3′; and reverse, 5′-GACGGTACCTCAAAGCTTCTTGTCATCGTC-3′. mCherry-FAK (pmCherry-C1-FAK-HA) and mCherry-FAK-V744G (pmCherry-C1-FAK-HA-V744G) were generated by PCR subcloning into the BglII and KpnI sites of the pmCherry-C1 vector (19) using the following primers: forward, 5′-GACAGATCTATGGCAGCTGCTTATCTTGACCCAAAC-3′; and reverse, 5′-GTAGGTACCTTATCTAGATCCGGTGGATC-3′. pMX-mCherry was generated by PCR subcloning mCherry into the NotI and SalI sites of the pMX-IRES-GFP vector (a gift from Clive Svendsen) from which internal ribosome entry site-GFP was excised. pMX-mCherry-FAK and pMX-mCherry-FAK-V744G were generated by PCR subcloning mCherry-FAK and mCherry-FAK-V744G into the BamHI and SalI sites of the pMX vector using the following primers: forward, 5′-ACAGGATCCGCCACCATGGTGAGCAAGGGCGAG-3′; and reverse, 5′-ATATGTATTCTAGATGATCGTCTGTCGACTCAGTGTGGCCGTGTCTGCCCTAGC-3′. FPGV-v-Src was provided by M. Frame (Edinburgh Cancer Research Centre, Edinburgh, Scotland, UK). GFP-talin1 has been described previously (16).
Cell Culture and Transfection
Human embryonic kidney (HEK) 293 cells were purchased from the American Tissue Culture Collection (Manassas, VA). Cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum, nonessential amino acids, and antibiotics. Transfections were performed by standard calcium phosphate precipitation methods. Cells were analyzed 24–48 h post-transfection. Wild-type and calpain 4 knock-out mouse embryonic fibroblasts have been described previously (20). NIH 3T3 control and calpain 2 small interfering RNA (siRNA) fibroblasts were maintained as described previously (16). v-Src-transformed NIH 3T3 cells were generated by retroviral infection of FPGV-v-Src. Briefly, cells were infected at 32 °C for 6 h and allowed to recover in growth medium for 24 h before selection with 1 mg/ml G418 (Invitrogen) for 7 days. HEK 293 cells stably expressing mCherry and mCherry-FAK constructs were generated by retroviral infection and sorted by fluorescence-activated cell sorting (University of Wisconsin-Madison Flow Cytometry Facility).
siRNA
HEK 293 control and calpain 2 siRNA cells were generated by retroviral infection of pSUPER.retro (Oligoengine, Seattle, WA) containing sequence encoding short hairpin RNA against a non-targeting sequence or human calpain 2 as described previously (21). The sense sequences for the Stealth siRNA oligonucleotides (Invitrogen) used are as follows: Controlsi, 5′-GAAUCUCAUCUAUUUCGUAACGGAC-3′; FAKsi-203, 5′-UGACAGAUACCUAGCAUCUAGCAAA-3′; FAKsi-385, 5′-GGGCCAGUAUUAUCAGGCAUGGAGA-3′; and FAKsi-2225, 5′-GCGGCCCAGGUUUACUGAACUUAAA-3′. Transfections of siRNA into HEK 293 cells were performed by calcium phosphate precipitation using 180 pmol of siRNA/6-cm dish containing 1.1 × 106 cells.
Expression and Purification of Recombinant FAK
The pFastBac-FAK-FLAG construct was transformed into DH10Bac Escherichia coli cells (Invitrogen) for recombination into bacmid DNA. High-titer viral stocks were used to infect Sf9 insect cells. Seventy-two hours after infection, cells were lysed in phosphate-buffered saline (10 mm sodium phosphate, 138 mm NaCl, and 2.7 mm KCl, pH 7.4) with 1% Triton X-100, 0.2 mm phenylmethylsulfonyl fluoride (PMSF), 1 μg/ml pepstatin, 2 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 μm E-64 (Sigma-Aldrich). Lysate was circulated over anti-FLAG M2-agarose beads (Sigma-Aldrich), washed with phosphate-buffered saline with 0.1% Triton X-100 and 0.2 mm PMSF, and eluted in phosphate-buffered saline with 100 μg/ml FLAG peptide (Sigma-Aldrich).
In Vitro Calpain Cleavage Assay
Baculovirus-purified FAK (5 μg) was incubated in cleavage buffer (50 mm Tris-HCl pH 7.4, 137 mm KCl, and 1 mm MgCl2) with the indicated concentrations of purified calpain 2 at 30 °C for 30 min in the absence or presence of CaCl2. Cleavage reactions were stopped by the addition of 6× SDS sample buffer.
Cleavage Site Mapping
FAK-FLAG was expressed in HEK 293 cells. 24–48 h later, cells were plated on dishes coated with 10 μg/ml fibronectin and incubated at 37 °C under 5% CO2 for 1 h. The medium was exchanged with complete medium containing a vehicle control or 1 μm ionomycin and 10 mm CaCl2, and cells were incubated for 10 min at 37 °C under 5% CO2. Cell lysates were incubated for 2 h with anti-FLAG M2-agarose beads, which were washed extensively with lysis buffer (20 mm Tris, pH 8.0, 1% Nonidet P-40, 10% glycerol, 137 mm NaCl, 0.2 mm PMSF, 1 μg/ml pepstatin, 2 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 mm sodium orthovanadate). The beads were boiled in nonreducing sample buffer to elute proteins, which were separated on SDS-polyacrylamide gels. Proteins were transferred to polyvinylidene difluoride membrane (Bio-Rad) and stained with Coomassie Blue, and an individual band was subjected to N-terminal sequencing (performed at the Baylor College of Medicine Protein Chemistry/Proteomics Core).
Immunocytochemistry
Glass coverslips were acid-washed, silanized, and coated with 10 μg/ml fibronectin. Cells were plated on coverslips in Dulbecco's modified Eagle's medium supplemented with 1% fetal bovine serum and nonessential amino acids and allowed to adhere for 4 h. Cells were then fixed, permeabilized, and stained as described previously (22).
