Abstract
Eukaryotic Holliday junction (HJ) resolvases have attracted much attention recently with the identification of at least three distinct proteins that can cleave model HJs in vitro. However, the specific DNA structure(s) that these proteins act upon in the cell is unknown. Here, we describe a system in budding yeast to directly and quantitatively monitor in vivo HJ resolution. We found that Yen1 acts redundantly with Mus81, but not Slx1, to resolve a model HJ in vivo. This functional overlap specifically extends to the repair/bypass of lesions that impede the progression of replication forks but not to the repair of double-strand breaks induced by ionizing radiation. Together, these results suggest a direct role for Yen1 in the response to DNA damage and implicate overlapping HJ resolution functions of Yen1 with Mus81 during replication fork repair.
Keywords: DNA, DNA Damage, DNA Enzymes, DNA Recombination, DNA Repair, DNA Replication, DNA Structure, DNA Topology, Holliday Junction, Holliday Junction Resolvase
Introduction
Homologous recombination (HR)2 is a conserved process for the repair of double-strand breaks (DSBs) that can arise directly as a result of genotoxins such as ionizing radiation or indirectly as a result of stalled replication forks that can collapse into a DSB. HR-mediated DSB repair is initiated by 5′–3′ DNA end resection to facilitate Rad51-mediated strand exchange and the generation of a D-loop (1). Following repair synthesis, the D-loop may be dismantled to facilitate synthesis-dependent strand annealing. Alternatively, nicks may be ligated resulting in the formation of Holliday junctions (HJs) (1). These four-way DNA structures are processed by one of two pathways. In the dissolution pathway, a hemicatenane generated by convergent branch migration of two HJs is unlinked by a topoisomerase (1). However, in the resolution pathway of HR, HJs are resolved by specialized nucleases known as HJ resolvases (1). HJ resolvases specifically cleave one of the two pairs of strands at the junction to resolve HJs into either crossover or non-crossover products and thus allow recombinant molecules to segregate during mitosis (2–7).
Eukaryotes do not possess an ortholog of the archetypal HJ resolvase RuvC found in Escherichia coli. However, at least two proteins in humans, GEN1 and SLX1, which acts in a heterodimeric complex with SLX4, have been shown to symmetrically cleave model HJs in vitro in a manner akin to RuvC (4–7). Whereas the Saccharomyces cerevisiae GEN1 ortholog, Yen1, can also symmetrically resolve model HJs, the S. cerevisiae Slx1-Slx4 complex cleaves HJs but does so in an asymmetrical manner (5, 8). Another conserved nuclease that has been implicated in HJ resolution is Mus81, which acts as a heterodimeric complex with EME1 in humans and Schizosaccharomyces pombe and with Mms4 in S. cerevisiae (3, 9–11). Mus81 is required for the formation of meiotic crossovers in S. pombe and, to a lesser degree, in S. cerevisiae (12). However, HJs are poor substrates for Mus81-Mms4, whereas Mus81-Eme1 asymmetrically cleaves HJs and has a preference for HJs that already contain a single nick or a stretch of single-stranded DNA (10, 11, 13). This has raised the possibility that Mus81 may act on HR intermediates that arise early in the DSB repair pathway prior to the formation of covalently closed HJs (12). Roles for Mus81 in the processing of branched structures that arise at stalled/collapsed replication forks have also been proposed (13, 14).
Although GEN1/Yen1, SLX1, and MUS81 can resolve model HJs in vitro, it has not been possible to unequivocally determine whether a particular protein is specifically required for in vivo HJ resolution or might otherwise be required for the processing of other non-HJ branched HR intermediates. Here, we have addressed the question as to whether any of the potential HJ nucleases identified to date can specifically resolve a model HJ in vivo. To do this, we generated a HJ-containing plasmid-based molecule, JM-HJ, that can be transformed into S. cerevisiae and resolved into selectable products. The ability to select for products of HJ resolution provides a quantitative measurement of in vivo HJ resolution efficiency. Moreover, this system directly measures HJ resolution because resolution of JM-HJ occurs without the need for the preceding steps and formation of intermediates that arise prior to HJ formation during HR-mediated DSB repair. Whereas Slx1 and Mus81 mutants have clear defects in the response to DNA damage, there is little evidence to suggest that Yen1 plays a significant role in HR or the maintenance of genome stability. However, using this system, we reveal that Yen1 acts redundantly with Mus81 to cleave a model HJ in vivo and that this genetic interaction is specifically functional during replication stress.