Immunoblot Analysis
Cells were scraped into lysis buffer on ice and clarified by centrifugation. Protein concentrations were determined using a bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Rockford, IL) according to the manufacturer's instructions. Equal amounts of total protein were loaded onto 6–20% gradient SDS-polyacrylamide gels and transferred to nitrocellulose. Western blots were imaged with an Odyssey infrared imaging system (LI-COR Biosciences, Omaha, NE).
Immunoprecipitation
For p130Cas co-immunoprecipitations, HEK 293 cells were transfected and lysed 24–48 h later in 50 mm Tris, pH 8.0, 1% Nonidet P-40, 150 mm NaCl, 0.5% sodium deoxycholate, 0.05% SDS, 0.2 mm PMSF, 1 μg/ml pepstatin, 2 μg/ml aprotinin, 1 μg/ml leupeptin, 10 mm sodium fluoride, and 1 mm sodium orthovanadate. For paxillin co-immunoprecipitations, cells were lysed in 50 mm HEPES, pH 7.4, 1% Nonidet P-40, 75 mm NaCl, 0.2 mm PMSF, 1 μg/ml pepstatin, 2 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 mm sodium orthovanadate. Cleared lysates (0.5–1 mg) were incubated with 2 μg of anti-GFP antibody (Invitrogen) for 1–2 h. Immune complexes were captured with GammaBind G-Sepharose beads (GE Healthcare), washed with lysis buffer, and analyzed by immunoblotting.
Live Fluorescence Microscopy
Fluorescence imaging of live cells was performed using a 60× objective on an Olympus IX-70 inverted microscope housed in a 37 °C closed system. Glass-bottom dishes were acid-washed and coated with 10 μg/ml fibronectin. Cells were plated in Dulbecco's modified Eagle's medium containing 1% fetal bovine serum, nonessential amino acids, and 25 mm HEPES, pH 7.4, and allowed to adhere for 1 h, after which the medium was replaced with Ham's F-12 supplemented with 1% fetal bovine serum, nonessential amino acids, and 25 mm HEPES, pH 7.4. Fluorescent images were then captured every 2 min for 2 h using MetaVue imaging software (Molecular Devices, Downingtown, PA).
Quantification of Adhesion Dynamics
Quantification of adhesion dynamics was performed as described previously (4, 17). Time-lapse sequences from live fluorescence imaging of GFP-talin1 were first subjected to high-pass filtration based on the “water” algorithm (23) to remove diffuse background fluorescence. Fluorescence intensities of individual adhesions from background-subtracted images were measured over time using MetaVue imaging software. Of the adhesions that formed and disassembled within the period of the movie, 10–15 randomly selected adhesions in six individual cells/condition over 3 separate days were analyzed. Rate constant measurements for assembly (increasing fluorescence intensity) and disassembly (decreasing fluorescence intensity) of individual adhesions were determined from the slopes of trend lines fitted to semilogarithmic plots of fluorescence intensity ratios over time as described previously (4, 17). The duration of an adhesion was determined by the time elapsed between the first and last frames in which an adhesion was observed.
RESULTS
FAK Regulates the Adhesion Dynamics of Talin
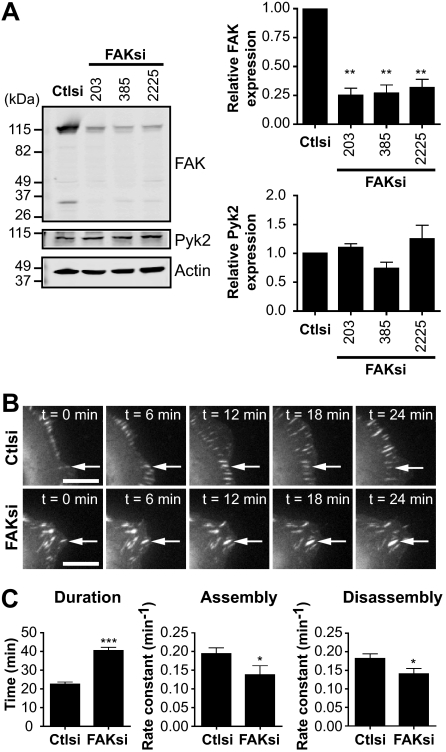
Previous studies have demonstrated that FAK and its phosphorylation at Tyr-397 are important for regulating focal adhesion turnover (4, 5). However, the contribution of calpain proteolysis of FAK to the regulation of adhesion dynamics has not been defined. To begin to address this, we first tested the effects of FAK depletion on the dynamics of the focal adhesion protein talin. We generated FAK-deficient HEK 293 cells by using siRNA to deplete endogenous levels of FAK by ∼80% without affecting the expression of the proline-rich tyrosine kinase Pyk2 (Fig. 1A) (24). Immunoblotting of endogenous FAK (∼115 kDa) with an antibody specific for the C terminus showed an additional band of ∼35 kDa in size. We visualized the dynamics of GFP-talin1 in control and FAK-deficient cells by time-lapse microscopy (Fig. 1B and supplemental Movies 1 and 2). Live fluorescence imaging demonstrated that talin-containing adhesions in FAK-depleted cells were extended in duration by ∼2-fold compared with control cells (Fig. 1C). From plots of GFP-talin1 fluorescence intensities over time, we generated rate constants for net adhesion assembly and disassembly rates. Interestingly, both adhesion assembly and disassembly rates were impaired in FAK-deficient cells. Thus, the extended duration of talin-containing adhesions in FAK-deficient cells resulted from the impaired ability of talin both to incorporate into adhesions and to disassemble from adhesions. Our results are consistent with findings in FAK-null bone marrow macrophages, which exhibit impaired adhesion turnover due to decreased assembly and disassembly rates (25). Taken together, these data indicate that FAK regulates talin dynamics in HEK 293 cells.
FIGURE 1.
FAK regulates adhesion turnover in HEK 293 cells. A, cell lysates from HEK 293 cells transiently transfected with control siRNA (Ctlsi) or FAK siRNA (FAKsi; FAKsi-203, FAKsi-385, FAKsi-2225) were analyzed by immunoblotting and probed for FAK and Pyk2. Actin was probed as a loading control. Immunoblots shown are from one of four separate experiments. Quantification of relative FAK or Pyk2 expression is defined as the ratio of FAK or Pyk2 to the expression in control siRNA. Relative FAK or Pyk2 expression is shown as the mean ± S.E. **, p < 0.01 (by one-way analysis of variance) compared with control siRNA. B, GFP-talin1 was transiently cotransfected with control or FAK (FAKsi-203) siRNA into HEK 293 cells. Cells were plated on fibronectin-coated glass-bottom dishes and analyzed by time-lapse fluorescence microscopy. Time-lapse montages demonstrate representative images of the dynamics of GFP-talin1 over a period of 24 min. Scale bars = 10 μm. Arrows indicate a representative adhesion. Representative movies are shown in supplemental Movies 1 and 2. C, duration was measured as the time elapsed between the appearance and dissolution of an observed adhesion. Rate constants for net adhesion assembly and disassembly were calculated from plots of fluorescence intensities of GFP-talin1 as described under “Experimental Procedures.” Data for each condition are the mean ± S.E. of a total of six cells over three independent experiments. *, p < 0.05; ***, p < 0.001 (by t test) compared with control siRNA.
FAK Is Cleaved by Calpain 2 in Vitro and in Vivo
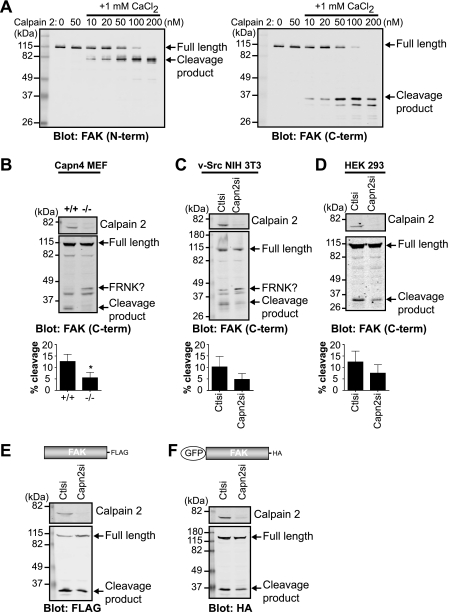
On the basis of previous studies (16, 26, 27), we hypothesized that the 35-kDa band present upon FAK immunoblotting was due to calpain proteolysis. However, the specific calpain-mediated FAK proteolytic site had not yet been characterized. We first wanted to confirm the ability of calpain 2 to cleave purified FAK using an in vitro cleavage assay (Fig. 2A). As expected, in the absence of calpain 2, FAK was not cleaved. The addition of calpain 2 without calcium also did not result in cleavage of FAK. In the presence of 1 mm calcium and low concentrations of calpain 2, immunoblot analysis with N and C terminus-specific antibodies showed a single predominant cleavage band at 80 kDa (Fig. 2A, left panel) and 35 kDa (right panel), respectively, indicating that calpain likely cleaves FAK in the C-terminal region of the protein. Higher concentrations of calpain resulted in limited cleavage and subsequent degradation of FAK. Therefore, these data demonstrate that FAK is a direct substrate of calpain 2 in vitro.
FIGURE 2.
FAK is cleaved by calpain 2 in vitro and in vivo. A, in vitro calpain cleavage assay of FAK was performed by incubating FAK in cleavage buffer alone or with calpain 2 or with 1 mm CaCl2 and increasing concentrations of calpain 2. Cleavage reactions were analyzed by immunoblotting and probed with anti-FAK antibodies specific to the N-terminal (N-term) and C-terminal (C-term) regions of FAK. Immunoblots shown are from one of two independent experiments. B–D, cell lysates from wild-type (+/+) and calpain 4 knock-out (−/−) mouse embryonic fibroblasts (Capn4 MEF) (B); v-Src-transformed NIH 3T3 control (Ctlsi) and calpain 2 (Capn2si) siRNA fibroblasts (C); and HEK 293 parental, control, and calpain 2 siRNA cells (D) were analyzed by immunoblotting and probed for calpain 2 and FAK. Immunoblots shown are from one of three independent experiments. Quantification of percent cleavage is defined as the ratio of cleaved FAK to total FAK (full-length + cleaved). Percent cleavage is shown as the mean ± S.E. *, p < 0.05 (by t test) compared with the control. E and F, cell lysates from HEK 293 control and calpain 2 siRNA cells transiently transfected with FAK-FLAG (E) or GFP-FAK (F) were analyzed by immunoblotting and probed with anti-calpain 2 and anti-FLAG (E) or anti-HA (F) antibodies. Immunoblots shown are from one of three independent experiments.
We next tested the capacity of several cell lines to generate the proteolytic fragment of FAK. Control mouse embryonic fibroblasts, v-Src-transformed NIH 3T3 fibroblasts, and HEK 293 cells were able to produce a 35-kDa C-terminal cleavage fragment, which was diminished in calpain 2-deficient cell lines (Fig. 2, B–D). In addition, upon exogenous expression of FAK-FLAG (Fig. 2E) or GFP-FAK (Fig. 2F), calpain 2-deficient HEK 293 cells displayed a reduction in the single proteolytic fragment compared with control cells. Similar to our results from in vitro calpain cleavage of FAK (Fig. 2A), a predominant single band was detected, although we did not observe FAK degradation, indicating that there are likely mechanisms in vivo that limit calpain activity and FAK degradation (reviewed in Ref. 14). Intriguingly, in calpain-deficient mouse embryonic fibroblasts and v-Src-transformed NIH 3T3 fibroblasts, immunoblot analysis revealed an increase in an additional band of higher molecular mass, which was similar in size to the FAK-related non-kinase (FRNK) (Fig. 2, B and C) (28, 29). Thus, to specifically analyze the effects of calpain-mediated cleavage of FAK separable from the calpain knockdown-mediated enhancement of the FRNK-like protein, we performed subsequent analyses in HEK 293 cells, which do not express FRNK. Taken together, these data indicate that calpain cleaves FAK both in vitro and in vivo, generating a single cleavage fragment in vivo. These findings also indicate that calpain 2-mediated proteolysis of FAK generates a fragment that is similar in size to FRNK.