EXPERIMENTAL PROCEDURES
Strains
The yeast strains used in this study are listed in Table 1. yen1EE and mus81DD alleles were constructed using the delitto perfetto methodology (15). JW2861-2 cells were obtained from the Coli Genetic Stock Center (Yale University), and RM40 cells were a kind gift from David Sherratt.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| Wild-type | BY4741 (Mata; his3Δ1, leu2Δ0, met15Δ0, ura3Δ0) | Open Biosystems |
| yen1Δ | BY4741 (YEN1::KanMX4) | Open Biosystems |
| mus81Δ | BY4741 (MUS81::KanMX4) | Open Biosystems |
| slx1Δ | BY4741 (SLX1::KanMX4) | Open Biosystems |
| mus81Δ yen1Δ | BY4741 (MUS81::KanMX4 YEN1::natR) | This study |
| mus81Δ yen1Δ rad1Δ | BY4741 (MUS81::KanMX4 YEN1::natR RAD1::HygR) | This study |
| mus81Δ yen1Δ rad1Δ slx1Δ | BY4741 (MUS81::KanMX4 YEN1::natR RAD1::Kl.URA3 SLX1::HygR) | This study |
| yen1EE | BY4741 (YEN1::YEN1(E193A/E195A)) | This study |
| mus81DD | BY4741 (MUS81::MUS81(D414A/D415A)) | This study |
| mus81DD yen1EE | BY4741 (MUS81::MUS81(D414A/D415A) YEN1::YEN1(E193A/E195A)) | This study |
Plasmids
pRS411, pRS313, and pRS415 were obtained from the American Type Culture Collection. pSD115 was a kind gift from David Sherratt. pGSHU and pGSKU were kind gifts from Francesca Storici and used to facilitate the creation of the yen1EE and mus81DD alleles.
Cloning of JM
The construction of JM and the subsequent purification of JM-HJ are described under supplemental “Experimental Procedures.”
In Vivo Resolution of JM-HJ Assays
350 ng of purified JM-HJ was cotransformed with 30 ng of pRS415 into yeast using the Frozen-EZ Yeast Transformation II KitTM (Zymo Research). Cells were plated onto the appropriate medium to select for resolution events (synthetic dextrose medium lacking His and Met) or pRS415 transformants (synthetic dextrose medium lacking Leu), and plates were incubated at 30 °C for 3 days. Resolution efficiencies were normalized against pRS415 transformation efficiency and expressed as a fraction of wild-type efficiencies.
RusA Reactions
Cleavage of 30 ng of JM-HJ by 100 nm RusA was performed using published procedures (16). RusA protein was a kind gift from Robert Lloyd.
Southern Analysis
Purified DNA or genomic DNA prepared from transformants of JM-HJ was subjected to Southern blot analysis using the cer sequence as a probe.
Analysis of JM-HJ Resolution Products
The cer1 and cer2 portions of JM-HJ resolution products were amplified by PCR from genomic DNA prepared from JM-HJ transformants using primer pairs G/H and I/J, respectively (supplemental Table 1). PCR fragments were purified and subjected to sequence analysis.
Drug Sensitivity Assays
10-Fold serial dilutions of mid-log phase yeast cells were plated onto drug-containing yeast/peptone/dextrose plates or exposed to 100 or 200 grays in a Cs-137 source and incubated at 30 °C for 3 days.
RESULTS AND DISCUSSION
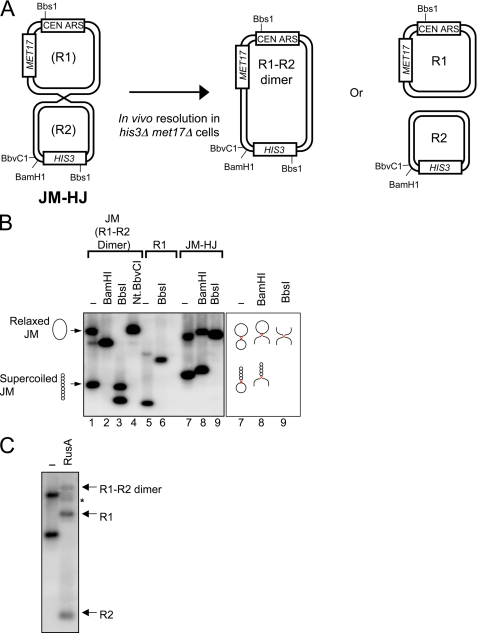
To analyze HJ resolution in an in vivo setting, we created a molecule, JM-HJ, that contains a single HJ that we could introduce into S. cerevisiae and select for resolution events. JM-HJ comprises two circular domains, R1 and R2, linked by a single HJ. R1 contains a MET17 marker and a CEN-ARS element to allow propagation in yeast, whereas R2 contains the selectable HIS3 marker but no origin of replication (Fig. 1A). Assuming that there is no bias in resolution orientation, 50% of resolution events of JM-HJ following transformation into yeast would be expected to generate a dimeric circular R1-R2 molecule in which the HIS3 marker and ARS would now be linked, allowing for selection of the HIS3 gene. To create JM-HJ, we exploited the XerC/D site-specific recombination system of E. coli to form a HJ in the plasmid JM via an intramolecular recombination event between direct repeats of the cer sequence (17). Details of the creation of JM and the subsequent induction and purification of JM-HJ are included under supplemental “Experimental Procedures.”
FIGURE 1.
System to directly analyze HJ resolution in vivo. A, depiction of the system used to analyze HJ resolution in vivo. Resolution of the HJ in JM-HJ into an R1-R2 dimer is detected as methionine and histidine prototrophs that arise following JM-HJ transformation into S. cerevisiae. See “Results and Discussion” for details. B, left panel, Southern blot analysis of purified JM, R1, and JM-HJ digested with various enzymes; right panel, representations of JM-HJ molecules present in lanes 7–9. The HJ in JM-HJ is shown in red. C, Southern blot analysis of purified JM-HJ digested with RusA. The asterisk indicates the linear R1-R2 dimer.
Southern blot analysis was used to confirm the structure of JM-HJ. Purified JM-HJ appeared as two species following gel electrophoresis: a fully relaxed molecule in which both the R1 and R2 domains of JM-HJ are relaxed and a partially relaxed molecule in which the R1 domain is supercoiled (Fig. 1B, lane 7). The electrophoretic mobilities of both of these species were distinct from either the supercoiled or relaxed form of JM, consistent with the presence of a HJ in JM-HJ (Fig. 1B, compare lanes 1 and 7). Linearization of both the R1 and R2 domains by BbsI or just the R2 domain by BamHI converted JM-HJ into species that had electrophoretic mobilities consistent with χ and α structures, respectively (Fig. 1, A and B, lanes 8 and 9). In contrast, BamHI and/or BbsI digestion of the parental JM or R1 molecule generated the predicted linear fragments (Fig. 1, A and B, lanes 2, 3, and 6). There was no evidence of these JM- or R1-derived fragments in the JM-HJ digestion products, indicating that the JM-HJ preparation was free of any contaminating JM or R1 (Fig. 1B, compare lanes 2, 3, and 6 with lanes 8 and 9). Further verification of the structure of JM-HJ was sought by treating JM-HJ with the HJ resolvase RusA, which should act to resolve the HJ in JM-HJ to form either the R1-R2 dimer or R1 and R2 monomers (Fig. 1A) (16). Indeed, treatment of JM-HJ with RusA resulted in 90% of the substrate being converted into either the R1-R2 dimer or monomeric circular R1 and R2 molecules (Fig. 1C). Approximately 10% of the RusA-generated products arose as a result of aberrant resolution events in which three strands at the junction must have been nicked, giving rise to a linear R1-R2 dimer. Overall, these data confirm the presence of a single HJ in JM-HJ, which can be resolved in vitro by a known HJ resolvase.