Mapping the Calpain Cleavage Site of FAK
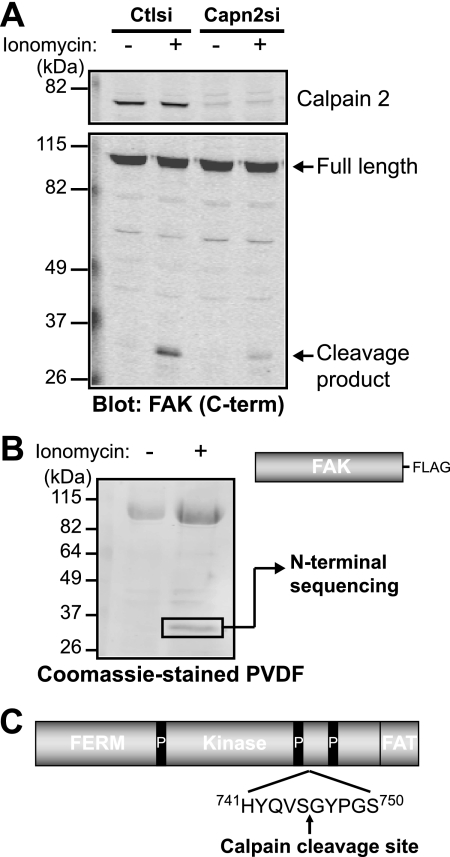
The site of calpain proteolysis of FAK had not been previously determined likely due to the lack of a consensus calpain cleavage site (30) and to the fact that only a small percentage (5–10%) of FAK is cleaved. To enhance calpain-mediated generation of the FAK cleavage fragment, we treated HEK 293 cells with the calcium ionophore ionomycin (Fig. 3A). The ionomycin-induced proteolysis of FAK was diminished in calpain 2-deficient cells. Likewise, treatment of HEK 293 cells ectopically expressing FAK-FLAG with ionomycin resulted in the enhanced generation of the proteolytic fragment. We were able to isolate the in vivo cleavage fragment, which was further analyzed by N-terminal sequencing (Fig. 3B). The cleavage site of FAK occurs after Ser-745, located between the two C-terminal proline-rich regions (Fig. 3C).
FIGURE 3.
Calpain cleaves FAK between the two C-terminal proline-rich regions. A, HEK 293 control (Ctlsi) or calpain 2 (Capn2si) siRNA cells were treated with vehicle (−) or 1 μm ionomycin + 1 mm CaCl2 (+). Cell lysates were analyzed by immunoblotting and probed for calpain 2 and FAK. Immunoblots shown are from one of two independent experiments. C-term, C-terminal. B, FAK-FLAG was transiently transfected into HEK 293 cells, which were treated with vehicle (−) or 1 μm ionomycin + 1 mm CaCl2 (+). Cell lysates were incubated with anti-FLAG beads, and protein complexes were transferred to polyvinylidene difluoride membrane. The 35-kDa band was analyzed by N-terminal sequencing. The Coomassie Blue-stained polyvinylidene difluoride (PVDF) membrane shown is from one of three independent experiments. C, a schematic of FAK domain organization (4.1, ezrin, radixin, moesin (FERM); kinase; focal adhesion targeting (FAT)) denotes the site of calpain cleavage after Ser-745, between the two C-terminal proline-rich (P) regions.
Generation of a Calpain-resistant Mutant of FAK
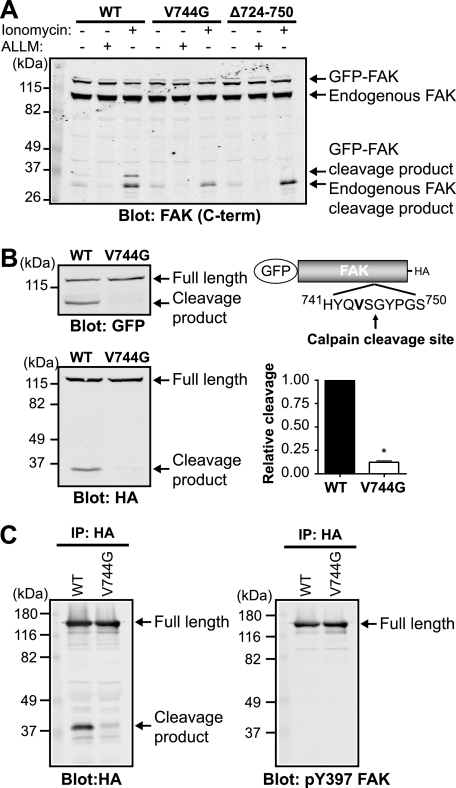
To specifically address the effects of calpain proteolysis on adhesion dynamics, we engineered a calpain-resistant mutant form of FAK. We performed site-directed mutagenesis of GFP-FAK in several residues proximal to the cleavage site (Fig. 4A). A 27-amino acid deletion (Δ724–750) surrounding the cleavage site abolished the cleavage fragment. Furthermore, analogous to the mutations introduced into talin and spectrin that rendered them calpain-resistant (17, 31), a point mutation of amino acid 744 at the P2 position from a valine to a glycine (V744G) significantly impaired calpain-mediated proteolysis of GFP-FAK. An additional point mutation (P748G) and a 6-amino acid deletion (Δ743–748) also impaired calpain proteolysis of GFP-FAK, whereas an R724L or S725F mutation did not affect the ability of FAK to be cleaved (data not shown). Furthermore, expression of GFP-FAK and calpain-resistant mutants did not affect the capacity of endogenous FAK to be cleaved. Quantification of the susceptibility to calpain proteolysis demonstrated that GFP-FAK-V744G was 10-fold resistant compared with wild-type GFP-FAK (Fig. 4B). Moreover, we confirmed that GFP-FAK-V744G retained the ability to be phosphorylated at Tyr-397 (Fig. 4C). These data demonstrate, for the first time, the site of calpain cleavage of FAK and the successful generation of a point mutant of FAK that is resistant to calpain proteolysis.