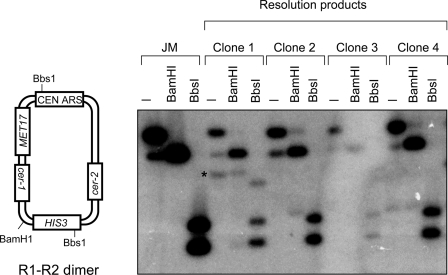
We next transformed JM-HJ into S. cerevisiae to determine whether yeast cells can resolve the HJ in JM-HJ in vivo. To monitor the in vivo resolution of JM-HJ, transformants of JM-HJ were screened for histidine and methionine prototrophy to select for resolution events giving rise to R1-R2 dimers (Fig. 1A). Plasmids recovered from these transformants had the predicted structure of a dimeric circular R1-R2 plasmid (Fig. 2), confirming that resolution of the HJ in JM-HJ had occurred as opposed to, for example, JM-HJ undergoing some aberrant rearrangement event or the HIS3 and MET17 markers ectopically integrating into the genome. We next analyzed the fidelity of the resolution events that had given rise to R1-R2 dimers. Resolution of the HJ in JM-HJ must occur within the cer sequences because the cer repeats in JM are flanked by heterologous sequences that prevent the HJ in JM-HJ branch-migrating outside these sequences (supplemental Fig. 2A). We therefore sequenced both cer sequences in the R1-R2 dimers recovered from 15 independent JM-HJ transformants. Only one of these products contained a mutation in the form of a dinucleotide GG-AA substitution. This mutation was located, however, 34 bp upstream of cer2, making it unlikely that it arose as a result of aberrant HJ resolution. Overall, these data indicate that the HJ in JM-HJ can be resolved in vivo to generate dimers and that resolution is done so in a faithful manner, preserving the nucleotide sequence.
FIGURE 2.
R1-R2 dimers can be detected as products of JM-HJ resolution. Left panel, schematic of the predicted R1-R2 dimer with the positions of relevant restriction sites; right panel, Southern analysis of purified JM and genomic DNA from four independent MET17 HIS3 clones arising from JM-HJ transformation digested with various enzymes as indicated. The asterisk indicates monomeric R1 that most likely arose through a post-JM-HJ resolution event through cer-mediated intramolecular recombination of the R1-R2 dimer product.
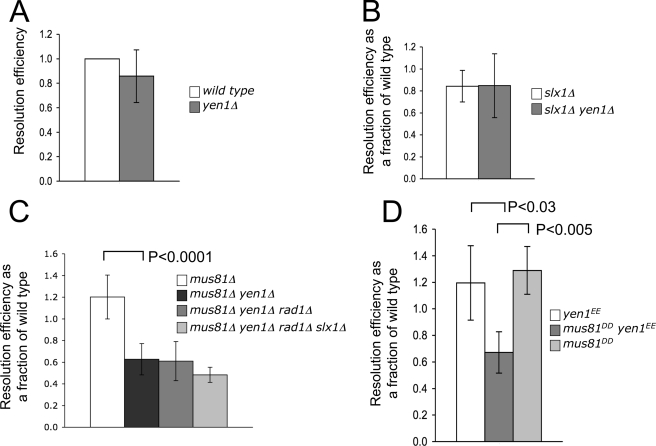
We next investigated if mutants in putative HJ resolvases are defective in resolving the HJ in JM-HJ. JM-HJ was cotransformed with a control plasmid, pRS415, to normalize resolution efficiencies against intersample variations in transformation efficiencies. yen1Δ mutants were found to be able to resolve JM-HJ with an efficiency that was equivalent to that of wild-type cells (Fig. 3A). To investigate if Yen1 might act redundantly with other nucleases, we transformed JM-HJ into slx1Δ yen1Δ and mus81Δ yen1Δ double mutants. We found that, in the absence of both Slx1 and Yen1, cells were still able to resolve JM-HJ with wild-type efficiencies (Fig. 3B). However, in contrast to wild-type cells, loss of Yen1 in a mus81Δ background resulted in an ∼2-fold decrease in JM-HJ resolution efficiency (Fig. 3C). Loss of Mus81 alone did not reduce JM-HJ resolution efficiency, indicating that Yen1 and Mus81 therefore possess redundant, overlapping functions required for the resolution of JM-HJ. We examined the possibility that Slx1 and/or Rad1 might be responsible for the resolution activity that persisted in the mus81Δ yen1Δ double mutants because Slx1 and Rad1 have overlapping substrate specificities with Mus81 in vitro (18). However, mus81Δ yen1Δ slx1Δ rad1Δ quadruple mutants were no more defective in JM-HJ resolution than the mus81Δ yen1Δ double mutants (Fig. 3C). The residual JM-HJ resolution activity in the quadruple mutant suggests the existence of additional nucleases that can also resolve the HJ in JM-HJ in vivo. We investigated the fidelity of JM-HJ resolution events in the absence of Yen1 and Mus81. No cer mutations were found in any of 20 independent JM-HJ resolution events isolated from mus81Δ yen1Δ double mutant cells, indicating that, in the absence of Yen1 and Mus81, JM-HJ is still resolved with absolute fidelity.
FIGURE 3.
Yen1 and Mus81 act redundantly to resolve JM-HJ in vivo. A–D, resolution efficiencies of JM-HJ in various strain backgrounds as indicated. Error bars are means ± S.D. from at least three independent experiments.
To further explore the synthetic resolution defect observed for yen1Δ and mus81Δ mutations, we went tested if the ability of Yen1 and Mus81 to resolve JM-HJ requires the nucleolytic activities of these proteins. To do this, we introduced, into the endogenous YEN1 and MUS81 genes, mutations that resulted in the substitution of amino acid residues that are essential for nuclease activity (5, 19). The resulting alleles were termed mus81DD, which contained substitutions D414A and D415A, and yen1EE, which contained substitutions E193A and E195A. Cells carrying either mus81DD or yen1EE alleles had resolution efficiencies comparable with that of wild-type cells (Fig. 3D). However, mus81DD yen1EE cells were as defective as mus81Δ yen1Δ double mutant cells in resolving JM-HJ, confirming that the resolution defect in mus81Δ yen1Δ double mutant cells is due to the loss of the nuclease activities of these proteins. Overall, these data demonstrate that JM-HJ resolution in vivo requires the nuclease activities of Yen1 and Mus81, which act in a redundant manner.
In vitro, Mus81 and Yen1 have distinct substrate preferences. Whereas Yen1 can efficiently and symmetrically cleave intact model HJs, Mus81-Mms4 displays poor activity toward HJs but has a preference for branched structures that contain nicks or gaps at the junction (5, 13, 20). The fact that Mus81 and Yen1 act redundantly to resolve JM-HJ suggests that Mus81 may indeed be able to cleave intact HJs in vivo in a manner akin to RuvC. This notion is consistent with recent findings that Mus81 can symmetrically cleave plasmid-borne palindromes that extrude into cruciform structures (21, 22).