FIGURE 4.
Point mutant of FAK is resistant to calpain proteolysis. A, HEK 293 cells were transfected with wild-type GFP-FAK (WT), GFP-FAK-V744G, or GFP-FAKΔ724–750. Cells were treated with vehicle (−), 50 μg/ml calpain inhibitor N-acetyl-Leu-Leu-Met (ALLM), or 1 μm ionomycin and 10 mm CaCl2. Cell lysates were analyzed by immunoblotting and probed for FAK. The immunoblot shown is from one of three independent experiments. C-term, C-terminal. B, cell lysates from HEK 293 cells transiently transfected with GFP-FAK or GFP-FAK-V744G were analyzed by immunoblotting and probed with anti-GFP and anti-HA antibodies. Immunoblots shown are from one of five independent experiments. Quantification of relative cleavage is defined as the ratio of cleaved GFP-FAK to total GFP-FAK (full-length + cleaved) relative to GFP-FAK. Relative cleavage is shown as the mean ± S.E. *, p < 0.05 (by t test) compared with wild-type GFP-FAK. C, HEK 293 cells were transiently transfected with GFP-FAK or GFP-FAK-V744G. Proteins were immunoprecipitated (IP) with anti-HA antibody, followed by immunoblotting with anti-HA and anti-phospho-Tyr-397 (pY397) FAK antibodies. Immunoblots shown are from one of three independent experiments.
Calpain-resistant FAK Retains Localization and Biochemical Interactions
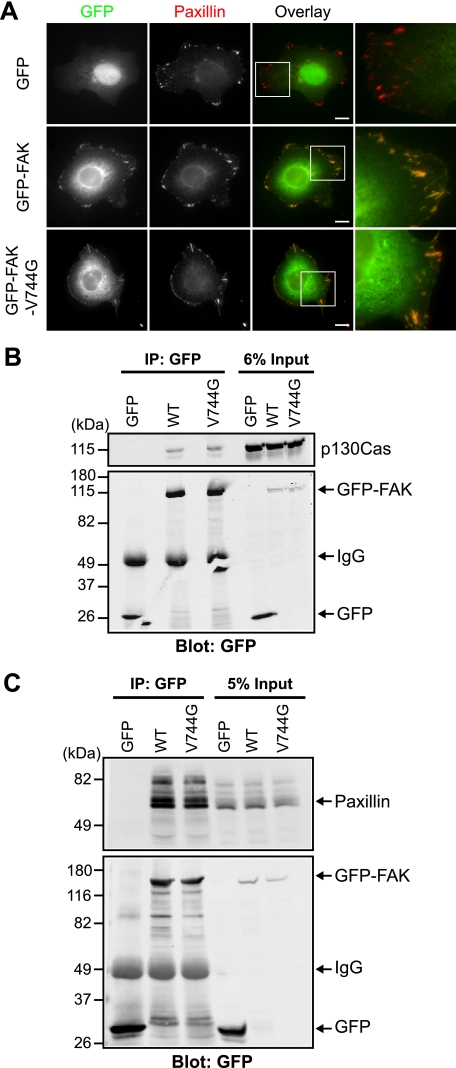
To further determine whether calpain-resistant GFP-FAK-V744G is functional, we examined the intracellular distribution of GFP-FAK-V744G. We expressed GFP-FAK and GFP-FAK-V744G in HEK 293 cells, followed by immunostaining with an antibody to the adhesion marker paxillin (Fig. 5A). We found that both GFP-FAK and GFP-FAK-V744G localized with paxillin-containing adhesions. Moreover, to determine whether wild-type and calpain-resistant GFP-FAK retained interactions with normal binding partners, we performed co-immunoprecipitation experiments and found that GFP-FAK and GFP-FAK-V744G were able to interact with p130Cas (Fig. 5B) and paxillin (Fig. 5C). Thus, these data indicate that calpain-resistant GFP-FAK-V744G is otherwise functional.
FIGURE 5.
Calpain-resistant FAK localizes to adhesions and retains biochemical properties. A, HEK 293 cells transiently transfected with GFP, GFP-FAK, or GFP-FAK-V744G were cultured on fibronectin-coated coverslips and stained with anti-paxillin antibody. Images shown are representative of three independent experiments. Scale bars = 10 μm. Regions outlined by boxes correspond to magnified images of adhesions. B and C, HEK 293 cells were transiently transfected with GFP, GFP-FAK, or GFP-FAK-V744G. Proteins were immunoprecipitated (IP) with anti-GFP antibody. Cell lysates and co-immunoprecipitations were analyzed by immunoblotting and probed with anti-GFP antibody and anti-p130Cas (B) or anti-paxillin (C) antibody. Immunoblots shown are from one of at least three independent experiments. WT, wild-type GFP-FAK.