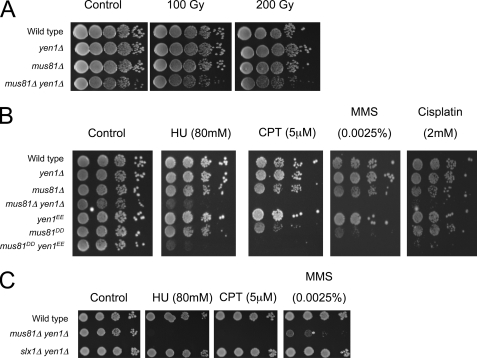
We next established if this novel genetic interaction between Yen1 and Mus81 has biological relevance outside the context of JM-HJ resolution. mus81Δ yen1Δ cells were not more sensitive to ionizing radiation than either of the single mutants, suggesting that the redundant functions of Yen1 and Mus81 in JM-HJ resolution are not required for HJ resolution during HR-mediated DSB repair (Fig. 4A). A central role for HR is the repair of collapsed replication forks that have encountered lesions in the DNA by facilitating break-induced replication (23). Unlike ionizing radiation-induced two-ended DSBs, which can be repaired by synthesis-dependent strand annealing or dissolution, processes that do not require HJ resolution, break-induced replication events are one-ended events that generate a single HJ that cannot be subjected to either synthesis-dependent strand annealing or dissolution but must be resolved prior to mitosis (1). We therefore reasoned that, in the absence of Yen1 and Mus81, the failure of a cell to resolve 50% of break-induced replication-induced HJs would result in extreme sensitivity to replication stress. As has been found previously, mus81 mutants show mild sensitivity to hydroxyurea, camptothecin, methyl methanesulfonate, and cisplatin at the doses used here. All of these agents perturb replication fork progression but through different mechanisms (Fig. 4B). In contrast to mus81 mutants, neither yen1Δ nor yen1EE cells were sensitive to any of the replication inhibitors tested here. However, the mus81Δ yen1Δ and mus81DD yen1EE double mutant cells were acutely sensitive to all replication inhibitors, consistent with the notion that Yen1 and Mus81 act redundantly in response to replication stress (Fig. 4B). In contrast, yen1Δ slx1Δ cells, which have wild-type JM-HJ resolution efficiencies, were not sensitive to replication stress (Figs. 3B and 4C). Overall, these results indicate that a compromised ability in mus81Δ yen1Δ cells to resolve JM-HJ in vivo is specifically associated with a defect in the ability to respond to replication stress.
FIGURE 4.
Yen1 and Mus81 are required for replication fork repair. A–C, DNA damage sensitivity assays of various strains as indicated. Gy, grays; HU, hydroxyurea; CPT, camptothecin; MMS, methyl methanesulfonate.
Conclusions
Because of the multipathway and multistep nature of HR repair, it has not been possible to specifically analyze the resolution step of HR in vivo. Here, we have described a system to analyze directly and in a quantitative manner HJ resolution in vivo that does not require the preceding steps that occur during HR-mediated DSB repair. The ability to genetically dissect in vivo HJ resolution reveals a redundant role for Yen1 with Mus81 in this process. Moreover, this novel genetic interaction between Yen1 and Mus81 is specifically relevant in the context of the repair/bypass of DNA lesions that can cause replication fork damage. The functional overlap between Yen1 and Mus81 is consistent with the recent finding that expression of the human homolog of Yen1, GEN1, can complement mus81Δ phenotypes in S. pombe (24).
Although mus81Δ yen1Δ cells displayed defective resolution of the HJ in JM-HJ, JM-HJ resolution still occurred at ∼50% of the levels seen in wild-type cells. This suggests that additional resolvases act in parallel to Yen1 and Mus81 and can do so in a faithful manner. Our results support the notion that these additional resolvases are neither Slx1 nor Rad1 and demonstrate that analysis of in vivo JM-HJ resolution can discriminate between nucleases that have similar biochemical activities in vitro. The system described in this work thus paves the way to identify those activities that can act in parallel to Yen1 and Mus81 to specifically resolve HJs in vivo.
Supplementary Material
Acknowledgments
We thank Peter McHugh and Ian D. Hickson for helpful comments on the manuscript; Francesca Storici, David Sherrat, and Robert Lloyd for reagents; Steve West for sharing results prior to publication; and Katsuhiro Hanada and members of the Chromosome Stability Group for useful discussions.
This work was supported by Cancer Research UK.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. 1–3, Table 1, and an additional reference.
- HR
- homologous recombination
- DSB
- double-strand break
- HJ
- Holliday junction.