Expression of Wild-type but Not Calpain-resistant FAK Rescues Impaired Adhesion Turnover of Talin in FAK-deficient Cells
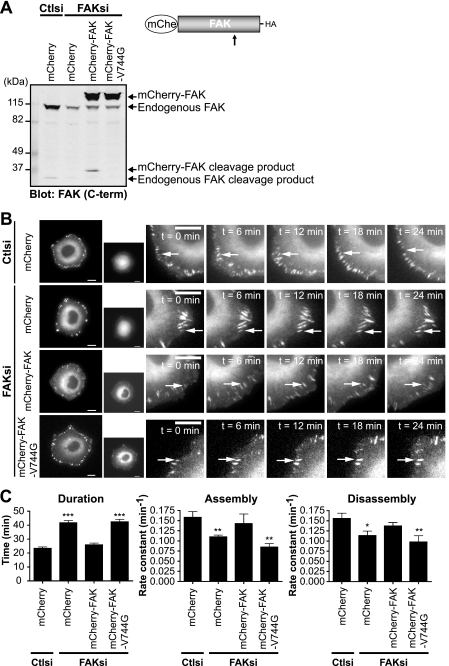
Given our findings that FAK regulates the adhesion dynamics of talin, we wanted to investigate the role of calpain cleavage of FAK in regulating talin dynamics. Accordingly, we generated mCherry fusions of wild-type and calpain-resistant FAK and were able to deplete cells of endogenous FAK without affecting the expression of exogenously expressed mCherry-FAK or mCherry-FAK-V744G (Fig. 6A). Using cell lines stably expressing mCherry, mCherry-FAK, or mCherry-FAK-V744G, we transiently depleted cells of endogenous FAK and visualized the dynamics of GFP-talin1 (Fig. 6B and supplemental Movies 3–6). Live fluorescence imaging showed that talin-containing adhesions in FAK-deficient mCherry-expressing cells were extended in duration compared with those in control cells (Fig. 6C). Quantification of adhesion dynamics demonstrated that the extended durations were due to significantly decreased rates of adhesion assembly and disassembly in FAK-deficient cells. Expression of wild-type mCherry-FAK was able to restore adhesion dynamics of talin in FAK-deficient cells. However, expression of calpain-resistant mCherry-FAK-V744G failed to rescue adhesion dynamics, indicating that calpain cleavage of FAK regulates talin dynamics. Taken together, these data indicate that FAK and its cleavage by calpain contribute to the regulation of adhesion turnover.
FIGURE 6.
Expression of wild-type but not calpain-resistant FAK rescues impaired adhesion turnover in FAK-deficient cells. A, HEK 293 cells were transiently cotransfected with control (Ctlsi) or FAK (FAKsi) siRNA and mCherry (mChe), mCherry-FAK, or mCherry-FAK-V744G. Cell lysates were analyzed by immunoblotting and probed for FAK. The immunoblot shown is from one of three independent experiments. C-term, C-terminal. B, HEK 293 cells stably expressing mCherry, mCherry-FAK, or mCherry-FAK-V744G were transiently cotransfected with control or FAK siRNA and GFP-talin1. Cells were plated on fibronectin-coated glass-bottom dishes and analyzed by time-lapse fluorescence microscopy. Time-lapse montages demonstrate representative images of the dynamics of GFP-talin1 over a period of 24 min. Scale bars = 10 μm. Arrows indicate a representative adhesion. Representative movies are shown in supplemental Movies 3–6. C, duration was measured as the time elapsed between the appearance and dissolution of an observed adhesion. Rate constants for net adhesion assembly and disassembly were calculated from plots of fluorescence intensities of GFP-talin1 as described under “Experimental Procedures.” Data for each condition are the mean ± S.E. from a total of six cells over three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (by t test) compared with control siRNA.
DISCUSSION
We showed previously that calpain-mediated proteolysis of talin regulates adhesion dynamics (17); however, the contribution of cleavage of other adhesion proteins has not been defined. Here, we have demonstrated that FAK regulates talin dynamics. We have identified the calpain cleavage site of FAK and have generated mutant FAK that is resistant to calpain proteolysis but retains proper localization and biochemical interactions. Expression of wild-type but not calpain-resistant FAK restores the adhesion dynamics of talin in FAK-deficient cells. Collectively, our data highlight a novel function for calpain proteolysis of FAK in regulating adhesion dynamics.
We found that the calpain cleavage site of FAK resides between consensus caspase cleavage sites (32), indicating that this region is particularly susceptible to proteolytic regulation. Caspases have been demonstrated to cleave FAK under conditions of apoptosis (32–35). Recent evidence has also shown the incorporation of caspase-8 into a complex containing FAK and calpain 2 to promote tumor cell migration and metastasis (36). Alternatively, it has been proposed that calpain cleavage may play a role in terminating FAK signaling by attenuating its kinase activity (27).
Our findings suggest that calpain proteolysis of FAK regulates the dynamic turnover of talin at adhesions. This is interesting in light of our previous study (17) that demonstrated that calpain proteolysis of talin is necessary for its own disassembly from focal adhesions as well as for affecting the dynamics of other adhesion components. We now provide evidence to suggest that calpain cleavage of FAK can also affect talin dynamics, indicating that proteolysis of multiple substrates rather than one specific substrate at adhesion sites is likely important for regulating the dynamic turnover of adhesions. However, how calpain cleavage of FAK regulates the adhesion dynamics of talin has yet to be defined. Also, at present, our data cannot determine whether the simultaneous or sequential cleavage of talin and/or FAK is required for regulating adhesion dynamics. Interestingly, similar to our findings with talin proteolysis (data not shown), we did not find a significant difference in migration in FAK-deficient cells re-expressing wild-type or calpain-resistant FAK, suggesting that proteolysis of multiple adhesion proteins may be necessary to affect cell migration. Future studies should address whether the inability to cleave both FAK and talin leads to a synergistic impairment of adhesion dynamics.
We cannot exclude the possibility that the effects of calpain-mediated proteolysis may depend on cellular context. Several oncogenic programs have been shown to exhibit differential cleavage of adhesion proteins (37). For example, in Src-transformed cells, FAK is extensively cleaved compared with other adhesion complex proteins (38, 39). In addition, human tumors overexpressing cyclin E have been associated with increased proteolysis of FAK (40). By contrast, in myoblasts overexpressing calpastatin, the proteolytic patterns of talin and FAK are unchanged, whereas an accumulation of MARCKS (myristoylated alanine-rich C kinase substrate) has been observed, indicating that calpain may preferentially cleave MARCKS to affect myoblast migration (41, 42).
It is intriguing that calpain proteolysis of FAK generates an FRNK-like fragment, suggesting that calpain proteolysis can generate a functional FRNK-like protein temporally and spatially within cells. Interestingly, we detected an increase in expression of an FRNK-like protein in calpain-deficient mouse embryonic fibroblasts and v-Src-transformed NIH 3T3 fibroblasts. FRNK expression is under the control of an alternative promoter that resides within an intron of FAK (29). These findings suggest that increased FRNK may compensate for the absence of calpain-mediated generation of the 35-kDa C-terminal fragment in calpain-deficient cells. Previous studies have shown that adhesion disassembly is disrupted but not abrogated in cells lacking calpain (15, 16). It is possible that, in cells deficient in calpain activity, the enhanced generation of FRNK functions as a compensatory mechanism to regulate adhesion turnover. Indeed, exogenously expressed FRNK has been demonstrated to function as a dominant-negative inhibitor of FAK (2, 43, 44). Intriguingly, a parallel mode of regulation has been observed in hippocampal neurons, in which the expression of protein kinase Mζ is directed by an intronic promoter within the protein kinase Cζ gene (45). The calpain-dependent proteolytic fragment of protein kinase C is similar in size to the protein generated from the alternative gene transcript and may be important in memory formation (46). Thus, it is very intriguing to speculate that calpain may play a role in mediating cross-talk between transcriptional and post-translational mechanisms of signaling.
In summary, our findings demonstrate a novel role for calpain-mediated limited proteolysis of FAK in affecting talin dynamics and support the notion that proteolysis of adhesion proteins is an important mechanism for regulating the turnover of adhesions. It is interesting to speculate that FAK may modulate proteolysis-mediated effects on adhesion dynamics both by functioning as an adaptor protein through its capacity to interact with calpain 2 (47) and by its direct proteolysis by calpain 2. With our successful generation of a calpain-resistant FAK mutant, future studies should investigate the interplay between FAK and calpain 2 binding and cleavage in modulating adhesion dynamics and cell migration. A challenge will be to determine how the proteolysis of different substrates temporally and spatially within cells affects adhesion turnover and cell motility.
Supplementary Material
Acknowledgment
We thank Christa Cortesio for useful discussions and critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA085862-08 (to A. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Movies 1–6.
- FAK
- focal adhesion kinase
- HA
- hemagglutinin
- GFP
- green fluorescent protein
- HEK
- human embryonic kidney
- siRNA
- small interfering RNA
- PMSF
- phenylmethylsulfonyl fluoride
- FRNK
- FAK-related non-kinase.
REFERENCES
- 1.Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Parsons J. T., Horwitz A. R. (2003) Science 302, 1704–1709 [DOI] [PubMed] [Google Scholar]
- 2.Sieg D. J., Hauck C. R., Schlaepfer D. D. (1999) J. Cell Sci. 112, 2677–2691 [DOI] [PubMed] [Google Scholar]
- 3.Schaller M. D., Hildebrand J. D., Shannon J. D., Fox J. W., Vines R. R., Parsons J. T. (1994) Mol. Cell. Biol. 14, 1680–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Webb D. J., Donais K., Whitmore L. A., Thomas S. M., Turner C. E., Parsons J. T., Horwitz A. F. (2004) Nat. Cell Biol. 6, 154–161 [DOI] [PubMed] [Google Scholar]
- 5.Schober M., Raghavan S., Nikolova M., Polak L., Pasolli H. A., Beggs H. E., Reichardt L. F., Fuchs E. (2007) J. Cell Biol. 176, 667–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamadi A., Bouali M., Dontenwill M., Stoeckel H., Takeda K., Rondé P. (2005) J. Cell Sci. 118, 4415–4425 [DOI] [PubMed] [Google Scholar]
- 7.Chan K. T., Cortesio C. L., Huttenlocher A. (2009) J. Cell Biol. 185, 357–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomar A., Lim S. T., Lim Y., Schlaepfer D. D. (2009) J. Cell Sci. 122, 1852–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ezratty E. J., Partridge M. A., Gundersen G. G. (2005) Nat. Cell Biol. 7, 581–590 [DOI] [PubMed] [Google Scholar]
- 10.Huttenlocher A., Palecek S. P., Lu Q., Zhang W., Mellgren R. L., Lauffenburger D. A., Ginsberg M. H., Horwitz A. F. (1997) J. Biol. Chem. 272, 32719–32722 [DOI] [PubMed] [Google Scholar]
- 11.Palecek S. P., Huttenlocher A., Horwitz A. F., Lauffenburger D. A. (1998) J. Cell Sci. 111, 929–940 [DOI] [PubMed] [Google Scholar]
- 12.Glading A., Chang P., Lauffenburger D. A., Wells A. (2000) J. Biol. Chem. 275, 2390–2398 [DOI] [PubMed] [Google Scholar]
- 13.Glading A., Lauffenburger D. A., Wells A. (2002) Trends Cell Biol. 12, 46–54 [DOI] [PubMed] [Google Scholar]
- 14.Franco S. J., Huttenlocher A. (2005) J. Cell Sci. 118, 3829–3838 [DOI] [PubMed] [Google Scholar]
- 15.Bhatt A., Kaverina I., Otey C., Huttenlocher A. (2002) J. Cell Sci. 115, 3415–3425 [DOI] [PubMed] [Google Scholar]
- 16.Franco S., Perrin B., Huttenlocher A. (2004) Exp. Cell Res. 299, 179–187 [DOI] [PubMed] [Google Scholar]