REFERENCES
- 1.Mimitou E. P., Symington L. S. (2009) Trends Biochem. Sci. 34, 264–272 [DOI] [PubMed] [Google Scholar]
- 2.Andersen S. L., Bergstralh D. T., Kohl K. P., LaRocque J. R., Moore C. B., Sekelsky J. (2009) Mol. Cell 35, 128–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boddy M. N., Gaillard P. H., McDonald W. H., Shanahan P., Yates J. R., 3rd, Russell P. (2001) Cell 107, 537–548 [DOI] [PubMed] [Google Scholar]
- 4.Fekairi S., Scaglione S., Chahwan C., Taylor E. R., Tissier A., Coulon S., Dong M. Q., Ruse C., Yates J. R., 3rd, Russell P., Fuchs R. P., McGowan C. H., Gaillard P. H. (2009) Cell 138, 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ip S. C., Rass U., Blanco M. G., Flynn H. R., Skehel J. M., West S. C. (2008) Nature 456, 357–361 [DOI] [PubMed] [Google Scholar]
- 6.Muñoz I. M., Hain K., Déclais A. C., Gardiner M., Toh G. W., Sanchez-Pulido L., Heuckmann J. M., Toth R., Macartney T., Eppink B., Kanaar R., Ponting C. P., Lilley D. M., Rouse J. (2009) Mol. Cell 35, 116–127 [DOI] [PubMed] [Google Scholar]
- 7.Svendsen J. M., Smogorzewska A., Sowa M. E., O'Connell B. C., Gygi S. P., Elledge S. J., Harper J. W. (2009) Cell 138, 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fricke W. M., Brill S. J. (2003) Genes Dev. 17, 1768–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X. B., Melchionna R., Denis C. M., Gaillard P. H., Blasina A., Van de Weyer I., Boddy M. N., Russell P., Vialard J., McGowan C. H. (2001) Mol. Cell 8, 1117–1127 [DOI] [PubMed] [Google Scholar]
- 10.Gaillard P. H., Noguchi E., Shanahan P., Russell P. (2003) Mol. Cell 12, 747–759 [DOI] [PubMed] [Google Scholar]
- 11.Osman F., Dixon J., Doe C. L., Whitby M. C. (2003) Mol. Cell 12, 761–774 [DOI] [PubMed] [Google Scholar]
- 12.Hollingsworth N. M., Brill S. J. (2004) Genes Dev. 18, 117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaliraman V., Mullen J. R., Fricke W. M., Bastin-Shanower S. A., Brill S. J. (2001) Genes Dev. 15, 2730–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doe C. L., Ahn J. S., Dixon J., Whitby M. C. (2002) J. Biol. Chem. 277, 32753–32759 [DOI] [PubMed] [Google Scholar]
- 15.Storici F., Resnick M. A. (2006) Methods Enzymol. 409, 329–345 [DOI] [PubMed] [Google Scholar]
- 16.Chan S. N., Harris L., Bolt E. L., Whitby M. C., Lloyd R. G. (1997) J. Biol. Chem. 272, 14873–14882 [DOI] [PubMed] [Google Scholar]
- 17.McCulloch R., Coggins L. W., Colloms S. D., Sherratt D. J. (1994) EMBO J. 13, 1844–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rouse J. (2009) Biochem. Soc. Trans. 37, 495–510 [DOI] [PubMed] [Google Scholar]
- 19.de los Santos T., Hunter N., Lee C., Larkin B., Loidl J., Hollingsworth N. M. (2003) Genetics 164, 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fricke W. M., Bastin-Shanower S. A., Brill S. J. (2005) DNA Repair 4, 243–251 [DOI] [PubMed] [Google Scholar]
- 21.Coté A. G., Lewis S. M. (2008) Mol. Cell 31, 800–812 [DOI] [PubMed] [Google Scholar]
- 22.Taylor E. R., McGowan C. H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3757–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Llorente B., Smith C. E., Symington L. S. (2008) Cell Cycle 7, 859–864 [DOI] [PubMed] [Google Scholar]
- 24.Lorenz A., West S. C., Whitby M. C. (2010) Nucleic Acids Res., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.