- 17.Franco S. J., Rodgers M. A., Perrin B. J., Han J., Bennin D. A., Critchley D. R., Huttenlocher A. (2004) Nat. Cell Biol. 6, 977–983 [DOI] [PubMed] [Google Scholar]
- 18.Ruoslahti E., Hayman E. G., Pierschbacher M., Engvall E. (1982) Methods Enzymol. 82, 803–831 [DOI] [PubMed] [Google Scholar]
- 19.Lokuta M. A., Senetar M. A., Bennin D. A., Nuzzi P. A., Chan K. T., Ott V. L., Huttenlocher A. (2007) Mol. Biol. Cell 18, 5069–5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dourdin N., Bhatt A. K., Dutt P., Greer P. A., Arthur J. S., Elce J. S., Huttenlocher A. (2001) J. Biol. Chem. 276, 48382–48388 [DOI] [PubMed] [Google Scholar]
- 21.Su L. T., Agapito M. A., Li M., Simonson W. T., Huttenlocher A., Habas R., Yue L., Runnels L. W. (2006) J. Biol. Chem. 281, 11260–11270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huttenlocher A., Ginsberg M. H., Horwitz A. F. (1996) J. Cell Biol. 134, 1551–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zamir E., Katz B. Z., Aota S., Yamada K. M., Geiger B., Kam Z. (1999) J. Cell Sci. 112, 1655–1669 [DOI] [PubMed] [Google Scholar]
- 24.Lim Y., Lim S. T., Tomar A., Gardel M., Bernard-Trifilo J. A., Chen X. L., Uryu S. A., Canete-Soler R., Zhai J., Lin H., Schlaepfer W. W., Nalbant P., Bokoch G., Ilic D., Waterman-Storer C., Schlaepfer D. D. (2008) J. Cell Biol. 180, 187–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owen K. A., Pixley F. J., Thomas K. S., Vicente-Manzanares M., Ray B. J., Horwitz A. F., Parsons J. T., Beggs H. E., Stanley E. R., Bouton A. H. (2007) J. Cell Biol. 179, 1275–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carragher N. O., Levkau B., Ross R., Raines E. W. (1999) J. Cell Biol. 147, 619–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooray P., Yuan Y., Schoenwaelder S. M., Mitchell C. A., Salem H. H., Jackson S. P. (1996) Biochem. J. 318, 41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaller M. D., Borgman C. A., Parsons J. T. (1993) Mol. Cell. Biol. 13, 785–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nolan K., Lacoste J., Parsons J. T. (1999) Mol. Cell. Biol. 19, 6120–6129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tompa P., Buzder-Lantos P., Tantos A., Farkas A., Szilágyi A., Bánóczi Z., Hudecz F., Friedrich P. (2004) J. Biol. Chem. 279, 20775–20785 [DOI] [PubMed] [Google Scholar]
- 31.Stabach P. R., Cianci C. D., Glantz S. B., Zhang Z., Morrow J. S. (1997) Biochemistry 36, 57–65 [DOI] [PubMed] [Google Scholar]
- 32.Gervais F. G., Thornberry N. A., Ruffolo S. C., Nicholson D. W., Roy S. (1998) J. Biol. Chem. 273, 17102–17108 [DOI] [PubMed] [Google Scholar]
- 33.Mian M. F., Kang C., Lee S., Choi J. H., Bae S. S., Kim S. H., Kim Y. H., Ryu S. H., Suh P. G., Kim J. S., Kim E. (2008) Chem. Biol. Interact. 171, 57–66 [DOI] [PubMed] [Google Scholar]
- 34.Wen L. P., Fahrni J. A., Troie S., Guan J. L., Orth K., Rosen G. D. (1997) J. Biol. Chem. 272, 26056–26061 [DOI] [PubMed] [Google Scholar]
- 35.Carragher N. O., Fincham V. J., Riley D., Frame M. C. (2001) J. Biol. Chem. 276, 4270–4275 [DOI] [PubMed] [Google Scholar]
- 36.Barbero S., Mielgo A., Torres V., Teitz T., Shields D. J., Mikolon D., Bogyo M., Barilà D., Lahti J. M., Schlaepfer D., Stupack D. G. (2009) Cancer Res. 69, 3755–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carragher N. O., Fonseca B. D., Frame M. C. (2004) Neoplasia 6, 53–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Westhoff M. A., Serrels B., Fincham V. J., Frame M. C., Carragher N. O. (2004) Mol. Cell. Biol. 24, 8113–8133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carragher N. O., Westhoff M. A., Riley D., Potter D. A., Dutt P., Elce J. S., Greer P. A., Frame M. C. (2002) Mol. Cell. Biol. 22, 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Libertini S. J., Robinson B. S., Dhillon N. K., Glick D., George M., Dandekar S., Gregg J. P., Sawai E., Mudryj M. (2005) Cancer Res. 65, 10700–10708 [DOI] [PubMed] [Google Scholar]
- 41.Dedieu S., Mazères G., Poussard S., Brustis J. J., Cottin P. (2003) Biol. Cell 95, 615–623 [DOI] [PubMed] [Google Scholar]
- 42.Dedieu S., Poussard S., Mazères G., Grise F., Dargelos E., Cottin P., Brustis J. J. (2004) Exp. Cell Res. 292, 187–200 [DOI] [PubMed] [Google Scholar]
- 43.Cooley M. A., Broome J. M., Ohngemach C., Romer L. H., Schaller M. D. (2000) Mol. Biol. Cell 11, 3247–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heidkamp M. C., Bayer A. L., Kalina J. A., Eble D. M., Samarel A. M. (2002) Circ. Res. 90, 1282–1289 [DOI] [PubMed] [Google Scholar]
- 45.Hernandez A. I., Blace N., Crary J. F., Serrano P. A., Leitges M., Libien J. M., Weinstein G., Tcherapanov A., Sacktor T. C. (2003) J. Biol. Chem. 278, 40305–40316 [DOI] [PubMed] [Google Scholar]
- 46.Bougie J. K., Lim T., Farah C. A., Manjunath V., Nagakura I., Ferraro G. B., Sossin W. S. (2009) J. Neurochem. 109, 1129–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carragher N. O., Westhoff M. A., Fincham V. J., Schaller M. D., Frame M. C. (2003) Curr. Biol. 13, 1442–1450 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.